Abstract
Four cases of central venous catheter-related Methylobacterium radiotolerans infection are presented here. The patients were all long-term catheter carriers with an underlying diagnosis of leukemia, and they mostly manifested fevers. The isolated bacterial strains all showed far better growth on buffered charcoal yeast extract agar during the initial isolation and/or subcultures than they did on sheep blood or chocolate agar. This microbiological feature may improve the culture recovery of this fastidious pink Gram-negative bacillus that has rarely been isolated in clinical microbiology laboratories.
TEXT
Methylobacterium species are fastidious aerobic Gram-negative bacilli (1). They are of environmental origin and are rarely isolated in clinical microbiology laboratories (2, 3). Of these pink-colored organisms, Methylobacterium mesophilicum has occasionally been reported as an opportunistic pathogen that may infect immunocompromised hosts, such as patients with malignancy, organ transplant, HIV infection, renal failure, or alcoholism (4–7). Methylobacterium radiotolerans is less known, however, with only two series of infections reported thus far (2, 8). In this study, we report four cases of central venous line-related M. radiotolerans infection in patients who had underlying leukemia. We paid attention to the culture features of the bacterial strains.
These cases occurred from 2009 to 2014 in patients at The University of Texas M. D. Anderson Cancer Center, a comprehensive cancer center with 600 beds in Houston, TX. Anticancer chemotherapy often required the long-term use of indwelling venous catheters. Cultures of the blood samples from patients with suspected bloodstream infection were performed using the Bactec 9240 automated system with Plus Aerobic/F bottles (BD Diagnostic Systems, Sparks, MD) and Isolator tubes (Wampole Laboratories, Princeton, NJ) (9). Typically, a 20-ml blood sample was divided equally into a Bactec bottle and an Isolator tube for cultures. The bottle was incubated for 7 days at 35°C with aeration in the system. The Isolator tube was processed by centrifugation to pellet the lysed cellular sludge, which was evenly divided and spread onto four agar plates. Before October 2009, culture involved two sheep blood agar plates and two chocolate agar plates; after this time, one buffered charcoal yeast extract plate (BCYE) was included in exchange for one sheep blood agar plate. All the plates were from BBL (BD Microbiology Systems, Cockeysville, MD). The plates were incubated for 4 days at 35°C in 5% CO2 to allow the bacterial or fungal colonies to be counted. About 30,000 blood cultures were performed annually at M. D. Anderson during the study period.
The fastidious nature of the M. radiotolerans strains required 16S rRNA gene sequencing for the initial identification, using a previously published method (10). Briefly, bacterial genomic DNA was extracted from cultured colonies and amplified by PCR for a 594-bp amplicon in the mid-region of the gene using two universal bacterial primers (5′-TGCCAGCAGCCGCGGTAATAC [forward, UBFO]; and 5′-CGCTCGTTGCGGGACTTAACC [reverse, UBRE]). The amplicon was sequenced. The sequence matches were made using the GenBank Basic Local Alignment Search Tool (BLAST), and the best match with a type or reference strain yielded species identification. During this study, the sequence of each strain was extended to near full length (∼1,420 bp) of the 16S gene for greater accuracy. Two additional sets of primers were used for the extension, including 5′-GCGTGCTTAACACATGCAAGTC (forward, AFBFO), 5′-GCACAAGCGGTGGAGCATGTG (reverse, 16R), primer UBFO as defined above, and 5′-AGGAGGTGATCCAACCGCA (reverse, 16R). Culture studies of all strains were also performed to determine microbiological features.
Case 1.
A 59-year-old female with relapsed acute myelogenous leukemia (AML) presented to the emergency room with fever in February 2009. She had been neutropenic for 2 weeks. Laboratory studies revealed no countable white blood cells. Two blood samples, one drawn from a peripheral vein and one from the peripherally inserted central catheter (PICC) that had been in place for 156 days, were cultured.
While the peripheral blood culture remained negative, the PICC line blood grew a Gram-negative rod in the Bactec bottle after an incubation period of 6 days. This bacillus with pink colonies was identified as M. radiotolerans by 16S gene sequencing. The patient was treated with levofloxacin and recovered.
Eleven days later, the patient experienced another febrile episode, and the PICC line blood also grew M. radiotolerans after 5 days of incubation in the Bactec bottle. The peripheral blood culture was negative. Again, 47 days later, the patient developed fever, and the same Gram-negative bacillus was cultured from the PICC line after an incubation of 6 days in the Bactec bottle.
For all three positive cultures from the Bactec bottles, concurrent Isolator plates, i.e., two sheep blood agar plates and two chocolate agar plates (before the use of buffered charcoal yeast extract plates) showed no growth each time. After these episodes, which spanned 58 days, the PICC line was removed.
Case 2.
A 12-year-old boy with a diagnosis of AML presented to the emergency room with low-grade fever in November 2010. Laboratory examination showed a white blood cell count of 1,900 × 106/liter with 1,290 × 106/liter of neutrophils (an improvement from the nadir of 0 neutrophils 2 weeks earlier). A blood culture drawn from the PICC line that had been in place for 59 days grew M. radiotolerans. The concurrent blood culture drawn from a peripheral vein was negative. The patient was treated with levofloxacin.
The positive PICC line blood showed growth only on the BCYE plate upon spread of the centrifuged sediment of the lysed blood, and 40 bacterial colonies were noted after an incubation of 5 days. The concurrent sheep blood plate and two chocolate plates that also contained the blood sediment did not show growth.
Five days later, blood samples drawn from the PICC line and a peripheral vein were cultured again due to low-grade fever in the patient. The PICC line blood showed growth, and the seven bacterial colonies were noted on BCYE only after 4 days of incubation. After this episode, the PICC line was removed, and the patient was cured of this M. radiotolerans infection. In these positive cultures, the Bactec bottles were negative.
Case 3.
An 87-year-old female with AML presented to the emergency room with a fever of 38.4°C in December 2013. The laboratory data revealed a white blood cell count of 700 × 106/liter with 63 × 106/liter of neutrophils. The patient had carried a PICC line for 7 months. Two blood cultures drawn from the PICC line 1 day apart grew M. radiotolerans. A peripheral blood sample was also drawn and remained negative. The patient was treated with levofloxacin and recovered.
Five days later, a third PICC line blood was also culture positive for the same organism. In these positive cultures, a few to many bacterial colonies were noted on the BCYE plates after incubations of 3 to 5 days, but far fewer colonies were seen on the chocolate agar plates and none on the sheep blood agar plates. The Bactec bottles of these cultures were all negative.
Case 4.
A 23-year-old woman presented to the emergency room for seizures in September 2014. The patient was a recipient of a cord blood hematopoietic stem cell transplant for acute lymphoblastic leukemia. She had suffered from graft-versus-host disease that was treated with prednisone. A central venous catheter (CVC) had been in place for 13 months.
Physical examination showed no fever. Laboratory tests revealed anemia (hemoglobin, 106 g/liter) but a normal white blood cell count (9,100 × 106/liter). A blood culture drawn from the CVC line grew M. radiotolerans, while a concurrent blood culture drawn from a peripheral vein was negative. The organism grew on the BCYE plate only, with 25 colonies noted. The patient was treated with ciprofloxacin and piperacillin-tazobactam.
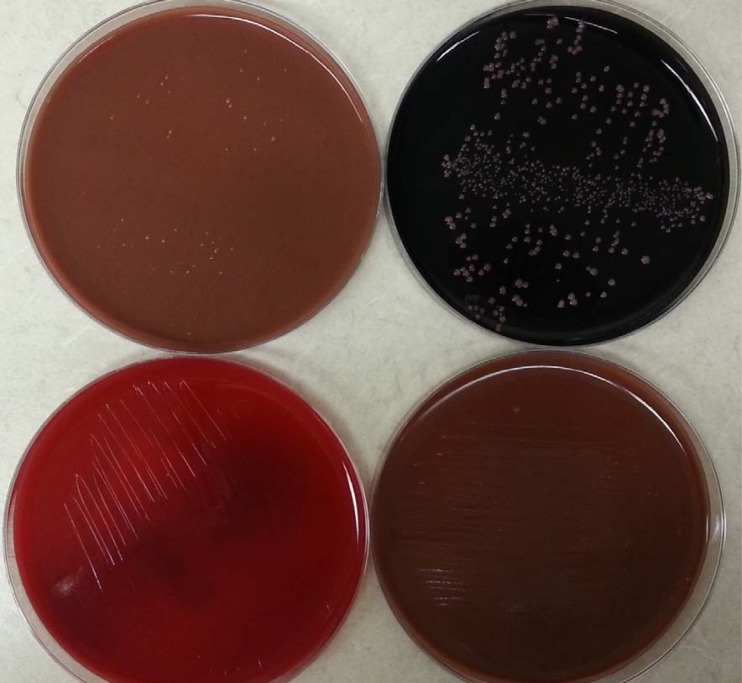
During the week, two additional CVC blood samples grew the same organism, and far more colonies were noted on the BCYE plates than on the chocolate plates. No growth was noted on the sheep blood plates. Figure 1 illustrates one such culture. In the third culture, the Bactec bottle also turned positive. The CVC line was removed after these positive cultures were obtained.
FIG 1.
Initial isolation of M. radiotolerans from the blood culture of a 23-year-old patient (case 4). The plates, upon spread of lysed blood sediment from the Isolator tube, were incubated for 5 days with 5% carbon dioxide. Numerous colonies grew on buffered charcoal yeast extract agar (top right), fewer and smaller colonies on the chocolate agar (top left and lower right), and none on blood agar (lower left).
Table 1 summarizes the culture and clinical features of these cases.
TABLE 1.
Clinical and microbiologic features of four patients with central venous line-related M. radiotolerans infection
| Feature | Case 1 | Case 2 | Case 3 | Case 4 |
|---|---|---|---|---|
| Age (yr), sexa | 59, F | 12, M | 87, F | 23, F |
| Underlying diseaseb | AML | AML | AML | ALL |
| ANC (×106/liter) at each culturec | 0, 180, 1,540 | 1,290, 560 | 60, 60, 460 | 1,790, 730, 3,330 |
| Symptom of infection episodes | Fever (38.9°C) × 2, afebrile | Fever (37.9°C), afebrile | Fever (38.4°C to 38.9°C) × 3 | Afebrile × 3 |
| Antimicrobial treatment | Levofloxacin | Clindamycin, levofloxacin, cefepime | Levofloxacin, linezolid | Ciprofloxacin, piper-tazod |
| Catheter type, days at 1st culturee | PICC, 156 | PICC, 59 | PICC, 217 | CVC, 385 |
| Growth of blood cultures | ||||
| Blood from CVC line | Positive | Positive | Positive | Positive |
| Peripheral blood | Negative | Negative | Negative | Negative |
| Growth medium(a) | Bactec bottle | Only BCYE agar | BCYE, chocolate | BCYE, chocolate, Bactec |
| Times of positivity | 3× over 58 days | 2× over 5 days | 3× over 6 days | 3× over 7 days |
F, female; M, male.
AML, acute myelogenous leukemia; ALL, acute lymphoblastic leukemia.
ANC, absolute neutrophil count.
Piper-tazo, piperacillin-tazobactam.
PICC, peripherally inserted central catheter; CVC, central venous catheter.
Additional microbiological studies.
The initial identification of these M. radiotolerans strains was based on the culture features (Gram-negative reaction, pink colonies, and positive reactions of catalase and oxidase) and the 594-bp 16S gene sequences. These sequences all matched 100% with those of the M. radiotolerans type strain JCM 2831 (GenBank accession no. CP001001 and others). To improve accuracy and discern minor strain differences, we further sequenced the 16S genes of all strains to nearly full length (1,398 bp to 1,421 bp). These sequences all best matched, at 100% to 99.8% (zero to two nucleotide mismatches), with those of the M. radiotolerans type strain and strain 7210 (GenBank accession no. GU294325). Therefore, these organisms are clearly identified. The second best matches were with M. mesophilicum, at up to 97.9% (1,387 of 1,417 bp), which allowed confidence in separating the two morphologically similar species.
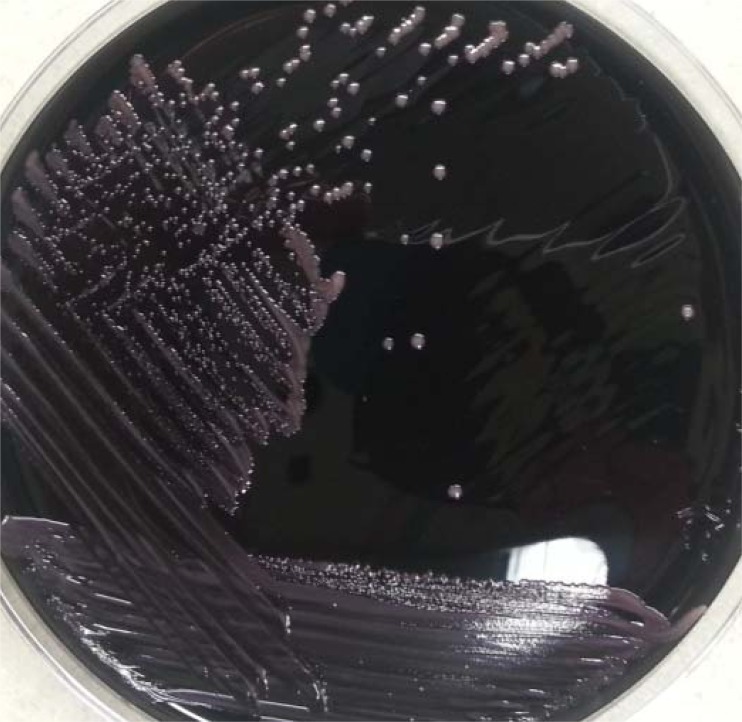
The features of these strains in culture were also compared side by side in subcultures on BCYE, sheep blood, and chocolate agars. All strains grew best on BCYE agar, with colonies reaching ∼1 mm after an incubation of 72 h. The colonies were light coral pink, smooth, raised, and entire, and they continued to grow to nearly 3 mm after 6 days. In contrast, all strains showed poor growth on sheep blood agar and chocolate agar, with the colony size at about half of that on BCYE. Even after 6 days of incubation, the colonies hardly exceeded 1 mm in diameter. None of the strains grew on MacConkey agar. Therefore, BCYE agar is the preferred medium for these M. radiotolerans strains. Figure 2 illustrates the colonies grown from the strain from case 4 on BCYE agar after 4 days of incubation.
FIG 2.
Subculture of M. radiotolerans on buffered charcoal yeast extract agar upon an incubation of 4 days (strain from case 4). Note the pink smooth shining colonies of ∼1.2 mm in diameter.
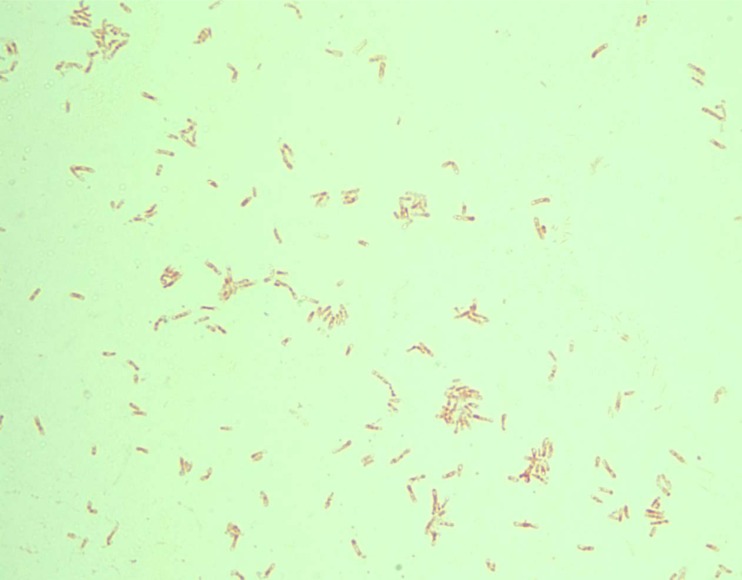
On Gram stain, the bacterial cells of all strains were poorly fixed on glass slides (i.e., washed away) but showed similar morphology microscopically (Fig. 3). The cells are bacillary (∼3 μm in length) and faintly Gram negative. They also showed vacuolated cytoplasm and faint cellular contours. These features fit the textbook description of Methylobacterium species (1, 3).
FIG 3.
Gram stain of M. radiotolerans strain from case 2. A 3-day-old culture on buffered charcoal yeast extract agar was stained to show faint safranin staining, cytoplasmic vacuoles, inconspicuous cellular contour, and poor fixation on glass slide.
In view of the growing popularity of mass spectrometry for rapid microbial identification, we also tested these M. radiotolerans strains with a 3-day-old culture from the BCYE plates using the Vitek MS instrument (bioMérieux, Durham, NC). Two strains were identified correctly (99.9% confidence, nonvalidated). Two strains were not identified, including the strain that was correctly identified 2 months earlier during the initial isolation with a 5-day BCYE culture. Thus, with 3 of the 4 strains identified, matrix-assisted laser desorption ionization–time of flight [MALDI-TOF] mass spectrometry works reasonably well with this species in spite of its rare isolation or being less known.
All strains grew poorly on Mueller-Hinton agar and on blood Mueller-Hinton agar, which made it difficult for us to perform antimicrobial susceptibility tests by the Etest method.
Discussion.
In this series of four cases with M. radiotolerans infection, a few consistent features are notable: all patients had leukemia, with profound neutropenia in three of them, a long-indwelling central venous catheter was the source of repeated positive blood cultures, and the M. radiotolerans strains grew solely or far better on BCYE plates. These results suggest that M. radiotolerans may colonize PICC or CVC lines. This was true in the recent report by Lai et al. (8) in Taiwan, in which the two cases with M. radiotolerans bacteremia were also CVC line related. In another report of patients with hemodialysis from Italy, CVC line colonization with M. radiotolerans was found to be the cause of repeated febrile episodes in the patients (2). To our knowledge, infection with M. radiotolerans has not been reported elsewhere.
The finding of the good growth of these M. radiotolerans strains on BCYE agar is novel. With BCYE in the initial cultures, colonies were noted upon incubation for 3 to 5 days of the Isolator tube blood sludge. Without this medium, two of the four cases would have gone undiagnosed. When the organism did grow in the Bactec bottle, as in the first case before our routine use of BCYE for blood cultures, extended incubation from 5 to 6 days was required. Therefore, our incorporation of BCYE agar in blood cultures might have contributed to the cultivation of this otherwise very fastidious organism. Conversely, the current practice of using automated blood cultures for 5 days and the rare use of the Isolator tube method elsewhere might have played a role in the scant reports of this infection in spite of the common use of CVC lines in hospitals.
In clinical microbiology laboratories, the differentiation of pink fastidious Gram-negative bacilli may be necessary at times. In our experience, Roseomonas species grow similarly on sheep blood agar, chocolate agar, and BCYE agar; the rosy-pink colonies are usually mucoid for commonly encountered species, such as Roseomonas mucosa and Roseomonas gilardii (11). These organisms are also coccobacillary on Gram stain. Thus, the better growth of M. radiotolerans on BCYE agar and its relatively long bacillary morphology on Gram stain with cytoplasmic vacuoles may offer presumptive identification of this organism. The 16S gene sequencing method should offer confident identification, and mass spectrometry is also helpful in this effort.
The sole manifestation of febrile episodes in our neutropenic patients and the negative peripheral blood cultures are also consistent with the low pathogenicity of M. radiotolerans. The lack of fever response of the fourth patient can be explained by the use of immunosuppressants. As an environmental Gram-negative rod, M. radiotolerans has a 6-Mbp genome (GenBank accession no. CP001001) that is relatively large and may be able to support complex growth needs. The reason it prefers BCYE agar is uncertain and may warrant further investigation. BCYE is also used to cultivate Legionella species, another group of environmental Gram-negative bacilli and a cause of pneumonia, from respiratory specimens. In addition, Legionella species do not grow on sheep blood agar, but, in our experience, they do grow considerably on chocolate agar; these features are also similar to those of the M. radiotolerans strains observed in this study.
ACKNOWLEDGMENTS
This work was supported in part by the National Institutes of Health grant CA16672 for our institutional DNA analysis core facility.
We thank the staff at our DNA analysis core facility and molecular diagnostic laboratory for DNA sequencing, the staff of microbiology laboratory for cultures, and the clinical staff for taking care of the patients.
We declare no conflicts of interest.
REFERENCES
- 1.Holt JG, Krieg NR, Sneath PHA, Staley JT, Williams ST. 1994. Bergey's manual of determinative bacteriology, 9th ed Williams & Wilkins, Baltimore, MD. [Google Scholar]
- 2.de Cal M, Cazzavillan S, Cruz D, Nalesso F, Brendolan A, Rassu M, Ronco C. 2009. Methylobacterium radiotolerans bacteremia in hemodialysis patients. G Ital Nefrol 26:616–620. (In Italian.) [PubMed] [Google Scholar]
- 3.Vaneechoutte M, Dijkshoorn L, Nemec A, Kampfer P, Wauters G. 2011. Acinetobacter, Chryseobacterium, Moraxella, and other nonfermentative Gram-negative rods, p 729–730. In Versalovic J, Carroll KC, Funke G, Jorgensen JH, Landry ML, Warnock DW (ed), Manual of clinical microbiology, 10th ed ASM Press, Washington, DC. [Google Scholar]
- 4.Fernandez M, Dreyer Z, Hockenberry-Eaton M, Baker CJ. 1997. Methylobacterium mesophilica [sic] as a cause of persistent bacteremia in a child with lymphoma. Pediatr Infect Dis J 16:1007–1008. doi: 10.1097/00006454-199710000-00023. [DOI] [PubMed] [Google Scholar]
- 5.Liu JW, Wu JJ, Chen HM, Huang AH, Ko WC, Chuang YC. 1997. Methylobacterium mesophilicum synovitis in an alcoholic. Clin Infect Dis 24:1008–1009. doi: 10.1093/clinids/24.5.1008. [DOI] [PubMed] [Google Scholar]
- 6.Truant AL, Gulati R, Giger O, Satishchandran V, Caya JG. 1998. Methylobacterium bacteremia in AIDS. Clin Microbiol Infect 4:112–113. doi: 10.1111/j.1469-0691.1998.tb00369.x. [DOI] [PubMed] [Google Scholar]
- 7.Sanders JW, Martin JW, Hooke M, Hooke J. 2000. Methylobacterium mesophilicum infection: case report and literature review of an unusual opportunistic pathogen. Clin Infect Dis 30:936–938. doi: 10.1086/313815. [DOI] [PubMed] [Google Scholar]
- 8.Lai CC, Cheng A, Liu WL, Tan CK, Huang YT, Chung KP, Lee MR, Hsueh PR. 2011. Infections caused by unusual Methylobacterium species. J Clin Microbiol 49:3329–3331. doi: 10.1128/JCM.01241-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tarrand JJ, Guillot C, Wenglar M, Jackson J, Lajeunesse JD, Rolston KV. 1991. Clinical comparison of the resin-containing Bactec 26 Plus and the Isolator 10 blood culturing systems. J Clin Microbiol 29:2245–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han XY, Pham AS, Tarrand JJ, Sood PK, Luthra R. 2002. Rapid and accurate identification of mycobacteria by sequencing hypervariable regions of the 16S ribosomal RNA gene. Am J Clin Pathol 118:796–801. doi: 10.1309/HN44-XQYM-JMAQ-2EDL. [DOI] [PubMed] [Google Scholar]
- 11.Han XY, Pham AS, Tarrand JJ, Rolston KV, Helsel LO, Levett PN. 2003. Bacteriologic characterization of 36 strains of Roseomonas species and proposal of Roseomonas mucosa sp. nov. and Roseomonas gilardii subsp. rosea subsp. nov. Am J Clin Pathol 120:256–264. doi: 10.1309/731VVGVCKK351Y4J. [DOI] [PubMed] [Google Scholar]