Abstract
Melioidosis, a disease of public health importance in Southeast Asia and northern Australia, is caused by the Gram-negative soil bacillus Burkholderia pseudomallei. Melioidosis is typically acquired through environmental exposure, and case clusters are rare, even in regions where the disease is endemic. B. pseudomallei is classed as a tier 1 select agent by the Centers for Disease Control and Prevention; from a biodefense perspective, source attribution is vital in an outbreak scenario to rule out a deliberate release. Two cases of melioidosis within a 3-month period at a residence in rural northern Australia prompted an investigation to determine the source of exposure. B. pseudomallei isolates from the property's groundwater supply matched the multilocus sequence type of the clinical isolates. Whole-genome sequencing confirmed the water supply as the probable source of infection in both cases, with the clinical isolates differing from the likely infecting environmental strain by just one single nucleotide polymorphism (SNP) each. For the first time, we report a phylogenetic analysis of genomewide insertion/deletion (indel) data, an approach conventionally viewed as problematic due to high mutation rates and homoplasy. Our whole-genome indel analysis was concordant with the SNP phylogeny, and these two combined data sets provided greater resolution and a better fit with our epidemiological chronology of events. Collectively, this investigation represents a highly accurate account of source attribution in a melioidosis outbreak and gives further insight into a frequently overlooked reservoir of B. pseudomallei. Our methods and findings have important implications for outbreak source tracing of this bacterium and other highly recombinogenic pathogens.
INTRODUCTION
Melioidosis is an underrecognized disease of significant public health burden in many tropical regions across the globe, especially northern Australia and Southeast Asia, where the greatest number of cases are reported annually (1). Melioidosis is caused by the environmental dwelling Gram-negative bacterium Burkholderia pseudomallei, an opportunistic pathogen that most commonly affects people with underlying disease or risk factors, particularly diabetes and hazardous alcohol use (2). Disease severity varies widely and depends on the strain, host immunity, and inoculum size. The highest case fatality rates exceed 90% in septic shock or untreated septic cases (3). Even when appropriate therapy is administered, mortality ranges from 13% in northern Australia (4, 5) to 50% in Southeast Asia (2). In October 2012, B. pseudomallei was upgraded to a tier 1 select agent by the Centers for Disease Control and Prevention (www.selectagents.gov) owing to fears of a deliberate release coupled with the high mortality rate, lack of a vaccine, intrinsic resistance to standard antimicrobial agents, and protean disease presentations that confound diagnosis, particularly in regions where the disease is not endemic.
B. pseudomallei infection primarily occurs via percutaneous inoculation; however, case reports associated with severe weather events and contaminated water supplies have highlighted the potentially important roles of inhalation and ingestion (2, 4). Bore (automated well) water supplies contaminated with B. pseudomallei have been linked to four and three deaths in the Northern Territory and western Australia, respectively (6, 7). Currently, there are an estimated 2,600 domestic water bores in the Darwin rural region of the Northern Territory, Australia (8), most of which are unchlorinated. Samples from 55 of these bores showed that one-third were culture positive for B. pseudomallei (9).
In early 2012, two nonfatal melioidosis cases were diagnosed in a single household supplied with B. pseudomallei-contaminated bore water in the Darwin rural region. Using multilocus sequence typing (MLST), we previously showed that clinical B. pseudomallei isolates from both cases were the same sequence type (ST 325) as an isolate from the bore water supply (8). The property's water supply was remediated with a UV filter, leading to undetectable levels of B. pseudomallei (8). ST 325 has also been recovered from other locations in the Darwin rural region, supporting the notion that MLST alone lacks resolution for source attribution. To obtain greater resolution, we performed whole-genome sequencing (WGS) on B. pseudomallei obtained from these two cases, from their water supply, and from other melioidosis patients and environmental samples from the surrounding region. PCR-based multilocus variable-number tandem repeat (VNTR) analysis (MLVA) of four loci (10) was incorporated to augment WGS short read data, which are unable to span paralogous or certain highly repetitive loci in B. pseudomallei (11, 12). As a novel aspect of this study, we performed genomewide phylogenetic analysis of small insertions/deletions (indels), which largely comprise short read-mappable VNTRs across the B. pseudomallei genome, to increase our resolution among closely related isolates. This work represents the most accurate account of source attribution in a melioidosis case to date and highlights the challenges of tracing a highly recombinogenic bacterium of public health importance, with ramifications that extend to forensic source tracing of this bacterium in the event of a nefarious release.
MATERIALS AND METHODS
Ethics approval.
Ethics approval for this study was detailed previously (4).
Genotyping.
MLVA-4 and MLST were performed as described elsewhere (10, 13).
Study site and B. pseudomallei isolates used in this study.
Details of the study site have been described (8). The 13 B. pseudomallei isolates used in the current study were all identified as ST 325 (Table 1). Six study site isolates, including the two human cases, and seven outgroup ST 325 isolates external to the study site were examined. These isolates were obtained either as part of the 25-year Darwin Prospective Melioidosis Study (4) or from B. pseudomallei environmental sampling efforts conducted by Menzies School of Health Research in the Darwin region over the past 2 decades (9, 14). Two outgroup isolates originated from bore water supplies on nearby properties (<9.7 km from the outbreak property), and the remaining five isolates were from human cases with patient exposure history linking them to probable infection in the Darwin rural region (Table 1). Outgroups were included to identify relatedness among isolates from the outbreak property.
TABLE 1.
Molecular typing results and sampling data of B. pseudomallei isolates
| Isolate no. | MLVA-4 type | MLST | Patient identification/water source | Source of sample | Isolation date | Sample origination |
|---|---|---|---|---|---|---|
| MSHR5990 | 223 | 325 | P741 | Clinical | Jan 2012 | Outbreak property |
| MSHR6955 | 402 | 325 | P811 | Clinical | Apr 2012 | Outbreak property |
| MSHR6137 | 223 | 325 | Water storage tank | Environmental | Jan 2012 | Outbreak property |
| MSHR7176 | 223 | 325 | Water storage tank | Environmental | May 2012 | Outbreak property |
| MSHR7406 | 223 | 325 | Shower | Environmental | Jun 2012 | Outbreak property |
| MSHR7446 | 244 | 325 | Bore head | Environmental | Jun 2012 | Outbreak property |
| MSHR1539 | 244 | 325 | Bore head | Environmental | Feb 2003 | Nearby location |
| MSHR3554 | 223 | 325 | Bore head | Environmental | Nov 2009 | Nearby location |
| MSHR270 | 438 | 325 | P92 | Clinical | Feb 1994 | Unrelated case |
| MSHR2037 | 437 | 325 | P429 | Clinical | Mar 2014 | Unrelated case |
| MSHR4182 | 244 | 325 | P646 | Clinical | Apr 2011 | Unrelated case |
| MSHR4438 | 400 | 325 | P690 | Clinical | Dec 2010 | Unrelated case |
| MSHR6354 | 430 | 325 | P785 | Clinical | Sep 2013 | Unrelated case |
B. pseudomallei culture detection from water (9) or soil (6) specimens was performed using established methods. Species confirmation was performed using a real-time PCR assay targeting a B. pseudomallei-specific 115-bp segment within the type three secretion system 1 (TTS1) gene (15).
Whole-genome sequencing (WGS) and de novo assembly.
Genomic DNA was extracted using the Qiagen DNeasy blood and tissue kit (Qiagen, Chadstone, Victoria, Australia) as previously described (16). Samples were sequenced at Macrogen, Inc. (Gasan-dong, Seoul, Republic of Korea), using the Illumina HiSeq2000 platform (Illumina, Inc., San Diego, CA). MSHR6137 was also subjected to WGS using the 454 GS FLX+ platform (454 Life Sciences, Branford, CT, USA). Genome assembly for this strain was performed as previously described (12). The final genome contains 65 contigs totaling 7,208,016 bp, with an N50 of 258,651 bp, and encodes a predicted 6,656 proteins.
Variant identification of outbreak-associated isolates.
Identification of orthologous core genome single-nucleotide polymorphism (SNP) and short read-mappable indel variants from Illumina WGS data were performed using SPANDx (17). MSHR6137 was isolated from the water storage tank in the same month as P741 presented with melioidosis and was therefore chosen as the reference genome for alignment. Following identification, genetic variants were visualized in Tablet v1.14.04.10 to ensure accuracy (18). MSHR6137 Illumina reads were included in the analyses as a control, allowing a small number of discrepant variant calls to be eliminated across all genomes. SNP and indel variants identified with SPANDx were used for phylogenetic reconstruction based on the maximum-parsimony module of PAUP v4.0. Phylogenetic trees were visualized and manipulated in FigTree v1.4 (http://tree.bio.ed.ac.uk/software/figtree/). Genetic loss was assessed using BedTools (19), which is incorporated into SPANDx.
Nucleotide sequence accession numbers.
The whole-genome shotgun project for MSHR6137 has been deposited into GenBank under the accession number AXDS00000000. The version described in this paper is version AXDS00000000.1.
RESULTS AND DISCUSSION
Two melioidosis cases (P741 and P811) from the same rural residential property presented within a 3-month period in early 2012 (8). Melioidosis is not generally considered communicable, and case clusters are rare; thus, the presentation of two cases from the same property over a short time frame prompted suspicion of a single B. pseudomallei-contaminated source. Our previous case study identified the likely point source of the two infections as a contaminated household bore water supply, as both clinical and environmental isolates from this property were ST 325 (8).
In certain microbes, a single genotyping technique may suffice for inferring relatedness; however, the high recombination rate of B. pseudomallei can confound attempts to trace the source of an infection (20). We recently reported two situations where B. pseudomallei strains from different countries in different hemispheres had identical STs but were highly divergent on a whole-genome level (21) and an instance where MLST failed to detect a polyclonal infection that was identifiable by WGS (22). Together, these studies demonstrate that MLST can suffer from a lack of discrimination power.
To further investigate the point source of the rural property outbreak, we first examined high-resolution tandem repeat loci using MLVA-4 (10). The MLVA-4 approach is inexpensive and thus useful in an outbreak scenario to rapidly analyze a large number of strains for additional genetic or genomic analyses. A predominant MLVA-4 type, 223, was found in 14 of 15 ST 325 isolates from the outbreak property's water supply. These isolates harbored an identical MLVA-4 type to MSHR5990, the isolate obtained from P741 (Table 1). Interestingly, the clinical isolate from P811 (MSHR6955) was MLVA-4 type 402, which differed from other strains isolated from the outbreak property by a single repeat deletion at the 2341k locus (Table 1). A single isolate from the water supply possessed a third MLVA-4 type (244) that differed from other outbreak ST 325 isolates at the 933k locus. These results suggest that all ST 325 isolates from the outbreak property were closely related. MLVA-4 types 223 and 244, but not 402, were also found in three ST 325 isolates: an unrelated case (MSHR4182) and two environmental samples from a nearby area (MSHR1539, MSHR3554) (Table 1). Given that the clinical isolate from P811 was the only instance of MLVA-4 type 402, it is possible that this MLVA variant arose as either a consequence of within-host evolution or via laboratory passage. This finding was not surprising given that MLVA loci are known to evolve rapidly, and multiple mutations have been documented in acute B. pseudomallei infections (11, 23). However, despite the MLVA differences of P741 and P811, no remarkable features were noted between the clinical presentation of these two cases, consistent with the intergenic nature of these MLVA loci, which are not expected to impart a selective advantage to the bacterium or to cause more severe disease (10, 24). It remains a possibility that the variant MLVA-4 type 402 is present in a lower abundance in the water supply, and therefore insufficient sampling precluded its isolation.
Following MLVA-4 analysis, 13 ST 325 B. pseudomallei isolates, including at least 1 from each known ST 325 MLVA-4 type, were selected for WGS (Table 1). Whole-genome SNP identification uncovered 111 SNPs among the ST 325 isolates (see Fig. S1 in the supplemental material). SNP variants were visualized using the Integrative Genomics Viewer 2.3.4 (25). SNPs were relatively evenly distributed, suggesting that the majority of variants were due to point mutation rather than to recombination (see Fig. S3 in the supplemental material). Fifty-seven of the SNPs resulted in a synonymous change, and 54 were nonsynonymous. None of the SNPs were associated with known virulence or antibiotic resistance genes. Phylogenetic reconstruction of these SNPs using maximum parsimony showed only minimal differences (one to four SNPs across the entire genome) among all outbreak isolates, including the two clinical isolates from patients 741 and 811. No evidence of genetic loss was observed in any ST 325 strain from the outbreak property relative to MSHR6137. However, one large-scale deletion (35.8 kb) was detected in outgroup isolate MSHR4438 (AXDS01000015, 120,323 to 156,123 kb). The two clinical isolates from the outbreak property differed from each other by two SNPs and from MSHR6137 by a single SNP each; thus, the environmental isolate MSHR6137 was more closely related to both clinical isolates than they were to each other (see Fig. S1 in the supplemental material). Based on these SNP results, we deduced that both melioidosis cases from the outbreak property were most likely caused by infection from this single point source.
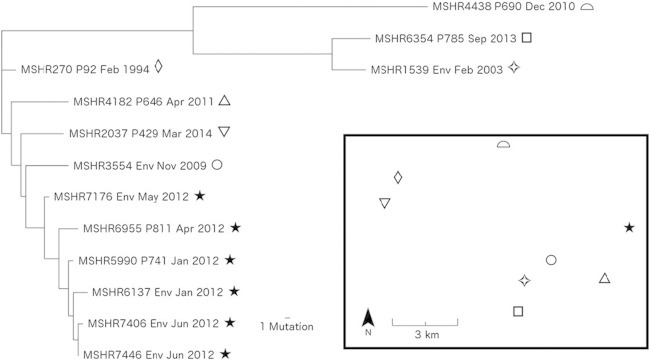
B. pseudomallei encodes a large number of predominantly intergenic tandem-repeat loci, with 609 such loci documented in the K96243 type strain (24). These loci provide a rich source of genetic variation in this organism. Genome-wide analysis of small (<30 bp) indel mutations in the ST 325 isolates, most of which encode VNTR loci, identified 77 variants; despite their lower number compared with SNPs, we hypothesized that these indels might be useful for phylogenetic analysis. Small indels can occur at highly mutable genomic positions, making them more prone to homoplasy, and are therefore conventionally considered problematic for phylogenetic reconstruction. However, in this comparison of closely related isolates, indel-based phylogenetic reconstruction yielded a topology very similar to the SNP data, with all isolates being placed into the same major clades (see Fig. S1 and S2 in the supplemental material). To provide higher resolution among ST 325 isolates, the SNP and indel data sets were combined into a single phylogeny (Fig. 1). This approach not only reduces homoplasy but allows further resolution within SNP-defined clades by comparison of indels within subclades. Four and nine indels were identified in P741 (MSHR5990) and P811 (MSHR6955), respectively, relative to MSHR6137. Fewer indels were identified between MSHR5990 and MSHR6137, which was expected given that both isolates were collected in the same month (Table 1). In contrast, P811 was diagnosed with melioidosis 3 months after MSHR6137 was collected. Thus, the time difference in sampling may explain the greater number of indels observed in MSHR6955. Importantly, all outbreak property isolates remained in the outbreak clade, and all outgroup isolates were distinct from this clade. Therefore, phylogenetic reconstruction of closely related bacterial populations using indels complements the findings of phylogenetic reconstruction using SNPs.
FIG 1.
Maximum parsimony whole-genome combined SNP and indel phylogeny of environmental and clinical isolates from a melioidosis outbreak in the Darwin rural region, northern Australia. P, patient; Env, environmental. The consistency index was 0.98. Inset, sampling locations for this study; solid stars, patient and environmental samples collected at the outbreak property; hollow shapes, isolates used as outgroups. Mutation refers to both SNPs and indels.
Closer examination of the indels identified between P741 and P811 isolates and MSHR6137 showed that 7 of the 9 indels were located in putative VNTRs. Interestingly, these indels did not fall within MLVA loci commonly screened in B. pseudomallei (24), including the four MLVA loci examined in this study. The additional indel detected with MLVA in MSHR6955 from P811 demonstrates that there are at least 10 indels separating this strain from MSHR6137. Environmental samples obtained from the outbreak property water supply in May 2012 (MSHR7176) and again in June 2012 (MSHRs 7406 and 7446) showed indel variations compared with MSHR6137, consistent with the SNP phylogeny (see Fig. S1 and S2 in the supplemental material). Both MSHR7176 and MSHR7406 were identical to MSHR6137 by MLVA, but the former differed by 9 indels and 1 SNP, and the latter differed by 3 indels and 2 SNPs. MSHR7446 differed from MSHR6137 by a single MLVA repeat, 3 indels, and a single SNP (see Fig. S1 and S2). Thus, based on the observed genetic diversity of ST 325 strains in the outbreak property water supply, we cannot rule out the possibility that the strains infecting P741 and P811 were present in the environment but not sampled.
Seven additional ST 325 B. pseudomallei isolates from the same geographic region as the outbreak property were also analyzed to provide closely related reference material for our outbreak findings. Five isolates were from clinical cases presenting between 1994 and 2014 (P92, MSHR270; P429, MSHR2037; P646, MSHR4182; P690, MSHR4438; and P785, MSHR6354), and two were environmental isolates obtained from surveillance of nearby domestic water supplies (MSHR1539 and MSHR3554) in 2003 and 2009, respectively (Table 1). Three of the seven nonoutbreak strains matched MLVA-4 genotypes found in the outbreak cluster (types 223 and 244), two strains differed at a single MLVA locus from the predominant type 223 (types 430 and 437), and two strains differed at two loci from the predominant type (types 400 and 430) (Table 1).
Based on the SNP-indel phylogeny (Fig. 1), the isolate obtained from P785 (MSHR6354) was strikingly similar to an isolate from a nearby property's water supply (MSHR1539), differing by just three SNPs. These two isolates were distinct from all other isolates by a comparatively large number of SNPs (n ≥ 45) and formed their own distinct clade. The residential address of patient 785 is 2.4 km from the bore water supply where MSHR1539 was isolated (Fig. 1). These data suggest that the bore at P785's property and the bore where MSHR1539 was isolated tap into a shared aquifer. Interestingly, a detailed clinical history indicated that P785 may also have contracted melioidosis from swimming in a nearby dam, reflecting the common uncertainties of attributing infection to a specific event or location and potentially implicating a separate untreated water source in P785's infection (4); however, sampling of this dam was not conducted. The tight relatedness of all isolates in this study suggests that the underground aqueous environment in this area contains a relatively nondiverse group of ST 325 strains, which is a stark contrast to B. pseudomallei populations in soil where several different STs can be present in a single sample (26). Of note, although clearly separated by WGS, MSHR1539 shared the same MLVA profile as MSHR7446, an environmental isolate from the outbreak property. Thus, these clade differences were only apparent at a whole-genome level and were not detected using MLVA. We conclude from this observation that temporal sampling is an important consideration for accurate source attribution, particularly when interpreting MLVA data, due to potential issues of homoplasy.
During the outbreak of a potentially deadly environmentally acquired pathogen, it is vital to identify the source of infection to prevent additional cases. In the current study, WGS combined with detailed epidemiological information was required to identify the probable source of the melioidosis outbreak. Although MLST was a good predictor of whole-genome similarity and successfully excluded distantly related B. pseudomallei isolates as an infection source, it is a relatively expensive technique that lacks resolving power among closely related isolates. MLVA-4 provided an inexpensive and higher-resolution approach that was informative but suffered from homoplasy; thus, MLVA-4 results should be interpreted with caution. In this study, we showed for the first time that a combination of SNP and indel variants into a single phylogeny provided the best fit with the epidemiological data and should be explored in future studies. Our study demonstrated confident source attribution of two melioidosis cases originating from the same water supply; however, an exact match was not found, possibly due to within-host evolution or laboratory passage or, alternatively, to insufficient sampling of the environmental reservoir. To our knowledge, this work constitutes the most accurate source attribution of a highly recombinogenic pathogen and highlights issues using traditional typing methods. These data will inform future source-tracing investigations of melioidosis and other pathogens in a naturally occurring outbreak or in the unlikely event of an intentional release.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by project grant 1046812 from the Australian National Health and Medical Research Council. WGS data were partially funded by a grant from the Northern Territory Research and Innovation Board and the U.S. Department of Homeland Security Science and Technology Directorate award HSHQDC-10-C-US. Evan McRobb is supported by a university postgraduate research scholarship provided by Charles Darwin University.
We thank Vanessa Theobald for assistance with B. pseudomallei culturing and molecular typing and Glenda and Ian Harrington for their expertise in environmental sampling.
We thank Roche Diagnostics Australia for provision of the 454 data used in this study.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.03453-14.
REFERENCES
- 1.Limmathurotsakul D, Wongratanacheewin S, Teerawattanasook N, Wongsuvan G, Chaisuksant S, Chetchotisakd P, Chaowagul W, Day NP, Peacock SJ. 2010. Increasing incidence of human melioidosis in northeast Thailand. Am J Trop Med Hyg 82:1113–1117. doi: 10.4269/ajtmh.2010.10-0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wiersinga WJ, Currie BJ, Peacock SJ. 2012. Melioidosis. N Engl J Med 367:1035–1044. doi: 10.1056/NEJMra1204699. [DOI] [PubMed] [Google Scholar]
- 3.Bossi P, Tegnell A, Baka A, Van Loock F, Hendriks J, Werner A, Maidhof H, Gouvras G. 2004. Bichat guidelines for the clinical management of glanders and melioidosis and bioterrorism-related glanders and melioidosis. Euro Surveill 9:E17–E18. http://www.eurosurveillance.org/images/dynamic/em/v09n12/0912-238.pdf. [PubMed] [Google Scholar]
- 4.Currie BJ, Ward L, Cheng AC. 2010. The epidemiology and clinical spectrum of melioidosis: 540 cases from the 20 year Darwin prospective study. PLoS Negl Trop Dis 4:e900. doi: 10.1371/journal.pntd.0000900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parameswaran U, Baird RW, Ward LM, Currie BJ. 2012. Melioidosis at Royal Darwin Hospital in the big 2009-2010 wet season: comparison with the preceding 20 years. Med J Aust 196:345–348. doi: 10.5694/mja11.11170. [DOI] [PubMed] [Google Scholar]
- 6.Currie BJ, Mayo M, Anstey NM, Donohoe P, Haase A, Kemp DJ. 2001. A cluster of melioidosis cases from an endemic region is clonal and is linked to the water supply using molecular typing of Burkholderia pseudomallei isolates. Am J Trop Med Hyg 65:177–179. [DOI] [PubMed] [Google Scholar]
- 7.Inglis TJ, Garrow SC, Henderson M, Clair A, Sampson J, O'Reilly L, Cameron B. 2000. Burkholderia pseudomallei traced to water treatment plant in Australia. Emerg Infect Dis 6:56–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McRobb E, Kaestli M, Mayo M, Price EP, Sarovich DS, Godoy D, Spratt BG, Currie BJ. 2013. Melioidosis from contaminated bore water and successful UV sterilization. Am J Trop Med Hyg 89:367–368. doi: 10.4269/ajtmh.13-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mayo M, Kaestli M, Harrington G, Cheng AC, Ward L, Karp D, Jolly P, Godoy D, Spratt BG, Currie BJ. 2011. Burkholderia pseudomallei in unchlorinated domestic bore water, tropical Northern Australia. Emerg Infect Dis 17:1283–1285. doi: 10.3201/eid1707.100614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Currie BJ, Haslem A, Pearson T, Hornstra H, Leadem B, Mayo M, Gal D, Ward L, Godoy D, Spratt BG, Keim P. 2009. Identification of melioidosis outbreak by multilocus variable number tandem repeat analysis. Emerg Infect Dis 15:169–174. doi: 10.3201/eid1502.081036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Limmathurotsakul D, Holden MT, Coupland P, Price EP, Chantratita N, Wuthiekanun V, Amornchai P, Parkhill J, Peacock SJ. 2014. Microevolution of Burkholderia pseudomallei during an acute infection. J Clin Microbiol 52:3418–3421. doi: 10.1128/jcm.01219-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Price EP, Sarovich DS, Mayo M, Tuanyok A, Drees KP, Kaestli M, Beckstrom-Sternberg SM, Babic-Sternberg JS, Kidd TJ, Bell SC, Keim P, Pearson T, Currie BJ. 2013. Within-host evolution of Burkholderia pseudomallei over a twelve-year chronic carriage infection. mBio 4(4):e00388–13. doi: 10.1128/mBio.00388-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Godoy D, Randle G, Simpson AJ, Aanensen DM, Pitt TL, Kinoshita R, Spratt BG. 2003. Multilocus sequence typing and evolutionary relationships among the causative agents of melioidosis and glanders, Burkholderia pseudomallei and Burkholderia mallei. J Clin Microbiol 41:2068–2079. doi: 10.1128/JCM.41.5.2068-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaestli M, Mayo M, Harrington G, Ward L, Watt F, Hill JV, Cheng AC, Currie BJ. 2009. Landscape changes influence the occurrence of the melioidosis bacterium Burkholderia pseudomallei in soil in northern Australia. PLoS Negl Trop Dis 3:e364. doi: 10.1371/journal.pntd.0000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Novak R, Glass M, Gee J, Gal D, Mayo M, Currie BJ, Wilkins PP. 2006. Development and evaluation of a real-time PCR assay targeting the type III secretion system of Burkholderia pseudomallei. J Clin Microbiol 44:85–90. doi: 10.1128/JCM.44.1.85-90.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Currie B, Gal D, Mayo M, Ward L, Godoy D, Spratt BG, LiPuma J. 2007. Using BOX-PCR to exclude a clonal outbreak of melioidosis. BMC Infect Dis 7:68. doi: 10.1186/1471-2334-7-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sarovich DS, Price EP. 2014. SPANDx: a genomics pipeline for comparative analysis of large haploid whole genome re-sequencing datasets. BMC Res Notes 7:618. doi: 10.1186/1756-0500-7-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Milne I, Bayer M, Cardle L, Shaw P, Stephen G, Wright F, Marshall D. 2010. Tablet–next generation sequence assembly visualization. Bioinformatics 26:401–402. doi: 10.1093/bioinformatics/btp666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pearson T, Giffard P, Beckstrom-Sternberg S, Auerbach R, Hornstra H, Tuanyok A, Price EP, Glass MB, Leadem B, Beckstrom-Sternberg JS, Allan GJ, Foster JT, Wagner DM, Okinaka RT, Sim SH, Pearson O, Wu Z, Chang J, Kaul R, Hoffmaster AR, Brettin TS, Robison RA, Mayo M, Gee JE, Tan P, Currie BJ, Keim P. 2009. Phylogeographic reconstruction of a bacterial species with high levels of lateral gene transfer. BMC Biol 7:78. doi: 10.1186/1741-7007-7-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Smit B, Sarovich DS, Price EP, Mayo M, Theobald V, Kham C, Heng S, Thong P, Holden MTG, Parkhill J, Peacock SJ, Spratt BG, Jacobs JA, Vandamme P, Currie BJ. 2015. Whole-genome sequencing confirms that Burkholderia pseudomallei multilocus sequence types common to both Cambodia and Australia are due to homoplasy. J Clin Microbiol 53:323–326. doi: 10.1128/JCM.02574-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Price EP, Sarovich DS, Viberg L, Mayo M, Kaestli M, Tuanyok A, Foster JT, Keim P, Pearson T, Currie BJ. 2015. Whole-genome sequencing of Burkholderia pseudomallei isolates from an unusual melioidosis case identifies a polyclonal infection with the same multilocus sequence type. J Clin Microbiol 53:282–286. doi: 10.1128/JCM.02560-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Price EP, Hornstra HM, Limmathurotsakul D, Max TL, Sarovich DS, Vogler AJ, Dale J, Ginther JL, Leadem B, Colman RE, Foster JT, Tuanyok A, Wagner DM, Peacock SJ, Pearson T, Keim P. 2010. Within-host evolution of Burkholderia pseudomallei in four cases of acute melioidosis. PLoS Pathog 6:e1000725. doi: 10.1371/journal.ppat.1000725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.U'Ren JM, Schupp JM, Pearson T, Hornstra H, Friedman CL, Smith KL, Daugherty R, Rhoton SD, Leadem B, Georgia S. 2007. Tandem repeat regions within the Burkholderia pseudomallei genome and their application for high resolution genotyping. BMC Microbiol 7:23. doi: 10.1186/1471-2180-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chantratita N, Wuthiekanun V, Limmathurotsakul D, Vesaratchavest M, Thanwisai A, Amornchai P, Tumapa S, Feil EJ, Day NP, Peacock SJ. 2008. Genetic diversity and microevolution of Burkholderia pseudomallei in the environment. PLoS Negl Trop Dis 2:e182. doi: 10.1371/journal.pntd.0000182. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.