Abstract
In order to limit the interference of HIV-1 cellular nucleic acids in estimating viral load (VL), the feasibility of leukodepletion of a small whole-blood (WB) volume to eliminate only leukocyte cell content was investigated, using a selection of filters. The efficacy of leukocyte filtration was evaluated by counting, CD45 quantitative PCR, and HIV-1 DNA quantification. Plasma HIV-1 was tested by real-time reverse transcription (RT)-PCR. A specific, miniaturized filter was developed and tested for leukocyte and plasma virus retention, WB sample dilution, and filtration parameters in HIV-1-spiked WB samples. This device proved effective to retain >99.9% of white blood cells in 100 μl of WB without affecting plasma VL. The Samba sample preparation chemistry was adapted to use a leukodepleted WB sample for VL monitoring using the point-of-care Samba-1 semiautomated system. The clinical performance of the assay was evaluated by testing 207 consecutive venous EDTA WB samples from HIV-1-infected patients attending a CD4 testing clinic. Most patients were on antiretroviral treatment (ART), but their VL status was unknown. Compared to the Roche Cobas AmpliPrep/Cobas TaqMan HIV-1 test, the new Samba assay had a concordance of 96.5%. The use of the Samba system with a VL test for WB might contribute to HIV-1 ART management and reduce loss-to-follow-up rates in resource-limited settings.
INTRODUCTION
As of 2010, there were a recorded 6,650,000 individuals receiving antiretroviral therapy (ART) in low- and middle-income countries (1). As diagnosis of HIV-1 infection and access to treatment increases, so does the need for effective monitoring. Monitoring results of ART to assess the development of resistance and the need for initiation of second-line ART can be done with clinical, immunological, or virological markers. Monitoring of viral suppression is necessary to be able to identify treatment failure and initiate a timely switch to second-line therapy. The WHO has recently reduced the threshold for defining virological failure from 5,000 copies/ml to 1,000 copies/ml in plasma samples (2).
Currently, the monitoring of HIV-1 by viral load (VL) testing in resource-limited settings is performed largely in centralized laboratories by PCR-based methods using plasma samples. Any delay in receiving the results increases loss-to-follow-up (LTFU) rates. Dried blood spots (DBS) have in many instances been employed as an alternative sample type to plasma. DBS are attractive for resource-limited settings because they are cheap, require minimal training for collection, can be stored easily, and are readily transportable, even in harsh environmental conditions. However, the appropriateness of DBS for VL testing has been questioned. Different studies suggest that DBS overquantify VL compared to plasma (3, 4), that there is a good correlation between DBS and plasma (5, 6), and also that DBS underquantify VL compared to plasma (7, 8). Depending on the assay used, the proviral DNA present in DBS samples interferes with RNA quantification, and RNA in DBS might not be sufficiently stable. In addition, DBS being tested centrally does little to improve LTFU rates (9).
The proposed solution for rural sites in resource-limited settings is to test patients at their point of care (POC). Although centralized testing is cost-effective, in areas with a high prevalence of HIV infections, decentralized testing is required to care for the greatest number of patients and to limit LTFU (10). In response to these issues, POC tests have been developed. The possibility of using whole blood (WB) for HIV-1 VL monitoring, as opposed to plasma, has been considered. The availability of a simpler method, including less equipment- and electricity-intensive centrifugation, would benefit POC sites. A blood sample could be collected using finger or heel prick and processed without additional instrumentation before use with a POC VL assay.
Whole-blood leukodepletion filters are widely used in blood transfusion centers and are designed to accommodate an input of ∼450 ml of WB, with filtration being completed under gravity. These filters typically deplete WB units of 3 to 4 logs of nucleated cells but retain around 10% of red blood cells (RBCs) and cause a 2-log reduction in platelets (11), although some improvements have recently been made.
The aim of this investigation was to assess the possibility of using a miniaturized version of WB filter systems to deplete a small WB sample of HIV-1-infected cells, so that the filtrate may be used to complete a VL assessment without the interference of intracellular proviral DNA or viral RNA. An investigation into the development and feasibility of such a methodology was conducted, keeping in mind usage at POC sites in resource-limited settings.
MATERIALS AND METHODS
Filter materials.
Filter materials were donated by Macopharma (Tourcoing, France) following the establishment of a material transfer agreement. Leukosorb medium was purchased as potential filter material from Pall Corporation (Port Washington, NY, USA). Filter sheets were cut using a Harris Unicore cutter (Redding, CA, USA) and cutting mat to fit the 8-mm internal diameter of the Samba plastic column. Layers of disks were then assembled and fitted into the Samba column by using the Samba column assembly jig that pushes the filter disks to the bottom of the column by air pressure and secured the material placement with a retaining o-ring (Fig. 1).
FIG 1.
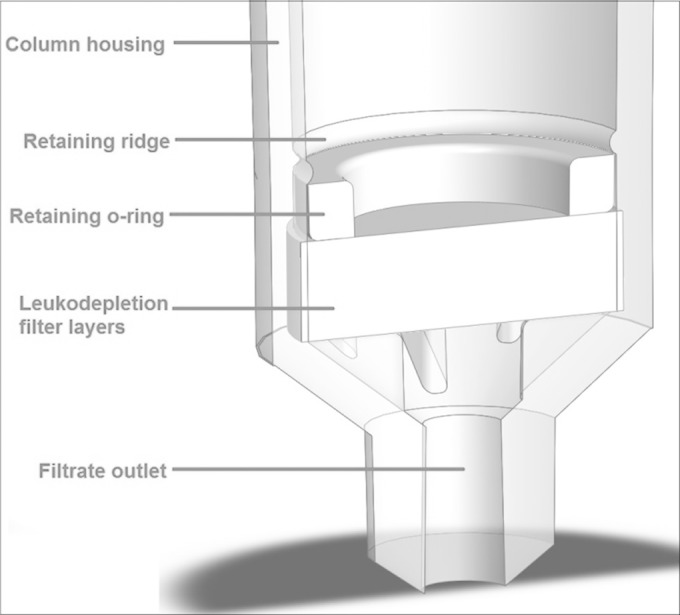
Cross section of the Samba filter column. The column was originally designed to house a silica solid phase for nucleic acid extraction but has been repurposed to hold the layers of leukodepletion materials for filtering WBCs from a WB sample. Diluted WB is passed through the filter using air pressure.
Macopharma supplied five different filter materials of different compositions and pore sizes, along with two recommended configurations. Membranes were either of polypropylene with a pore size of 16 μm or of polybutylene terephthalate with a pore size of either 9 or 6 μm. Filters assembled from Pall's Leukosorb medium consisted of a single type of material, packed into the column in up to 10 layers. The Samba platform uses syringe pumps to apply air pressure to move liquids through a solid phase. The air pressure employed to move WB samples through the fitted column was low, as was the pipetting speed of the instruments, so as to minimize cell lysis during sample handling.
Separation and quantification of WBC.
Several methods were jointly used to quantify the effect of filtration: WBC count by flow cytometry, CD45 gene quantification by real-time PCR (qPCR), and quantification of HIV-1 proviral DNA by qPCR.
WBC count.
WBC counts were performed on a small number of selected samples using a flow cytometric assay, the BD Leucocount kit (Becton Dickinson, Oxford, United Kingdom). Testing was completed by the staff of the Components Development Laboratory (National Health Service Blood & Transplant, Brentwood, United Kingdom).
CD45+ cell detection by reverse transcription (RT) and qPCR.
For evaluations of relative leukocyte retention, CD45 mRNA was quantified by qPCR. This method detected both white blood cells (WBC) in WB and spiked HIV-1-infected cells of the 8E5 cell line. The introduction of additional cells was performed so that cell numbers were at detectable levels even in greatly leukodepleted samples.
Relative CD45+ cell concentrations were assessed using the TaqMan gene expression PCR assay (Life Technologies, Paisley, United Kingdom; primer/probe set Hs04189704_m1) according to the instructions supplied by the manufacturer for a 50-μl reaction volume. The RT step required for this assay was completed using the SuperScript III first-strand synthesis system (Life Technologies) according to the manufacturer's specifications. Following RT, reaction samples were diluted 1:4 by the addition of 80 μl of water. The CD45 qPCR was completed using the Stratagene Mx3000 instrument (Agilent, La Jolla, CA, USA).
Standards and samples.
The study was conducted in three phases: in vitro spiking of calibrated reagents into normal WB, testing of calibrated panel samples, and comparative testing of clinical venous WB samples in the field. For these purposes, human WB negative for viral markers was obtained from Sera Laboratories International (Haywards Heath, United Kingdom) or Innovative Research (Novi, USA). Negative pooled human plasma was obtained from the Diagnostics Development Unit (DDU; Department Hematology, University of Cambridge), from residual samples collected during a trial in 2009.
To study the relative retention of HIV-1-infected cells, viral-marker-negative WB or plasma samples were spiked with 8E5 cells (purchased from American Type Culture Collection [ATCC], Manassas, VA, USA), a cell line containing one copy of proviral HIV-1 DNA per cell (12). Cells remaining in samples following treatment were quantified by qPCR targeting the 5′ noncoding region of the proviral DNA. For HIV-1 RNA studies, negative WB or plasma samples were spiked with either culture supernatant of HIV-1 subtype B LAI or the WHO 3rd HIV International Standard (subtype B) (National Institute of Biological Standards & Controls [NIBSC], Potter's Bar, United Kingdom). The reference standard for quantifying CD45+ cells was a known number of 8E5 cells. Cell concentrations from HIV-1 culture supernatants were calculated using a hemocytometer, and then the volume corresponding to 1 × 106 cells was aliquoted, the cells were pelleted, and the supernatant was removed prior to freezing. One pellet was resuspended in 1 ml of cell-free negative plasma as the standard sample. The reference HIV-1 RNA standard was a secondary subtype B viral standard calibrated by the Artus HIVirus-1 RG RT-PCR kit (testing completed by the Public Health Laboratory [PHL], Cambridge, United Kingdom) in copies/milliliter. HIV-1 proviral DNA was quantified against a calibrated plasmid-derived standard generated in-house.
HIV-1 proviral DNA detection by qPCR.
The 50-μl qPCR mixture consisted of 10 μl template nucleic acid in 1× buffer with 4 mM MgCl2, 0.8 mM deoxynucleoside triphosphates (dNTPs), 0.4 μM each target-specific primer, 0.2 μM probe, and 0.5 μl SureStart Taq polymerase. The reaction mixtures were incubated at 50°C for 10 min and 95°C for 10 min, followed by 40 cycles of 95°C for 30 s and 60°C for 1 min. Assays were performed using the Stratagene Mx3000 instrument. The standard was used as described above.
RT-qPCR for HIV-1 RNA quantification.
For HIV-1 RNA quantification, in-house reverse transcription (RT)-qPCR was used, as previously described (13).
Samba semiquantitative assay.
The Samba assay was used as previously described using 200 μl of filtered, diluted WB as the input instead of plasma (14). All other steps and reagents were identical to the published method. For internal purposes, the dipstick test line signal was visually semiquantified on a scale from 0 to 5 printed on a reference card. A signal of 0.5 was doubtfully reactive, signals of 1 and 2 were weak, and signals above 2 were strong. It was recommended to repeat testing when the Samba test line signal was weak, but sample volume was limited in this study to the extent that systematic repeat testing was not permitted.
Clinical samples.
HIV-1-infected blood donor plasma samples (subtype C) were donated by the Namibia National Blood Centre (Windhoek, Namibia) and were used to spike WB with a known concentration of HIV-1 in plasma (14). These samples were diluted in WB to reach a viral load ranging between 102 and 104 copies/ml in order to test mimicked infected patient blood in six replicates for each dilution, constituting a panel of 120 aliquots to be tested for VL with the Roche Amplicor VL assay and the semiquantitative Samba-1 VL using a cutoff of 3 ± 0.3 log copies/ml (14).
The efficacy of the leukodepletion method in assessing VL in patient samples was determined by comparing data generated by the Roche Cobas AmpliPrep/Cobas TaqMan HIV-1 assay applied to plasma and Samba applied to filtered WB on the same random WB patient samples. Clinical WB venous samples were obtained from patients attending routine CD4/viral load monitoring services at the support center of the KEMRI/CDC Research and Public Health Collaboration (Kisumu, Kenya) and tested with the Samba WB semiquantitative assay at the KEMRI/CDC HIV-R laboratory. WB samples were leftover venous blood samples. Aliquots were used as WB for Samba testing before being centrifuged to produce plasma for VL testing using the KEMRI/CDC standard protocol (2,200 × g, 5 min) for Roche CAP/CTM assay version 2.0 (550-μl plasma input). Testing results were mutually blinded. At the end of the study, discrepant samples were identified and, when sample volume permitted, the Samba test was repeated and tested by the Abbott RealTime HIV-1 test, the result of which acted as a tiebreaker between Roche and Samba results.
Three HIV-1-positive clinical samples with undetectable VL (from consented patients on ART) were purchased from Sera Laboratories International (Haywards Heath, United Kingdom). VL status was confirmed by testing at the PHL, Cambridge, United Kingdom, as indicated above.
Statistical analysis.
Statistical analysis of data was completed using Microsoft Office Excel 2007 software tools and PRISM software. The ninety-five-percent confidence interval was derived using binomial exact proportion. Categorical variables were compared using Fisher's exact test and, for continuous variables, the nonparametric Mann-Whitney test. In all statistical analyses, a P value of <0.05 was regarded as significant. A Kappa coefficient was used to determine statistical significance. The 95% confidence intervals (CIs) were calculated using VassarStats (http://vassarstats.net).
RESULTS
Leukodepletion filters and optimization of filtration conditions.
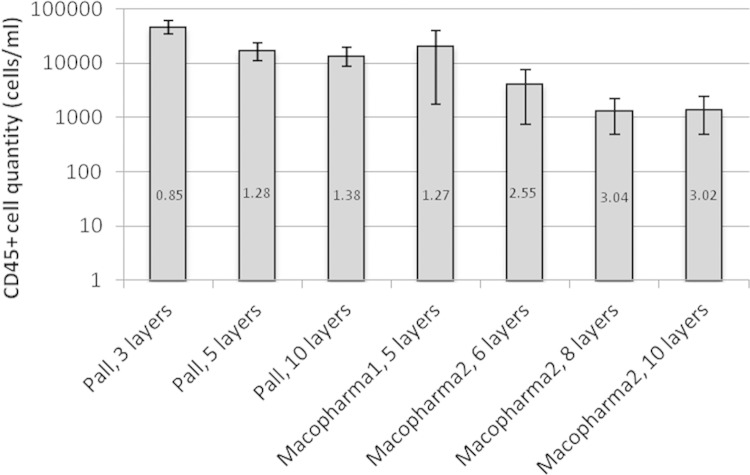
Macopharma filter materials were supplied with a recommended filter composition by the manufacturer. Pall filters were comprised of only one material (Leukosorb). Preliminary testing revealed that an increased number of filter layers resulted in an increased efficacy of CD45+ cell removal. The necessity for a tradeoff between the number of filter layers and the amount of sample lost to the dead volume of the filter also became apparent. To maximize the volume of recovered sample, blood samples were diluted 1:2 to 1:4 with phosphate-buffered saline (PBS). The three dilutions tested did not significantly affect WBC retention or HIV-1 RNA recovery. A 300-μl final volume (100 μl WB and 200 μl PBS) was selected as the limiting sample dilution with acceptable dead volume retention. It resulted that a filter configuration consisting of eight layers of Macopharma materials gave the highest level of leukodepletion (Fig. 2) with an acceptable dead volume (≤50 μl). All subsequent testing was done with this filter format. An air pressure of 50 μl/s was optimal to pass the sample through the filter, with faster speeds reducing filtration efficacy and lower speeds increasing the amount of sample lost to the dead volume of the filter.
FIG 2.
Leukoreduction performance of seven filter configurations as assessed by CD45 qPCR. The number given in each bar is the log reduction of WBCs for each filter type compared to diluted WB spiked with 100,000 8E5 cells/ml. For each filter type, data were collected from three experiments. Error bars represent the standard deviations. Macopharma 1 and Macopharma 2 are two different arrangements of filters.
Assessment of efficacy.
The above-given conditions were used to test the performance of the filter in terms of removal of WBCs and retention of plasma viral particles. The level of WBC reduction in WB samples spiked with 100,000 8E5 cells/ml was approximately 3 logs when assessed with quantification of CD45 genes or counting by flow cytometry (Table 1). Results obtained with HIV-1 DNA quantification were significantly lower than with the other two methods (P < 0.01) due to substantial breakage of 8E5 cells during filtration, since HIV-1 DNA was detected in the filtrate.
TABLE 1.
Efficacy of diluted whole-blood cell removal by centrifugation and filtration, assessed by three different methods
| WBC removal method | Reduction in WBC concna |
|||||
|---|---|---|---|---|---|---|
| RT and CD45 qPCR |
HIV-1 qPCR |
WBC count |
||||
| Log difference | % | Log difference | % 8E5 | Log difference | % | |
| Centrifugation | 2.74 | 99.66 | 1.70 | 98.02 | 3.06 | 99.91 |
| Filtration | 3.15 | 99.93 | 1.86 | 98.63 | 3.48 | 99.97 |
PCR data were collected over three experiments for each type. Flow cytometric analysis was performed on two experiments, averaged in this table. Reduction in WBC concentration is expressed in log difference and percentage of initial values.
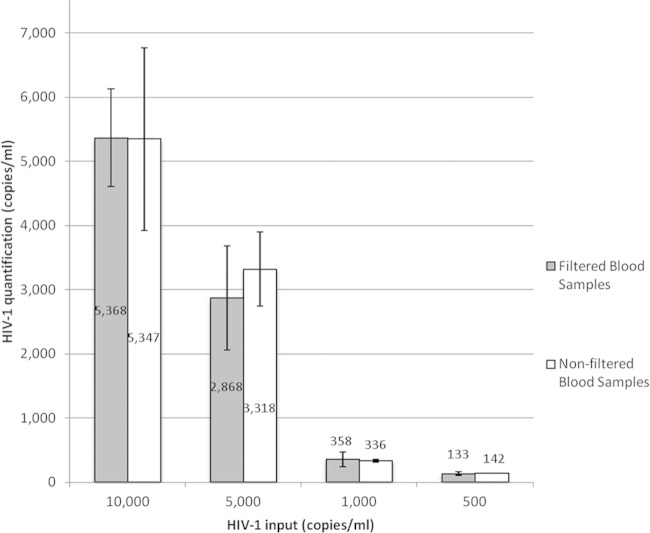
The level of retention of plasma HIV-1 viral particles was assessed to determine the impact of filtration on the VL. WB control samples were spiked with a concentration range of 500 to 10,000 copies/ml of LAI culture supernatant containing HIV-1 subtype B. The HIV-1 content was assessed pre- and postfiltration by RT-qPCR. Filtration did not retain a significant amount of virus over the range tested (Fig. 3).
FIG 3.
Comparison of free virus levels between filtered and nonfiltered blood samples. HIV-1 concentrations tested in the range of 500 to 10,000 copies/ml, assessed by HIV-1 RT-qPCR. Data are averaged from three experiments. Error bars represent the standard deviation. Numbers inside the bars indicate the VL calculated from the in-house RT-qPCR results.
Inclusion of filtration into the VL Samba-1 assay.
The leukodepletion filter was used as a sample preparation step preceding the Samba point-of-care nucleic acid-based assay 0 (14, 15). The Samba-1 platform had been previously used to monitor HIV-1 RNA load from WB with a cutoff of 103 ± 0.3 log copies/ml as described (Samba-1 semiquantitative HIV-1 test for WB) (14). The 120-member panel of WB spiked with subtype C-infected plasma samples calibrated with CAP/CTM assay was tested with the semiquantitative Samba-1 assay after filtration. The analytical accuracy of the protocol was 99.2%, as 1/120 samples was incorrectly identified. This sample was diluted to contain 303 copies/ml but was identified as >1,000 copies/ml by the Samba-1 test with a weak reactive signal. The other five replicates of this dilution were correctly identified as <1,000 copies/ml. In addition, three HIV-1-infected donor WB samples antibody positive but RNA negative were tested with and without filtration followed by the Samba-1 assay. HIV-1 VL was confirmed as being <50 copies/ml with RT-qPCR. Three replicates from each sample were tested using the Samba-1 semiquantitative HIV-1 test for WB, before and after filtration. All unfiltered samples were Samba positive (>1,000 copies/ml) but negative after WB filtration, suggesting the presence of HIV-1 proviral DNA in the unfiltered samples. This result was confirmed with qPCR for HIV-1 DNA.
Clinical evaluation.
Of the 207 clinical samples tested, 198 were carried forward to analysis; nine were excluded based on there being insufficient sample available or inconclusive testing. In all instances where the VL was undetectable by the Roche assay, the Samba assay identified the corresponding blood sample as containing <1,000 copies/ml. Considering the accepted accuracy of the test as 3 ± 0.3 log10 copies/ml, 30/33 samples with a VL of >500 copies/ml by CAP/CTM assay were correctly identified by Samba-1 (Table 2). Eleven samples were discrepant, two of them with strong Samba signal (VL 450 and 263 IU/ml). The remaining nine samples had weak signal, among which six had sufficient volume to repeat Samba testing. Four of them were negative on repeat testing and were reclassified as concordant. The other two samples with weak signal were repeat tested and were classified as true discrepant together with the five samples with weak or strong signal that could not be repeat tested. These seven samples had VLs ranging between 78 and 396 copies/ml and were confirmed to contain <500 copies/ml by the Abbott test (Table 3). In all cases where tiebreaker testing was done, the Abbott and Roche assays were in agreement, although for the same sample the quantification given by the Abbott test was lower than that given by Roche. As expected, samples with a VL between 500 and 200 copies/ml were distributed below or above the 1,000-copy cutoff. All samples of >2,000 copies/ml with the CAP/CTM assay were reactive with Samba. Overall, the Samba-1 semiquantitative HIV-1 test for WB showed a concordance with the CAP/CTM assay of 96.5% (95% confidence interval [CI] of 92.2 to 97.9).
TABLE 2.
Comparison of Samba-filtered WB and Roche CAP/CTM assays for evaluation of patient viral load
| Parameter | Viral load (copies/ml) | Roche CAP/CTM result by viral load (copies/ml) |
|||
|---|---|---|---|---|---|
| Negative | <500 | 500–2,000 | >2,000 | ||
| Samba-filtered WB | <1,000 | 115 | 43 | 3 | |
| >1,000 | 0 | 7 | 6 | 24 | |
| Total no. of patients | 115 | 50 | 9 | 24 | |
| % concordant results | 100 | 86 | 100 | 100 | |
| % discordant results | 0 | 14 | 0 | 0 | |
TABLE 3.
Detailed results of the seven confirmed discordant samples between Samba-1-filtered, Roche CAP/CTM, and Abbott qPCR
| Sample ID | VL Roche result (copies/ml) | SAMBA signal strengtha |
Abbott VL result (copies/ml) | |
|---|---|---|---|---|
| Initial | Repeat | |||
| S7 | 78 | 1 | ND | Fail |
| S8 | 450 | 5 | ND | Neg |
| S39 | 128 | 2 | ND | Neg |
| S51 | 262 | 3 | ND | <40 |
| S148 | 168 | 0.5 | ND | Fail |
| S164 | 396 | 2 | 2 | <40 |
| S174 | 372 | 1 | 0.5 | <40 |
Samba signal strength is estimated within a range between 0 and 5 grades determined against a scale card. A grade of 0.5 is barely visible, grades 1 and 2 are weak, and grades above 2 are clearly visible.
DISCUSSION
The use of leukodepletion filters on a miniature scale (Fig. 1) proved to be a simple and effective method of removing WBCs from a venous WB sample (Fig. 2). The chosen filter configuration achieved a reduction in WBCs of >99.9% by two separate methods (Table 1). Assessing the efficacy of filtration with the spiked 8E5 cell WB was inappropriate since after both filtration and centrifugation, cell breakage resulted in HIV-1 proviral DNA found in plasma and filtrate. The leukoreduction filter did not appear to retain any significant amounts of free virus (Fig. 3), thus meeting the objectives of the study. However, it is noted that the whole development experiments were conducted with EDTA whole blood spiked with either HIV-1 culture supernatant or infected plasma samples when the intended use is for capillary blood. Clinical samples were also EDTA venous blood. Although in both types of situations similar results were obtained, the efficacy of the method applied to finger/heel prick-collected EDTA blood remains to be ascertained. However, a study using CAP/CTM reported that there was a significant difference in VL quantification between venous and capillary whole blood (16).
The Samba semiquantitative HIV-1 test for WB has been shown to correlate well with the commercial gold-standard VL monitoring assay, with 96.5% concordance with the Roche Cobas AmpliPrep/Cobas TaqMan assay. The Samba test had 100% accuracy with samples with undetectable VL by the Roche assay, confirming that the leukoreduction step employed does not leave enough HIV-1 DNA to perceptibly interfere with Samba. For the Samba platform using the NASBA system, both HIV-1 proviral DNA and viral RNA are detected, requiring cell depletion for accurate VL assessment. Where DBS have been trialed as an alternative sample for VL monitoring, it was suspected that proviral DNA was being coamplified with viral particle RNA. One study on the feasibility of using DBS with the Abbott RealTime VL assay found that in 9% of samples with a plasma VL of <40 copies/ml, the corresponding DBS sample had a detectable nucleic acid content (17). Furthermore, this study demonstrated with a specific RT reaction that these DBS samples had undetectable RNA levels, strongly suggesting that proviral DNA was the source of reactivity. More details regarding the downsides of DBS usage for VL measurement have been previously published by our group (14).
There is limited literature on the use of WB for VL monitoring, except with DBS. However, data on the performance of the Alere total HIV-1/2 RNA test, an assay intended for POC monitoring of VL from a very small DBS sample, were recently published. These data revealed that for samples with a plasma VL of ≤5,000 copies/ml, the test had poor correlation with the Roche CAP/CTM plasma VL assay (18). In this case, even though WB volume was small, some samples appeared to contain contaminating proviral DNA, which, added to intracellular RNA, contributed to an overestimation of VL. In a study comparing the performance of CAP/CTM for VL quantification in plasma and venous WB or DBS, it was shown that there was no correlation in VL values between the two sample types below a VL of 3,000 copies/ml, and 60% of plasma VL-negative samples were positive with WB (16). These differences were attributed to the interference of proviral DNA that remained significant with relatively low VL levels. In another study comparing plasma and DBS for VL quantification with CAP/CTM, cellular DNA interference was also shown (19). Not unexpectedly, the same problems with VL quantification in WB were found in liquid WB and DBS. These data sets further suggest that removing the cellular HIV-1 nucleic acids is important to obtain accurate VL estimation with current commercial assays. In addition, overestimation of VL in patients on ART might lead to unnecessary changes of therapeutic regimens to second-line ART.
Despite the good performance in samples with undetectable VL, the Samba assay was reactive (≥1E3 ± 0.3 log copies/ml), with several samples containing <500 copies/ml according to both Roche and Abbott VL assays (Table 2). In most cases, these discrepant samples showed a weak signal, and following the general recommendation of repeating the assay in such circumstances, several samples repeated negative. The Samba test line signal strength does not strictly correlate to the nucleic acid content of the sample. Weak test lines are undesirable because of the increased frequency of misinterpretation by the user, especially from staff with inadequate training (20). Although a number of clinical samples tested were from patients attending a baseline CD4 assessment (and therefore not yet on ART), the majority of samples were from individuals already on ART. The Samba chemistry has been assessed for inhibition or interference from ARV compounds in clinical sample testing, and no evidence of such interference was observed (14). Two hypothetical explanations can then be offered. First is that not all cellular nucleic acids are being eliminated from these samples and may contribute to an overestimation of the VL. This is unlikely, since none of the samples from infected patients VL negative with the CAP/CTM assay were reactive with Samba-filtered WB. Alternatively, the detuning of Samba semiquant to set up the 1,000-copy cutoff established for plasma samples might need to be slightly adjusted when using WB samples (14). The latter hypothesis is currently being examined in the DDU laboratory.
The aim of this project was to develop a simple method of assessing HIV-1 VL in WB samples. The simplicity of the result above or below a single working cutoff appears to be an advantage for clinicians or health workers in the field who do not have to agonize over borderline results of a quantitative test. We conclude that leukodepletion by filtration of the blood sample is a feasible method to reduce the CD4+ cell content to a level sufficiently low to play a negligible role in the quantification of a patient's VL. As the leukodepletion filter used in this study has been easily incorporated into the current Samba instrumentation, showing that no overtly complex technology is required, the method is applicable in a POC context.
The method has been demonstrated only in the Samba-1 instrument and should be transferred to the Samba-2 instrument upon its release. The lack of available cartridge space in the Samba-1 Sambaprep cartridge results in the requirement for manual steps, which would increase the risk of contamination and user error. The overestimation of the VL in some clinical samples may be remedied upon transfer to the Samba-2 instrument, when the cutoff should be readjusted. It is a weakness of this preliminary study that the WB clinical data presented here were obtained by venipuncture, and so the performance must in the future be confirmed in capillary blood samples obtained through finger prick in adults and heel prick in infants.
In conclusion, commercial WB leukodepletion filters can be miniaturized for the effective removal of >99.9% of WBCs from small anticoagulated WB samples. We have shown this to be applicable to the quantification of HIV-1 VL by making negligible interference from cellular nucleic acids. The resultant development of a Samba assay adapted for use with leukodepleted blood has been shown to be comparable to the performance of the gold-standard plasma VL assay. The implementation of this assay in resource-limited settings could increase access to POC VL testing, which improves HIV treatment management through earlier identification of virological failure and a reduction in loss-to-follow-up rates.
ACKNOWLEDGMENTS
We thank the team of the NHSBT Components Development Laboratory, Brentwood, headed by R. Cardigan, for performing the flow cytometric testing. Thanks to S. Parmar of the Public Health Laboratory, Cambridge, for completion of the VL quantifications by Qiagen.
This project has been funded in part with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contract no. HHSN272200800014C and HHSN2722009000028C.
T.V. is head of research and development for Macopharma, which provided filters. H.L. is CEO of diagnostics for Real World, which produces the Samba assay.
REFERENCES
- 1.WHO. 2011. Global HIV/AIDS response: epidemic update and health sector progress towards universal access: progress report 2011. WHO, Geneva, Switzerland. [Google Scholar]
- 2.WHO. 2013. Consolidated guidelines on the use of antiretroviral drugs for treating and preventing HIV infection: recommendations for a public health approach. WHO, Geneva, Switzerland. [PubMed] [Google Scholar]
- 3.Monleau M, Montavon C, Laurent C, Segondy M, Montes B, Delaporte E, Boillot F, Peeters M. 2009. Evaluation of different RNA extraction methods and storage conditions of dried plasma or blood spots for human immunodeficiency virus type 1 RNA quantification and PCR amplification for drug resistance testing. J Clin Microbiol 47:1107–1118. doi: 10.1128/JCM.02255-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waters L, Kambugu A, Tibenderana H, Meya D, John L, Mandalia S, Nabankema M, Namugga I, Quinn TC, Gazzard B, Reynolds SJ, Nelson M. 2007. Evaluation of filter paper transfer of whole blood and plasma samples for quantifying HIV RNA in subjects on antiretroviral therapy in Uganda. J Acquir Immune Defic Syndr 46:590–593. doi: 10.1097/QAI.0b013e318159d7f4. [DOI] [PubMed] [Google Scholar]
- 5.Kane CT, Ndiaye HD, Diallo S, Ndiaye I, Wade AS, Diaw PA, Gaye-Diallo A, Mboup S. 2008. Quantitation of HIV-1 RNA in dried blood spots by the real-time Nuclisens EasyQ HIV-1 assay in Senegal. J Virol Methods 148:291–295. doi: 10.1016/j.jviromet.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 6.Rottinghaus EK, Ugbena R, Dialo K, Bassey O, Azeez A, Devos J, Zhang G, Aberle-Grasse J, Nkengasong J, Yang C. 2012. Dried blood spot specimens are suitable alternative sample type for HIV-1 viral load measurement and drug resistance genotyping in patients receiving first-line antiretroviral therapy. Clin Infect Dis 54:1187–1195. doi: 10.1093/cid/cis015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garrido C, Zahonero N, Cprral A, Arredondo M, Soriano V, de Mendoza C. 2009. Correlation between human immunodeficiency virus type 1 (HIV-1) RNA measurements obtained with dried blood spots and those obtained with plasma by use of Nuclisens EasyQ HIV-1 and Abbott RealTime HIV load tests. J Clin Microbiol 47:1031–1036. doi: 10.1128/JCM.02099-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johannessen A, Garrido C, Zahonero N, Sandvik L, Naman E, Kivuyo SL, Kasubi MJ, Gendersen SG, Bruun JN, de Mendoza C. 2009. Dried blood spots perform well in viral load monitoring of patients who receive antiretroviral treatment in rural Tanzania. Clin Infect Dis 49:976–981. [DOI] [PubMed] [Google Scholar]
- 9.Lofgren SM, Morrisey AB, Chevallier CC, Malabeja AI, Edmonds S, Amos B, Sifuna DJ, von Seidlein L, Schimana W, Stevens WS, Bartlett JA, Crump JA. 2009. Evaluation of a dried blood spot HIV-1 RNA program for early infant diagnosis and viral load monitoring at rural and remote healthcare facilities. AIDS 23:2459–2466. doi: 10.1097/QAD.0b013e328331f702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerlach J, Boyle D, Domingo G, Weigl B, Free M. 2008. Increased access to diagnostic tests for HIV case management. PATH Publications, Seattle, WA. [Google Scholar]
- 11.Prowse CV, Hornsey VS, Drummond O, Mac Gregor IR, Pepper DS, Barclay GR, Bethel H, Walker B, Barnard G, Kirby L, Hope J. 1999. Preliminary assessment of whole-blood, red-cell and platelet-leucodepleting filters for possible induction of prion release by leucocyte fragmentation during room temperature processing. Br J Haematol 106:240–247. doi: 10.1046/j.1365-2141.1999.01530.x. [DOI] [PubMed] [Google Scholar]
- 12.Lightfoote MM, Coligan JE, Folks TM, Fauci AS, Martin MA, Venkatesan S. 1986. Structural characterization of reverse transcriptase and endonuclease polypeptides of the acquired immunodeficiency syndrome virus. J Virol 60:771–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Candotti D, Temple J, Owusu-Ofori S, Allain JP. 2004. Multiplex real-time quantitative RT-PCR assay for hepatitis B virus, hepatitis C virus, and human immunodeficiency virus type 1. J Virol Methods 118:39–47. doi: 10.1016/j.jviromet.2004.01.017. [DOI] [PubMed] [Google Scholar]
- 14.Ritchie AV, Ushiro-Lumb I, Edemaga D, Joshi HA, De Ruiter A, Szumilin E, Jendrulek I, McGuire M, Goel N, Sharma PI, Allain JP, Lee HH. 2014. Samba HIV semiquantitative test, a new point of care viral load monitoring assay for resource-limited settings. J Clin Microbiol 52:3377–3383. doi: 10.1128/JCM.00593-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee HH, Dineva MA, Chua YL, Ritchie AV, Ushiro-Lumb I, Wisnievski CA. 2010. Simple amplification-based assay: a nucleic acid-based point-of-care platform for HIV-1 testing. J Infect Dis 201(Suppl 1):S65–S72. doi: 10.1086/650385. [DOI] [PubMed] [Google Scholar]
- 16.Steinmetzer K, Seidel T, Stallmach A, Ermantraut E. 2010. HIV load testing with small samples of whole blood. J Clin Microbiol 48:2786–2792. doi: 10.1128/JCM.02276-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arredondo M, Garrido C, Parkin N, Zahonero N, Bertagniolio S, Soriano V, de Mendoza C. 2012. Comparison of HIV-1 RNA measurements obtained by using plasma and dried blood spots in the automated Abbott real-time viral load assay. J Clin Microbiol 50:569–572. doi: 10.1128/JCM.00418-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jani IV, Meggi B, Mabunda N, Vubil A, Sitoe NE, Tobaiwa O, Quevedo JI, Lehe JD, Loquiha O, Vojnov L, Peter TF. 2014. Accurate early infant HIV diagnosis in primary health clinics using a point of care nucleic acid test. J Acquir Immune Defic Syndr 67:e1–e4. doi: 10.1097/QAI.0000000000000250. [DOI] [PubMed] [Google Scholar]
- 19.Ouma KN, Basavaraju SV, Okonji JA, Williamson J, Thomas TK, Mills LA, Nkengasong JN, Zeh C. 2013. Evaluation of quantification of HIV-1 RNA viral load in plasma and dried blood spots by use of the semiautomated Cobas Amplicor assay and the fully automated Cobas Ampliprep/Taqman assay, version 2.0, in Kisumu, Kenya. J Clin Microbiol 51:1208–1218. doi: 10.1128/JCM.03048-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiu YH, Ong J, Walker S, Kumlawati J, Gartinah T, McPhee DA, Dax EM. 2011. Photographed rapid HIV test results pilot novel quality assessment and training schemes. PLoS One 6:e18294. doi: 10.1371/journal.pone.0018294. [DOI] [PMC free article] [PubMed] [Google Scholar]