Abstract
The discrimination of Leishmania species from patient samples has epidemiological and clinical relevance. In this study, different gene target PCR-restriction fragment length polymorphism (RFLP) protocols were evaluated for their robustness as Leishmania species discriminators in 61 patients with cutaneous leishmaniasis. We modified the hsp70-PCR-RFLP protocol and found it to be the most reliable protocol for species identification.
TEXT
Human infections by Leishmania spp. produce a pleiomorphic syndrome in which symptomatology depends on the parasite species and the immunological stage of the host. The symptoms range from completely asymptomatic to cutaneous, mucocutaneous, and visceral (1). Several authors have reported differences in the treatment outcomes linked to the parasite species (2–5). Furthermore, mucocutaneous leishmaniasis (MCL) is a belated complication associated with specific parasite species (3, 6) most commonly occurring in infections caused by the Leishmania (Viannia) subgenus. American cutaneous leishmaniasis (ACL) cases are usually the result of infections produced by this subgenus, and species identification is useful for treatment and prognosis. Molecular techniques may become a routine way to confirm suspected cases of ACL (7–10); the present study describes the best PCR-restriction fragment length polymorphism (RFLP) gene target for determining the species of Leishmania present in clinical samples from ACL lesions in a set of Colombian patients.
The study was approved by the boards of ethical conduct of the Hospital Militar Central-Bogota-Colombia (HOMIC) and Centro Dermatologico Federico Lleras Acosta Bogota-Colombia (CDFLL) in accordance with national (resolution 008430 of the Colombian Health Ministry) and international (Declaration of Helsinki and amendments, World Medical Association, South Korea, 2008) guidelines. DNA was extracted from skin biopsy specimens from the internal border of the lesions from 42 adult patients with a clinical diagnosis of ACL. The diagnosis was confirmed microscopically in 35 patients and by PCR detection of the parasite in 7 patients. All patients voluntarily participated in the study and signed an informed consent.
The CDFLL biobank provided 19 Giemsa-stained slide smears from cutaneous lesions. In 17 of them, the presence of Leishmania sp. amastigotes was microscopically confirmed, and in 2 smears, the detection of the parasite was established by PCR. DNA was recovered from the Giemsa-stained smears.
All PCRs performed included DNA from 2 negative-control patients from CDFLL (with confirmed diagnoses of sporotrichosis and ecthyma gangrenosum) and from three healthy volunteers. The entire group of patients had been infected within the Colombian borders.
We selected genes and sequences previously reported to be useful markers for species identification by PCR-RFLP of Leishmania species for further evaluation. We analyzed zinc-metalloprotease (gp63) (11), spliced leader (SL) (12), cysteine protease B1 (cpb) (13), and heat shock protein 70 (hsp70) (14, 15) for their ability to discriminate the L. (Viannia) subgenus in clinical samples. Using the PCR-RFLP protocols previously reported for each gene, we extracted DNA from the reference strains of Leishmania species commonly associated with ACL in Colombia [Leishmania (Viannia) panamensis, L.(V.) braziliensis, L.(V.) guyanensis, Leishmania (Leishmania) amazonensis, and L.(L.) mexicana] (5, 16, 17). Other Leishmania species circulating in South America were not considered in the present study.
gp63 PCR amplification of DNA from the reference strains produced an expected fragment of 870 bp, in agreement with previous reports (11) (data not shown). However, when the fragments were digested with SalI and ApaI, only the L. (L.) amazonensis amplicon digestion behaved as described previously (11).
Amplification of the SL sequence was performed with organisms of the Leishmania (Viannia) and Leishmania (Leishmania) subgenera, as described previously (12). Using genomic DNA from L. (L) amazonensis and L. (L) mexicana reference strains, products of 300 and 320 bp were identified, respectively. For species belonging to the Leishmania (Viannia) subgenus, an expected 226-bp fragment was obtained. Further digestion of those fragments using HaeIII allowed for the identification of L. (V.) braziliensis but could not discriminate between L. (V.) guyanensis and L. (V.) panamensis. We successfully amplified the genomic DNA of cultured promastigotes; however, when we amplified up to 200 ng of genomic DNA from various clinical samples, the results were negative.
Amplification of the cysteine protease B (cpb) gene from the genomic DNA derived from parasite cultures, according to the protocol reported elsewhere (13), was successful. When patient samples were used, the amplification of human DNA was also obtained. A fragment of about 1 kb was present in all human DNA negative controls and in clinical samples (data not shown). We isolated and cultured parasites from patient 8 and amplified the cpb gene. This amplification yielded a unique 1.3-kb band of the expected size. However, when DNA was extracted directly from the patient biopsy sample, we detected a light specific band and a strong band of about 1 kb.
Using DNA from reference strains and 10 clinical samples randomly selected from this patient's cohort, the hsp70 gene was PCR amplified and digested with HaeIII and BccI, as previously reported (14, 15). This protocol was applied to the entire set of clinical samples, and the hsp70 gene was chosen for further evaluation.
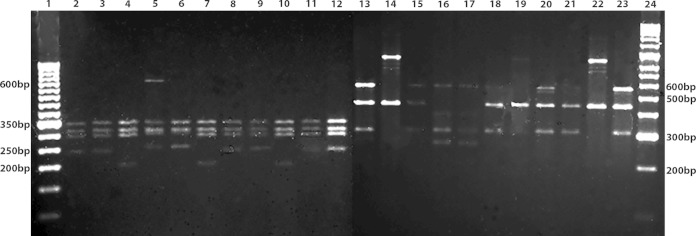
In 39 out of 61 samples with clinically suspected ACL, no obvious amplification products of the hsp70 gene fragment were observed. A nested-PCR protocol was designed to improve the yield of hsp70 DNA. After cleaning the PCR product, amplification yielded the original hsp70 fragment, as previously described (14, 15), and this product was used as the DNA template for a second round of amplification. After determining the ideal amount of DNA (ranging from 1.9 ng to 19.2 ng), nested-PCR was performed, using the described conditions for the first round of amplification (15). The following internal primers were used for the second round of amplification: Fw (5′-ACTTCAACGACTCGCAGCGCCA-3′) and Rv (5′-ATCGGGTTGCATGTGCTCTCCA-3′). The amplification products were digested with HaeIII and BccI (14, 15) (Fig. 1).
FIG 1.
hsp70-nested-PCR-RFLP for Leishmania species identification in clinical samples of ACL. SYBR-safe-stained 2% agarose gels showing HaeIII (lanes 2 to 12) and BccI (lanes 13 to 23) enzymatic digestions of hsp70-nested-PCR products, using as a template the genomic DNA of the following reference strains: lane 2 and 13, L. (V.) guyanensis (MHOM/GF/79/LEM85); lanes 3 and 14, L. (V.) panamensis (MHOM/PA/71/LS94); lanes 4 and 15, L. (V.) braziliensis (MHOM/BR/75/M2903); lanes 5 and 16, L. (L.) amazonensis (MHOM/BR/73/M2269); lanes 6 and 17, L. (L.) mexicana (MHOM/BZ/82/BEL 21); or, using as a template the genomic DNA from different types of clinical samples: lanes 7 and 18, biopsy specimen from patient 21 identified as L. (V.) braziliensis; lanes 8 and 19, Giemsa-stained smear from patient 23 identified as L. (V.) panamensis; lanes 9 and 20, Giemsa-stained smear from patient 15 identified as L. (V.) guyanensis; lanes 10 and 21, biopsy specimen from patient 60 identified as L. (V.) braziliensis; lanes 11 and 22, Giemsa-stained smear from patient 24 identified as L. (V.) panamensis; lanes 12 and 23, biopsy specimen from patient 63 identified as L. (V.) guyanensis; lane 1, size ruler, 50 bp; lane 24, size ruler, 100 bp.
The approach described in this report allowed for the identification of Leishmania species in clinical samples (Table 1). The predominance of L. (V.) braziliensis associated with ACL in our patients contrasts with previous reports that describe L. (V.) panamensis as being the main species producing ACL in Colombia (5, 16, 17). This might be a result of a bias of the present study in the selection of patients, given that most of them come from eastern Colombia, where L.(V) braziliensis is predominant.
TABLE 1.
Distribution of Leishmania species identified by hsp70-RFLP and hsp70-nested-PCR-RFLP
| Leishmania species identifieda | No. of patients | % |
|---|---|---|
| L. braziliensis | 46 | 75.4 |
| L. panamensis | 9 | 14.8 |
| L. guyanensis | 2 | 3.3 |
| Undefined pattern | 3 | 4.9 |
| No amplification of hsp70 | 1 | 1.6 |
From a total of 61 samples, the method improved in the present study allowed species identification in 93.5% of the samples. The undefined patterns in 4.9% might correspond to mixed infections or rare gene polymorphisms. A DNA sample from a patient whose direct skin smear was positive for microscopy did not amplify using either the hsp70 or the hsp70-nested-PCR.
The identification of Leishmania species of the subgenus Leishmania (Viannia) in clinical samples has been challenging with the current PCR-based protocols (18). Even with rigorous approaches, such as stepwise PCR, samples with a weak-positive target gene signal after diagnostic PCR have not enabled identification at the species level (19). The nested-PCR for hsp70 implemented in the present study successfully identifies Leishmania species in clinical samples, even from low concentrations of parasite DNA, as was the case for the direct smears. This is a significant contribution for species differentiation in cases with little parasite DNA in the specimen, such as samples obtained by noninvasive diagnostic sampling.
ACKNOWLEDGMENTS
This work was supported by the Universidad Nacional de Colombia grant HERMES-18994 from División de Investigación Sede Bogota and CDFLL grant 4000-16-1S. Mónica L. Cruz-Barrera and Viviana Ortegon-Vergara were funded by the Colciencias young researchers program.
REFERENCES
- 1.Bañuls AL, Bastien P, Pomares C, Arevalo J, Fisa R, Hide M. 2011. Clinical pleiomorphism in human leishmaniasis, with special mention of asymptomatic infection. Clin Microbiol Infect 17:1451–1461. doi: 10.1111/j.1469-0691.2011.03640.x. [DOI] [PubMed] [Google Scholar]
- 2.Llanos-Cuentas A, Tulliano G, Araujo-Castillo R, Miranda-Verastegui C, Santamaria-Castrellon G, Ramirez L, Lazo M, De Doncker S, Boelaert M, Robays J, Dujardin JC, Arevalo J, Chappuis E. 2008. Clinical and parasite species risk factors for pentavalent antimonial treatment failure in cutaneous leishmaniasis in Perú. Clin Infect Dis 46:223–231. doi: 10.1086/524042. [DOI] [PubMed] [Google Scholar]
- 3.Croft S, Sundar S, Fairlamb A. 2006. Drug resistance in leishmaniasis. Clin Microbiol Rev 19:111–126. doi: 10.1128/CMR.19.1.111-126.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rojas R, Valderrama L, Valderrama M, Varona MX, Ouellette M, Saravia NG. 2006. Resistance to antimony and treatment failure in human Leishmania (Viannia) infection. J Infect Dis 193:1375–1383. doi: 10.1086/503371. [DOI] [PubMed] [Google Scholar]
- 5.Fernández OL, Diaz-Toro Y, Ovalle C, Valderrama L, Muvdi S, Rodríguez I, Gomez MA, Saravia NG. 2014. Miltefosine and antimonial drug susceptibility of Leishmania Viannia species and populations in regions of high transmission in Colombia. PLoS Negl Trop Dis 8:e2871. doi: 10.1371/journal.pntd.0002871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saravia NG, Segura I, Holguin AF, Santrich C, Valderrama L, Ocampo C. 1998. Epidemiologic, genetic, and clinical associations among phenotypically distinct populations of Leishmania (Viannia) in Colombia. Am J Trop Med Hyg 59:86–94. [DOI] [PubMed] [Google Scholar]
- 7.Boggild AK, Ramos AP, Espinosa D, Valencia BM, Veland N, Miranda-Verastegui C, Arevalo J, Low DE, Llanos-Cuentas A. 2010. Clinical and demographic stratification of test performance: a pooled analysis of five laboratory diagnostic methods for American cutaneous leishmaniasis. Am J Trop Med Hyg 83:345–350. doi: 10.4269/ajtmh.2010.09-0414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Valencia BM, Veland N, Alba M, Adaui V, Arevalo J, Low DE, Llanos-Cuentas A, Boggild AK. 2012. Non-invasive cytology brush PCR for the diagnosis and causative species identification of American cutaneous leishmaniasis in Peru. PLoS One 7:e49738. doi: 10.1371/journal.pone.0049738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boggild AK, Valencia BM, Espinosa D, Veland N, Ramos AP, Arevalo J, Llanos-Cuentas A, Low DE. 2010. Detection and species identification of Leishmania DNA from filter paper lesion impressions for patients with American cutaneous leishmaniasis. Clin Infect Dis 50:e1–6. doi: 10.1086/648730. [DOI] [PubMed] [Google Scholar]
- 10.Shirian S, Oryan A, Hatam GR, Panahi S, Daneshbod Y. 2014. Comparison of conventional, molecular, and immunohistochemical methods in diagnosis of typical and atypical cutaneous leishmaniasis. Arch Pathol Lab Med 138:235–240. doi: 10.5858/arpa.2013-0098-OA. [DOI] [PubMed] [Google Scholar]
- 11.Victoir K, De Doncker S, Cabrera L, Alvarez E, Arevalo J, Llanos-Cuentas A, Le Ray D, Dujardin J. 2003. Direct identification of Leishmania species in biopsies from patients with American tegumentary leishmaniasis. Trans R Soc Trop Med Hyg 97:80–87. doi: 10.1016/S0035-9203(03)90031-9. [DOI] [PubMed] [Google Scholar]
- 12.Marfurt J, Niederwieser I, Makia ND, Beck HP, Felger I. 2003. Diagnostic genotyping of Old and New World Leishmania species by PCR-RFLP. Diagn Microbiol Infect Dis 46:115–124. doi: 10.1016/S0732-8893(03)00040-3. [DOI] [PubMed] [Google Scholar]
- 13.López L, Robayo M, Vargas M, Vélez ID. 2012. Thermotherapy. An alternative for the treatment of American cutaneous leishmaniasis. Trials 13:58. doi: 10.1186/1745-6215-13-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia L, Kindt A, Bermudez H, Llanos-Cuentas A, De Doncker S, Arevalo J, Tintaya KWQ, Dujardin J-C. 2004. Culture independent species typing of neotropical Leishmania for clinical validation of a PCR-based assay targeting heat shock protein 70 genes. J Clin Microbiol 42:2294–2297. doi: 10.1128/JCM.42.5.2294-2297.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Montalvo A, Fraga J, Monzote L, Montano I, De Doncker S, Dujardin J, Van der Auwera G. 2010. Heat-shock protein 70 PCR-RFLP: a universal simple tool for Leishmania species discrimination in the New and Old World. Parasitology 137:1159–1168. doi: 10.1017/S0031182010000089. [DOI] [PubMed] [Google Scholar]
- 16.Corredor A, Kreutzer RD, Tesh RB, Boshell J, Palau MT, Caceres E, Duque S, Pelaez D, Rodriguez G, Nichols S, Hernandez C, Morales A, Young D, Ferro C. 1990. Distribution and etiology of leishmaniasis in Colombia. Am J Trop Med Hyg 42:206–214. [DOI] [PubMed] [Google Scholar]
- 17.Ovalle C, Porras L, Rey M, Ríos M, Camargo Y. 2006. Distribución geográfica de especies de Leishmania aisladas de pacientes consultantes al Instituto Nacional de Dermatología Federico Lleras Acosta, E.S.E., 1995–2005. Biomédica 26:145–151. [PubMed] [Google Scholar]
- 18.Rocha MN, Margonari C, Presot IM, Soares RP. 2010. Evaluation of 4 polymerase chain reaction protocols for cultured Leishmania spp. typing. Diagn Microbiol Infect Dis 68:401–409. doi: 10.1016/j.diagmicrobio.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 19.Veland N, Boggild AK, Valencia C, Valencia BM, Llanos-Cuentas A, Van der Auwera G, Dujardin JC, Arevalo J. 2011. Leishmania (Viannia) species identification on clinical samples from cutaneous leishmaniasis patients in Peru: assessment of a molecular stepwise approach. J Clin Microbiol 50:495–498. doi: 10.1128/JCM.05061-11. [DOI] [PMC free article] [PubMed] [Google Scholar]