Abstract
Statutory surveillance of bovine spongiform encephalopathy (BSE) indicates that cattle are susceptible to both classical BSE (C-BSE) and atypical forms of BSE. Atypical forms of BSE appear to be sporadic and thus may never be eradicated. A major challenge for prion surveillance is the lack of sufficiently practical and sensitive tests for routine BSE detection and strain discrimination. The real-time quaking-induced conversion (RT-QuIC) test, which is based on prion-seeded fibrillization of recombinant prion protein (rPrPSen), is known to be highly specific and sensitive for the detection of multiple human and animal prion diseases but not BSE. Here, we tested brain tissue from cattle affected by C-BSE and atypical L-type bovine spongiform encephalopathy (L-type BSE or L-BSE) with the RT-QuIC assay and found that both BSE forms can be detected and distinguished using particular rPrPSen substrates. Specifically, L-BSE was detected using multiple rPrPSen substrates, while C-BSE was much more selective. This substrate-based approach suggests a diagnostic strategy for specific, sensitive, and rapid detection and discrimination of at least some BSE forms.
INTRODUCTION
Transmissible spongiform encephalopathies (TSEs), or prion diseases, such as Creutzfeldt-Jakob disease (CJD) in humans and bovine spongiform encephalopathy (BSE) in cattle, are fatal neurodegenerative disorders characterized by spongiosis, neuronal loss, gliosis, and abnormal deposition of host-encoded prion protein (PrP) in the brain. The infectious agent responsible for TSEs appears to be largely composed of a misfolded and multimeric form of prion protein (PrPSc), which is able to induce polymerization and conformational conversion of normal protease-sensitive prion protein (PrPSen) into PrPSc and its partially protease-resistant forms (PrPRes) (1).
BSE and its link to variant Creutzfeldt-Jakob disease (vCJD) in humans have raised important food safety issues. The incidence of classical BSE (C-BSE) has decreased as a result of disease-control programs, such as the ruminant feed ban (2). However, atypical forms of bovine prion disease have been identified in several countries (3). These emerging bovine prion strains can be categorized as H-type BSE (H-BSE) or L-type BSE (L-BSE or bovine amyloidotic spongiform encephalopathy [BASE] when amyloid plaques are present in the brain). These atypical BSE strains are differentiated biochemically by the electrophoretic mobility and glycoform pattern of PrPRes after proteinase K (PK) digestion (4–7). Currently, both of these atypical bovine prion strains, which mainly affect older animals (8), are thought to be sporadic forms of bovine prion diseases (9) and are still rare (less than 100 cases identified worldwide to date).
It is suspected that one of these atypical L-BSE or H-BSE strains may have instigated the C-BSE epidemic, as suggested, for example, by the fact that the L-BSE strain can convert to C-BSE following serial passage in mice (5, 6, 10). Further concerns about the atypical BSE strains have arisen from observations that nonhuman primates and transgenic mice expressing human PrPSen show differing susceptibilities to C-BSE and L-BSE (11–16). Although controversial (16), some findings suggest that L-BSE has greater pathogenicity than C-BSE in transgenic mice and Syrian golden hamsters (10, 17, 18). Furthermore, L-BSE prions have been shown to be experimentally transmissible to cattle (19, 20) and transgenic mice overexpressing bovine or human prion protein (PrP) (21–23) with shorter incubation periods and more severe spongiform changes than C-BSE prions. Although there is no evidence of atypical bovine prion transmission to humans as yet, these findings collectively suggest that, even as the incidence of C-BSE wanes, it remains important to be able to monitor and discriminate among prion strains in cattle.
In pursuit of these goals, recent studies have described in vitro detection of C-BSE prions in brain tissue and blood by protein misfolding cyclic amplification (PMCA), which is much more sensitive than the direct immunoblotting that is required for the biochemical analyses of PrPRes (24). Furthermore, PMCA can selectively detect C-BSE in mixtures of brain homogenates (BHs) from C-BSE- and scrapie-infected sheep with 100% specificity and 97% sensitivity (25). Still, it remains important to develop highly sensitive methods that can discriminate C-BSE from other prion strains in cattle. Such methods should also be rapid and practical enough for routine screening and surveillance applications. Development of these methods would reduce the risk of prion-contaminated meat entering the human or animal food supply.
Real-time quaking-induced conversion (RT-QuIC) assays can rapidly detect subinfectious doses of prion seeding activity and have been used successfully to detect multiple human, cervid, ovine, hamster, and mouse prion strains in a variety of biological tissues, such as cerebrospinal fluid, saliva, blood, and nasal fluid (26–36). In RT-QuIC reactions, prion-associated seeds induce amyloid fibril formation of bacterially expressed recombinant PrPSen (rPrPSen) in multiwell plates. The resulting rPrP amyloid fibrils are then detected by the enhanced fluorescence of the amyloid-sensitive dye thioflavin T (ThT). Here, we adapted RT-QuIC assays to the sensitive detection and discrimination of the C-BSE and L-BSE prion strains of cattle.
MATERIALS AND METHODS
Western blotting of cattle brain homogenates.
Analyses of brain tissue collected from two healthy cattle and nine Italian BSE-infected field cases (Table 1), four classified as C type and five as L type, were done by Western blotting on 150-mg (±20 mg) equivalents of each sample as described previously (37). Briefly, each sample was separated by SDS-PAGE on 14% polyacrylamide handmade minigels and then transferred onto polyvinylidene difluoride (PVDF) membranes (Immobilon P; Millipore, Billerica, MA) for 2 h at 60 V. The membranes were blocked with 5% bovine serum albumin (BSA) in Tris-buffered saline (TBS) for 1 h and incubated at 4°C overnight with monoclonal antibody 6H4 (0.9 μg/ml, epitope 144-152, DYEDRYYRE; Prionics, Schlieren-Zurich, Switzerland). Immunodetection was carried out with an alkaline phosphatase conjugated goat anti-mouse IgG (Western 2 Ab-AP; Prionics), revealed by a chemiluminescent substrate (Lumi-Phos; Thermo Scientific, Waltham, MA, USA) and visualized on Hyperfilm ECL (GE-Healthcare Ltd., St. Giles, United Kingdom) with a 15-min exposure.
TABLE 1.
Epidemiological data and biochemical results from cattle
| Cattle identification | Breed | Age (yr) | Cause of death | Western blot PrPRes glycoform profilea | PrPSc distributionb | Main PrPSc deposition pattern |
|---|---|---|---|---|---|---|
| L-BSE 1 | Alpine Brown | 11 | Slaughtered | D ≤ M | FC > B | Amyloid plaques |
| L-BSE 2 | Piedmontese | 15 | Fallen stock | D ≤ M | FC > B | Amyloid plaques |
| L-BSE 3 | Piedmontese | 14 | Fallen stock | D ≤ M | FC > B | Amyloid plaques |
| L-BSE 4 | Holstein Friesian | 13 | Slaughtered | D ≤ M | NAc | NA |
| L-BSE 5 | Alpine Brown | 14 | Slaughtered | D ≤ M | FC > B | Small aggregates, granular |
| C-BSE 1 | Holstein Friesian | 7 | Slaughtered | D > M | FC < B | Granular, linear tract, glial |
| C-BSE 2 | Holstein Friesian | 7 | Slaughtered | D > M | FC < B | Granular, linear tract, glial |
| C-BSE 3 | Holstein Friesian | 6 | Slaughtered | D > M | FC < B | Granular, linear tract, glial |
| C-BSE 4 | Pezzata Rossa | 6 | Slaughtered | D > M | FC < B | Granular, linear tract, glial |
D, diglycosylated band; M, monoglycosylated band.
FC, frontal cortex; B, brainstem.
NA, not available.
Recombinant prion protein purification.
Syrian golden hamster (residues 23 to 231 [hamster 23-231] [GenBank accession no. K02234] or residues 90 to 231 [hamster 90-231]), human (residues 23 to 231 [human 23-231] [accession no. M13899.1]), and chimeric hamster-sheep (Ha-S; Syrian hamster residues 23 to 137 followed by sheep residues 141 to 234 of the R154Q171 polymorph [accession no. AY907689]) prion protein genes were ligated into the pET41 vector (EMD Biosciences). Escherichia coli carrying this vector was grown in Luria broth (LB) medium in the presence of kanamycin and chloramphenicol. rPrPSen expression was induced using Overnight Express Autoinduction system 1 (Novagen) and Bug Buster master mix (Novagen) to isolate inclusion bodies. Following solubilization of the inclusion bodies in 8 M guanidinium-HCl, the denatured protein was purified under 6 M guanidinium-HCl denaturing conditions using nickel nitrilotriacetic acid (Ni-NTA) superflow resin (Qiagen) with an AKTA fast protein liquid chromatography instrument (GE Healthcare). The rPrPSen was subjected to on-column refolding using a linear gradient into phosphate buffer and then eluted using an imidazole gradient as previously described (26). The purified protein was extensively dialyzed into 10 mM sodium phosphate buffer (pH 5.8). Then, following filtration (0.22-μm syringe filter; Fisher), a concentration measurement by absorbance at 280 nm was performed and the rPrPSen was stored at −80°C.
Brain homogenate preparation and RT-QuIC protocol.
Normal (n = 2), C-BSE-infected (n = 4), and L-BSE-infected (n = 5) bovine brain homogenates (BHs) (10%, wt/vol) were prepared as previously described (38) and stored at −80°C. For RT-QuIC analysis, BHs were serially diluted in 0.1% SDS (Sigma)–N2 (Gibco)–PBS as previously reported (26). The RT-QuIC reaction mix was composed of 10 mM phosphate buffer (pH 7.4), 300 mM NaCl, 10 μM ThT, 1 mM EDTA, and 0.1 mg/ml of rPrPSen. Aliquots of this mix (98 μl) were loaded into each well of a black 96-well plate with a clear bottom (Nunc) and seeded with 2 μl of 10−4 to 10−9 brain tissue dilutions. Normal bovine BH dilutions were used as negative controls (as shown in Fig. 2 and 3), and 10−5 brain tissue dilutions from hamsters with clinical scrapie were initially included as positive controls when bovine brain samples were tested for the first time (not shown). The plate was then sealed with a plate-sealer film (Nalgene Nunc International) and incubated for 55 h at 42°C in a BMG Labtech FLUOstar plate reader with cycles of 1 min of shaking (700 rpm double orbital) and 1 min of rest throughout the incubation. ThT fluorescence measurements (excitation, 450 ± 10 nm; emission, 480 ± 10 nm; bottom read) were recorded every 45 min. RT-QuIC reactions were deemed acceptable when the negative controls remained negative for at least 55 h and the positive scrapie (strain 263K) controls were positive within 5 h according to the criteria described in the next paragraph.
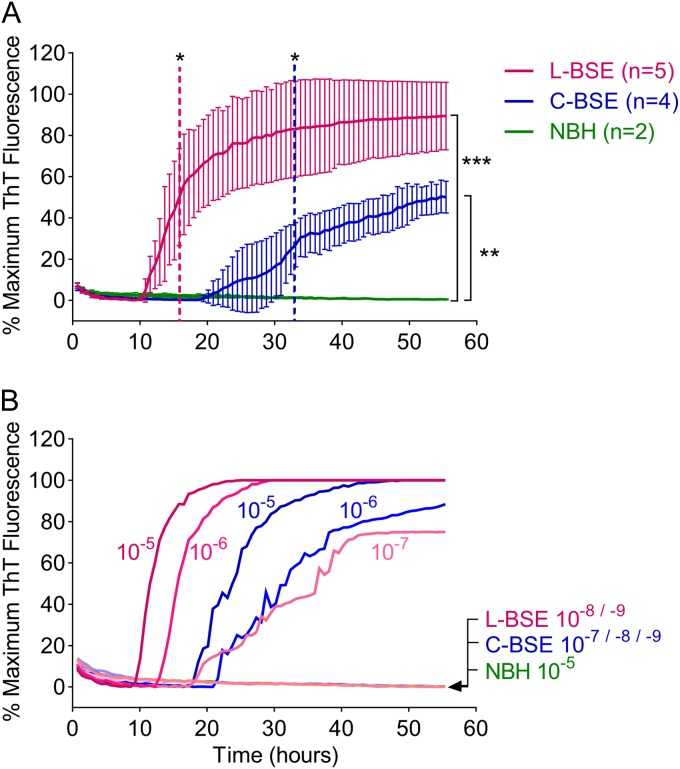
FIG 2.
RT-QuIC sensitivity for C-BSE and L-BSE detection. (A) L-BSE-infected (magenta), C-BSE-infected (blue), or normal negative control (NBH, green) 10−5 brain tissue dilutions were used to seed quadruplicate RT-QuIC reactions using the Ha-S rPrPSen substrate. Average ThT fluorescence readings from the quadruplicate reactions for each case were determined and then, in turn, averaged (±SD) over the designated number of cattle in each group. Color-matched dashed lines indicate the time at which the signal from a given sample became significantly greater than the control. Brackets signify the difference between the test groups and the control at the 55-h endpoint. *, P < 0.05; **, P < 0.005; ***, P < 0.0005. Comparable data were obtained in two additional independent RT-QuIC tests done on different days and plate readers (28). (B) Serial dilutions (10−5 to 10−9) of C-BSE-infected or L-BSE-infected brain tissue or a 10−5 dilution of uninfected brain tissue were used to seed quadruplicate RT-QuIC reactions with Ha-S rPrPSen as the substrate. The data show the average ThT fluorescence of 4 replicate wells. Similar results were obtained from two additional C-BSE-infected and two additional L-BSE-infected brain specimens (not shown). Each ThT reading is indicated as the percentage of the maximum value achievable by the plate readers (see Materials and Methods) as a function of reaction time.
FIG 3.
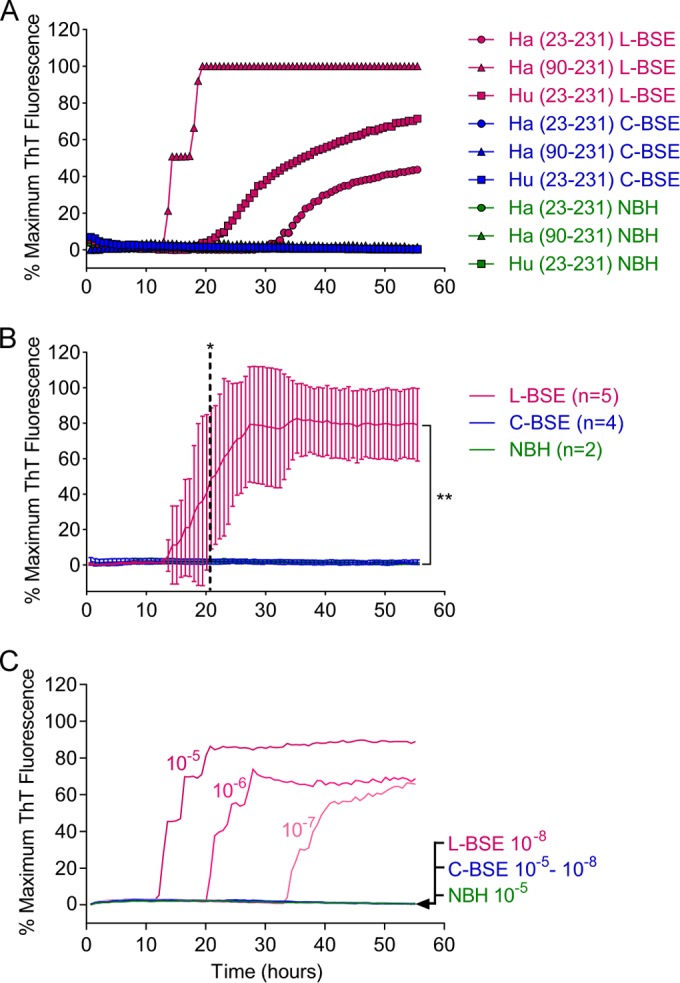
RT-QuIC for L-BSE versus C-BSE strain discrimination. (A) L-BSE-infected, C-BSE-infected, and uninfected (NBH) 10−5 brain tissue dilutions were used to seed quadruplicate RT-QuIC reactions. Hamster 23-231, hamster 90-231, or human 23-231 rPrPSen were used as the substrates. (B) Averaged (±SD) RT-QuIC responses from 10−5 brain tissue dilutions of all of the L-BSE- and C-BSE-infected brain samples using hamster 90-231 PrPSen as a substrate. Dashed lines, the time at which the signals from L-BSE- and C-BSE-infected samples became significantly different; brackets, the difference between the L-BSE and C-BSE test groups at the 55-h endpoint. *, P < 0.05; **, P < 0.005. As in Fig. 2A, the data points are averages of the average readings from quadruplicate reactions run on each specimen with normalization as described in Materials and Methods. Similar results were obtained in three additional repeat experiments performed with these samples. (C) Serial dilutions (10−5 to 10−8) of C-BSE- or L-BSE-infected brain tissue or a 10−5 dilution of uninfected brain tissue were used to seed RT-QuIC reactions using Ha (90-231) rPrPSen as the substrate. Each data point shows the ThT fluorescence average of 4 replicate wells. All ThT readings are indicated as a percentage of the maximum value achievable by the plate readers.
Data analysis.
RT-QuIC fluorescence readings were analyzed as previously described (28). Briefly, to compensate for differences between the fluorescence plate readers, data sets were normalized to a percentage of the maximal fluorescence response of the instrument, and the obtained values were plotted against the reaction times. Samples were judged to be RT-QuIC positive using criteria similar to those previously described for RT-QuIC analyses of brain specimens (26, 28), except for the use of baseline-adjusted normalized fluorescence values. A 55-h time point was chosen based on multiple (n = 20) repeat runs in which no spontaneous conversions of the substrate in negative-control seeded reactions were observed up to the experimentally designated time point. Data are displayed as the average of four technical replicates except where indicated. Where multiple biological replicates are displayed, they are represented as the mean ± the standard deviation. At pertinent time points, the signals were analyzed for statistical significance using a two-tailed unpaired t test with Welch's correction.
RESULTS
Immunoblot profile of PrPRes in brains from Italian C-BSE- and L-BSE-infected cattle.
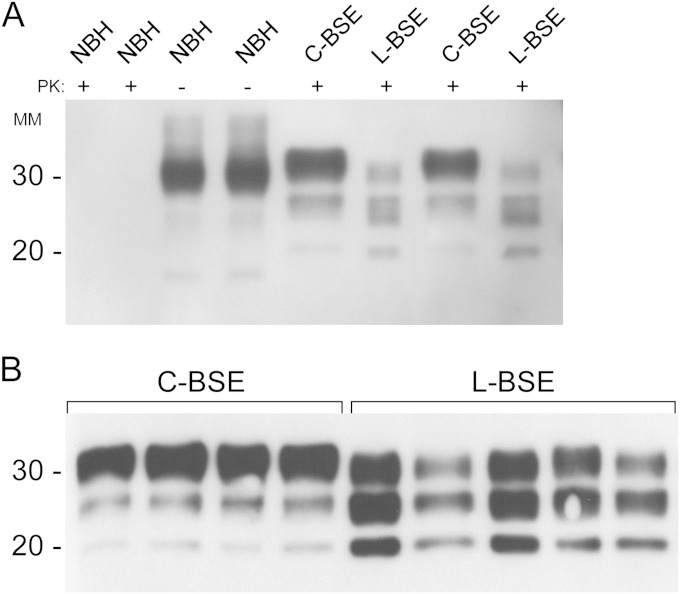
To develop RT-QuIC conditions for the detection of C-BSE and L-BSE, we used brainstem and frontal cortex brain samples, respectively, from four C-BSE-infected and five L-BSE-infected Italian cattle. To first confirm that the C-BSE- and L-BSE-infected samples differed as expected in the PrPRes banding pattern after proteinase K (PK) treatment, we performed Western blot analysis on all of these samples (Fig. 1A and B). Consistent with previous reports (4), C-BSE-infected brain samples showed the typical predominance of the highest-molecular-mass glycoform, while the L-BSE glycotype profile had a more prominent monoglycosylated (middle) band and an expected overall ∼2-kDa downward shift of the un-, mono-, and diglycosylated bands.
FIG 1.
Western blots of brain stem and frontal cortex samples from normal, C-BSE-infected, and L-BSE-infected cattle. (A) Two brain samples from normal cattle (NBH) were subjected to immunoblotting without proteinase K (PK) digestion (NBH −PK lanes). The same two brain samples from healthy cattle along with two brainstem specimens from C-BSE-positive and two frontal cortex samples from L-BSE-positive cattle were PK digested as described in Materials and Methods (NBH, C-BSE, and L-BSE +PK lanes). (B) Homogenized brainstem specimens from four C-BSE-positive and frontal cortex samples from five L-BSE-positive cattle were treated with proteinase K (PK) and subjected to immunoblotting using monoclonal antibody 6H4 (epitope within PrP residues 144 to 152). MM, molecular mass (in kilodaltons). The data are representative of multiple immunoblots of these specimens.
Detection and differentiation of C-BSE and L-BSE.
Because C-BSE causes vCJD in humans and shares most, if not all, of its strain characteristics, we initially tried an rPrPSen substrate that works well for the detection of human vCJD (33), namely, a chimeric form comprising an N-terminal hamster PrP domain and a C-terminal sheep domain (Ha-S rPrPSen). C-BSE- and L-BSE-infected brain homogenates were used as a source of prion seeds, and equivalent dilutions of two uninfected brain homogenates were used as specificity controls. Figure 2A shows the results from RT-QuIC reactions seeded with 10−5 dilutions of five L-BSE-infected and four C-BSE-infected brain samples, each of which gave positive reactions in all of the quadruplicate reactions within 55 h. The average fluorescence increase was stronger and faster for the L-BSE-infected samples, showing positive signals (P < 0.05) over the control samples as early as 16 h, compared to the 33 h it took for the BSE-infected samples to show positive signals. Although the two strains exhibited distinct RT-QuIC seeded polymerization kinetics, our results show that Ha-S rPrPSen supports detection of C-BSE and L-BSE seeding activity (P < 0.005 and 0.0005, respectively, at 55 h). Next, we investigated the RT-QuIC sensitivities for C-BSE and L-BSE using Ha-S rPrPSen. Figure 2B shows the RT-QuIC endpoint dilution analysis of two representative C-BSE- and L-BSE-positive brain samples. We saw positive reactions with C-BSE-infected and L-BSE-infected brain tissue dilutions down to 10−6 and 10−7, respectively.
To explore the relative abilities of C-BSE and L-BSE prion strains to initiate polymerization of other rPrPSen substrates, we tested human 23-231, hamster 23-231, and hamster 90-231 rPrPSen substrates (Fig. 3A). RT-QuIC reactions were seeded with 10−5 C-BSE- or L-BSE-infected brain tissue dilutions and incubated for 55 h using the same experimental conditions described for Ha-S rPrPSen. All three substrates resulted in positive RT-QuIC reactions with the L-BSE seed, with Ha rPrPSen 90-231 giving the fastest response. Conversely, C-BSE was unable to seed conversion of any of these three rPrPSen substrates under these conditions.
Because stronger and faster RT-QuIC responses were seen with Ha rPrPSen 90-231 (Fig. 3A), we used this substrate to test all four C-BSE-infected and five L-BSE-infected brain samples previously analyzed with Ha-S rPrPSen substrate (Fig. 2A). Our results confirmed that all of the L-BSE-infected brain samples induced fibrillization of Ha rPrPSen 90-231 with an average lag phase of ∼13 h, while none of the C-BSE-infected brain samples gave positive reactions within 55 h (Fig. 3B). To investigate the minimum difference in sensitivity for the C-BSE- and L-BSE-infected brain tissues using this substrate, we seeded RT-QuIC reactions with 10−4 to 10−8 dilutions of L-BSE- or C-BSE-infected brain tissue. Whereas we observed the L-BSE strain in reactions with as little as 10−7 dilutions of brain tissue, we failed to detect the C-BSE strains even at the most concentrated homogenates tested (i.e., 10−4 [data not shown, and Fig. 3C]). These findings indicate that Ha rPrPSen 90-231 is at least 10,000-fold more sensitive for detecting L-BSE seeding activity than for detecting C-BSE, and L-BSE can be distinguished from C-BSE within 21 h (P < 0.05). Taken together, these findings indicate that by using appropriate substrates, the RT-QuIC assay can rapidly detect and discriminate C-BSE and L-BSE strains.
DISCUSSION
Although active and passive surveillance programs implemented by the European Union have led to significant decreases in the incidence of C-BSE, the recent identifications of atypical bovine (4, 7) and ovine prion strains (39) pose new concerns for the safety of livestock and the food supply. Current knowledge of the origins, pathogenesis, and environmental spreading mechanisms of these atypical BSE strains is limited. Thus, there are concerns that precautions that are presently taken to minimize the risk of prion contamination of the food supply might not be as effective at preventing the spread of these recently recognized strains. Consequently, as stated recently by the European Food Safety Authority (EFSA), there is a need for sensitive, fast, practical, and strain-specific prion detection techniques. Furthermore, regulation (EC) no. 999/20015 was amended to require that, beginning in July 2014, samples from BSE-positive cattle be submitted for TSE classification by Western blotting and immunohistochemistry (40).
Our findings indicate that by using the Ha-S rPrPSen substrate, RT-QuIC assays can sensitively detect both C-BSE- and L-BSE-associated seeding activity in less than 48 h. A systematic comparison of the European Union-approved rapid tests used for surveillance showed that the IDEXX HerdChek BSE-scrapie short assay is the most sensitive test for all BSE forms (41). However, our studies indicate that using Ha-S rPrPSen as the substrate, the RT-QuIC test is at least 104-fold more sensitive than the IDEXX enzyme-linked immunosorbent assay (ELISA) for the detection of C-BSE- and L-BSE-infected brain tissue. Our detection of 106-fold dilutions of C-BSE-infected brain tissue and 107-fold dilutions of L-BSE-infected brain tissue makes our RT-QuIC test at least as sensitive as infectivity bioassays (42). Furthermore, our observation that human 23-231, hamster 23-231, and hamster 90-231 rPrPSen substrates selectively allowed L-BSE but not C-BSE detection might be exploited as a sensitive and practical approach to prion strain discrimination. However, while detection with these substrates would allow the exclusion of C-BSE, it may not discriminate between L-BSE and H-BSE based upon the choice of substrate. Further studies will be required to assess this possibility. In any case, the ability to discriminate between C-BSE and L-BSE based on RT-QuIC substrate provides a prototypic example of how prion strains can have different seeding activities that may, in turn, help with strain identification. This new strategy may have important implications for TSE surveillance and might in the long run eliminate the need for follow-up strain identification by immunoblotting.
The reasons behind the differential RT-QuIC seeding performance of C-BSE and L-BSE strains remain unclear. Presumably, differences in their molecular conformation and/or aggregation state influence their relative abilities to interact with specific prion protein sequences (i.e., Ha-S rPrPSen) and not with others (i.e., Ha 90-231 rPrPSen). We note that the ability of L-BSE to induce fibrillization of multiple types of prion proteins (e.g., human and hamster) is reminiscent of the relatively promiscuous sporadic CJD RT-QuIC seeding capacity that has been described by us and others (27–29, 31, 33, 43). This sporadic CJD-like RT-QuIC seeding competence of the L-BSE strain is also consistent with the molecular similarities between type 2 sporadic CJD and L-BSE PrPRes that have been described by Casalone et al. (4). Additional studies on a larger number of animals are necessary to confirm the practical value and applicability of the assay. Furthermore, testing of other atypical bovine prion strains (e.g., H-type BSE) and ovine prion strains (e.g., Nor 98) would be of interest. Nevertheless, our findings so far show promise for the application of the RT-QuIC assay to the fast and sensitive detection and discrimination of prion seeding activity from different bovine strains.
ACKNOWLEDGMENTS
This study was funded in part by the Intramural Research Program of the NIAID and grants from the Italian Ministry of Health to Cristina Casalone and Cristiano Corona (grants RF-2009-1474758 and IZS-PLV05/11RC).
We thank Jay Carroll, Roger Moore, and Allison Kraus for critical review of the manuscript. We also thank Lynne Raymond for providing bacterial expression vectors for these studies and Gregory Raymond for assisting with sample shipping.
REFERENCES
- 1.Kraus A, Groveman BR, Caughey B. 2013. Prions and the potential transmissibility of protein misfolding diseases. Annu Rev Microbiol 67:543–564. doi: 10.1146/annurev-micro-092412-155735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ducrot C, Arnold M, de Koeijer A, Heim D, Calavas D. 2008. Review on the epidemiology and dynamics of BSE epidemics. Vet Res 39:15. doi: 10.1051/vetres:2007053. [DOI] [PubMed] [Google Scholar]
- 3.Jacobs JG, Langeveld JP, Biacabe AG, Acutis PL, Polak MP, Gavier-Widen D, Buschmann A, Caramelli M, Casalone C, Mazza M, Groschup M, Erkens JH, Davidse A, van Zijderveld FG, Baron T. 2007. Molecular discrimination of atypical bovine spongiform encephalopathy strains from a geographical region spanning a wide area in Europe. J Clin Microbiol 45:1821–1829. doi: 10.1128/JCM.00160-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casalone C, Zanusso G, Acutis P, Ferrari S, Capucci L, Tagliavini F, Monaco S, Caramelli M. 2004. Identification of a second bovine amyloidotic spongiform encephalopathy: molecular similarities with sporadic Creutzfeldt-Jakob disease. Proc Natl Acad Sci U S A 101:3065–3070. doi: 10.1073/pnas.0305777101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baron T, Vulin J, Biacabe AG, Lakhdar L, Verchere J, Torres JM, Bencsik A. 2011. Emergence of classical BSE strain properties during serial passages of H-BSE in wild-type mice. PLoS One 6:e15839. doi: 10.1371/journal.pone.0015839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Torres JM, Andreoletti O, Lacroux C, Prieto I, Lorenzo P, Larska M, Baron T, Espinosa JC. 2011. Classical bovine spongiform encephalopathy by transmission of H-type prion in homologous prion protein context. Emerging Infect Dis 17:1636–1644. doi: 10.3201/eid1709.101403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biacabe AG, Laplanche JL, Ryder S, Baron T. 2004. Distinct molecular phenotypes in bovine prion diseases. EMBO Rep 5:110–115. doi: 10.1038/sj.embor.7400054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Windl O, Dawson M. 2012. Animal prion diseases. Subcell Biochem 65:497–516. doi: 10.1007/978-94-007-5416-4_18. [DOI] [PubMed] [Google Scholar]
- 9.Brown P, McShane LM, Zanusso G, Detwiler L. 2006. On the question of sporadic or atypical bovine spongiform encephalopathy and Creutzfeldt-Jakob disease. Emerging Infect Dis 12:1816–1821. doi: 10.3201/eid1212.060965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Capobianco R, Casalone C, Suardi S, Mangieri M, Miccolo C, Limido L, Catania M, Rossi G, Di Fede G, Giaccone G, Bruzzone MG, Minati L, Corona C, Acutis P, Gelmetti D, Lombardi G, Groschup MH, Buschmann A, Zanusso G, Monaco S, Caramelli M, Tagliavini F. 2007. Conversion of the BASE prion strain into the BSE strain: the origin of BSE? PLoS Pathog 3:e31. doi: 10.1371/journal.ppat.0030031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nicot S, Bencsik A, Morignat E, Mestre-Frances N, Perret-Liaudet A, Baron T. 2012. Differentiation of prions from L-type BSE versus sporadic Creutzfeldt-Jakob disease. Emerging Infect Dis 18:2028–2031. doi: 10.3201/eid1812.120342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barria MA, Ironside JW, Head MW. 2014. Exploring the zoonotic potential of animal prion diseases: in vivo and in vitro approaches. Prion 8:85–91. doi: 10.4161/pri.28124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Comoy EE, Casalone C, Lescoutra-Etchegaray N, Zanusso G, Freire S, Marce D, Auvre F, Ruchoux MM, Ferrari S, Monaco S, Sales N, Caramelli M, Leboulch P, Brown P, Lasmezas CI, Deslys JP. 2008. Atypical BSE (BASE) transmitted from asymptomatic aging cattle to a primate. PLoS One 3:e3017. doi: 10.1371/journal.pone.0003017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ono F, Tase N, Kurosawa A, Hiyaoka A, Ohyama A, Tezuka Y, Wada N, Sato Y, Tobiume M, Hagiwara K, Yamakawa Y, Terao K, Sata T. 2011. Atypical L-type bovine spongiform encephalopathy (L-BSE) transmission to cynomolgus macaques, a non-human primate. Jpn J Infect Dis 64:81–84. [PubMed] [Google Scholar]
- 15.Beringue V, Herzog L, Reine F, Le Dur A, Casalone C, Vilotte JL, Laude H. 2008. Transmission of atypical bovine prions to mice transgenic for human prion protein. Emerging Infect Dis 14:1898–1901. doi: 10.3201/eid1412.080941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilson R, Hart P, Piccardo P, Hunter N, Casalone C, Baron T, Barron RM. 2012. Bovine PrP expression levels in transgenic mice influence transmission characteristics of atypical bovine spongiform encephalopathy. J Gen Virol 93:1132–1140. doi: 10.1099/vir.0.040030-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beringue V, Andreoletti O, Le Dur A, Essalmani R, Vilotte JL, Lacroux C, Reine F, Herzog L, Biacabe AG, Baron T, Caramelli M, Casalone C, Laude H. 2007. A bovine prion acquires an epidemic bovine spongiform encephalopathy strain-like phenotype on interspecies transmission. J Neurosci 27:6965–6971. doi: 10.1523/JNEUROSCI.0693-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nicot S, Baron T. 2011. Strain-specific barriers against bovine prions in hamsters. J Virol 85:1906–1908. doi: 10.1128/JVI.01872-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fukuda S, Iwamaru Y, Imamura M, Masujin K, Shimizu Y, Matsuura Y, Shu Y, Kurachi M, Kasai K, Murayama Y, Onoe S, Hagiwara K, Sata T, Mohri S, Yokoyama T, Okada H. 2009. Intraspecies transmission of L-type-like bovine spongiform encephalopathy detected in Japan. Microbiol Immunol 53:704–707. doi: 10.1111/j.1348-0421.2009.00169.x. [DOI] [PubMed] [Google Scholar]
- 20.Lombardi G, Casalone C, D'Angelo A, Gelmetti D, Torcoli G, Barbieri I, Corona C, Fasoli E, Farinazzo A, Fiorini M, Gelati M, Iulini B, Tagliavini F, Ferrari S, Caramelli M, Monaco S, Capucci L, Zanusso G. 2008. Intraspecies transmission of BASE induces clinical dullness and amyotrophic changes. PLoS Pathog 4:e1000075. doi: 10.1371/journal.ppat.1000075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buschmann A, Gretzschel A, Biacabe AG, Schiebel K, Corona C, Hoffmann C, Eiden M, Baron T, Casalone C, Groschup MH. 2006. Atypical BSE in Germany–proof of transmissibility and biochemical characterization. Vet Microbiol 117:103–116. doi: 10.1016/j.vetmic.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 22.Masujin K, Shu Y, Yamakawa Y, Hagiwara K, Sata T, Matsuura Y, Iwamaru Y, Imamura M, Okada H, Mohri S, Yokoyama T. 2008. Biological and biochemical characterization of L-type-like bovine spongiform encephalopathy (BSE) detected in Japanese black beef cattle. Prion 2:123–128. doi: 10.4161/pri.2.3.7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kong Q, Zheng M, Casalone C, Qing L, Huang S, Chakraborty B, Wang P, Chen F, Cali I, Corona C, Martucci F, Iulini B, Acutis P, Wang L, Liang J, Wang M, Li X, Monaco S, Zanusso G, Zou WQ, Caramelli M, Gambetti P. 2008. Evaluation of the human transmission risk of an atypical bovine spongiform encephalopathy prion strain. J Virol 82:3697–3701. doi: 10.1128/JVI.02561-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lacroux C, Comoy E, Moudjou M, Perret-Liaudet A, Lugan S, Litaise C, Simmons H, Jas-Duval C, Lantier I, Beringue V, Groschup M, Fichet G, Costes P, Streichenberger N, Lantier F, Deslys JP, Vilette D, Andreoletti O. 2014. Preclinical detection of variant CJD and BSE prions in blood. PLoS Pathog 10:e1004202. doi: 10.1371/journal.ppat.1004202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gough KC, Bishop K, Maddison BC. 2014. Highly sensitive detection of small ruminant bovine spongiform encephalopathy within transmissible spongiform encephalopathy mixes by serial protein misfolding cyclic amplification. J Clin Microbiol 52:3863–3868. doi: 10.1128/JCM.01693-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilham JM, Orrú CD, Bessen RA, Atarashi R, Sano K, Race B, Meade-White KD, Taubner LM, Timmes A, Caughey B. 2010. Rapid end-point quantitation of prion seeding activity with sensitivity comparable to bioassays. PLoS Pathog 6:e1001217. doi: 10.1371/journal.ppat.1001217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Atarashi R, Satoh K, Sano K, Fuse T, Yamaguchi N, Ishibashi D, Matsubara T, Nakagaki T, Yamanaka H, Shirabe S, Yamada M, Mizusawa H, Kitamoto T, Klug G, McGlade A, Collins SJ, Nishida N. 2011. Ultrasensitive human prion detection in cerebrospinal fluid by real-time quaking-induced conversion. Nat Med 17:175–178. doi: 10.1038/nm.2294. [DOI] [PubMed] [Google Scholar]
- 28.Orrù CD, Bongianni M, Tonoli G, Ferrari S, Hughson AG, Groveman BR, Fiorini M, Pocchiari M, Monaco S, Caughey B, Zanusso G. 2014. A test for Creutzfeldt-Jakob disease using nasal brushings. N Engl J Med 371:519–529. doi: 10.1056/NEJMoa1315200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peden AH, McGuire LI, Appleford NE, Mallinson G, Wilham JM, Orrù CD, Caughey B, Ironside JW, Knight RS, Will RG, Green AJ, Head MW. 2012. Sensitive and specific detection of sporadic Creutzfeldt-Jakob disease brain prion protein using real-time quaking induced conversion. J Gen Virol 93:438–449. doi: 10.1099/vir.0.033365-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orrù CD, Hughson AG, Race B, Raymond GJ, Caughey B. 2012. Time course of prion seeding activity in cerebrospinal fluid of scrapie-infected hamsters after intratongue and intracerebral inoculations. J Clin Microbiol 50:1464–1466. doi: 10.1128/JCM.06099-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McGuire LI, Peden AH, Orrù CD, Wilham JM, Appleford NE, Mallinson G, Andrews M, Head MW, Caughey B, Will RG, Knight RSG, Green AJE. 2012. RT-QuIC analysis of cerebrospinal fluid in sporadic Creutzfeldt-Jakob disease. Ann Neurol 72:278–285. doi: 10.1002/ana.23589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vascellari S, Orrù CD, Hughson AG, King D, Barron R, Wilham JM, Baron GS, Race B, Pani A, Caughey B. 2012. Prion seeding activities of mouse scrapie strains with divergent PrPSc protease sensitivities and amyloid plaque content using RT-QuIC and eQuIC. PLoS One 7:e48969. doi: 10.1371/journal.pone.0048969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Orrù CD, Wilham JM, Raymond LD, Kuhn F, Schroeder B, Raeber AJ, Caughey B. 2011. Prion disease blood test using immunoprecipitation and improved quaking-induced conversion. MBio 2:e00078–11. doi: 10.1128/mBio.00078-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sano K, Satoh K, Atarashi R, Takashima H, Iwasaki Y, Yoshida M, Sanjo N, Murai H, Mizusawa H, Schmitz M, Zerr I, Kim YS, Nishida N. 2013. Early detection of abnormal prion protein in genetic human prion diseases now possible using real-time QUIC assay. PLoS One 8:e54915. doi: 10.1371/journal.pone.0054915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henderson DM, Manca M, Haley NJ, Denkers ND, Nalls AV, Mathiason CK, Caughey B, Hoover EA. 2013. Rapid antemortem detection of CWD prions in deer saliva. PLoS One 8:e74377. doi: 10.1371/journal.pone.0074377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elder AM, Henderson DM, Nalls AV, Wilham JM, Caughey BW, Hoover EA, Kincaid AE, Bartz JC, Mathiason CK. 2013. In vitro detection of prionemia in TSE-infected cervids and hamsters. PLoS One 8:e80203. doi: 10.1371/journal.pone.0080203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mazza M, Iulini B, Vaccari G, Acutis PL, Martucci F, Esposito E, Peletto S, Barocci S, Chiappini B, Corona C, Barbieri I, Caramelli M, Agrimi U, Casalone C, Nonno R. 2010. Co-existence of classical scrapie and Nor98 in a sheep from an Italian outbreak. Res Vet Sci 88:478–485. doi: 10.1016/j.rvsc.2009.11.015. [DOI] [PubMed] [Google Scholar]
- 38.Saa P, Castilla J, Soto C. 2006. Ultra-efficient replication of infectious prions by automated protein misfolding cyclic amplification. J Biol Chem 281:35245–35252. doi: 10.1074/jbc.M603964200. [DOI] [PubMed] [Google Scholar]
- 39.Pirisinu L, Nonno R, Esposito E, Benestad SL, Gambetti P, Agrimi U, Zou WQ. 2013. Small ruminant nor98 prions share biochemical features with human Gerstmann-Straussler-Scheinker disease and variably protease-sensitive prionopathy. PLoS One 8:e66405. doi: 10.1371/journal.pone.0066405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.European Food Safety Authority. 2014. Protocol for further laboratory investigations into the distribution of infectivity of atypical BSE. EFSA J 12:3798. doi: 10.2903/j.efsa.2014.3798. [DOI] [Google Scholar]
- 41.Meloni D, Davidse A, Langeveld JP, Varello K, Casalone C, Corona C, Balkema-Buschmann A, Groschup MH, Ingravalle F, Bozzetta E. 2012. EU-approved rapid tests for bovine spongiform encephalopathy detect atypical forms: a study for their sensitivities. PLoS One 7:e43133. doi: 10.1371/journal.pone.0043133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buschmann A, Groschup MH. 2005. Highly bovine spongiform encephalopathy-sensitive transgenic mice confirm the essential restriction of infectivity to the nervous system in clinically diseased cattle. J Infect Dis 192:934–942. doi: 10.1086/431602. [DOI] [PubMed] [Google Scholar]
- 43.Cramm M, Schmitz M, Karch A, Zafar S, Varges D, Mitrova E, Schroeder B, Raeber A, Kuhn F, Zerr I. 9 May 2014. Characteristic CSF prion seeding efficiency in humans with prion diseases. Mol Neurobiol doi: 10.1007/s12035-014-8709-6. [DOI] [PMC free article] [PubMed] [Google Scholar]