Abstract
The EasyNAT assay was evaluated for the detection of tuberculosis in sputum smears from presumptive pulmonary tuberculosis (TB) patients in an African high-TB and high-HIV setting. The sensitivity of the EasyNAT assay was 66.7%, and the specificity and positive predictive value were 100% for the culture-positive patients. The sensitivity was only 10% in the smear-negative and culture-positive patients.
TEXT
Tuberculosis (TB), caused by the Mycobacterium tuberculosis complex, was reported in about 8.9 million cases and led to 1.3 million TB-related deaths in 2012 (1). The early and accurate diagnosis of TB and treatment are the mainstays of TB control (2, 3). Smear microscopy is still the sole TB diagnostic tool used in most resource-limited settings, where a high prevalence of HIV disease reduces its sensitivity from 55% to 39% because of paucibacillary disease (3, 4). The use of TB culture is restricted by the slow growth of the pathogen, high operational costs, and comprehensive biosafety requirements. Hence, a point-of-care test (POCT) for the diagnosis of active pulmonary TB is urgently needed (5).
Nucleic acid amplification tests (NAATs) for TB are high-performing tests for detecting TB RNA and DNA in clinical samples (6). The EasyNAT tuberculosis isothermal nucleic acid amplification diagnostic kit by Ustar Biotechnologies Co., Ltd. uses isothermal cross-priming amplification technology for the qualitative detection of M. tuberculosis. Six to eight primers target the gyrB gene of M. tuberculosis, amplifying it at a constant temperature of 63 or 65°C (7). Isothermal amplification is considered a promising platform for the point-of-care molecular detection of TB, because the technology does not need an initial denaturation step or the addition of a nicking enzyme (8), and it has a short hands-on time (9).
The recent clinical evaluation of the EasyNAT TB kit at four country-level TB clinics in China included 2,200 presumptive TB patients. Lowenstein-Jensen (LJ) culture was used as the reference standard. The overall sensitivity of the kit was 84.1%, and the specificity was 97.8%. The positive and negative predictive values (PPV and NPV, respectively) were 83.4% and 97.9%, respectively. In the smear-negative patients, the sensitivity was 59.8% (10).
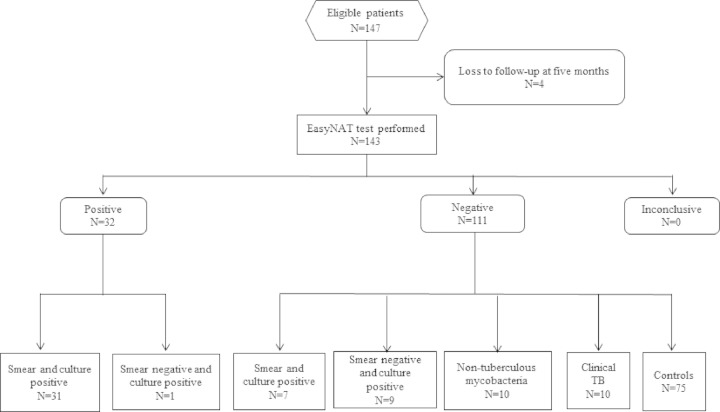
In this substudy, the diagnostic performance of the EasyNAT assay was evaluated for the first time in an African high-TB-burden setting. We evaluated the detection of M. tuberculosis from 1 ml of frozen untreated morning or spot sputum samples from a clinically and microbiologically well-characterized cohort of presumptive pulmonary TB patients in Bagamoyo, Tanzania. Of the 147 participants, four were excluded because information was not available at the 5-month follow-up (Fig. 1). The EasyNAT assay was performed on one sputum sample from each of the 143 eligible participants who were assigned to classification groups, as described in Table 1. In classification groups A and B, the EasyNAT assay was applied only to samples that were also culture positive.
FIG 1.
Patient flow and EasyNAT test results by patient classification.
TABLE 1.
Classification of study population according to clinical and microbiological data
| No. (%) of samples | HIV prevalence (no. [%]) | Description |
|---|---|---|
| 38 (26.6) | 13 (34.2) | Smear-positive/culture-positive, M. tuberculosis |
| 10 (7.0) | 10 (100) | Smear-negative/culture-positive, M. tuberculosis |
| 10 (7.0) | 7 (70.0) | Smear-negative or smear-positive/culture-positive, NTMa |
| 10 (7.0) | 4 (40.0) | All cultures negative, chest radiograph and clinical symptoms very suspect for pulmonary TB (clinically diagnosed TB) |
| 75 (52.4) | 32 (42.7) | All smears and cultures negative, sustained recovery up to 5 mo after antibiotic therapy (controls) |
NTM, nontuberculous mycobacteria.
The mycobacteriological reference included the results of smear microscopy for acid-fast bacilli, the Bactec Mycobacterium Growth Indicator Tube (MGIT) 960 system, and LJ culture, as well as subsequent MPT64 antigen or molecular testing (GenoType MTBC or Mycobacterium CM/AS assay; Hain Lifescience, Nehren, Germany).
The study was approved by the Ifakara Health Institute (IHI) institutional review board and the Medical Research Coordinating Committee of Tanzania. Informed consent was obtained from all study participants.
The frozen stored sputum samples were thawed, and DNA extraction, amplification, hybridization, and detection using the EasyNAT assay were performed as previously described (10). The appearance of both the control and test lines on the EasyNAT kits indicated the presence of M. tuberculosis DNA. A single control line indicated the absence of M. tuberculosis DNA or that DNA copies were below the detection limit of the assay (7, 10). All tests were done at the TB research laboratory in Bagamoyo, Tanzania. The laboratory personnel were blinded to the classification of the patients and the microbiological results other than those of the EasyNAT assay. Parallel rapid HIV testing was done, as per the research protocol.
The mean patient age was 40.5 years (standard deviation, 15.3 years), and 54.6% of the patients were male. The overall HIV prevalence was 46.2% (95% confidence interval [CI], 37.8% to 54.7%). The HIV prevalences between the culture-confirmed TB patients and controls were 47.9% and 42.7%, respectively (P = 0.568, chi-square test).
The sensitivity of the EasyNAT assay with culture as the reference standard was 66.7% (95% CI, 51.6% to 79.6%). All patients who were classified as controls were negative by the EasyNAT assay (specificity, 100.0%; 95% CI, 95.2% to 100.0%). The PPV and NPV for the culture-positive patients were 100.0% (95% CI, 89.1% to 100.0%) and 82.4% (95% CI, 73.0% to 89.6%), respectively (Table 2).
TABLE 2.
Performance of EasyNAT kit with MGIT and LJ combined, smear microscopy, MGIT, and LJ alone versus controls as reference standards
| Performance parameter | Estimate ([95% CI]) for reference standards in: |
|||
|---|---|---|---|---|
| Main analysis | Subanalysis |
|||
| Culture MGIT and LJ vs controls | Smear microscopy vs controls | Culture MGIT alone vs controls | Culture LJ alone vs controls | |
| Sensitivity (%) | 66.7 (51.6–79.6) | 81.6 (65.7–92.3) | 66.7 (51.6–79.6) | 69.2 (52.4–83.0) |
| Specificity (%) | 100 (95.2–100.0) | 100 (95.2–100.0) | 100 (95.2–100.0) | 100 (95.2–100.0) |
| Positive predictive value (%) | 100.0 (89.1–100.0) | 100 (88.8–100.0) | 100.0 (89.1–100.0) | 100.0 (87.2–100.0) |
| Negative predictive value (%) | 82.4 (73.0–89.6) | 91.50 (83.2–96.5) | 82.40 (73.0–89.6) | 86.2 (77.1–92.7) |
| Positive likelihood ratioa | NC | NC | NC | NC |
| Negative likelihood ratio | 0.3 (0.2–0.5) | 0.19 (0.1–0.4) | 0.3 (0.2–0.5) | 0.3 (0.2–0.5) |
The positive likelihood ratio could not be computed, since it is given by sensitivity/(1 − specificity). In all cases, the specificity was 1 (or 100%). NC, not calculated.
The EasyNAT assay identified 31 of 38 smear- and culture-positive patients, respectively (sensitivity, 81.6%; 95% CI, 65.7% to 92.3%). One of the 10 smear-negative and culture-positive TB patients was positive by the EasyNAT assay (sensitivity, 10%; 95% CI, 0.3% to 44.5%). The sensitivity of the EasyNAT assay was 81.6% (95% CI, 65.7% to 92.3%) compared to that with Ziehl-Neelsen (ZN) staining, and 66.7% (95% CI, 51.6% to 79.6) and 69.2% (95% CI, 52.4% to 83.0%), respectively, compared to that with the MGIT system or LJ alone. No M. tuberculosis was detected by the EasyNAT assay in 10 patients categorized as having clinically diagnosed TB and in 10 patients who had the following Mycobacterium species and strains: M. fortuitum strain 1, M. fortuitum strain 2/M. mageritense, M. malmoense/M. haemophilum/M. pasture, M. celatum I+III, M. simiae, M. celatum, M. intracellulare, M. asiaticum, M. scrofulaceum, or M. smegmatis. One patient with both M. intracellulare and M. tuberculosis had a positive EasyNAT assay result.
In this cohort of presumptive TB patients from sub-Saharan Africa, the diagnostic accuracy for smear- and culture-positive TB patients was lower than that in previously studies on the EasyNAT assay (7, 10) and other isothermal NAATs (11). However, the sensitivity of the EasyNAT assay in the rather small group of smear-negative, culture-positive participants was considerably lower than that in the two evaluation studies from Asia (10% versus 59.8% and 87.5, respectively) (7, 10). Since all patients (n = 10) in this classification group were HIV positive, the low detection rate was most likely caused by a paucibacillary TB disease (12) with DNA copies below the detection limit of the assay. This might also explain why none of the clinically diagnosed TB patients, who presumably also had a low bacillary load, were positive by the EasyNAT assay. The assay did not cross-react with nontuberculous mycobacteria.
The estimated cost per EasyNAT test is $4 to 5 (10), which may be considered low. However, the utility of the EasyNAT assay in primary health care settings might be limited, because the sample preparation techniques are fairly complicated and may require a biological safety cabinet (10). Also, the EasyNAT assay does not concurrently detect drug resistance in TB, which is a disadvantage in high-burden settings.
In conclusion, the EasyNAT assay detected M. tuberculosis with an excellent specificity and positive predictive value. The sensitivity was acceptable in the smear-positive patients. However, the low detection rate for the smear-negative, culture-positive sputum samples might be a limitation for wider clinical use and requires further evaluation in larger study populations from different regions that are endemic for TB.
ACKNOWLEDGMENTS
The Ifakara Health Institute purchased the EasyNAT kits at a trial price for research purposes. Ustar Biotechnologies Co., Ltd. was not involved in the study design, analysis, or writing of the manuscript. None of the authors received financial compensation for this work.
This study was part of the Ph.D. project of M.B. financed by the Swiss National Science Foundation grant 32EC30_131192/1 to Hans Peter Beck of the Swiss TPH through EDCTP, in the framework of the TB CHILD Consortium focus on “Evaluation of new and emerging diagnostics for childhood tuberculosis in high burden countries” (IP.2009.32040.007). The TB cohort in Bagamoyo was funded by the Rudolf Geigy Foundation, Basel, Switzerland.
REFERENCES
- 1.WHO. 2013. Global tuberculosis report 2013. World Health Organization, Geneva, Switzerland: http://apps.who.int/iris/bitstream/10665/91355/1/9789241564656_eng.pdf. [Google Scholar]
- 2.Chiang CY, Van Weezenbeek C, Mori T, Enarson DA. 2013. Challenges to the global control of tuberculosis. Respirology 18:596–604. doi: 10.1111/resp.12067. [DOI] [PubMed] [Google Scholar]
- 3.Steingart KR, Henry M, Ng V, Hopewell PC, Ramsay A, Cunningham J, Urbanczik R, Perkins M, Aziz MA, Pai M. 2006. Fluorescence versus conventional sputum smear microscopy for tuberculosis: a systematic review. Lancet Infect Dis 6:570–581. doi: 10.1016/S1473-3099(06)70578-3. [DOI] [PubMed] [Google Scholar]
- 4.Whitelaw A, Peter J, Sohn H, Viljoen D, Theron G, Badri M, Davids V, Pai M, Dheda K. 2011. Comparative cost and performance of light-emitting diode microscopy in HIV–tuberculosis-co-infected patients. Eur Respir J 38:1393–1397. doi: 10.1183/09031936.00023211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pai NP, Vadnais C, Denkinger C, Engel N, Pai M. 2012. Point-of-care testing for infectious diseases: diversity, complexity, and barriers in low- and middle-income countries. PLoS Med 9:e1001306–e1001306. doi: 10.1371/journal.pmed.1001306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niemz A, Boyle DS. 2012. Nucleic acid testing for tuberculosis at the point-of-care in high-burden countries. Expert Rev Mol Diagn 12:687–701. doi: 10.1586/erm.12.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fang R, Li X, Hu L, You Q, Li J, Wu J, Xu P, Zhong H, Luo Y, Mei J, Gao Q. 2009. Cross-priming amplification for rapid detection of Mycobacterium tuberculosis in sputum specimens. J Clin Microbiol 47:845–847. doi: 10.1128/JCM.01528-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu G, Hu L, Zhong H, Wang H, Yusa S-I, Weiss TC, Romaniuk PJ, Pickerill S, You Q. 2012. Cross priming amplification: mechanism and optimization for isothermal DNA amplification. Sci Rep 2:246. doi: 10.1038/srep00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boehme CC, Nabeta P, Henostroza G, Raqib R, Rahim Z, Gerhardt M, Sanga E, Hoelscher M, Notomi T, Hase T, Perkins MD. 2007. Operational feasibility of using loop-mediated isothermal amplification for diagnosis of pulmonary tuberculosis in microscopy centers of developing countries. J Clin Microbiol 45:1936–1940. doi: 10.1128/JCM.02352-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ou X, Song Y, Zhao B, Li Q, Xia H, Zhou Y, Pang Y, Wang S, Zhang Z, Cheng S, Liu C, Zhao Y. 2014. A multicenter study of cross-priming amplification for tuberculosis diagnosis at peripheral level in China. Tuberculosis 94:428–433. doi: 10.1016/j.tube.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 11.Mitarai S, Okumura M, Toyota E, Yoshiyama T, Aono A, Sejimo A, Azuma Y, Sugahara K, Nagasawa T, Nagayama N, Yamane A, Yano R, Kokuto H, Morimoto K, Ueyama M, Kubota M, Yi R, Ogata H, Kudoh S, Mori T. 2011. Evaluation of a simple loop-mediated isothermal amplification test kit for the diagnosis of tuberculosis. Int J Tuberc Lung Dis 15:1211–1217. doi: 10.5588/ijtld.10.0629. [DOI] [PubMed] [Google Scholar]
- 12.Sharma SK, Kadhiravan T, Banga A, Goyal T, Bhatia I, Saha PK. 2004. Spectrum of clinical disease in a series of 135 hospitalised HIV-infected patients from north India. BMC Infect Dis 4:52–52. doi: 10.1186/1471-2334-4-52. [DOI] [PMC free article] [PubMed] [Google Scholar]