Abstract
Background & objectives:
Shiga toxin producing Escherichia coli (STEC) is an important zoonotic foodborne pathogen, capable of causing haemorrhagic colitis (HC) and haemolytic uremic syndrome (HUS). As data from India on human infections caused by STEC are limited, this study was carried out for hospital based surveillance for STEC as a causative agent of diarrhoea, bloody diarrhoea and HUS at a tertiary care centre and to study the virulence gene profile and strain relatedness by multi locus variable tandem repeat analysis (MLVA).
Methods:
A total of 600 stool samples were studied. Stool samples of every fifth patient presenting with non-bloody diarrhoea, all cases of bloody diarrhoea and diarrhoea associated HUS (D+HUS) were collected from October 2009 to September 2011. Stool samples were cultured for STEC and characterization of STEC was done by serogrouping, virulence genes analysis, and MLVA typing.
Results:
STEC were isolated as a sole pathogen from 11 stool samples [5 of 290 (1.7%) non-blood diarrhoea and 5 of 300 (1.6%) blood diarrhoea cases]. STEC was also isolated from one fatal case of HUS who was an eight month old child. Only six of 11 isolates were positive for stx2 gene, whereas stx1 was present in all 11 isolates. Only one isolate was positive for eae. Other adhesion genes present were iha in five isolates, followed by toxB and efa1 in two each and saa gene in one, isolate. Among the plasmid encoded genes, espP, hly and etpD were each present in one isolate each. In the MLVA typing, diverse profiles were obtained except two untypeable isolates from different patients shared the same MLVA profile. Both these isolates were not epidemiologically linked.
Interpretation & conclusions:
This study demonstrated that STEC could be a causative agent of diarrhoea, bloody diarrhoea and sporadic HUS. However, further work needs to be done to study and explore the prevalence of these organisms in the food chain in this region.
Keywords: Bloody diarrhoea, haemolytic uremic syndrome, human, Shiga-toxigenic Escherichia coli
Shiga-toxigenic Escherichia coli (STEC) is the most common food-borne zoonotic pathogen implicated in causing gastrointestinal illnesses, haemorrhagic colitis (HC) and haemolytic uremic syndrome (HUS). Though O157:H7 is the most common serotype implicated in human illness, other serotypes are also emerging. The Centers for Disease Control and Prevention (CDC) data show six non-O157 serotypes [O111:H8 or Non-motile (NM); O103:H2, H11, H25; O26:H11 or NM; O45:H2 or NM; O121:H19 or H7; and O145:NM] to be responsible for 71 per cent of non-O157 STEC illnesses in the USA1. STEC infections are a cause of concern particularly for elderly and paediatric patients because of the higher risk of HUS in these populations2. CDC has updated guidelines in October 2009 for the detection of STEC in acute community-acquired diarrhoea, and recommended tests for detection for shiga toxins or stx genes in addition to conventional culture3 (http://www.cdc.gov/mmwr/preview/mmwrhtml/rr5812a1.html).
In contrast to the developed world, little is known about STEC epidemiology in humans in India, though multiple reservoirs for STEC have been described, including healthy and diarrhoeagenic cattle, calves, goats and yaks4,5,6,7. STEC were isolated from 1.4 and 0.6 per cent of human bloody and watery diarrhoea samples in a study from Kolkata5. A study from Vellore has reported 2.3 per cent isolation in preschool-aged children admitted to the hospital with diarrhoea and an equivalent (2%) asymptomatic population8. However, from India no outbreaks of human illness are reported and no data are available supporting the role of STEC in HUS. Therefore, we conducted a hospital based surveillance for these pathogens in human cases of diarrhoea and HUS presenting to a tertiary case centre in north India. The virulence profile and the genetic inter-relatedness of these organisms were also studied using multi locus variable tandem repeat analysis (MLVA).
Material & Methods
Sample collection: Stool samples (n=600) were collected from every fifth patient presenting with non-bloody diarrhoea (290 of 1438), all cases of bloody diarrhoea and diarrhoea associated HUS (D+HUS) (n=310) in patients from October 2009 to September 2011 in Postgraduate Institute of Medical Education and Research (PGIMER), Chandigarh. The samples were submitted to the Enteric Laboratory of department of Medical Microbiology at the PGIMER, Chandigarh. A total of 1270 cases were excluded from the study. Acute non-bloody diarrhoea was defined as the sudden onset of diarrhoea with frequent loose stool >3 times/day without the presence of visible blood. Bloody diarrhoea was defined as the presence of visible blood in loose stools, and diarrhoea associated HUS was defined as the occurrence of haemolytic anaemia, acute renal failure and thrombocytopenia after an episode of bloody diarrhoea. Patients presenting with diarrhoea at the time of OPD of both pediatric and adult patients (Medicine and Gastroenterology OPDs) visit/admission or developing diarrhoea within 48 h of admission were included while patients who developed diarrhoea after 48 h of admission to the hospital were excluded. The study protocol was approved by the Institute Ethics committee. Written informed consent was obtained from patients.
Culture of stool samples: About 5 ml of fresh stool was collected in a wide mouth container and a small portion of the stool specimen was transferred to Cary-Blair medium (Difco, USA). The stool sample was grossly examined for presence of blood and mucus. All stool samples were plated onto the MacConkey agar (Hi-Media Ltd., Mumbai, India), xylose lysine deoxycholate agar (XLD) (Hi-Media Ltd., Mumbai, India), thiosulphate citrate bile salt sucrose agar (TCBS) (Difco, USA), blood agar (Difco, USA) and inoculated into selenite F (Hi-Media Ltd., Mumbai, India) and alkaline peptone water (Hi Media Ltd., Mumbai, India) for isolation of Salmonella, Shigella, Vibrio cholerae and Aeromonas. A multiplex PCR assay was used to characterize the biochemically confirmed E. coli for enteroaggregative (EAEC), enterotoxigenic (ETEC) and enteropathogenic E. coli (EPEC)9.
Processing of samples for STEC: For STEC screening, enrichment of stool specimens was performed by inoculating. 5 ml of stool samples into 3 ml of enriched culture (EC) medium (Difco, USA) and incubated overnight at 37˚C with constant shaking. DNA was extracted from one ml of the broth culture by boiling method10. Supernatant was used in the PCR for the detection of stx1 and stx2 genes by using primers and PCR conditions as described previously11. Broth cultures positive for either stx1 and/or stx2 by PCR, were serially diluted in 10 mM phosphate-buffered saline (PBS) (pH 7.0) and 100 μl volume of each dilution was spread on Sorbitol MacConkey agar (SMAC) (Difco, USA), in duplicate. Isolated presumptive E. coli colonies including non-sorbitol fermenting colonies were picked up randomly from dilution plates and subjected to biochemical analysis for confirmation. The E.coli colonies were further spot-inoculated on master Luria agar plates. After division into rows and columns, colonies from each row and each column were pooled into fresh EC broths. After overnight incubation at 37˚C, DNA was extracted by boiling method. Duplex PCR, using primers for stx1 and stx2 was performed. When positive results were obtained in one column and one row, the shared colony was considered as STEC positive. A stx-positive colony was preserved in brain heart infusion broth (BHI, Difco, USA) containing 15 per cent glycerol at -80°C, till further use.
Phenotypic characterization
Serogrouping: On the basis of O antigen, isolates were serogrouped by the National Salmonella and Escherichia Centre at the Central Research Institute at Kasauli, India.
Analysis of virulence genes: PCR assays were carried out targeting the STEC virulence genes stx1, stx2, eae, hly, katP, espP and adhesion genes iha, saa, efa1 and toxB as described previously12,13,14,15,16,17. The following standard strains of STEC were used as controls in the virulence gene detection: EDL933, VT3 and Wani 1. EDL933 was gifted from Pasteur Institute, Paris, France. VT3 strain was obtained from National Institute of Cholera and Enteric Diseases (NICED), Kolkata and Wani 1, was obtained from Dr Shakil Wani, S.K University of Agriculture Sciences and Technology of Kashmir, Srinagar. All three belonged to serogroup O157 and were characterized for stx1 and stx2 as well as various other virulence genes.
Antimicrobial drug susceptibility testing: Isolates were tested for antimicrobial drug resistance by Kirby Bauer disc diffusion method18 for nalidixic acid (30 μg), norfloxacin (10 μg), ofloxacin (5μg), amikacin (30 μg), ceftriaxone (30 μg), cefoperazone (30 μg), gentamicin (30 μg), co-trimoxazole (25 μg), azithromycin (30 μg), and ciprofloxacin (5 μg) (Oxoid Limited, Hampshire, UK) according to CLSI (Clinical and Laboratory Standards Institute) guidelines19. E.coli ATCC 25922 was used as the control strain.
MLVA typing: MLVA typing was performed by using five VNTR loci CVN001, CVN004, CVN007, CVN014 and CVN015 as described previously20. These loci were chosen on the basis of reproducibility and discriminatory indices and PCR was performed as described20. EDL933 was used as the control strain. After amplification, products were separated on a 6 per cent denaturing polyacrylamide gel (PAGE) by using SequiGen gel apparatus 38 × 50 × 0.4 cm (BioRad Laboratories Inc., USA). After electrophoresis, gels were fixed overnight in 10 per cent acetic acid and then silver stained using the Promega silver staining kit (Promega, USA). On the basis of amplicon size, allele numbers were assigned to each locus20 and allele string was made for each isolate with an order: CVN001-CVN004-CVN007-CVN014-CVN015.
The dendrogram was constructed by using UPGMA clustering method with START vs. 2.0.5 software21. Minimum spanning tree (MST) was constructed by using the online software (http://pubmlst.org/analysis.).
Results
Table I shows the age and sex distribution along with results of stool culture. Of the 600 cases, 56 per cent were males (mean age 4.7 yr; range from 2 months to 60 yr) and 44 per cent were females (mean age 5.2 yr; range from 40 days to 67 yr). From 600 stool samples (290 from patients with acute non-bloody diarrhoea, 300 from bloody diarrhoea and 10 from HUS) screened by PCR, STEC were isolated as a sole pathogen from 11 (1.8%) samples. Of these 11 isolates, five isolates each were obtained each from acute non-bloody diarrhoea (1.7%) and bloody diarrhoea (1.6%) cases and one was isolated from a case of HUS.
Table I.
Age, sex and type of disease distribution of human patients whose stool samples grew STEC
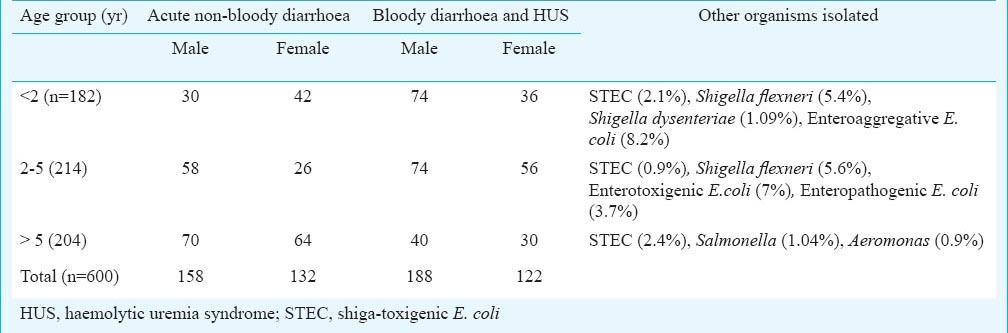
Table II shows details of 11 patients from whom STEC were isolated. Of these, eight were admitted to the hospital while the remaining three were out-door patients. Their age ranged from <2 months to 60 yr. All patients were admitted to different wards of the hospital. Stool sample of only one HUS case, an eight month old male infant, was positive for STEC. HUS developed after a week of bloody diarrhoea. The patient had the typical triad of acute renal failure (urea 164 mg/dl, creatinine 7.9 mg/dl), anaemia (6.4 g/dl) and thrombocytopenia (platelet count 99000/μl). There was an epidemiological evidence of contact with animals and the case was fatal22. The STEC isolate was positive for stx1, stx2, eae and iha. Thus enterohaemorrhagic E.coli (EHEC) which is a subset of STEC (defined as stx1+/stx2+ and eae+) was isolated from stool sample of only one patient.
Table II.
Demographic details of STEC positive acute non-bloody and bloody diarrhoea cases and virulence factors harboured

Characterization of virulence genes: The stx1 gene was the most common virulence gene, present in 11 (100%) isolates followed by stx2 in six (54.5%) isolates. The eae gene was present in HUS isolate only. Among the plasmid encoded genes, espP, hly and etpD were each present in one (9%) isolate. The most common adhesion gene was iha, present in five (45.4%) isolates, followed by toxB and efa1 each in two (18%) isolates and saa gene in one (9%), isolate (Table II).
Serogrouping: Isolates belonged to serogroup O85 (2 isolates), O95 (one isolate) and O102 (one isolate). Six isolates were found to be untypable with somatic (O) antiserum and one was found to be rough.
Antimicrobial drug susceptibility testing & MLVA: All isolates were found to be sensitive to all the antimicrobials tested. In the dendrogram analysis, 11 MLVA genotypes were observed (Fig. 1); 10 genotypes were represented by one isolate each and one genotype was shared by two isolates (9,10). Two main clades were identified (clade I and clade II). Clade I included four isolates (including EDL933) and clade II included eight isolates. In the MST analysis, HUS and two other isolates were found to be closely related to EDL933 control STEC strain (Fig. 2).
Fig. 1.
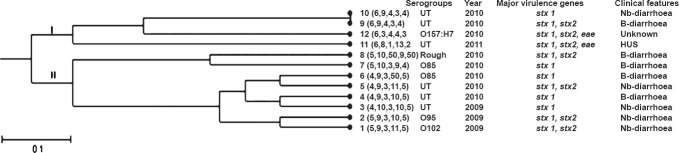
Dendrogram based on MLVA profiles of 11 non-O157 STEC isolates. Dendrogram based on allelic profiles of MLVA for 11 isolates including reference strain 12 (EDL933) was constructed by using START v2. Nb, non-bloody diarrhoea; B, bloody.
Fig. 2.
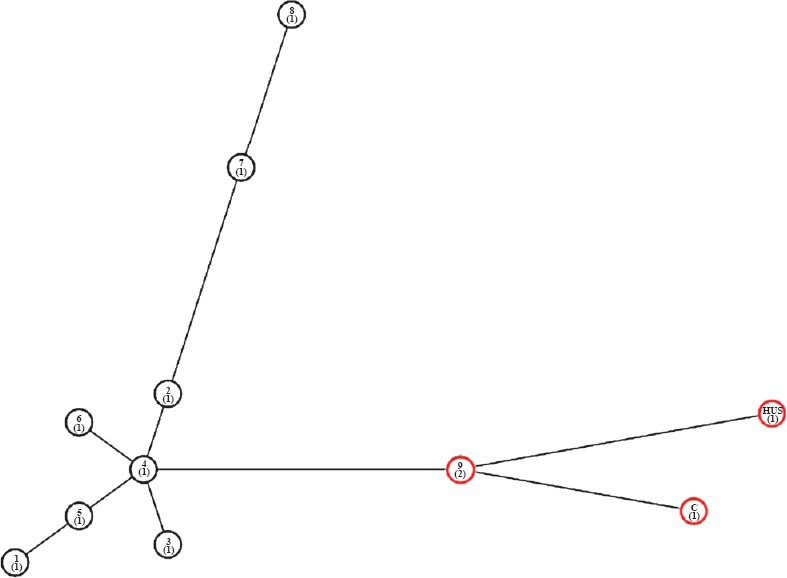
Minimum spanning tree (MST). MST based on the MLVA profiles of STEC isolates was constructed. Profile 9 in red circle represents the two isolates (H9, H10) sharing the profile. C-EDL933 control.
Discussion
This study was carried out in a tertiary care centre of north India. The geographic region around this centre is a major milk producing area of India and animal rearing is one of the main sources of income. In the present study, STEC were isolated from five cases non-bloody diarrhoea (1.7%) and five bloody diarrhoea (1.6%) cases. These rates were similar to those reported from the Netherlands (1.7% from cases of bloody diarrhoea)23 and higher than those reported from Alberta, Canada (1.4% in stool samples suspected for viral gastroenteritis)2. Although STEC reservoirs exist and are clearly present in the food chain and environment in India, yet, the human illness appears to be infrequent. In a study from Kolkata, STEC were isolated from 1.4 and 0.6 per cent of human bloody and watery diarrhoea samples5. Other pathogens like Salmonella, Shigella flexneri, S. dysentriae, Aeromonas, EPEC, ETEC and EAEC were also isolated from the stool samples in our study. Isolation rate for STEC as a causative agent in our hospital-based surveillance for community acquired diarrhoea was slightly higher than Salmonella (1.04%), indicating that these organisms should also be routinely looked for in the diagnostic microbiology laboratories, especially in areas where animals are reared.
In the present study, all STEC were sensitive to all 10 antimicrobials tested. Khan et al24 demonstrated antimicrobial resistance in 49.2 per cent of the STEC isolated from cow stool, beef and human stool samples collected from Kolkata, and resistance was commonly observed to ampicillin, tetracycline, and streptomycin and less frequently to cephalothin, co-trimoxazole, nalidixic acid, and neomycin. A high level of antimicrobial resistance has been reported in other pathotypes of diarrhoeagenic E. coli (enteroaggregative E coli, enterotoxigenic E. coli and enteropathogenic E. coli) from our region9. The antimicrobial sensitivity of STEC may be due to less antimicrobial pressure on these bacteria.
It has been shown that a large spectrum of serologically different STEC types can infect humans25. The serogroups found in this study have also been reported from animal stool and food samples from India4,6,7. The serogroup O157 was conspicuously absent.
The stx1 gene was present in all the isolates whereas stx2 was present in six of 11 isolates. Strains carrying eae and stx2 are considered to be more pathogenic to humans26. The low prevalence of eae positivity indicates the absence of LEE (locus for enterocyte attaching and effacing lesions) or presence of eae variants/other adhesins in these isolates. Though many studies have shown that LEE is essential for host colonization and virulence27,28, others have demonstrated that some STEC isolates without LEE, such as STEC O113:H21, O1:H7, NT:H7, O76:H7, O128:H2, O91:H, O113:H21, O116:H21, O130:H11 and O5:H- are associated with sporadic cases and outbreaks of HUS29,30,31. Other adhesins described for STEC include efaI (EHEC factor for adherence), chromosomal Iha (iron-regulated gene A homologue)32, STEC autoagglutinating adhesin (Saa)33, and plasmid-encoded ToxB34. In our study, the most common adhesion gene was iha, followed by toxB and efa1; saa gene was present in one isolate only. Other toxins such as espP, etpD, hly and katP present on the large plasmid may also contribute to the severity of STEC illness. These genes were found to be variably present in our isolates. Though, the interplay between these genes is not well understood, but these factors provide additional traits to increase the virulence in STEC.
Diverse MLVA profiles were obtained from patients except for two showing the same MLVA profile. Both of these isolates were not epidemiologically linked. Small number of HUS cases was the main limitation of our study. To conclude, this study demonstrates the presence of STEC isolates as causative agents of diarrhoea and bloody diarrhoea and sporadic HUS. Further work needs to be done to study the epidemiological and evolutionary relationships between STEC present in the environment, cattle and/or the food chain, and STEC ultimately acquired by humans.
Acknowledgment
Authors acknowledge the Indian Council of Medical Research (ICMR), New Delhi, India for financial support and the Director, National Salmonella and Escherichia Centre, Central Research Institute, Kasauli, India for serogrouping of the isolates.
References
- 1.Bosilevac JM, Koohmaraie M. Prevalence and characterization of non-O157 shiga toxin-producing Escherichia coli isolates from commercial ground beef in the United States. Appl Environ Microbiol. 2011;77:2103–12. doi: 10.1128/AEM.02833-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Couturier MR, Lee B, Zelyasand N, Chui L. Shiga-toxigenic Escherichia coli detection in stool samples screened for viral gastroenteritis in Alberta, Canada. J Clin Microbiol. 2011;49:574–8. doi: 10.1128/JCM.01693-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. [accessed on October 12, 2012]. Available from: http://www.cdc.gov/mmwr /preview/mmwrhtml/rr5812a1.html .
- 4.Bandyopadhyay S, Lodh C, Rahaman H, Bhattacharya D, Bera AK, Ahmed FA, et al. Characterization of shiga toxin producing (STEC) and enteropathogenic Escherichia coli (EPEC) in raw yak (Poephagus grunniens) milk and milk products. Res Vet Sci. 2012;93:604–10. doi: 10.1016/j.rvsc.2011.12.011. [DOI] [PubMed] [Google Scholar]
- 5.Khan A, Yamasaki S, Sato T, Ramamurthy T, Pal A, Datta S, et al. Prevalence and genetic profiling of virulence determinants of non-O157 Shiga toxin-producing Escherichia coli isolated from cattle, beef, and humans, Calcutta, India. Emerg Infect Dis. 2002;8:54–62. [PubMed] [Google Scholar]
- 6.Wani SA, Hussain I, Fayaz I, Mirand MA, Nishikawa Y. Subtype analysis of stx1, stx2 and eae genes in Shiga toxin-producing Escherichia coli (STEC) and typical and atypical enteropathogenic E. coli (EPEC) from lambs in India. Vet J. 2009;182:489–90. doi: 10.1016/j.tvjl.2008.07.017. [DOI] [PubMed] [Google Scholar]
- 7.Wani SA, Samanta I, Munshi ZH, Bhatand MA, Nishikawa Y. Shiga toxin-producing Escherichia coli and enteropathogenic Escherichia coli in healthy goats in India: occurrence and virulence properties. J Appl Microbiol. 2006;100:108–13. doi: 10.1111/j.1365-2672.2005.02759.x. [DOI] [PubMed] [Google Scholar]
- 8.Rajendran P, Rajan DP, Kangand G, Thorpe CM. Shiga toxin-producing Escherichia coli infection in South India. J Med Microbiol. 2009;58:1525–6. doi: 10.1099/jmm.0.010900-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aranda KRS, Fagundes-Neto U, Scaletsky ICA. Evaluation of multiplex PCRs for diagnosis of infection with diarrheagenic Escherichia coli and Shigella spp. J Clin Microbiol. 2004;42:5849–53. doi: 10.1128/JCM.42.12.5849-5853.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Queipo-Ortuño MI, De Dios Colmenero J, Macias M, Bravo MJ, Morata P. Preparation of bacterial DNA template by boiling and effect of immunoglobulin Gas an inhibitor in real-time PCR for serum samples from patients with brucellosis. Clin Vaccine Immunol. 2008;15:293–6. doi: 10.1128/CVI.00270-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar A, Taneja N, Kumarand Y, Sharma M. Detection of Shiga toxin variants among Shiga toxin-forming Escherichia coli isolates from animal stool, meat and human stool samples in India. J Appl Microbiol. 2012;113:1208–16. doi: 10.1111/j.1365-2672.2012.05415.x. [DOI] [PubMed] [Google Scholar]
- 12.Paton AW, Paton JC. Direct detection and characterization of Shiga toxigenic Escherichia coli by multiplex PCR for stx1, stx2, eae, ehxA, and saa. J Clin Microbiol. 2002;40:271–4. doi: 10.1128/JCM.40.1.271-274.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paton AW, Paton JC. Direct detection and characterization of Shiga toxigenic Escherichia coli by multiplex PCR for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157. J Clin Microbiol. 1998;36:598–602. doi: 10.1128/jcm.36.2.598-602.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmidt H, Henkeland B, Karch H. A gene cluster closely related to type II secretion pathway operons of Gram-negative bacteria is located on the large plasmid of enterohemorrhagic Escherichia coli O157 strains. FEMS Microbiol Lett. 1997;148:265–72. doi: 10.1111/j.1574-6968.1997.tb10299.x. [DOI] [PubMed] [Google Scholar]
- 15.Brunder W, Schmidtand H, Karch H. EspP, a novel extracellular serine protease of enterohaemorrhagic Escherichia coli O157:H7 cleaves human coagulation factor V. Mol Microbiol. 1997;24:767–78. doi: 10.1046/j.1365-2958.1997.3871751.x. [DOI] [PubMed] [Google Scholar]
- 16.Tarr CL, Large TM, Moeller CL, Lacher DW, Tarr PI, Acheson DW, et al. Molecular characterization of a serotype O121:H19 clone, a distinct Shiga toxin-producing clone of pathogenic Escherichia coli. Infect Immun. 2002;70:6853–59. doi: 10.1128/IAI.70.12.6853-6859.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brunder W, Schmidtand H, Karch H, Kat P. a novel catalaseperoxidase encoded by the large plasmid of enterohaemorrhagic Escherichia coli O157:H7. Microbiology. 1996;142:3305–15. doi: 10.1099/13500872-142-11-3305. [DOI] [PubMed] [Google Scholar]
- 18.Bauer AW, Kirby WMM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966;45:493–6. [PubMed] [Google Scholar]
- 19.Wayne,PA: CLSI; 2008. Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing. 18th Informational Supplement. M100-S18. [Google Scholar]
- 20.Lindstedt BA, Brandal LT, Aas L, Vardundand T, Kapperud G. Study of polymorphic variable-number of tandem repeats loci in the ECOR collection and in a set of pathogenic Escherichia coli and Shigella isolates for use in a genotyping assay. J Microbiol Methods. 2007;69:197–205. doi: 10.1016/j.mimet.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 21.Joley KA, Feil EJ, Chanand MS, Maiden MC. Sequence type analysis and recombinational tests (START) Bioinformatics. 2001;17:1230–31. doi: 10.1093/bioinformatics/17.12.1230. [DOI] [PubMed] [Google Scholar]
- 22.Kumar A, Taneja N, Singhi S, Shahand R, Sharma M. Haemolytic uraemic syndrome in India due to Shiga toxigenic Escherichia coli. J Med Microbiol. 2013;62:157–60. doi: 10.1099/jmm.0.044131-0. [DOI] [PubMed] [Google Scholar]
- 23.van Duynhoven YT, Friesema IH, Schuurman T, Roovers A, van Zwet AA, Sabbe LJ, et al. Prevalence, characterisation and clinical profiles of Shiga toxin-producing Escherichia coli in The Netherlands. Clin Microbiol Infect. 2008;14:437–45. doi: 10.1111/j.1469-0691.2008.01963.x. [DOI] [PubMed] [Google Scholar]
- 24.Khan A, Das SC, Ramamurthy T, Sikdar A, Khanam J, Yamasaki S, et al. Antibiotic resistance, virulence gene, and molecular profiles of Shiga toxin-producing Escherichia coli isolates from diverse sources in Calcutta, India. J Clin Microbiol. 2002;40:2009–15. doi: 10.1128/JCM.40.6.2009-2015.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boerlin P, McEwen SA, Boerlin-Petzold F, Wilson JB, Johnsonand RP, Gyles CL. Associations between virulence factors of Shiga toxin-producing Escherichia coli and disease in humans. J Clin Microbiol. 1999;37:497–503. doi: 10.1128/jcm.37.3.497-503.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ostroff SM, Tarr PI, Neill MA, Lewis JH, Hargrett-Bean N, Kobayashi JM. Toxin genotypes and plasmid profiles as determinants of systemic sequelae in Escherichia coli O157:H7 infections. J Infect Dis. 1989;160:994–8. doi: 10.1093/infdis/160.6.994. [DOI] [PubMed] [Google Scholar]
- 27.Marches O, Nougayrede JP, Boullier S, Mainil J, Charlier G, Raymond I, et al. Role of tir and intimin in the virulence of rabbit enteropathogenic Escherichia coli serotype O103:H2. Infect Immun. 2000;68:2171–82. doi: 10.1128/iai.68.4.2171-2182.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McKee ML, Melton-Celsa AR, Moxley RA, Francis DH, O’Brien AD. Enterohemorrhagic Escherichia coli O157:H7 requires intimin to colonize the gnotobiotic pig intestine and to adhere to HEp-2 cells. Infect Immun. 1995;63:3739–44. doi: 10.1128/iai.63.9.3739-3744.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bonnet R, Souweine B, Gauthier G, Rich C, Livrelli V, Sirot J, et al. Non-O157:H7 Stx2-producing Escherichia coli strains associated with sporadic cases of hemolytic-uremic syndrome in adults. J Clin Microbiol. 1998;36:1777–80. doi: 10.1128/jcm.36.6.1777-1780.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elliott EJ, Robins-Browne RM, O’Loughlin EV, Bennett-Wood V, Bourke J, Henning P, et al. Nationwide study of haemolytic uraemic syndrome: clinical, microbiological, and epidemiological features. Arch Dis Childhood. 2001;85:125–31. doi: 10.1136/adc.85.2.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Newton HJ, Sloan J, Bulach DM, Seemann T, Allison CC, Tauschek M, et al. Shiga toxin-producing Escherichia coli strains negative for locus of enterocyte effacement. Emerg Infect Dis. 2009;15:372–80. doi: 10.3201/eid1502.080631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tarr PI, Bilge SS, Vary JC, Jr, Jelacic S, Habeeb RL, Ward TR, et al. Iha: a novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a recently acquired chromosomal island of conserved structure. Infect Immun. 2000;68:1400–7. doi: 10.1128/iai.68.3.1400-1407.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paton AW, Srimanote P, Woodrowand MC, Paton JC. Characterization of Saa, a novel autoagglutinating adhesin produced by locus of enterocyte effacement-negative Shiga-toxigenic Escherichia coli strains that are virulent for humans. Infect Immun. 2001;69:6999–7009. doi: 10.1128/IAI.69.11.6999-7009.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tatsuno I, Horie M, Abe H, Miki T, Makino K, Shinagawa H, et al. ToxB gene on pO157 of enterohemorrhagic Escherichia coli O157:H7 is required for full epithelial cell adherence phenotype. Infect Immun. 2001;69:6660–9. doi: 10.1128/IAI.69.11.6660-6669.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]


