Abstract
Background
Recent studies have demonstrated that DNA polymorphisms in the solute carrier family 30 member 8 (SLC30A8) gene confer the risk susceptibility to type 2 diabetes (T2D). The present study aimed to analyze DNA methylation levels of this gene in T2D and diabetic nephropathy (DN).
Results
We confirmed the genetic association study of SLC30A8 in 992 Malay subjects with normal glucose tolerance and T2D patients with and without DN. Genotyping was conducted with TaqMan allelic discrimination. SNP rs11558471(A/G) in the SLC30A8 gene was strongly associated with T2D (P = 0.002, OR = 1.334, 95% CI = 1.110 to 1.602) and moderately associated with DN (P = 0.041, OR = 1.399, 95% CI = 1.013 to 1.932). We further performed DNA methylation analysis of six CpG sites in the SLC30A8 gene promoter with bisulfite pyrosequencing protocol. The average DNA methylation levels of the SLC30A8 gene in all Malay subjects were at approximately 81.4%. DNA methylation levels of the SLC30A8 gene in T2D patients were higher compared to non-diabetic subjects (82.9% vs. 80.1%, P = 0.014). But no significant difference of DNA methylation levels of the SLC30A8 gene between T2D patients with and without DN was observed.
Conclusion
The present study thus provides the first evidence that increased DNA methylation of the SLC30A8 gene promoter is associated with T2D but not DN in a Malay population.
Keywords: DNA methylation, SLC30A8, Type 2 diabetes
Background
The solute carrier family 30 member 8 (SLC30A8) gene is encoded for a zinc efflux transporter and highly expressed in the pancreas, particularly in alpha, beta, and PP cells of the islets of Langerhans. Functionally, this transporter is essential for zinc flux into beta-cell insulin-secretory granules and the subsequent crystallization of hexameric insulin [1]. Pathological studies have demonstrated that the SLC30A8 gene expression levels are downregulated in pancreatic islets of diabetic mice [2]. The downregulation of the SLC30A8 gene expression results in the reduction of insulin content and glucose-inducible insulin secretion [3].
Genome-wide association studies (GWASs) have identified a number of susceptibility variants for type 2 diabetes (T2D). The common alleles of single nucleotide polymorphisms (SNPs), rs13266634(C/T, Arg276Trp), and rs11558471(A/G) in the SLC30A8 gene are found to confer the risk susceptibility in T2D [4-9]. The genes including SLC30A8 identified by GWAS, however, can only explain approximately 10% of the overall heritable risk of T2D, which challenges our expectations to translate genetic information into clinical practice [10-12]. The missing information on heritability in T2D includes the impact of rare variants and epigenetic factors. The latter is involved in the complex interplay between genes and the environment [13-15]. To further understand the genetic effects of SLC30A8, Flannick et al. have recently conducted a mutation screening study for rare variants in the gene (the minor allele frequency is less 1%) and suggested that the rare variants with loss of function may protect against T2D [16].
Epigenetic factors mainly including DNA methylation changes have been considered to be involved in the pathogenesis of T2D [13-15]. The methylation of the 5′-carbon of cytosine, often in a gene promoter, is a form of epigenetic modification that does not affect the primary DNA sequences but affects secondary interactions that play a critical role in the regulation of gene expression [17,18]. DNA methylation levels are commonly analyzed at clusters of CpG methylation sites in the gene promoter and used for indication of epigenetic effects. We have recently reported that DNA methylation changes of the insulin-like growth factor 1 (IGF1) and its binding proteins (IGFBP1 and IGFBP7) are associated with T2D [19-22]. But whether the SLC30A8 gene is subject to epigenetic effects in T2D is unknown.
Malaysia is a country with a high prevalence of T2D at 14.9% for adults aged 30 years and above according to the latest National Health and Morbidity Surveys in 2006 [23]. Moreover, diabetic nephropathy (DN) is the most common cause of end-stage renal disease (ESRD). In this country, DN contributes to 57% of patients with T2D [24-26]. We have currently collected a cohort of Malay subjects including normal glucose tolerance (NGT) and T2D patients with and without DN for genetic studies. In the present study, we conducted genetic association study and further analyzed DNA methylation alteration of the SLC30A8 gene in this Malay population. The results may provide useful information to evaluate the genetic and epigenetic effects of the SCL30A8 gene in T2D and DN.
Results
Association of SLC30A8 genetic polymorphisms with T2D and DN
A total of 992 Malay subjects including NGT subjects (n = 476) and T2D patients with or without DN (516) were included in the genotyping experiments. Clinical parameters in all Malay subjects selected for genetic association study are summarized in Table 1.
Table 1.
Clinical parameters of Malay subjects in genetic association study
| Group | NGT | T2D a | T2D-DN | T2D + DN | P values b/c |
|---|---|---|---|---|---|
| N (men/women) | 476 (224/252) | 516 (260/256) | 239 (103/136) | 124 (66/58) | |
| Age (years) | 42 ± 15 | 56 ± 10 | 55 ± 10 | 57 ± 10 | <0.001/NS |
| Diabetes duration (years) | - | 14 ± 8 | 13 ± 8 | 14 ± 7 | -/NS |
| BMI (kg/m2) | 26 ± 7.0 | 28 ± 5 | 29 ± 5 | 27 ± 5 | <0.001/0.059 |
| SBP (mm Hg) | 127 ± 19 | 144 ± 25 | 139 ± 22 | 155 ± 31 | <0.001/0.001 |
| DBP (mm Hg) | 80 ± 11 | 81 ± 12 | 80 ± 12 | 82 ± 13 | NS/NS |
| FPG (mmol/l) | 4.87 ± 1.02 | 5.0 ± 1.22 | 4.76 ± 1.00 | 5.6 ± 1.05 | 0.028/0.035 |
| HbA1c (%) | 5.4 ± 0.48 | 8.1 ± 2.17 | 7.6 ± 2.0 | 8.6 ± 2.4 | <0.001/<0.001 |
| Serum creatinine (μmol/l) | 67.8 ± 33.7 | 122.3 ± 114.4 | 91.0 ± 72.9 | 207.3 ± 178.7 | <0.001/<0.001 |
| Urine creatinine (mmol/l) | 12.3 ± 7.4 | 10.0 ± 6.8 | 11.4 ± 6.2 | 5.0 ± 3.52 | <0.001/<0.001 |
| ACR (mg/mmol) | 3.32 ± 16.1 | 57.3 ± 175.4 | 1.10 ± 0.819 | 249.2 ± 311.5 | <0.001/<0.001 |
| eGFR (ml/min/1.73 m2) | 112.9 ± 45.2 | 71.4 ± 38.2 | 81.0 ± 29.9 | 48.3 ± 35.2 | <0.001/<0.001 |
Data are expressed as means ± SD.
NGT normal glucose tolerance, T2D type 2 diabetes, DN diabetic nephropathy, BMI body mass index, SBP systolic blood pressure, DBP diastolic blood pressure, FPG fasting plasma glucose, HbA1c glycosylated hemoglobin, ACR albumin creatinine ratio, eGFR estimated glomerular filtration rate.
aThe patients with microalbuminuria were included.
b P values were from tests of NGT vs. all T2D.
c P values were from tests of T2D without DN vs. T2D with DN.
The SLC30A8 gene is located on chromosome 8q24.11. Both SNPs rs13266634(C/T) and rs11558471(A/G) reside within exon 9 of the gene (Figure 1). The first SNP is non-synonymous, in which the amino acid arginine is changed to tryptophan, while the second one is located at 3′-UTR of SLC30A8. We genotyped these two SNPs in the Malay subjects with NGT and T2D, and data showed the genotype distributions of these two SNPs in Hardy-Weinberg equilibrium (HWE). Figure 2 illustrates that the frequencies of the major alleles in both SNPs , that is, allele C of rs13266634(C/T) and A of rs11558471(A/G) were increased from NGT to T2D without DN and to T2D with DN. Analyses for single marker association indicated that the A allele of rs11558471(A/G) was strongly associated with T2D (the A allele frequencies between NGT and all T2D were 0.552 vs. 0.620, P = 0.002, OR = 1.334, 95% CI = 1.110 to 1.602) and moderately associated with DN (T2D without and with DN, 0.593 vs. 0.671, P = 0.041, OR = 1.399, 95% CI = 1.013 to 1.932). The association of SNP rs13266634 (C/T) with T2D and DN was not significant (P = 0.053, OR = 1.200, 95% CI = 0.997 to 1.443; and P = 0.098, OR = 1.313, 95% CI = 0.950 to 1.815). Information of genotype distribution and allele frequency comparison analyses is summarized in Table 2.
Figure 1.
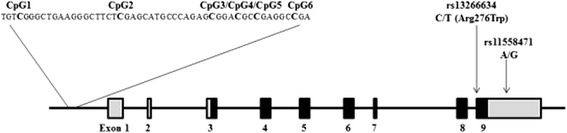
The CpG sites and single nucleotide polymorphisms in the SLC30A8 gene. The total length of the SLC30A8 gene is 226,442 bases. RefSeq DNA sequence at NCBI GenBank: NC_000008.10, NC_018919.2, NT_008046.17.
Figure 2.
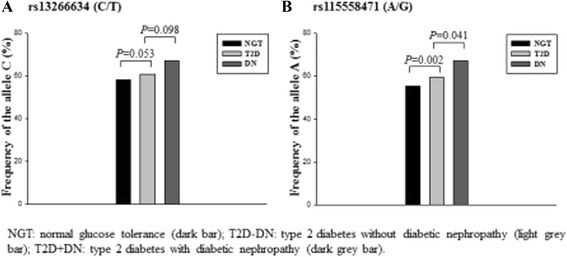
Risk allele frequencies of the SLC30A8 polymorphisms in Malay subjects. Risk allele frequencies (allele C of rs13266634(C/T) (A) and A of rs11558471(A/G) (B)) of the SLC30A8 polymorphisms in Malay subjects with normal glucose tolerance, type 2 diabetes, and diabetic nephropathy. NGT normal glucose tolerance (dark bar), T2D-DN type 2 diabetes without diabetic nephropathy (light grey bar), and T2D + DN type 2 diabetes with diabetic nephropathy (dark grey bar).
Table 2.
Association of the SLC30A8 genetic polymorphisms with type 2 diabetes and diabetic nephropathy in a Malay population
| SNP ID | Group | N | Genotypes | RAF | P value | Odds ratio | ||
|---|---|---|---|---|---|---|---|---|
| rs13266634 | CC | CT | TT | |||||
| NGT | 438 | 153 | 202 | 83 | 0.580 | 0.053 | 1.200 (0.997 to 1.443) | |
| All T2Da | 506 | 208 | 215 | 83 | 0.620 | |||
| T2D-DN | 236 | 90 | 107 | 39 | 0.608 | 0.098 | 1.313 (0.950 to 1.815) | |
| T2D + DN | 123 | 59 | 47 | 17 | 0.670 | |||
| rs11558471 | AA | AG | GG | |||||
| NGT | 441 | 139 | 209 | 93 | 0.552 | 0.002 | 1.334 (1.110 to 1.602) | |
| All T2Da | 509 | 204 | 225 | 80 | 0.620 | |||
| T2D-DN | 237 | 87 | 107 | 43 | 0.593 | 0.041 | 1.399 (1.013 to 1.932) | |
| T2D + DN | 123 | 57 | 51 | 15 | 0.671 | |||
Tests were conducted with an additive model.
NGT normal glucose tolerance, T2D type 2 diabetes, DN diabetic nephropathy, RAF risk allele frequency.
aThe patients with microalbuminuria were included.
Association of SLC30A8 DNA methylation with T2D and DN
To avoid the error caused by ages, the NGT subjects and T2D patients with age match were selected in DNA methylation analyses (Table 3). In the promoter region of the SLC30A8 gene, there is a cluster of neighboring six CpG sites (Figure 1). We analyzed SLC30A8 DNA methylation levels not only at each CpG site but also in total means of all CpG sites. In this Malay cohort, the average DNA methylation levels of all six CpG sites in the SLC30A8 gene promoter were high (81.4%). Figure 3A demonstrates the DNA methylation patterns of CpG sites in the SLC30A8 gene promoter region among NGT subjects and T2D patients with and without DN in the Malay cohort. DNA methylation levels at five CpG sites of the gene (except CpG2) in T2D patients were found to be higher than those in NGT subjects, respectively (CpG1 83.9% vs. 81.9%, P = 0.031; CpG3 82.1% vs. 84.8%, P = 0.003; CpG4 69.6% vs. 66.3%, P = 0.001; CpG5 86.2% vs. 83.7%, P = 0.004; and CpG6 79.8% vs. 78.1%, P = 0.001) (Figure 3A). Combining all six CpG sites together, total mean values of SLC30A8 DNA methylation levels were significantly increased in T2D patients compared with NGT subjects (82.9%, 95% CI = 79.2% to 80.5% vs. 80.1%, 95% CI = 75.4% to 78.6%, P = 0.014) (Figure 3B). A linear regression model was used to estimate the association between SLC30A8 DNA methylation levels in blood and phenotypes of T2D, and no significant association was found. Furthermore, the analyses of SLC30A8 DNA methylation levels between the patients with and without DN or according to the genotypes of SNPs rs13266634(C/T) and rs11558471(A/G) were done and no significant difference of the levels was found.
Table 3.
Clinical parameters of Malay subjects in epigenetic study
| Group | NGT | T2D a | T2D-DN | T2D + DN | P values b/c |
|---|---|---|---|---|---|
| N (men/women) | 27 (19/8) | 161 (81/80) | 109 (49/60) | 43 (25/18) | |
| Age (years) | 57 ± 9 | 56 ± 9 | 55 ± 10 | 56 ± 10 | NS/NS |
| Diabetes duration (years) | - | 12 ± 7 | 11 ± 8 | 11 ± 7 | -/NS |
| BMI (kg/m2) | 24 ± 4.0 | 28 ± 5 | 29 ± 5 | 27 ± 5 | <0.001/NS |
| SBP (mm Hg) | 132 ± 18 | 139 ± 24 | 135 ± 20 | 150 ± 33 | NS/NS |
| DBP (mm Hg) | 84 ± 14 | 80 ± 12 | 80 ± 12 | 83 ± 15 | NS/NS |
| HbA1c (%) | 5.7 ± 1.0 | 8.2 ± 2.0 | 8.1 ± 2.0 | 8.6 ± 2.4 | <0.001/NS |
| Serum creatinine (μmol/l) | 70.6 ± 17.1 | 126.4 ± 119.5 | 92.7 ± 72.9 | 215.8 ± 183.0 | <0.005/<0.001 |
| Urine creatinine (mmol/l) | 12.3 ± 7.4 | 8.1 ± 6.8 | 9.7 ± 6.2 | 5.0 ± 3.52 | <0.001/<0.001 |
| ACR (mg/mmol) | 0.86 ± 0.75 | 46.2 ± 127.1 | 1.19 ± 0.84 | 195.1 ± 206.1 | <0.001/<0.001 |
| eGFR (ml/min/1.73 m2) | 102.8 ± 28.6 | 67.1 ± 28.0 | 76.5 ± 23.4 | 44.1 ± 26.0 | <0.001/<0.001 |
| SLC30A8 DNA methylation levels (95% CI) | 80.1% (79.4 to 81.4) | 82.9% (81.2 to 83.5) | 82.7% (80.9 to 82.9) | 83.6% (79.8 to 84.4) | 0.014/0.632 |
Data are expressed as means ± SD or 95% CI.
NGT normal glucose tolerance, T2D type 2 diabetes, DN diabetic nephropathy, BMI body mass index, SBP systolic blood pressure, DBP diastolic blood pressure, FPG fasting plasma glucose, HbA1c glycosylated hemoglobin, ACR albumin creatinine ratio, eGFR estimated glomerular filtration rate.
aThe patients with microalbuminuria were included.
b P values were from tests of NGT vs. all T2D.
c P values were from tests of T2D without DN vs. T2D with DN.
Figure 3.
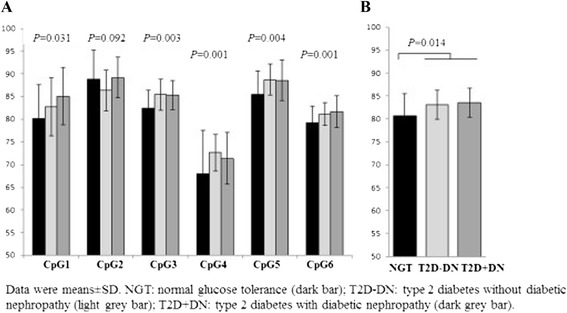
DNA methylation changes of the SLC30A8 gene in Malay subjects. DNA methylation changes of the SLC30A8 gene in Malay subjects with normal glucose tolerance, type 2 diabetes, and diabetic nephropathy. DNA methylation levels at five CpG sites of the gene (except CpG2) in T2D patients were found to be higher than those in NGT subjects, respectively (A). Combining all six CpG sites together, total mean values of SLC30A8 DNA methylation levels were significantly increased in T2D patients compared with NGT subjects (B). Data were means with 95% CI. NGT: normal glucose tolerance (dark bar); T2D-DN: type 2 diabetes without diabetic nephropathy (light grey bar) and T2D + DN: type 2 diabetes with diabetic nephropathy (dark grey bar).
Discussion
We have investigated DNA methylation levels of the SLC30A8 gene in a Malay population. Our main findings include: first, a cluster of CpG sites in the promoter region of the SLC30A8 gene is hypermethylated. Second, increased DNA methylation levels of this gene are associated with T2D but not with DN.
DNA methylation analysis can be performed in the scales of global genome or specific gene region and in peripheral blood with mixed cell types or specific tissues [27]. Dayeh et al. have recently performed human pancreatic islet tissue-specific DNA methylation analyses at the genomic regions of SNPs that are predicted to be associated with T2D by GWAS [28]. In the SLC30A8 gene, three CpG sites are located nearby SNP rs13266634. According to the genotypes of this polymorphism, DNA methylation changes at one of CpG sites are found to be significantly different. However, the DNA methylation levels at this CpG site are very low (<5%). Hall E et al. have further investigated the effects of palmitate on genome-wide mRNA expression and DNA methylation patterns in human pancreatic islets. With the palmitate treatment, the SLC30A8 mRNA expression levels in human pancreatic islets are found to be decreased. But whether DNA methylation levels of this gene are affected by palmitate is unclear [29]. A recent report has indicated that both approaches of whole-blood DNA methylation profiling and adipose tissue-specific methylation analysis for study of epigenetic changes are related to body mass index (BMI) and suggested that the analysis of blood DNA methylation is worthwhile and can reflect changes in relevant tissues for a phenotype [30]. The present study is a clinical observation. Although we have no tissue sample of pancreatic islets available for analysis, epigenetic study with blood samples is clinically accessible. Our findings are unlikely false positive because the average of DNA methylation of the SLC30A8 gene is high. We have analyzed the SLC30A8 gene DNA methylation levels according to gender and smoking factors; no significant change of the levels between NGT and T2D was found. Furthermore, the increased DNA methylation of the SLC30A8 gene in T2D is consistent with the downregulation of the gene expression in pancreatic islets of diabetic animals as previously reported [2,3]. Except age, gender, and smoking, the confounding factor of ethnic group has been taken into our study. Malaysian populations include Malays, Chinese, Indians, and indigenous Sabahans and Sarawakians. To avoid the error caused by this confounding factor, only Malay subjects are selected for this study. However, the replication study in other ethnic groups or populations may be necessary to ascertain the SLC30A8 epigenetic effect in T2D.
In the recent years, several groups including ours have begun to investigate the epigenetic effects in T2D. From our studies [19-22] and other reports [27-30], we have learnt that DNA methylation levels between non-diabetic control subjects and T2D patients differ significantly but the difference is small (often around 2%). The standard deviation of detected DNA methylation values is generally much less compared with the data from serum or plasma protein analyses. Compared with an epigenetic study in tumor, it may be more difficult to perform the same study in T2D. There are a number of genes contributing towards genetic and epigenetic effects to the disease, and the contribution of each gene may be minor. In this case, it is of importance to analyze the accumulating genetic and epigenetic effects in T2D. Data from the present study demonstrate that the SLC30A8 gene DNA methylation is high and implicate that the association of increased SLC30A8 DNA methylation levels with T2D should be included into the accumulating analyses.
Compared with genetic study of T2D, the progress of genetic study of DN has been slowed down mainly due to the difficulty of collection and characterization of subjects [31]. Although genetic association studies of the SLC30A8 genetic polymorphisms with T2D are well documented [4-9], no association of the genetic polymorphisms with DN has been reported. In the present study, we have replicated the association of the SLC30A8 genetic polymorphisms with T2D and also shown a moderate association between SNP rs1155847(A/G) in the gene and DN. The data provide the basic information for further epigenetic analysis of the SLC30A8 gene in the present study. However, the sample sizes of cases (T2D patients with DN) and controls (T2D without DN) are relatively small. Additional investigation with large cohorts is necessary to confirm the association of the SLC30A8 genetic polymorphisms with DN.
Conclusion
In conclusion, the present study provides the first evidence that increased DNA methylation of the SLC30A8 gene is associated with T2D but not DN in a Malay population.
Methods
Subjects
Malaysia is a country with multi-cultures and multi-ethnic populations. We collected the subjects with NGT and T2D from the collaborating centers all over Malaysia. The ethnic distribution of our cohort was 67.6% Malays, 15.3% Indians, 14.8% Chinese, and 2.3% indigenous Sabahans and Sarawakians. To avoid the error caused by ethnic stratification, Indian, Chinese, and indigenous Sabahans and Sarawakians were excluded in the present study. A total of 950 Malay subjects with NGT (n = 441) and T2D (509) were included in the genetic association study. The patients with T2D were diagnosed based on medical history by the medical officer or by oral glucose tolerance test (OGTT). All diagnoses were done based on World Health Organization (WHO) criteria [32]. The patients with T2D and normoalbuminuria (ACR <3.5 mg/mmol) were considered as control subjects without DN, while the patients with macroalbuminuria (ACR ≥35 mg/mmol) and ESRD who needed dialysis were included as the cases with DN. The diagnoses of DN were based on urine albumin-to-creatinine ratio (ACR) suggested by ADA [33]. There were 131 T2D patients with microalbuminuria (ACR 3.5 to 35 mg/mmol). Finally, 237 patients without DN and 123 patients with DN were included in the study. Clinical characteristics in all Malay subjects selected for genetic association study and epigenetic analysis are summarized in Tables 1 and 3, respectively.
The informed consent was given from all subjects, and the study was approved by the local ethical committees. All subjects need to answer a questionnaire and underwent a medical examination guided by the research staff. Data and material transfer agreement from the Institute for Medical Research, Malaysia to Karolinska Institutet, Sweden was completed prior to the study.
Genomic DNA extraction
Genomic DNA extraction was isolated from fresh peripheral blood samples using a DNeasy blood and tissue extraction kit according to the manufacturer’s protocol (Qiagen, Hilden, Germany) and quantified using a spectrophotometry (Biophotometer Plus, Eppendorf, Germany).
SNP selection and genotyping
Based upon information from the recent genetic association studies, two SNPs, that is, rs13266634(Arg276Trp C/T) and rs11558471(A/G) in the SLC30A8 gene, were selected for our study [4-9]. The first SNP is non-synonymous, in which the amino acid arginine is changed to tryptophan, while the second one is a synonymous SNP at the 3′-UTR. The SLC30A8 gene structure and location of these two polymorphisms are shown in Figure 1. Genotyping these two SNPs were done using TaqMan SNP genotyping assays purchased from Applied Biosystems (Applied Biosystems, Foster City, USA). Genotyping experiments were performed in ABI 7300 sequence detector with a Taqman allelic discrimination protocol (Applied Biosystems). DNA samples were distributed randomly across plates with cases and controls for genotyping quality control. All PCR reactions were run in 20 μl volumes using 10 to 20 ng genomic DNA. Millipore water was used as negative controls (blanks) on each plate.
Bisulfite treatment and pyrosequencing
Epigenetic analysis was performed by bisulfite pyrosequencing, which is a sensitive and accurate protocol [34,35]. DNA was treated with sodium bisulfite using EpiTect Bisulfite kit (Qiagen) and cleanup of bisulfite-converted DNA was done. PCR amplification was then carried out using PyroMark CpG assay (Qiagen) and PyroMark Gold Q96 Reagent kits (Qiagen) in a PyroMark Q96 system (Biotage AB, Uppsala Sweden). PyroMark PCR master mix includes HotStarTaq DNA polymerase and optimized PyroMark reaction buffer containing 3 mM MgCl2 and dNTPs, 10x CoralLoad Concentrate, 5x Q-Solution, 25 mM MgCl2, and RNase-free water. The PCR amplicon covers the sequence in human chromosome 8: 117962434 to 117962479 (version 37.56). There are four CpG sites in the SLC30A8 gene promoter region as indicated with the bold letter ‘C’ and recorded as CpG1-4 (Figure 1). Finally, methylation levels of these CpG sites were detected by using the PyroMark Gold 96 Reagent kit (Qiagen) and a PyroMark Q96 ID pyrosequencing system (Biotage). The unmethylated and unconverted DNA samples (Qiagen) were used for control of conversion efficiency in bisulfite treatment and accuracy in methylation analyses. PyroQ-CpG software (Biotage) was used for methylation data analysis.
Statistical analyses
Allele frequency and genotype distribution of the studied SNPs were tested for HWE using the χ2 statistic. For difference between NGT subjects and T2D patients, two models were tested comparing either allele frequencies in 2 × 2 contingency tables (dominant) or genotypes in 3 × 2 contingency tables (additive). Odd ratios (OD) and 95% confidence intervals (CI) were calculated to test the relative risk for association. Statistical power for genetic association study was calculated by using the software of PowerSampleSize (PS version 2.1.31). Since the major alleles of the studied SLC30A8 polymorphisms were risk for T2D, and their frequencies were high (55% to 67%), the sample sizes of cases and controls included in our studies were sufficient to detect an association with T2D at 85% powers. But the power to detect the association with DN is low (approximately 55%). Tests for comparison of continuous variables between groups were assessed by unpaired t-test or one-way ANOVA followed with Tukey’s post hoc test. Data in non-normally distributed traits were transformed to the natural logarithm for obtaining a normal distribution before performing statistical analysis. To estimate the association between SLC30A8 DNA methylation levels in blood and phenotypes of T2D, the linear regression model was used. Data were given as the means ± SD. P value less than 0.05 was considered significant. Statistical calculations were performed by using PASW Statistic Base 18 (SPSS Inc, Chicago, IL, USA).
Acknowledgements
The authors are grateful for all subjects who volunteered to participate in this study and for all Medical Laboratory Technologists in Malaysia who assisted with the sample collection. We also thank Drs Julien Pelletier, Zuheng Ma, Tianwei Gu, and Sara Babiker for constructive discussion and laboratory assistance. The study was supported by the Stig and Gunborg Westmans Foundation, Family Erling-Persson Foundation, and Fund of Karolinska Institutet. NAS is supported by a scholarship from Ministry of Science, Technology and Innovation, Malaysia.
Abbreviations
- T2D
Type 2 diabetes
- SLC30A8
Salute carrier family 30
- SNP
Single nucleotide polymorphism
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
All proposed the project; HFG and NAS designed the study; WNWM and NAS collected clinical data; NAS collected experimental data; NAS and HFG analyzed the experimental data; HFG and NAS wrote the manuscript. All contributed to data interpretation, discussion, and revision of the manuscript. All authors read and approved the final manuscript.
Contributor Information
Norhashimah Abu Seman, Email: norhashimah.abu.seman@ki.se.
Wan Nazaimoon Wan Mohamud, Email: nazaimoon@imr.gov.my.
Claes-Göran Östenson, Email: claes.ostensson@karolinska.se.
Kerstin Brismar, Email: kerstin.brismar@ki.se.
Harvest F Gu, Email: harvest.gu@ki.se.
References
- 1.Chimienti F, Devergnas S, Favier A, Seve M. Identification and cloning of a beta-cell specific zinc transporter, ZnT-8, localized into insulin secretory granules. Diabetes. 2004;53(9):2330–7. doi: 10.2337/diabetes.53.9.2330. [DOI] [PubMed] [Google Scholar]
- 2.Tamaki M, Fujitani Y, Uchida T, Hirose T, Kawamori R, Watada H. Downregulation of ZnT8 expression in pancreatic β-cells of diabetic mice. Islets. 2009;1(2):124–8. doi: 10.4161/isl.1.2.9433. [DOI] [PubMed] [Google Scholar]
- 3.Fu Y, Tian W, Pratt EB, Dirling LB, Shyng SL, Meshul CK, et al. Down-regulation of ZnT8 expression in INS-1 rat pancreatic beta cells reduces insulin content and glucose-inducible insulin secretion. PLoS One. 2009;4(5):e5679. doi: 10.1371/journal.pone.0005679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lyssenko V, Jonsson A, Almgren P, Pulizzi N, Isomaa B, Tuomi T, et al. Clinical risk factors, DNA variants, and the development of type 2 diabetes. N Engl J Med. 2008;359(21):2220–32. doi: 10.1056/NEJMoa0801869. [DOI] [PubMed] [Google Scholar]
- 5.Cauchi S, Del Guerra S, Choquet H, D’Aleo V, Groves CJ, Lupi R, et al. Meta-analysis and functional effects of the SLC30A8 rs13266634 polymorphism on isolated human pancreatic islets. Mol Genet Metab. 2010;100(1):77–82. doi: 10.1016/j.ymgme.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 6.Kanoni S, Nettleton JA, Hivert MF, Ye Z, van Rooij FJ, Shungin D, et al. Total zinc intake may modify the glucose-raising effect of a zinc transporter (SLC30A8) variant: a 14-cohort meta-analysis. Diabetes. 2011;60(9):2407–16. doi: 10.2337/db11-0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445(7130):881–5. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 8.Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316(5829):1341–5. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University, Novartis Institutes of BioMedical Research. Saxena R, Voight BF, Lyssenko V, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316(5829):1331–6. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 10.Billings LK, Florez JC. The genetics of type 2 diabetes: what have we learned from GWAS? Ann N Y Acad Sci. 2010;1212:59–77. doi: 10.1111/j.1749-6632.2010.05838.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahlqvist E, Ahluwalia TS, Groop L. Genetics of type 2 diabetes. Clin Chem. 2011;57(2):241–54. doi: 10.1373/clinchem.2010.157016. [DOI] [PubMed] [Google Scholar]
- 12.Imamura M, Maeda S. Genetics of type 2 diabetes: the GWAS era and future perspectives. Endocr J. 2011;58(9):723–39. doi: 10.1507/endocrj.EJ11-0113. [DOI] [PubMed] [Google Scholar]
- 13.Ling C, Groop L. Epigenetics: a molecular link between environmental factors and type 2 diabetes. Diabetes. 2009;58(12):2718–25. doi: 10.2337/db09-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drong AW, Lindgren CM, McCarthy MI. The genetic and epigenetic basis of type 2 diabetes and obesity. Clin Pharmacol Ther. 2012;92(6):707–15. doi: 10.1038/clpt.2012.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kirchner H, Osler ME, Krook A, Zierath JR. Epigenetic flexibility in metabolic regulation: disease cause and prevention? Trends Cell Biol. 2013;23(5):203–9. doi: 10.1016/j.tcb.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 16.Flannick J, Thorleifsson G, Beer NL, Jacobs SB, Grarup N, Burtt NP, et al. Loss-of-function mutations in SLC30A8 protect against type 2 diabetes. Nat Genet. 2014;46(4):357–63. doi: 10.1038/ng.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13(7):484–92. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 18.Branco MR, Ficz G, Reik W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat Rev Genet. 2011;13(1):7–13. doi: 10.1038/ni.2192. [DOI] [PubMed] [Google Scholar]
- 19.Gu T, Gu HF, Hilding A, Sjöholm LK, Ostenson CG, Ekström TJ, et al. Increased DNA methylation levels of the insulin-like growth factor binding protein 1 gene are associated with type 2 diabetes in Swedish men. Clin Epigenetics. 2013;5(1):21. doi: 10.1186/1868-7083-5-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gu HF, Gu T, Hilding A, Zhu Y, Kärvestedt L, Ostenson CG, et al. Evaluation of IGFBP-7 DNA methylation changes and serum protein variation in Swedish subjects with and without type 2 diabetes. Clin Epigenetics. 2013;5(1):20. doi: 10.1186/1868-7083-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gu T, Falhammar H, Gu HF, Brismar K. Epigenetic analyses of the insulin-like growth factor binding protein 1 gene in type 1 diabetes and diabetic nephropathy. Clin Epigenetics. 2014;6(1):10. doi: 10.1186/1868-7083-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gu T, Gu HF, Hilding A, Ostenson CG, Brismar K. DNA Methylation Analysis of the Insulin-like Growth Factor-1 (IGF1) Gene in Swedish Men with Normal Glucose Tolerance and Type 2 Diabetes. J Diabetes Metab. 2014;5:8. [Google Scholar]
- 23.Letchuman GR, Wan Nazaimoon WM, Wan Mohamad WB, Chandran LR, Tee GH, Jamaiyah H, et al. Prevalence of diabetes in the Malaysian National Health Morbidity Survey III 2006. Med J Malaysia. 2010;65(3):180–6. [PubMed] [Google Scholar]
- 24.Lim YN, Lim TO, Lee DG, Wong HS, Ong LM, Shaariah W, et al. A report of the Malaysian dialysis registry of the National Renal Registry, Malaysia. Med J Malaysia. 2008;63(Suppl C):5–8. [PubMed] [Google Scholar]
- 25.Shaza AM, Rozina G, Izham MI, Azhar SS. Dialysis for end stage renal disease: a descriptive study in Penang Hospital. Med J Malaysia. 2005;60(3):320–7. [PubMed] [Google Scholar]
- 26.Hooi LS, Wong HS, Morad Z. Prevention of renal failure: the Malaysian experience. Kidney Int Suppl. 2005;94:S70–4. doi: 10.1111/j.1523-1755.2005.09418.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu X, Su S, Barnes VA, De Miguel C, Pollock J, Ownby D, et al. A genome-wide methylation study on obesity: differential variability and differential methylation. Epigenetics. 2013;8(5):522–33. doi: 10.4161/epi.24506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dayeh TA, Olsson AH, Volkov P, Almgren P, Rönn T, Ling C. Identification of CpG-SNPs associated with type 2 diabetes and differential DNA methylation in human pancreatic islets. Diabetologia. 2013;56(5):1036–46. doi: 10.1007/s00125-012-2815-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hall E, Volkov P, Dayeh T, Bacos K, Rönn T, Nitert MD, et al. Effects of palmitate on genome-wide mRNA expression and DNA methylation patterns in human pancreatic islets. BMC Med. 2014;12:103. doi: 10.1186/1741-7015-12-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dick KJ, Nelson CP, Tsaprouni L, Sandling JK, Aïssi D, Wahl S, et al. DNA methylation and body-mass index: a genome-wide analysis. Lancet. 2014;S0140-6736(13):62674–4. doi: 10.1016/S0140-6736(13)62674-4. [DOI] [PubMed] [Google Scholar]
- 31.Gu HF, Brismar K. Genetic association studies in diabetic nephropathy. Curr Diabetes Rev. 2012;8(5):336–44. doi: 10.2174/157339912802083522. [DOI] [PubMed] [Google Scholar]
- 32.Alberti KG, Zimmet PZ. Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabet Med. 1998;15(7):539–53. doi: 10.1002/(SICI)1096-9136(199807)15:7<539::AID-DIA668>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 33.Molitch ME, DeFronzo RA, Franz MJ, Keane WF, Mogensen CE, Parving HH, et al. American Diabetes Association. Nephropathy in diabetes. Diabetes Care. 2004;27(Suppl 1):S79–83. doi: 10.2337/diacare.27.2007.s79. [DOI] [PubMed] [Google Scholar]
- 34.Colella S, Shen L, Baggerly KA, Issa JP, Krahe R. Sensitive and quantitative universal pyrosequencing methylation analysis of CpG sites. Biotechniques. 2003;35(1):146–50. doi: 10.2144/03351md01. [DOI] [PubMed] [Google Scholar]
- 35.Tost J, Dunker J, Gut IG. Analysis and quantification of multiple methylation variable positions in CpG islands by pyrosequencing. Biotechniques. 2003;35(1):152–6. doi: 10.2144/03351md02. [DOI] [PubMed] [Google Scholar]


