Abstract
It is well established that salicylic acid (SA) plays a critical role in the transcriptional reprograming that occurs during the plant defense response against biotic and abiotic stress. In the course of the defense response, the transcription of different sets of defense genes is controlled in a spatio-temporal manner via SA-mediated mechanisms. Interestingly, different lines of evidence indicate that SA interplays with reactive oxygen species (ROS) and glutathione (GSH) in stressed plants. In this review we focus on the evidence that links SA, ROS, and GSH signals to the transcriptional control of defense genes. We discuss how redox modifications of regulators and co-regulators involved in SA-mediated transcriptional responses control the temporal patterns of gene expression in response to stress. Finally, we examine how these redox sensors are coordinated with the dynamics of cellular redox changes occurring in the defense response to biotic and abiotic stress.
Keywords: glutathione, glutaredoxin GRXC9/GRX480, NPR1, reactive oxygen species, salicylic acid, thioredoxin TRXh5, TGA transcription factors
Interplay between Salicylic Acid (SA) and Redox Signals in the Defense Response to Stress
A feed-forward loop between salicylic acid (SA) and reactive oxygen species (ROS) production in the defense response to stress was first reported at the early 1990s (Chen et al., 1993). This early report was followed by a controversy on whether H2O2 was downstream or upstream of SA in the pathway for induction of Pathogenesis-Related 1 (PR1) expression (Neuenschwander et al., 1995; Chamnongpol et al., 1996). Later on, it was demonstrated that ROS signals are involved both upstream and downstream SA signaling in response to stress. Interestingly, the evidence indicates that SA does not only play a pro-oxidant role, but it also has an antioxidant role in concert with glutathione (GSH) in the response to stress. In this first section we present a comprehensive picture of the relationships between SA, ROS, and GSH in the response to stress signaling (Figure 1).
FIGURE 1.
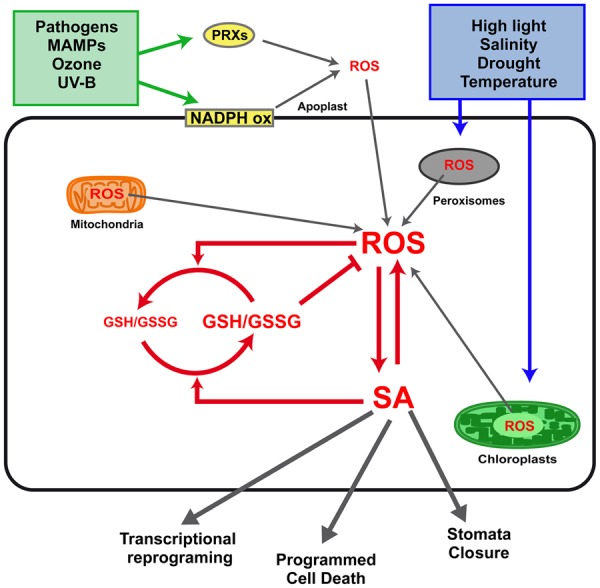
Interplay between salicylic acid (SA), reactive oxygen species (ROS), and glutathione (GSH) in defense responses to biotic and abiotic stress. Stress conditions such as infection with pathogens, exposure to microbe-associated molecular patterns (MAMPs), ozone, and UV-B treatments, trigger ROS production mainly at the apoplast. This production of ROS is mediated by plasma membrane NADPH oxidases (NADPH ox) and cell wall peroxidases (PRXs). Other stresses, such as high light radiation, salinity, drought, and temperature, trigger ROS production mainly at the chloroplasts and peroxisomes. Mitochondria have been also described as an important source of ROS during defense responses (Lam et al., 2001). A feed-forward loop between H2O2 and SA synthesis occurs in response to stress, as described in the text. SA also has an antioxidant role, increasing GSH levels and reducing power, which in turn is involved in ROS scavenging. Finally, the interplay between intracellular levels of SA, H2O2, and GSH determines transcriptional reprogramming, programmed cell death, and stomata closure, the three main outputs of the defense responses.
ROS Bursts Trigger SA Signaling
It is well known that activation of SA signaling in stressed plants is preceded by oxidative bursts originating in different cellular compartments (Wrzaczek et al., 2013). In the case of basal (PTI) and induced (ETI) defense responses against pathogens infection, it has been extensively reported that increases in SA levels are preceded by apoplastic H2O2 bursts mediated by NADPH oxidases and extracellular peroxidases (PRXs; Mackerness et al., 2001; Torres et al., 2002; Joo et al., 2005; Tsuda et al., 2008; O’Brien et al., 2012; Mammarella et al., 2014). Although PTI and ETI responses are activated in the plant by recognition of different pathogens molecules, they share several signals including ROS and SA. Differences in the timing and levels at which these signals are produced in PTI and ETI determine differences in the speed and strength at which these immune reactions are established to be effective in counteracting potential pathogens with low cost on fitness (Tsuda et al., 2008; Katagiri and Tsuda, 2010).
Apoplastic H2O2 bursts also precede SA signaling in plant responses to exposure to ozone and UV-B (Grant and Loake, 2000; Mackerness et al., 2001; Torres et al., 2002; Joo et al., 2005; Ogawa et al., 2007; Garcion et al., 2008; O’Brien et al., 2012). Pharmacological evidences supports that increases in apoplastic H2O2 levels after UV-B trigger SA biosynthesis (Mackerness et al., 2001). Noteworthy, in the case of ozone, ROS signaling starts at guard-cells chloroplasts and then it propagates to the apoplast of neighbor cells (Joo et al., 2005).
Salicylic acid also functions as a signal of other types of abiotic stresses such as high light exposure, salinity, drought, and temperature (Mateo et al., 2006; Lee and Park, 2010; Wan et al., 2012; Miura and Tada, 2014). In contrast to the above mentioned stresses, these conditions generate ROS accumulation in chloroplasts and peroxisomes (Apel and Hirt, 2004; Holuigue et al., 2007). Although involvement of SA in these cases has been demonstrated in SA-deficient and overproducer plants (Mateo et al., 2006; Lee and Park, 2010; Wan et al., 2012; Miura and Tada, 2014), direct evidence of increased SA levels has been only reported in oat plants exposed to drought (Sánchez-Martín et al., 2014). Interestingly, increased levels of SA have been detected in plants with sustained ROS production in peroxisomes (catalase 2 knockout, cat2; Chaouch et al., 2010) and in chloroplast (thylakoidal ascorbate PRX gene silencing, tAPX RNAi; Maruta et al., 2012; Noshi et al., 2012). The evidence obtained using these models indicate that H2O2 originated in chloroplasts and peroxisomes triggers SA biosynthesis, which is essential for main outputs of the defense response: transcriptional reprogramming, cell death, and stomatal closure (Figure 1).
The mechanisms by which H2O2 generated in the apoplast, chloroplasts, and peroxisomes triggers SA biosynthesis remains unknown. ICS1 and ICS2 are the two Arabidopsis genes coding for isochorismate synthase, the key enzyme controlling SA biosynthesis (Garcion et al., 2008). ICS1 upregulation was detected in the ETI response to pathogens, in response to UV-B, ozone, and drought stress (Wildermuth et al., 2001; Ogawa et al., 2007; Zhang et al., 2010; Wan et al., 2012), as well as in cat2 plants (Chaouch et al., 2010). In contrast, upregulation of ICS2 but not of ICS1 was detected in tAPX RNAi plants (Noshi et al., 2012). Transcription factors that regulate ICS1 expression, such as CBP60, SARD1, and WRKY8/28/48 (Zhang et al., 2010; van Verk et al., 2011; Gao et al., 2013), or upstream PAD4/EDS1 genes expression, such as CAMTA3/SR1 and ZAT6 (Du et al., 2009; Shi et al., 2014) represent potential candidates for ROS-mediated regulation of SA biosynthesis.
Remarkably, it has recently been proposed that Ca+2 signaling regulate SA production (Seyfferth and Tsuda, 2014), based on evidence that the activity of CBP60, WRKY8/28/48, and CAMTA3/SR1 factors are modulated by calcium dependent protein kinases (CDPKs) and calmodulin (CaM; Du et al., 2009; Gao et al., 2013; Truman et al., 2013). Indeed, intracellular increase of cytosolic Ca+2 was first described as an upstream signal that controls apoplastic ROS production through the modification of NADPH oxidase by CDPKs (Dubiella et al., 2013; Gao et al., 2013). Recently, Ca+2 has been also proposed to act downstream ROS signaling (Wrzaczek et al., 2013), based on previous evidence that exogenous treatments with H2O2 promote Ca+2 influxes (Price et al., 1994; Pei et al., 2000). Therefore, the possibility that a Ca+2 signal mediates activation of SA production triggered by ROS, represents an interesting aspect to explore.
SA Modulates Redox Homeostasis
An ambivalent effect of SA in promoting ROS accumulation (prooxidant) and ROS scavenging (antioxidant), has being reported in several stress models, including the ETI response to pathogens and responses to high light, drought, salinity, and cold stress (Mou et al., 2003; Mateo et al., 2006; Miura and Tada, 2014). On one hand, SA promotes ROS production during early events of signaling, being these ROS essential for defense responses (Garreton et al., 2002; Lee et al., 2010; Khokon et al., 2011). Furthermore, high concentrations of SA (>100 μM) promote ROS production, inducing oxidative stress, and reducing tolerance to drought and salinity (Lee et al., 2010; Miura and Tada, 2014). How can SA promote ROS accumulation? Early reports showed SA-mediated inhibition of catalase and cytosolic ascorbate PRX, two main H2O2 detoxifying enzymes (Chen et al., 1993; Durner and Klessig, 1995). Then, SA-promoted production of ROS by extracellular PRXs was identified in stomatal closure control in drought response (Khokon et al., 2011; Miura et al., 2013).
On the contrary, the available evidence supports that SA promotes ROS scavenging being essential for the antioxidant response that constrains ROS bursts in responses to avirulent bacteria (Grant and Loake, 2000), high light (Mateo et al., 2006), ozone (Yoshida et al., 2009), salinity (Lee et al., 2010), and in cat2 mutants (Chaouch et al., 2010). Recent studies show that SA and GSH interplay as redox signals, fostering a role for SA in the antioxidant response (Dubreuil-Maurizi et al., 2011; Foyer and Noctor, 2011; Dubreuil-Maurizi and Poinssot, 2012; Han et al., 2013). Plants that over accumulate SA show increased GSH levels and reducing power (ratio GSH/GSSG; Mateo et al., 2006) while abolishment of SA accumulation in a cat2 background (cat2 sid2) reduces the GSH/GSSG ratio (Chaouch et al., 2010; Noctor et al., 2014). Conversely, plants deficient in GSH biosynthesis (phytoalexin-deficient mutant, pad2-1) have decreased levels of SA and ICS1 transcripts (Dubreuil-Maurizi et al., 2011). This suggests that SA can play an antioxidant role by modulating GSH levels and reducing power (Figure 1), through still unknown mechanisms.
The dual redox effect of SA is reflected by a biphasic redox dynamics in plants treated with SA or INA (Mou et al., 2003; Mateo et al., 2006). A first oxidative phase, characterized by a transient increase in ROS levels and decline in GSH reducing power, is followed by a reductive phase characterized by an increase in GSH levels and reducing power. This temporal dynamics determines a sequential activation of the redox-regulated processes involved in the transcription of defense genes.
Redox-Modulated Processes in the SA-Mediated Control of Gene Expression
Salicylic acid plays a pivotal role in the genetic reprogramming, being responsible for transcriptional control of 100s of defense genes that are sequentially turned on/off (Maleck et al., 2000; Wang et al., 2006; Blanco et al., 2009). Interestingly, several redox-regulated processes have been discovered in the transcription of SA-regulated genes (Mou et al., 2003; Koornneef et al., 2008; Tada et al., 2008). The evidence suggests that cellular redox changes occurring in response to stress are translated into transcriptional responses, through redox modifications of master regulators and co-regulators (Moore et al., 2011). Here, we focus in the redox-modulated processes mediated by SA that control the expression of three Arabidopsis model genes: PR1, GRXC9 (glutaredoxin C9 or GRX480), and ORA59 (Octadecanoid-Responsive AP2/ERF domain protein 59; Figure 2). These genes have been studied in greater detail and they respond to SA with particular temporal patterns and mechanisms, being therefore good models for different classes of SA-regulated genes.
FIGURE 2.
Redox-modulated processes in the SA-mediated control of gene expression. Model for the transcriptional control of genes representing three main groups of SA-regulated genes: SA-induced non-expressor of pathogenesis-related (PR) genes 1 (NPR1) -dependent late genes (PR1, Left); SA-induced NPR1-independent early genes [glutaredoxin C9 (GRXC9), Medium]; and JA/ET-induced SA-repressed genes [Octadecanoid-Responsive AP2/ERF domain protein 59 (ORA59), Right]. The temporal dynamics of the redox changes (Δ Redox) occurring during the defense response to stress are represented by the bar at the left, where blue indicates reductive states and red indicates oxidative states. The temporal dynamics in the formation of transcriptionally active and inactive complexes in the promoter of PR1, GRXC9, and ORA59, according to redox changes dynamics, are included in each panel. The places where ROS/SA, and JA/ET signals act in these pathways, is indicated by red arrows. The components identified (or suspected) as redox sensors in these pathways, whose mechanisms of action are discussed in the text, are indicated in color. TGA factors (red) are involved in the three pathways. Homodimers or heterodimers of TGA2 and TGA3 (T2T3) or TGA2 and TGA5 (T2T5) factors act as platforms for the formation of transcriptionally inactive and active complexes. Active complexes promote recruitment of RNA polymerase II (RNAPII) and gene transcription (red arrows at promoters). NPR1 (yellow) is the master co-activator for SA-inducible NPR1-dependent pathway and is redox-regulated by oxido-reduction of Cys residues. TRXh5 and GRXC9 (green) are oxidoreductases coded by SA-inducible genes, which catalyze reduction of NPR1 and of a still unknown component in GRXC9 and ORA59 promoters. Other transcriptional factors and co-factors not directly involved in redox regulation are shown in gray tones.
Members of the TGA and WRKY transcription factor families, that recognize the TGA box (TGACGTCA) and the W box (TTGACT), respectively, have been involved in SA-mediated transcriptional regulation (Pandey and Somssich, 2009; Gatz, 2013). Furthermore, co-regulators including non-expressor of PR genes 1 (NPR1), SCL14, and Med 18 control transcription of different groups of SA-regulated genes (Fu and Dong, 2013). In this second section we will focus our attention on NPR1, TGA factors, and two oxidoreductases, to discuss evidence that point them as redox sensors in the expression of SA-regulated genes (Figure 2).
Non-Expressor of PR Genes 1 (NPR1), a Master Redox Sensor
Non-Expressor of PR genes is the master co-activator for PR1 and most SA-induced genes, and was the first redox sensor described for SA-regulated genes (Mou et al., 2003). Particularly at the PR1 promoter, SA stimulates NPR1 interaction with TGA2 and TGA3, which enhances its binding to TGA boxes, forming a trans-activating complex for RNA polymerase II (RNAPII) recruitment (Figure 2, left panel; Lebel et al., 1998; Kesarwani et al., 2007; Pape et al., 2010). Current knowledge indicates that SA-promoted redox modification of Cys residues in NPR1 determines the levels of the active, reduced, and monomeric form of NPR1 in the nucleus (Kinkema et al., 2000; Mou et al., 2003; Tada et al., 2008; Lindermayr et al., 2010). The levels of nuclear NPR1 are also regulated by other SA-mediated mechanisms, such as proteasome-mediated degradation and phosphorylation (Pajerowska-Mukhtar et al., 2013).
Salicylic acid is essential for NPR1 redox modification, but how it controls this process is still not well understood. NPR1 reduction is catalyzed by thioredoxin TRXh5 (Tada et al., 2008; Kneeshaw et al., 2014), coded by the only member of TRXh gene class transcriptionally induced by SA and oxidative stress (Laloi et al., 2004; Tada et al., 2008; Belin et al., 2014). Whether NPR1 monomerization also occurs under oxidative stress, has not been explored yet. Furthermore, evidence indicates that both, oligomerization and monomerization of NPR1 involves S-nitrosoglutathione (GSNO) mediated S-nitrosylation (Feechan et al., 2005; Rusterucci et al., 2007; Lindermayr et al., 2010).
Non-Expressor of PR genes 1 reduction and therefore induction of NPR1-dependent genes, including WRKYs and PR1, correlate with the reductive phase of the defense response (Mou et al., 2003). Based on the evidence summarized here, we propose a model for SA-mediated NPR1 redox control and its influence on PR1 induction (Figure 2, left panel).
Interestingly, the discovery of the direct binding of SA to NPR1 (Wu et al., 2012), and also to NPR3, and NPR4, which control NPR1 degradation (Fu et al., 2012), suggests the existence of a direct mechanism by which nuclear NPR1 levels and activity can be regulated according to the levels of SA, that in turn reflects the cellular redox state.
TGA Factors, a Potential Node for Integrative Cellular Redox Regulation?
TGA factors have been postulated as redox sensors (Spoel and Loake, 2011), based on evidence showing that modification of Cys residues in TGA1 and TGA4 modulate their binding to NPR1 and to DNA (Despres et al., 2003; Lindermayr et al., 2010). TGA1 and TGA4 compose class I TGA and their function is not critical for the expression of SA-regulated genes (Kesarwani et al., 2007; Shearer et al., 2012; Wang and Fobert, 2013; Herrera-Vásquez et al., 2014). In contrast, the evidence supports that class II TGAs (TGA2, TGA5, and TGA6), and to a lesser extent TGA3, are the essential factors for SA-regulated expression of defense genes (Johnson et al., 2003; Zhang et al., 2003; Kesarwani et al., 2007; Herrera-Vásquez et al., 2014). Intriguingly, there is still no direct evidence of regulation of these factors through redox modification. Nevertheless, a potential for TGA2/5/6 as a node for general redox regulation in response to stress, is supported by the evidence described below.
TGA2 represses PR1 basal expression but can also activate it upon SA-mediated stress challenge by interacting with negative and positive TGA boxes at the PR1 promoter (Johnson et al., 2003; Zhang et al., 2003; Kesarwani et al., 2007; Pape et al., 2010). The essential role of TGA2/5/6 in PR1 expression can be extrapolated to the group of NPR1-dependent genes with overrepresentation of the TGA box (Maleck et al., 2000).
We have shown that TGA2/5/6 are also essential for early SA-dependent and NPR1-independent induction of a set of genes with antioxidant and detoxifying activities (Blanco et al., 2009). GRXC9, which codes for a glutaredoxin of the plant-specific CC subfamily, is used here as a model for this pathway (Figure 2, medium panel; Ndamukong et al., 2007; Blanco et al., 2009; Herrera-Vásquez et al., 2014). SA-induced expression of GRXC9 requires two as-1 promoter elements that constitutively bind TGA2 and TGA3 factors (Herrera-Vásquez et al., 2014). as-1 elements, consisting of two TGA boxes separated by four base pairs (Krawczyk et al., 2002), confer early and transient induction by SA through ROS (Qin et al., 1994; Johnson et al., 2001; Garreton et al., 2002). Two as-1 elements were also found in the TRXh5 promoter, although its functionality has not been explored yet (Laloi et al., 2004). We propose that early induction of GRXC9, and probably of TRXh5 also, occurs during the oxidative phase of the defense response mediated by ROS signals (Figure 2; Mou et al., 2003; Herrera-Vásquez et al., 2014). TGA2/5/6 are also essential for the induction of as-1-containing genes involved in chemical detoxification (Mueller et al., 2008; Stotz et al., 2013).
Furthermore, the well-recognized antagonistic effect of SA on JA/ET-mediated transcriptional responses (Pieterse et al., 2009), is also mediated by class II TGAs (Ndamukong et al., 2007; Zander et al., 2010). SA inhibits expression of a group of JA/ET-induced genes, including PDF1.2, through repression of ORA59, which codes for a master transcription factor from the AP2/ERF family (Zander et al., 2010; Van der Does et al., 2013). Interestingly, ACC-induced and SA-repressed ORA59 expression depend of TGAs class II factors, through their binding to a TGA box present in the ORA59 promoter (Figure 2, right panel; Zander et al., 2014). Kinetic and pharmacological studies indicate that SA suppresses JA-responsive genes only within a specific time frame requiring SA-mediated increase in GSH levels (Koornneef et al., 2008). Therefore, SA-mediated ORA59 repression occurs in the reductive phase of the defense response, which is consistent with evidence indicating that NPR1 is required for SA-mediated repression of JA/ET-induced genes (Spoel, 2003).
Taken together, we can conclude that class II TGAs (particularly TGA2) are essential in different mechanisms of transcriptional control mediated by SA and ROS signals, which operate at different times in the defense response to stress (Figure 2). Accordingly, a strong phenotype of stress sensitivity is detected in tga2/5/6 triple mutant plants (Zhang et al., 2003; Mueller et al., 2008). The question is how TGA2 activity is controlled by SA and ROS signals? The only clue for a redox control of TGA2 is that it interacts with GRXC9 in the nucleus (Ndamukong et al., 2007). Interestingly, GRXC9 overexpression represses the expression of its own gene and of ORA59 while GRXC9 forms part of the complex bound to the as-1-containing region of the GRXC9 promoter (Herrera-Vásquez et al., 2014; Zander et al., 2014). These findings are integrated in the model shown in Figure 2. This model shows that SA, by inducing expression of GRXC9, controls the expression of antioxidant genes and at the same time represses JA/ET-mediated responses. We speculate that GRXC9 catalyzes the reduction of a protein from the transactivating complex in both genes, triggering their inactivation. Although evidence for functional associations of TGA factors and CC-type GRXs suggests that TGAs can be redox-modified (Murmu et al., 2010), there is still no evidence of this modification.
Oxidoreductases as Redox Sensors in the SA-Mediated Control of Gene Expression
The involvement of TRX/GRX oxidoreductases in SA-mediated transcription was first proposed some years ago (Fobert and Despres, 2005). As described above, two Cys-containing oxidoreductases, TRXh5, and GRXC9, were later on recognized as important elements for redox control in SA-mediated transcriptional responses. TRXh5 and GRXC9 genes are induced by SA during the oxidative phase of the defense response. TRXh5 reduces NPR1, which is essential for NPR1-dependent transcriptional responses (Tada et al., 2008). Instead, GRXC9 probably reduces a still unknown protein that represses the expression of genes from SA-dependent NPR1-independent as well as JA/ET-dependent SA-repressed pathways. These processes occur during the reductive phase of the defense response (Figure 2). Considering that TRXh5 and GRXC9 are in turn reduced and regenerated at the expense of the reducing power of NADPH and GSH, respectively (Meyer et al., 2012), these enzymes become key redox sensors that coordinate transcription and the cellular redox state.
Conclusion and Future Directions
The evidence discussed here indicates that redox-modulated processes are critical for the fine-tune regulation of gene expression mediated by SA. These processes occur in a temporaly controlled manner, coordinated with the cellular redox changes occurring during the defense response. Although important advances have occurred during the last years, we still have a fragmented knowledge of the network of redox processes that allows a coordinated transcriptional response to stressful conditions. Focusing on SA–ROS interplay, one important challenge is to understand how ROS generated in different cell compartments and cell types triggers SA biosynthesis. Furthermore, considering that all stress conditions generate oxidative bursts, but not all lead to SA accumulation, how is the specificity of ROS signals for triggering SA biosynthesis established? A point of convergence of the responses to different stresses mediated by SA, such as the PAD4/EDS1/SAG101 complex located upstream in the SA-signaling pathway (Wiermer et al., 2005), can be explored as a node for redox regulation of SA biosynthesis in response to stress.
In relation to the redox mechanisms that control the SA-mediated transcriptional response, the evidence discussed here supports the involvement of NPR1, TGA factors, and the oxidoreductases TRXh5 and GRXC9 as redox sensors. Several intriguing aspects about these sensors are pending, such as the promiscuous and essential role of TGA2 in the control of genes that respond to oxidant and reducing cellular redox states. Whether TGA2 itself can be redox modified, particularly reduced by GRXC9 to trigger gene repression during the reductive phase of the defense response, is a critical point that still needs to be answered. In this context, an interesting target to explore for redox regulation is MED18. The MED18 protein is a member of the Mediator Complex that interacts with the Ying Yang 1 transcription factor (YY1; Lai et al., 2014). This complex co-represses three genes coding for oxidoreductases involved in defense: GRXC9, TRXh5, and GRXS13 (La Camera et al., 2011; Laporte et al., 2012).
Finally, an important challenge for the future is to incorporate the temporal and spatial perspective in the analysis of the redox processes associated to transcriptional activity. New technical approaches that allow to record cell-specific changes in ROS levels, the redox state of GSH and new markers for gene expression will help in unraveling the sequential events occurring in different groups of cells exposed to stress during the time course of the defense response.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Commission for Science and Technology CONICYT (FONDECYT grant N∘ 1141202) and the Millennium Science Initiative (Nucleus for Plant Functional Genomics, grant N∘ P10-062-F).
References
- Apel K., Hirt H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55 373–399 10.1146/annurev.arplant.55.031903.141701 [DOI] [PubMed] [Google Scholar]
- Belin C., Bashandy T., Cela J., Delorme-Hinoux V., Riondet C., Reichheld J. P. (2014). A comprehensive study of thiol reduction gene expression under stress conditions in Arabidopsis thaliana. Plant Cell Environ. 38 299–314 10.1111/pce.12276 [DOI] [PubMed] [Google Scholar]
- Blanco F., Salinas P., Cecchini N. M., Jordana X., Van Hummelen P., Alvarez M. E., et al. (2009). Early genomic responses to salicylic acid in Arabidopsis. Plant Mol. Biol. 70 79–102 10.1007/s11103-009-9458-9451 [DOI] [PubMed] [Google Scholar]
- Chamnongpol S., Willekens H., Langebartels C., Van Montagu M., Inzé D., Van Camp W. (1996). Transgenic tobacco with a reduced catalase activity develops necrotic lesions and induces pathogenesis-related expression under high light. Plant J. 10 491–503 10.1046/j.1365-313X.1996.10030491.x [DOI] [Google Scholar]
- Chaouch S., Queval G., Vanderauwera S., Mhamdi A., Vandorpe M., Langlois-Meurinne M., et al. (2010). Peroxisomal hydrogen peroxide is coupled to biotic defense responses by ISOCHORISMATE SYNTHASE1 in a daylength-related manner. Plant Physiol. 153 1692–1705 10.1104/pp.110.153957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Silva H., Klessig D. F. (1993). Active oxygen species in the induction of plant systemic acquired resistance by salicylic acid. Science 262 1883–1886 10.1126/science.8266079 [DOI] [PubMed] [Google Scholar]
- Despres C., Chubak C., Rochon A., Clark R., Bethune T., Desveaux D., et al. (2003). The Arabidopsis NPR1 disease resistance protein is a novel cofactor that confers redox regulation of DNA binding activity to the basic domain/leucine zipper transcription factor TGA1. Plant Cell 15 2181–2191 10.1105/tpc.012849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Ali G. S., Simons K. A., Hou J., Yang T., Reddy A. S., et al. (2009). Ca(2+)/calmodulin regulates salicylic-acid-mediated plant immunity. Nature 457 1154–1158 10.1038/nature07612 [DOI] [PubMed] [Google Scholar]
- Dubiella U., Seybold H., Durian G., Komander E., Lassig R., Witte C. P., et al. (2013). Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc. Natl. Acad. Sci. U.S.A. 110 8744–8749 10.1073/pnas.1221294110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubreuil-Maurizi C., Poinssot B. (2012). Role of glutathione in plant signaling under biotic stress. Plant Signal. Behav. 7 210–212 10.4161/psb.18831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubreuil-Maurizi C., Vitecek J., Marty L., Branciard L., Frettinger P., Wendehenne D., et al. (2011). Glutathione deficiency of the Arabidopsis mutant pad2-1 affects oxidative stress-related events, defense gene expression, and the hypersensitive response. Plant Physiol. 157 2000–2012 10.1104/pp.111.182667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durner J., Klessig D. F. (1995). Inhibition of ascorbate peroxidase by salicylic acid and 26-dichloroisonicotinic acid, two inducers of plant defense responses. Proc. Natl. Acad. Sci. U.S.A. 92 11312–11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feechan A., Kwon E., Yun B. W., Wang Y., Pallas J. A., Loake G. J. (2005). A central role for S-nitrosothiols in plant disease resistance. Proc. Natl. Acad. Sci. U.S.A. 102 8054–8059 10.1073/pnas.0501456102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fobert P. R., Despres C. (2005). Redox control of systemic acquired resistance. Curr. Opin. Plant Biol. 8 378–382 10.1016/j.pbi.2005.05.003 [DOI] [PubMed] [Google Scholar]
- Foyer C. H., Noctor G. (2011). Ascorbate and glutathione: the heart of the redox hub. Plant Physiol. 155 2–18 10.1104/pp.110.167569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z. Q., Dong X. (2013). Systemic acquired resistance: turning local infection into global defense. Annu. Rev. Plant Biol. 64 839–863 10.1146/annurev-arplant-042811-105606 [DOI] [PubMed] [Google Scholar]
- Fu Z. Q., Yan S., Saleh A., Wang W., Ruble J., Oka N., et al. (2012). NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486 228–232 10.1038/nature11162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Chen X., Lin W., Chen S., Lu D., Niu Y., et al. (2013). Bifurcation of Arabidopsis NLR immune signaling via Ca(2)(+)-dependent protein kinases. PLoS Pathog. 9:e1003127 10.1371/journal.ppat.1003127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcion C., Lohmann A., Lamodiere E., Catinot J., Buchala A., Doermann P., et al. (2008). Characterization and biological function of the ISOCHORISMATE SYNTHASE2 gene of Arabidopsis. Plant Physiol. 147 1279–1287 10.1104/pp.108.119420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garreton V., Carpinelli J., Jordana X., Holuigue L. (2002). The as-1 promoter element is an oxidative stress-responsive element and salicylic acid activates it via oxidative species. Plant Physiol. 130 1516–1526 10.1104/pp.009886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatz C. (2013). From pioneers to team players: TGA transcription factors provide a molecular link between different stress pathways. Mol. Plant Microbe Interact. 26 151–159 10.1094/MPMI-04-12-0078-IA [DOI] [PubMed] [Google Scholar]
- Grant J. J., Loake G. J. (2000). Role of reactive oxygen intermediates and cognate redox signaling in disease resistance. Plant Physiol. 124 21–29 10.1104/pp.124.1.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Chaouch S., Mhamdi A., Queval G., Zechmann B., Noctor G. (2013). Functional analysis of Arabidopsis mutants points to novel roles for glutathione in coupling H(2)O(2) to activation of salicylic acid accumulation and signaling. Antioxid. Redox Signal. 18 2106–2121 10.1089/ars.2012.5052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera-Vásquez A., Carvallo L., Blanco F., Tobar M., Villarroel-Candia E., Vicente-Carbajosa J., et al. (2014). Transcriptional control of glutaredoxin GRXC9 expression by a salicylic acid-dependent and NPR1-independent pathway in Arabidopsis. Plant Mol. Biol. Rep. 10.1007/s11105-014-0782-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holuigue L., Salinas P., Blanco F., Garreton V. (2007). “Salicylic acid and reactive oxygen species in the activation of stress defense genes,” in Salicylic Acid – A Plant Hormone eds Hayat S., Ahmad A. (Dordrecht: Springer Science & Business Media) 197–246. [Google Scholar]
- Johnson C., Boden E., Arias J. (2003). Salicylic acid and NPR1 induce the recruitment of trans-activating TGA factors to a defense gene promoter in Arabidopsis. Plant Cell 15 1846–1858 10.1105/tpc.012211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C., Glover G., Arias J. (2001). Regulation of DNA binding and trans-activation by a xenobiotic stress-activated plant transcription factor. J. Biol. Chem. 276 172–178 10.1074/jbc.M005143200 [DOI] [PubMed] [Google Scholar]
- Joo J. H., Wang S., Chen J. G., Jones A. M., Fedoroff N. V. (2005). Different signaling and cell death roles of heterotrimeric G protein alpha and beta subunits in the Arabidopsis oxidative stress response to ozone. Plant Cell 17 957–970 10.1105/tpc.104.029603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri F., Tsuda K. (2010). Understanding the plant immune system. Mol. Plant Microbe Interact. 23 1531–1536 10.1094/MPMI-04-10-0099 [DOI] [PubMed] [Google Scholar]
- Kesarwani M., Yoo J., Dong X. (2007). Genetic interactions of TGA transcription factors in the regulation of pathogenesis-related genes and disease resistance in Arabidopsis. Plant Physiol. 144 336–346 10.1104/pp.106.095299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khokon A. R., Okuma E., Hossain M. A., Munemasa S., Uraji M., Nakamura Y., et al. (2011). Involvement of extracellular oxidative burst in salicylic acid-induced stomatal closure in Arabidopsis. Plant Cell Environ. 34 434–443 10.1111/j.1365-3040.2010.02253.x [DOI] [PubMed] [Google Scholar]
- Kinkema M., Fan W., Dong X. (2000). Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell 12 2339–2350 10.1105/tpc.12.12.2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneeshaw S., Gelineau S., Tada Y., Loake G. J., Spoel S. H. (2014). Selective protein denitrosylation activity of Thioredoxin-h5 modulates plant Immunity. Mol. Cell 56 153–162 10.1016/j.molcel.2014.08.003 [DOI] [PubMed] [Google Scholar]
- Koornneef A., Leon-Reyes A., Ritsema T., Verhage A., Den Otter F. C., Van Loon L. C., et al. (2008). Kinetics of salicylate-mediated suppression of jasmonate signaling reveal a role for redox modulation. Plant Physiol. 147 1358–1368 10.1104/pp.108.121392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawczyk S., Thurow C., Niggeweg R., Gatz C. (2002). Analysis of the spacing between the two palindromes of activation sequence-1 with respect to binding to different TGA factors and transcriptional activation potential. Nucleic Acids Res. 30 775–781 10.1093/nar/30.3.775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Camera S., L’haridon F., Astier J., Zander M., Abou-Mansour E., Page G., et al. (2011). The glutaredoxin ATGRXS13 is required to facilitate Botrytis cinerea infection of Arabidopsis thaliana plants. Plant J. 68 507–519 10.1111/j.1365-313X.2011.04706.x [DOI] [PubMed] [Google Scholar]
- Lai Z., Schluttenhofer C. M., Bhide K., Shreve J., Thimmapuram J., Lee S. Y., et al. (2014). MED18 interaction with distinct transcription factors regulates multiple plant functions. Nat. Commun. 5 3064 10.1038/ncomms4064 [DOI] [PubMed] [Google Scholar]
- Laloi C., Mestres-Ortega D., Marco Y., Meyer Y., Reichheld J. P. (2004). The Arabidopsis cytosolic thioredoxin h5 gene induction by oxidative stress and its W-box-mediated response to pathogen elicitor. Plant Physiol. 134 1006–1016 10.1104/pp.103.035782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam E., Kato N., Lawton M. (2001). Programmed cell death, mitochondria and the plant hypersensitive response. Nature 411 848–853 10.1038/35081184 [DOI] [PubMed] [Google Scholar]
- Laporte D., Olate E., Salinas P., Salazar M., Jordana X., Holuigue L. (2012). Glutaredoxin GRXS13 plays a key role in protection against photooxidative stress in Arabidopsis. J. Exp. Bot. 63 503–515 10.1093/jxb/err301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebel E., Heifetz P., Thorne L., Uknes S., Ryals J., Ward E. (1998). Functional analysis of regulatory sequences controlling PR-1 gene expression in Arabidopsis. Plant J. 16 223–233 10.1046/j.1365-313x.1998.00288.x [DOI] [PubMed] [Google Scholar]
- Lee S., Kim S. G., Park C. M. (2010). Salicylic acid promotes seed germination under high salinity by modulating antioxidant activity in Arabidopsis. New Phytol. 188 626–637 10.1111/j.1469-8137.2010.03378.x [DOI] [PubMed] [Google Scholar]
- Lee S., Park C. M. (2010). Modulation of reactive oxygen species by salicylic acid in Arabidopsis seed germination under high salinity. Plant Signal. Behav. 5 1534–1536 10.4161/psb.5.12.13159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindermayr C., Sell S., Muller B., Leister D., Durner J. (2010). Redox regulation of the NPR1-TGA1 system of Arabidopsis thaliana by nitric oxide. Plant Cell 22 2894–2907 10.1105/tpc.109.066464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackerness S., John C. F., Jordan B., Thomas B. (2001). Early signaling components in ultraviolet-B responses: distinct roles for different reactive oxygen species and nitric oxide. FEBS Lett. 489 237–242 10.1016/S0014-5793(01)02103-2 [DOI] [PubMed] [Google Scholar]
- Maleck K., Levine A., Eulgem T., Morgan A., Schmid J., Lawton K. A., et al. (2000). The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat. Genet. 26 403–410 10.1038/82521 [DOI] [PubMed] [Google Scholar]
- Mammarella N. D., Cheng Z., Fu Z. Q., Daudi A., Bolwell G. P., Dong X., et al. (2014). Apoplastic peroxidases are required for salicylic acid-mediated defense against Pseudomonas syringae. Phytochemistry 10.1016/j.phytochem.2014.07.010 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruta T., Noshi M., Tanouchi A., Tamoi M., Yabuta Y., Yoshimura K., et al. (2012). H2O2-triggered retrograde signaling from chloroplasts to nucleus plays specific role in response to stress. J. Biol. Chem. 287 11717–11729 10.1074/jbc.M111.292847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateo A., Funck D., Muhlenbock P., Kular B., Mullineaux P. M., Karpinski S. (2006). Controlled levels of salicylic acid are required for optimal photosynthesis and redox homeostasis. J. Exp. Bot. 57 1795–1807 10.1093/jxb/erj196 [DOI] [PubMed] [Google Scholar]
- Meyer Y., Belin C., Delorme-Hinoux V., Reichheld J. P., Riondet C. (2012). Thioredoxin and glutaredoxin systems in plants: molecular mechanisms, crosstalks, and functional significance. Antioxid. Redox Signal. 17 1124–1160 10.1089/ars.2011.4327 [DOI] [PubMed] [Google Scholar]
- Miura K., Okamoto H., Okuma E., Shiba H., Kamada H., Hasegawa P. M., et al. (2013). SIZ1 deficiency causes reduced stomatal aperture and enhanced drought tolerance via controlling salicylic acid-induced accumulation of reactive oxygen species in Arabidopsis. Plant J. 73 91–104 10.1111/tpj.12014 [DOI] [PubMed] [Google Scholar]
- Miura K., Tada Y. (2014). Regulation of water, salinity, and cold stress responses by salicylic acid. Front. Plant Sci. 5:4 10.3389/fpls.2014.00004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J. W., Loake G. J., Spoel S. H. (2011). Transcription dynamics in plant immunity. Plant Cell 23 2809–2820 10.1105/tpc.111.087346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou Z., Fan W., Dong X. (2003). Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113 935–944 10.1016/s0092-8674(03)00429-x [DOI] [PubMed] [Google Scholar]
- Mueller S., Hilbert B., Dueckershoff K., Roitsch T., Krischke M., Mueller M. J., et al. (2008). General detoxification and stress responses are mediated by oxidized lipids through TGA transcription factors in Arabidopsis. Plant Cell 20 768–785 10.1105/tpc.107.054809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murmu J., Bush M. J., Delong C., Li S., Xu M., Khan M., et al. (2010). Arabidopsis basic leucine-zipper transcription factors TGA9 and TGA10 interact with floral glutaredoxins ROXY1 and ROXY2 and are redundantly required for anther development. Plant Physiol. 154 1492–1504 10.1104/pp.110.159111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndamukong I., Abdallat A. A., Thurow C., Fode B., Zander M., Weigel R., et al. (2007). SA-inducible Arabidopsis glutaredoxin interacts with TGA factors and suppresses JA-responsive PDF1.2 transcription. Plant J. 50 128–139 10.1111/j.1365-313X.2007.03039.x [DOI] [PubMed] [Google Scholar]
- Neuenschwander U., Vernooij B., Friedrich L., Uknes S., Kessmann H., Ryals J. (1995). Is hydrogen peroxide a second messenger of salicylic acid in systemic acquired resistance? Plant J. 8 227–233 10.1046/j.1365-313X.1995.08020227.x [DOI] [Google Scholar]
- Noctor G., Lelarge-Trouverie C., Mhamdi A. (2014). The metabolomics of oxidative stress. Phytochemistry 10.1016/j.phytochem.2014.09.002 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Noshi M., Maruta T., Shigeoka S. (2012). Relationship between chloroplastic H2O2 and the salicylic acid response. Plant Signal. Behav. 7 944–946 10.4161/psb.20906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien J. A., Daudi A., Finch P., Butt V. S., Whitelegge J. P., Souda P., et al. (2012). A peroxidase-dependent apoplastic oxidative burst in cultured Arabidopsis cells functions in MAMP-elicited defense. Plant Physiol. 158 2013–2027 10.1104/pp.111.190140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa D., Nakajima N., Tamaoki M., Aono M., Kubo A., Kamada H., et al. (2007). The isochorismate pathway is negatively regulated by salicylic acid signaling in O3-exposed Arabidopsis. Planta 226 1277–1285 10.1007/s00425-007-0556-555 [DOI] [PubMed] [Google Scholar]
- Pajerowska-Mukhtar K. M., Emerine D. K., Mukhtar M. S. (2013). Tell me more: roles of NPRs in plant immunity. Trends Plant Sci. 18 402–411 10.1016/j.tplants.2013.04.004 [DOI] [PubMed] [Google Scholar]
- Pandey S. P., Somssich I. E. (2009). The role of WRKY transcription factors in plant immunity. Plant Physiol. 150 1648–1655 10.1104/pp.109.138990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape S., Thurow C., Gatz C. (2010). The Arabidopsis PR-1 promoter contains multiple integration sites for the coactivator NPR1 and the repressor SNI1. Plant Physiol. 154 1805–1818 10.1104/pp.110.165563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Z. M., Murata Y., Benning G., Thomine S., Klusener B., Allen G. J., et al. (2000). Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 406 731–734 10.1038/35021067 [DOI] [PubMed] [Google Scholar]
- Pieterse C. M., Leon-Reyes A., Van Der Ent S., Van Wees S. C. (2009). Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 5 308–316 10.1038/nchembio.164 [DOI] [PubMed] [Google Scholar]
- Price A. H., Taylor A., Ripley S. J., Griffiths A., Trewavas A. J., Knight M. R. (1994). Oxidative signals in tobacco increase cytosolic calcium. Plant Cell 6 1301–1310 10.1105/tpc.6.9.1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X. F., Holuigue L., Horvath D. M., Chua N. H. (1994). Immediate early transcription activation by salicylic acid via the cauliflower mosaic virus as-1 element. Plant Cell 6 863–874 10.1105/tpc.6.6.863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusterucci C., Espunya M. C., Diaz M., Chabannes M., Martinez M. C. (2007). S-nitrosoglutathione reductase affords protection against pathogens in Arabidopsis, both locally and systemically. Plant Physiol. 143 1282–1292 10.1104/pp.106.091686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Martín J., Heald J. I. M., Kingston-Smith A., Winters A. N. A., Rubiales D., Sanz M., et al. (2014). A metabolomic study in oats (Avena sativa) highlights a drought tolerance mechanism based on salicylate signalling pathways and the modulation of carbon, antioxidant and photo-oxidative metabolism. Plant Cell Environ. 10.1111/pce.12501 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Seyfferth C., Tsuda K. (2014). Salicylic acid signal transduction: the initiation of biosynthesis, perception and transcriptional reprogramming. Front. Plant Sci. 5:697 10.3389/fpls.2014.00697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearer H. L., Cheng Y. T., Wang L., Liu J., Boyle P., Despres C., et al. (2012). Arabidopsis clade I TGA transcription factors regulate plant defenses in an NPR1-independent fashion. Mol. Plant Microbe Interact. 25 1459–1468 10.1094/MPMI-09-11-0256 [DOI] [PubMed] [Google Scholar]
- Shi H., Wang X., Ye T., Chen F., Deng J., Yang P., et al. (2014). The Cysteine2/Histidine2-Type Transcription Factor ZINC FINGER OF Arabidopsis thaliana6 Modulates Biotic and Abiotic Stress Responses by Activating Salicylic Acid-Related Genes and C-REPEAT-BINDING FACTOR Genes in Arabidopsis. Plant Physiol. 165 1367–1379 10.1104/pp.114.242404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel S. H. (2003). NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 15 760–770 10.1105/tpc.009159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel S. H., Loake G. J. (2011). Redox-based protein modifications: the missing link in plant immune signalling. Curr. Opin. Plant Biol. 14 358–364 10.1016/j.pbi.2011.03.007 [DOI] [PubMed] [Google Scholar]
- Stotz H. U., Mueller S., Zoeller M., Mueller M. J., Berger S. (2013). TGA transcription factors and jasmonate-independent COI1 signalling regulate specific plant responses to reactive oxylipins. J. Exp. Bot. 64 963–975 10.1093/jxb/ers389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada Y., Spoel S. H., Pajerowska-Mukhtar K., Mou Z., Song J., Wang C., et al. (2008). Plant immunity requires conformational changes [corrected] of NPR1 via S-nitrosylation and thioredoxins. Science 321 952–956 10.1126/science.1156970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres M. A., Dangl J. L., Jones J. D. (2002). Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. U.S.A. 99 517–522 10.1073/pnas.012452499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman W., Sreekanta S., Lu Y., Bethke G., Tsuda K., Katagiri F., et al. (2013). The CALMODULIN-BINDING PROTEIN60 family includes both negative and positive regulators of plant immunity. Plant Physiol. 163 1741–1751 10.1104/pp.113.227108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K., Sato M., Glazebrook J., Cohen J. D., Katagiri F. (2008). Interplay between MAMP-triggered and SA-mediated defense responses. Plant J. 53 763–775 10.1111/j.1365-313X.2007.03369.x [DOI] [PubMed] [Google Scholar]
- Van der Does D., Leon-Reyes A., Koornneef A., van Verk M. C., Rodenburg N., Pauwels L., et al. (2013). Salicylic acid suppresses jasmonic acid signaling downstream of SCFCOI1-JAZ by targeting GCC promoter motifs via transcription factor ORA59. Plant Cell 25 744–761 10.1105/tpc.112.108548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Verk M. C., Bol J. F., Linthorst H. J. (2011). WRKY transcription factors involved in activation of SA biosynthesis genes. BMC Plant Biol. 11:89 10.1186/1471-2229-11-89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan D., Li R., Zou B., Zhang X., Cong J., Wang R., et al. (2012). Calmodulin-binding protein CBP60g is a positive regulator of both disease resistance and drought tolerance in Arabidopsis. Plant Cell Rep. 31 1269–1281 10.1007/s00299-012-1247-7 [DOI] [PubMed] [Google Scholar]
- Wang D., Amornsiripanitch N., Dong X. (2006). A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog. 2:e123 10.1371/journal.ppat.0020123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Fobert P. R. (2013). Arabidopsis clade I TGA factors regulate apoplastic defences against the bacterial pathogen Pseudomonas syringae through endoplasmic reticulum-based processes. PLoS ONE 8:e77378 10.1371/journal.pone.0077378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiermer M., Feys B. J., Parker J. E. (2005). Plant immunity: the EDS1 regulatory node. Curr. Opin. Plant Biol. 8 383–389 10.1016/j.pbi.2005.05.010 [DOI] [PubMed] [Google Scholar]
- Wildermuth M. C., Dewdney J., Wu G., Ausubel F. M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414 562–565 10.1038/35107108 [DOI] [PubMed] [Google Scholar]
- Wrzaczek M., Brosche M., Kangasjarvi J. (2013). ROS signaling loops – production, perception, regulation. Curr. Opin. Plant Biol. 16 575–582 10.1016/j.pbi.2013.07.002 [DOI] [PubMed] [Google Scholar]
- Wu Y., Zhang D., Chu J. Y., Boyle P., Wang Y., Brindle I. D., et al. (2012). The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep. 1 639–647 10.1016/j.celrep.2012.05.008 [DOI] [PubMed] [Google Scholar]
- Yoshida S., Tamaoki M., Ioki M., Ogawa D., Sato Y., Aono M., et al. (2009). Ethylene and salicylic acid control glutathione biosynthesis in ozone-exposed Arabidopsis thaliana. Physiol. Plant. 136 284–298 10.1111/j.1399-3054.2009.01220.x [DOI] [PubMed] [Google Scholar]
- Zander M., La Camera S., Lamotte O., Metraux J. P., Gatz C. (2010). Arabidopsis thaliana class-II TGA transcription factors are essential activators of jasmonic acid/ethylene-induced defense responses. Plant J. 61 200–210 10.1111/j.1365-313X.2009.04044.x [DOI] [PubMed] [Google Scholar]
- Zander M., Thurow C., Gatz C. (2014). TGA transcription factors activate the salicylic acid-suppressible branch of the ethylene-induced defense program by regulating ORA59 expression. Plant Physiol. 165 1671–1683 10.1104/pp.114.243360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Tessaro M. J., Lassner M., Li X. (2003). Knockout analysis of Arabidopsis transcription factors TGA2 TGA5 and TGA6 reveals their redundant and essential roles in systemic acquired resistance. Plant Cell 15 2647–2653 10.1105/tpc.014894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Xu S., Ding P., Wang D., Cheng Y. T., He J., et al. (2010). Control of salicylic acid synthesis and systemic acquired resistance by two members of a plant-specific family of transcription factors. Proc. Natl. Acad. Sci. U.S.A. 107 18220–18225 10.1073/pnas.1005225107 [DOI] [PMC free article] [PubMed] [Google Scholar]



