Abstract
The objectives of this study were to use non-equilibrium gravitational field-flow fractionation (GrFFF), an immunotag-less method of sorting mesenchymal stem cells (MSCs), to sort equine muscle tissue-derived mesenchymal stem cells (MMSCs) and bone marrow-derived mesenchymal stem cells (BMSC) into subpopulations and to carry out assays in order to compare their osteogenic capabilities. Cells from 1 young adult horse were isolated from left semitendinosus muscle tissue and from bone marrow aspirates of the fourth and fifth sternebrae. Aliquots of 800 × 103 MSCs from each tissue source were sorted into 5 fractions using non-equilibrium GrFFF (GrFFF proprietary system). Pooled fractions were cultured and expanded for use in osteogenic assays, including flow cytometry, histochemistry, bone nodule assays, and real-time quantitative polymerase chain reaction (qPCR) for gene expression of osteocalcin (OCN), RUNX2, and osterix. Equine MMSCs and BMSCs were consistently sorted into 5 fractions that remained viable for use in further osteogenic assays. Statistical analysis confirmed strongly significant upregulation of OCN, RUNX2, and osterix for the BMSC fraction 4 with P < 0.00001. Flow cytometry revealed different cell size and granularity for BMSC fraction 4 and MMSC fraction 2 compared to unsorted controls and other fractions. Histochemisty and bone nodule assays revealed positive staining nodules without differences in average nodule area, perimeter, or stain intensity between tissues or fractions. As there are different subpopulations of MSCs with different osteogenic capacities within equine muscle- and bone marrow-derived sources, these differences must be taken into account when using equine stem cell therapy to induce bone healing in veterinary medicine.
Résumé
Les objectifs de la présente étude étaient d’utiliser une méthode non-équilibrée de fractionnement par flot sous champs gravitationnel (GrFFF), une méthode sans marquage immunologique de séparation des cellules souches mésenchymateuses (MSCs), afin de séparer les cellules souches mésenchymateuses dérivées du tissu musculaire équin (MMSCs) et les cellules souches mésenchymateuses provenant de la moelle osseuse (BMSCs) en sous-populations et de réaliser des essais afin de comparer leurs capacités ostéogéniques. Des cellules provenant d’un jeune cheval adulte furent isolées du muscle semi-tendineux gauche et d’aspirations de la moelle osseuse de la quatrième et cinquième strernèbre. Des aliquotes de 800 × 103 MSCs provenant de chaque source de tissu furent séparés en 5 fractions par GrFFF non-équilibré (système breveté GrFFF). Des fractions regroupées ont été mises en culture afin de proliférer pour utilisation dans des essais ostéogéniques, incluant la cytométrie en flux, l’histochimie, des essais de nodules osseux, et l’amplification en chaine quantitative par la polymérase (qPCR) pour l’expression des gènes de l’ostéocalcine (OCN), RUNX2, et osterix. Les MMSCs et BMSCs équins étaient séparés de manière constante en 5 fractions qui demeuraient viables pour utilisation dans des essais ostéogéniques additionnels. Les analyses statistiques ont confirmé une régulation à la hausse très significative pour OCN, RUNX2 et osterix pour la fraction 4 des BMSC (P < 0,00001). La cytométrie en flux a révélé une taille et une granularité différente pour la fraction 4 des BMSCs et la fraction 2 des MMSCs comparativement aux témoins non-séparés et aux autres fractions. L’histochimie et les essais de nodules osseux ont révélé des nodules se colorant positivement sans différence pour les tissus ou les fractions dans la moyenne de la surface du nodule, du périmètre, ou de l’intensité de la coloration. Étant donné qu’il y a différentes sous-populations de MSCs avec différentes capacités ostéogéniques parmi les sources dérivées du muscle et de la moelle osseuse, ces différences doivent être prises en compte lors de l’utilisation thérapeutique en médecine vétérinaire des cellules souches pour induire la guérison osseuse.
(Traduit par Docteur Serge Messier)
Introduction
There is a constant need to expedite the healing of fractures in horses due to the inherent risks of contralateral limb laminitis, cyclic loading, and fatigue failure of implants (1) and nonunions due to disturbed blood supply to the injured bone (2). The most common method used to accelerate bone healing in horses is autogenous, cancellous bone graft. Bone tissue engineering methods, including the use of mesenchymal stem cells (MSCs) to enhance bone healing, have been evaluated in research studies in many species. The injection of MSCs into the distraction gap of rats has been shown to have encouraging results in preventing a nonunion (3). Several in-vitro studies (4,5) have addressed the healing of equine musculoskeletal injuries through MSC-based treatments.
While cell therapies show promise, they need to be standardized and refined before clinical use to increase the rate of equine bone healing. Part of that refinement can be addressed by attempting to use the purest cultures of stem cells that are optimal at healing the tissue of interest. It is well-known that mesenchymal stem cells (MSCs) derived from a single source are made up of a heterogeneous population (6,7). Subpopulations of MSCs are documented within many tissue types and many species (8), including mouse bone (9), mouse blood (10), mouse muscle (11), rat bone marrow (12), rat calvarial bone (13), human heart (14), human adipose (15), equine adipose (16), equine umbilical cord (17), and equine muscle, periosteum, adipose, and bone marrow (18). It has been found that these subpopulations have different shapes, proliferation, and differentiation abilities (12,19). It is therefore important to be able to isolate the fraction of MSCs that proliferates and differentiates optimally for the application of interest.
Identifying the source and subpopulation of MSCs with the best osteogenic potential may prove vital for moving basic science research toward clinical cell-based treatments to promote bone healing (20). Traditionally, cells are sorted into their respective subpopulations using fluorescence-activated cell sorting (FACS) (21). A more “cell-friendly” method is required, however, as this method is complicated, expensive, relies on clearly elucidated cell surface markers (22), and alters the function of cells (23). Field-flow fractionation (FFF) is a group of bioanalysis techniques that can be used to separate bioanalytes ranging from proteins and nucleic acids to viruses, organelles, and whole cells. Non-equilibrium gravitational field-flow fractionation (GrFFF) is a flowthrough type of FFF technique that relies on the earth’s gravity to sort MSCs (24). The cells settle into different velocity regions of the fluid-filled channel based on differences in their molar mass, size, and surface antigens. Due to these variances in biophysical properties, the cells are carried downstream at different speeds, exiting the channel after different retention times. The division of the cells into the various resulting fractions reveals differences in the cells’ molar mass, size, and surface antigens (25).
It has been shown that GrFFF-based methods are useful for cellular applications. Gravitational field-flow fractionation (GrFFF) has been used to sort human stem cells (26) and non-equilibrium GrFFF has been used to isolate, purify, and sort human MSCs derived from different sources (24). The different elution profiles may help elucidate important distinctions between MSC sources of donor tissue. The resulting fractions will have an array of commitment potentials that correlate with their differing degrees of stemness (21).
Previous work in our lab has validated the use of a non-equilibrium gravitational field-flow fractionation (GrFFF) MSC sorting system for use with equine MSCs (18) and has shown that muscle and bone marrow may be ideal tissue sources for promoting bone healing based on osteogenic capacity when compared to periosteum and adipose tissue (27). Based on these findings, we used the non-equilibrium GrFFF system to sort cells into their respective subpopulations.
We hypothesized that equine muscle- and bone marrow-derived MSCs would consist of subpopulations that did not have equal osteogenic capabilities. The purpose of the study reported here was to use a non-equilibrium GrFFF system to sort equine muscle-derived mesenchymal stem cells (MMSCs) and bone marrow-derived mesenchymal stem cells (BMSCs) into distinct fractions and to compare the osteogenic capacities of the resulting subpopulations.
Materials and methods
Samples
Bone marrow and skeletal muscle were collected from 1 young adult horse postmortem. The horse was donated to the Atlantic Veterinary College for reasons other than this study and was euthanized in accordance with protocols approved by the University of Prince Edward Island Animal Care Committee (27). The horse was first sedated with intravenous (IV) xylazine [1.1 mg/kg body weight (BW)] (Xylamax; Bimeda, Cambridge, Ontario) and then euthanized with pentabarbitol sodium administered IV (10 mL/50 kg BW) (Euthanyl Forte; Bimeda).
Tissue collection and cell isolation
Tissue collection and cell isolation techniques were carried out as described in an earlier study (27). Briefly, immediately after euthanasia, bone marrow was aseptically collected from the sternebrae using an Illinois Bone Marrow Biopsy Needle (Carefusion, San Diego, California, USA) and 9 cm3 of muscle was aseptically collected from the left semitendinosus/membranosus muscle. The aspirate (9.5 mL) of bone marrow was collected from the fourth sternebrae into a 12-mL syringe that had been pre-loaded with 2.5 mL of 1000 IU/mL heparin (Leo Pharma, Thornhill, Ontario). Another sample was immediately drawn from the fifth sternebra in the same fashion and transported to the laboratory. Cells were isolated from bone marrow using a centrifugation gradient technique. The tissues collected were placed in alpha minimal essential media (αMEM; Invitrogen, Toronto, Ontario) and transported to the laboratory. Cells were isolated from muscle by means of an enzyme digestion technique (27).
Cell cryopreservation
Cells were divided into 2.5 million cell aliquots with 1.8 mL of freezing media [10 mL dimethyl sulfoxide (DMSO)] in 90 mL of fetal bovine serum (FBS) in cryo-vials (Corning, Corning, New York, USA). They were kept at −80°C for a minimum of 24 h and then placed in a liquid nitrogen tank until removed for cell culture (17,18,27–29). Viable cells were plated in T-75 flasks (Corning) at a cell density of 33 × 103 cells/cm2 in standard media (SM) [αMEM supplemented with 10% FBS (PAA Laboratories, Etobicoke, Ontario), L-glutamine (2 mM) (Invitrogen), 10 000 U penicillin, 10 mg streptomycin/mL (Invitrogen), and 250 μg/mL amphotericin B (Invitrogen)].
Cell preparation
Cultured and expanded cells from passage 2 of both bone marrow and muscle tissue were used for the GrFFF. Cells were washed with phosphate-buffered saline (PBS) (Invitrogen) and then incubated for 30 min in a humidified 5% carbon dioxide (CO2) and 95% air atmosphere incubator at 37°C with 5 parts versene (Invitrogen) to 1 part trypsin (Invitrogen). The reaction was stopped with an equal amount of SM. The cell suspension was spun at 377 × g for 10 min and the supernatant removed. The pellet was vortexed and resuspended in 3 mL of mobile phase solution [1 g BSA (Bovine serum albumin; Fischer Scientific, Fair Lawn, New Jersey, USA) in 1 L PBS made with ultrapure water, 5000 U penicillin, and 5 mg streptomycin/mL]. Aliquots of 800 × 103 cells from each sample were seeded into 6 Eppendorf vials, spun down at 377 × g for 7 min and the supernatant was removed. Fifty microliters of mobile phase solution was added to each Eppendorf vial and cells were resuspended.
GrFFF system
The GrFFF system was purchased from byFlow s.r.l. (Bologna, Italy) and assembled and operated following the manufacturer’s instructions. Bleach solution [50% ultrapure water and 50% sodium hypochlorite solution (Sigma, Oakville, Ontario)] was used to sterilize the fractionation system and the 100-μL high-performance liquid chromatography (HPLC) syringe (Hamilton, Reno, Nevada, USA), to be used for sample loading, at the beginning of each working day as described and schematized in previous studies (21,26). The fractionation system was then flushed with sterile water and loaded with mobile phase solution. Each aliquot of 800 × 103 cells was then individually injected into the GrFFF system (21,24) and sorted into 5 fractions by changing the collection tube every 5 min. This timing was based on human MSC sorting work done with the GrFFF system (21) and validation work done in our laboratory that graphed the absorbency readings at different intervals and adjusted the collection times until fraction absorbencies (optical density), characterized by spectrophotometric analysis (LKB Biochrom Ultrospec II 4050 UV/Vis Spectrophotometer; Biochrom, Holliston, Massachusetts, USA) at a wavelength of 600 nm, were consistently different from 1 group to the next (18). The 5 fractions from each of the 6 aliquots and 1 unsorted group were combined to provide pooled fractions that were assessed by hemocytometric analysis. Pooled fractions were plated in T-75 flasks (Corning) at a cell density of 33 × 103 cells/cm2 in SM and expanded until there were appropriate numbers for osteogenic assays.
Osteoblastic cell differentiation
Sorted cells and unsorted control cells from both bone marrow and muscle tissue were seeded at a density of 1300 cells/cm2 into 30-mm dishes (Corning) and induced to differentiate in parallel with 1 control in SM as previously described. Cells were supplemented with an osteogenic induction medium (OM) [α-MEM, 5% FCS, 10 000 U penicillin and 10 mg streptomycin/mL, 250 μg/mL amphotericin B, 50 μg/mL ascorbic acid, dexamethazone 10−8 M, and 10 mM β-glycerophosphate (Sigma)]. Cultures were maintained for 7 d and then fixed for 20 min in 10% neutral buffered formalin at room temperature (27). Cultures were then stained for calcium with von Kossa stain (30) and with the substrate naphthol AS MX-PO4 and Red Violet LB salt for alkaline phosphatase (31) to confirm mineralization of bone nodules and osteoblastic differentiation. Light microscopy (Axiovert 40 CFL; Carl Zeiss Canada, Toronto, Ontario) digital images (Power Shot G5; Canon, Mississauga, Ontario) were taken on day 7 to document bone nodule formation for quantification of average area, average perimeter, and intensity (Sigma Scan Pro 5; Systat Software, San Jose, California, USA).
Flow cytometric analysis of MSC surface markers
Cultured and expanded cells from the second passage of each of the 5 fractions and the 1 unsorted control from both bone marrow and muscle tissue were used for the flow cytometric analysis. The amount of antibody was optimized with a cytometer (FACSAria Flow Cytometer; BD Biosciences, Mississauga, Ontario).
Cell preparation
Cells were washed with PBS solution and then incubated for 15 min in a humidified incubator at 5% CO2 and 95% air at 37°C with a mixture of versene (Invitrogen) and trypsin (5:1). This detachment method yielded the highest values for viability (up to 95% after 8 h). The reaction was stopped with an equal amount of standard medium. The cell suspension was centrifuged (377 × g for 10 min) and the pellet was then resuspended and washed in ice-cold 1% BSA in PBS solution. The cell suspension was again centrifuged (377 × g for 10 min) and the resulting pellet was resuspended in ice-cold 1% BSA in PBS solution, stained with trypan blue to determine viability, and counted for flow cytometric analysis.
Cell labeling
One million cells per sample were labeled. For each of the 5 fractions and 1 unsorted control group, 1 sample was unstained and served as a negative control sample. Successive samples were labeled with validated (22,32,33) antibodies (CD45, CD44, CD90, CD34, CD29, CD14, CD79, and MHCII respectively) and their isotypes for an internal negative control. Cells were centrifuged (377 × g for 10 min) and primary antibodies were added in 1% BSA in PBS solution (Table I). Samples were placed on ice and incubated for 45 min; samples were then washed in ice-cold 1% BSA in PBS solution and centrifuged (377 × g for 10 min). The washing and centrifugation steps were repeated 3 times. Cells were stored at 4°C until flow cytometric analysis. The secondary fluorescein isothiocyanate (FITC)-labeled antibody for CD90 was diluted in 1% BSA in PBS solution and incubated on ice for 30 min and then washed in ice-cold 1% BSA in PBS solution and centrifuged (377 × g for 10 min). The washing and centrifugation steps were repeated 3 times. Cells were stored at 4°C until flow cytometric analysis (FACSAria flow cytometer, BD Biosciences).
Table I.
Characteristics and source of antibodies used for flow cytometric analysis of mesenchymal stem cell (MSC) surface markers
| CD marker | Fluorochrome | Emission wavelength (nm) | Excitation wavelength (nm) | Source |
|---|---|---|---|---|
| CD34 | Allophycocyanin | 660 | 650 | AbD Seroteca |
| CD44 | Phycoerythrin | 667 | 496 | Biolegendb |
| CD45 | Allophycocyanin | 660 | 650 | AbD Seroteca |
| CD90 | NA | NA | NA | Accurate Chemical & Scientificc |
| CD14 | R Phycoerythrin-cyanine 5.1 | 693 | 565 | Beckman Coulter Canadad |
| CD29 | R Phycoerythrin-cyanine 5.1 | 693 | 565 | Biolegendb |
| MHCII | Fluorescein isothiocyanate (FITC) | 519 | 495 | AbD Seroteca |
| CD79 | Alexa fluor 647 | 665 | 650 | AbD Seroteca |
| IgG2a (mouse) | R Phycoerythrin-cyanine 5.1 | 693 | 565 | Beckman Coulter Canadad |
| IgG1k (mouse) | Phycoerythrin | 667 | 496 | Biolegendb |
| IgG1 (mouse) | Allophycocyanin | 660 | 650 | AbD Seroteca |
| IgG2bk (rat) | Phycoerythrin | 667 | 496 | Biolegendb |
| IgG2b | Phycoerythrin | 667 | 496 | AbD Seroteca |
| IgG1 (mouse) | Alexa fluor 647 | 665 | 650 | AbD Seroteca |
| IgM (mouse) | FITC | 519 | 495 | AbD Seroteca |
| IgG1 (mouse) | FITC | 519 | 495 | AbD Seroteca |
Raleigh, North Carolina, USA.
San Diego, California, USA.
Westbury, New York, USA.
Mississauga, Ontario.
NA = Not applicable.
Real-time quantitative PCR
Cultured and expanded cells from the second passage of each of the 5 fractions and the 1 unsorted control from both bone marrow and muscle tissue were seeded in 6-well plates at 200 cells/cm2 in triplicate. Half of the wells were induced with osteogenic medium and the other half were maintained in standard medium to serve as control cultures. Growth of the paired cultures was stopped on day 7 and total ribonucleic acid (RNA) was extracted (Aurum Total RNA Mini Kit, Bio-Rad Laboratories, Hercules, California, USA) from the cells. The complementary deoxyribonucleic acid (cDNA) was synthesized from total RNA via a cDNA synthesis kit (iScript cDNA Synthesis Kit; Bio-Rad Laboratories).
Primers derived from the coding regions of osteocalcin (OCN) were as follows: forward, 5′-CTGGGCCAGGACTCCGCATCT-3′ and reverse, 5′-AGCCAGCTCGTCACAGTCTGGGTTG-3′. Primers derived from the coding regions of RUNX2 were as follows: forward, 5′-CAGACCAGCAGCACTCCATA-3′ and reverse, 5′-CAGCGTCAACACCATCATTC-3′. Primers derived from the coding regions of osterix were as follows: forward, 5′-GGCTATGCCAATGACTACCC-3′ and reverse, 5′-GGTGAGATGCCTGCATGGA-3′. Expression of the OCN, RUNX2, and osterix genes were quantified via real-time PCR assay with a mix (iQ SYBR Green Supermix; Bio-Rad Laboratories). The PCR assay was done on a thermal cycler (Rotorgene-6 RG 3000; Corbett Research, Montreal, Quebec). Cycling conditions were as follows: 95°C for 5 min; 35 cycles of 95°C for 15 s; 56°C for 30 s for OCN (57.7°C for 30 s for RUNX2 and osterix); 72°C for 45 s; and melting from 55° to 99°C. Nuclease-free water instead of cDNA was used as a negative control sample. The housekeeping gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used to normalize the expression of each gene of interest (34,35).
Quantitative PCR statistical analysis
The data comprised gene expressions across 5 fractions and 1 unsorted control obtained from 2 tissues of 1 horse, with 3 replicate samples taken from each tissue. The study design has split-plot character, with samples within tissues representing whole-plots and fractions within each sample corresponding to sub-plots. Gene expression was quantified with real-time quantitative PCR. The outcome was computed using the comparative CT method (36,37) or 2 to the power of the negative difference of housekeeping gene, glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and gene of interest (GOI) threshold cycle (CT) values to a baseline medium (SM). Outcomes for 3 genes (OCN, RUNX2, and osterix) were analyzed separately. The analysis used linear-mixed models with random effects of samples (within tissues) after cubic root transformation of the outcome. The linear-mixed model assumptions were validated by residual analysis and found to be acceptable for all genes with this particular transformation. The linear-mixed model included fixed effects of tissues and fractions as well as their interaction. Least squares means were back-transformed to the scale of the outcome as estimated medians with 95% confidence intervals. The statistical analysis was carried out using SAS (proc mixed) software (Version 9.2; SAS Institute, Cary, North Carolina, USA) and the significance level was set at P < 0.05.
Results
Microscopic analysis of each fraction
Both MMSCs and BMSCs had cells in sorted fractions 1 to 5 as well as in unsorted fraction 6. Cells from all cell sources and fractions adhered to plastic. The cell recovery from the GrFFF sorting system was poor overall at about 50%. The highest number of cells in both tissues was found in fractions 2, 3, and 4. Muscle-derived and bone marrow-derived MSCs were sorted by GrFFF while maintaining sterility and viability.
Nodule quantification
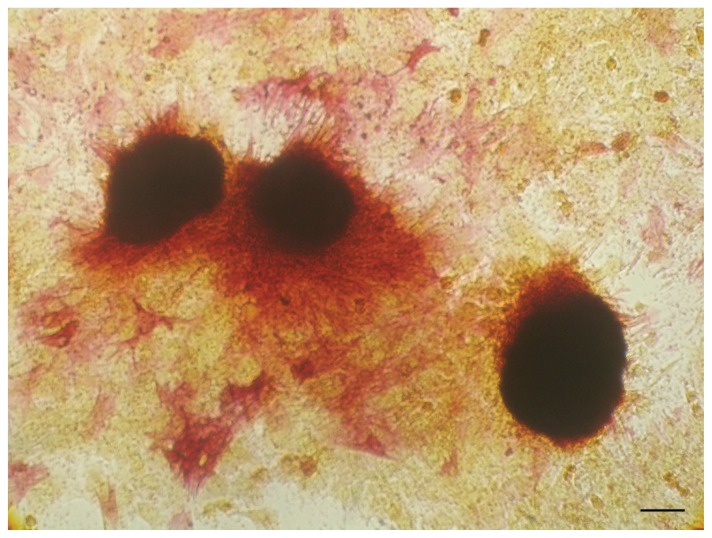
Bone nodules were assessed using light microscopy digital images of osteogenically induced cells from the second passage of each of the 5 fractions and the 1 unsorted control from both bone marrow and muscle tissue. Bone marrow-derived mesenchymal stem cell (BMSC) fractions 2, 4, and 5 had more von Kossa and alkaline phosphatase positive nodules present than BMSC fractions 1, 3, and the unsorted control BMSC population. Each of the 5 muscle-derived mesenchymal cell (MMSC) fraction cultures had more von Kossa and alkaline phosphatase positive nodules present than the unsorted control MMSC population. No differences in average nodule area, perimeter, or stain intensity were noted between tissues or fractions. A representative photomicrograph of von Kossa and alkaline phosphatase positive nodules is shown in Figure 1.
Figure 1.
Representative photomicrograph of von Kossa and alkaline phosphatase positive histochemical staining for muscle-derived mesenchymal stem cells (MMSCs) cultured from fraction 1. Magnification 10×. Bar = 200 μm.
Real-time quantitative PCR
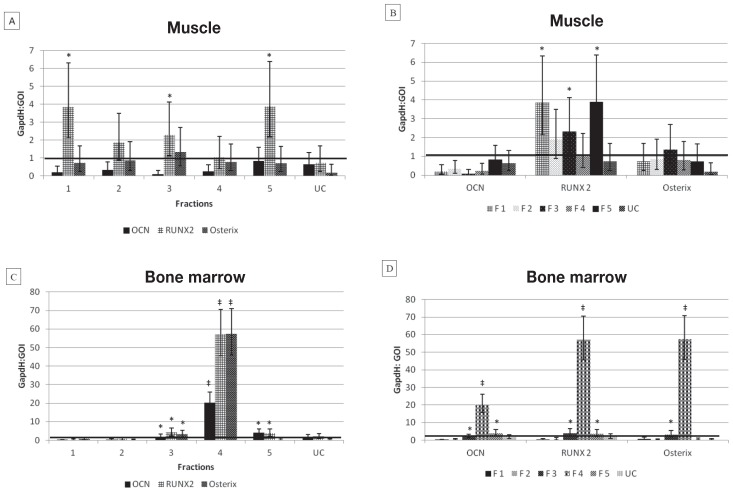
Osteogenic capacity was determined on the basis of gene expression of OCN, RUNX2, and osterix measured in sorted fractions 1 to 5, as well as in unsorted control, from both bone marrow and muscle tissue. Muscle-derived mesenchymal stem cell (MMSC) fractions 1, 3, and 5 had significantly higher (P < 0.05) RUNX2 expression after differentiation with osteogenic medium than did the control samples cultured in standard medium. There were no significant differences in OCN or osterix expression between differentiated and nondifferentiated cultures of MMSCs. Bone marrow-derived mesenchymal stem cell (BMSC) fraction 3 had significantly higher (P < 0.05) OCN, RUNX2, and osterix expression after differentiation with osteogenic medium than did the control samples cultured in standard medium. Bone marrow-derived mesenchymal stem cell (BMSC) fraction 5 had significantly higher (P < 0.05) OCN and RUNX2 expression after differentiation with osteogenic medium than did the control samples cultured in standard medium. Bone marrow-derived mesenchymal stem cell (BMSC) fraction 4 had highly significant (P ≤ 0.00000) OCN, RUNX2, and osterix expression after differentiation with osteogenic medium compared to the control samples cultured in standard medium. None of the unsorted control MMSCs or BMSCs showed significant differences in OCN, RUNX, or osterix expression between differentiated and nondifferentiated cultures of MMSCs (Figure 2).
Figure 2.
Graphical representation of quantitative polymerase chain reaction (qPCR) data. Mean ± standard deviation (SD) results of nontransformed data for real-time PCR assay of OCN, RUNX2, and osterix gene expression for 5 sorted fractions (F1–5) of mesenchymal stem cells (MSCs) and 1 unsorted control (UC) derived from muscle (A and B) and bone marrow (C and D). Note the extremely high expression of all 3 osteogenic markers in bone marrow fraction 4 compared to other tissue and fractions. Also note the difference in x-axis scales between muscle and bone marrow graphs.
* Significant (P ≤ 0.05) expression of gene of interest compared to cultures in standard medium (horizontal black line).
‡ Highly significant (P ≤ 0.00000) expression of gene of interest compared to cultures in standard medium.
GAPDH: GOI = Ratio of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene to the gene of interest (GOI).
Flow cytometric analysis
Cells isolated from the second passage of each of the 5 fractions and the 1 unsorted control from both bone marrow and muscle tissue were cultured via standard conditions and each positively expressed CD90, CD44, and CD29 and lacked expression of CD45, CD34, CD14, CD79, and MHCII as determined on the basis of flow cytometric data.
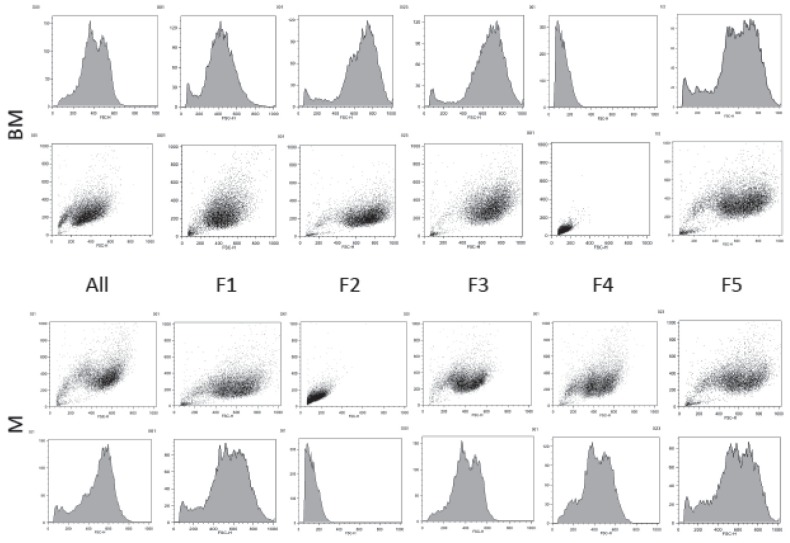
Triple-stained combinations were highly positive for expression of CD29-CD44, CD29-CD90, and CD44-CD90. Triple-stained combinations were highly negative for expression of CD14-CD79, CD14-MHCII, and CD79-MHCII. Results were similar across tissues, fractions, and unsorted controls. The unstained flow cytometric analysis for assessing the granularity and size of cells in the fraction subpopulations and unsorted whole population revealed a distinctly different population in bone marrow fraction 4 and muscle fraction 2 (Figure 3).
Figure 3.
Results of flow cytometric analysis of mesenchymal stem cells (MSCs) cultured from 5 sorted fractions (F1 to 5) and 1 unsorted control (UC) derived from equine muscle (M), bone marrow (BM), and unstained for assessing the population granularity (x-axis) and size (y-axis). Dot plot and corresponding histogram graphical representation of the subpopulations depict a distinctly different population in bone marrow fraction 4 and muscle fraction 2. The fluorescence intensity (arbitrary units) is depicted.
Discussion
This is the first study to compare the osteogenic capabilities of subpopulations of equine-derived MMSCs and BMSCs sorted using non-equilibrium GrFFF or any other sorting system. As expected, several subpopulations with varying degrees of osteogenic potential were present in each tissue. The fractions of cells had different osteogenic gene expression based on qPCR and different cell size and granularity based on flow cytometry. In a previous study (18), these subpopulations were found to have trilineage differentiation capabilities and therefore were not altered by the sorting process. Across all tissues, fractions, and unsorted controls, surface markers of all cells were consistent with what would be expected for MSCs (22,33). Sorting systems that rely on the cell surface markers alone would therefore have missed the differences found in subpopulations in this study. Evidence from this study indicates that cells must be sorted by properties other than cell surface markers when the phenotype has not been completely elucidated.
Interestingly, most of the fractionated BMSC and MMSC cultures had von Kossa and alkaline phosphatase positive nodules present and the unsorted control BMSC and MMSC populations had fewer nodules present. This revealed how very differently the same population of cells acts when separated into subpopulations. In this case, the subpopulations displayed more osteogenic activity than the population as a whole. We believe this finding is explained by the concept of an MSC microenvironment (38–41) that is a niche made up of other cells, local extracellular matrix, soluble molecules, and other naive MSCs (41). When this niche is altered by sorting cell populations into subpopulations, as was done in this study, it is reasonable to expect the subpopulations to behave differently from one another and from the original unsorted population.
The main limitation of this study was that only 1 horse was used. Inter-horse variation can be significant and could affect the results. We compensated for this limitation by carrying out all cultures, controls, and assays in triplicate. While equine MSC research is often subject to a low “n,” the published findings are nevertheless important (17,42). The main drawback of using this sorting system is the low number of cells that can be fractionated per run. We increased sorting throughput by pooling fractions collected at the same retention times from repeated runs (18). We also found that an expansion step after the sorting process easily made up for the low throughput. Others researchers have set up 2 GrFFF channels in parallel to increase sorting throughput (26) and placed a coating on the PVC in the GrFFF system, which improved the efficiency of the system (43).
In equine regenerative medicine, identifying the source of MSCs with the optimum osteogenic capability has been the focus of much evaluation (20,27,28,44,45). It is evident that the variation of MSCs within each source must also be taken into consideration (17,18). Techniques for sorting and enriching MSCs appear to be vital to the isolation of the equine MSC phenotypes that are ideal for bone healing.
In conclusion, subpopulations sorted from equine MMSCs and BMSCs with GrFFF are not created equally when it comes to osteogenic potential. Bone marrow-derived MSCs from fraction 4 may have more bone-healing potential because they express high levels of osteogenic gene markers. Further clarification of MSC subpopulation phenotypes is necessary in order to determine the optimum ultrapure population for the intended application.
Acknowledgments
The authors thank Glenda M. Wright, PhD for her assistance with this manuscript. This study was supported by the Atlantic Canada Opportunities Agency and Innovation PEI.
Footnotes
This manuscript represents a portion of a thesis to be submitted by Dr. Radtke to the University of Prince Edward Island, Department of Health Management, as a partial fulfillment of the requirements for a Doctor of Philosophy degree.
References
- 1.Brunner H. Fatigue fracture of bone plates. Injury. 1980;11:203–207. doi: 10.1016/s0020-1383(80)80044-1. [DOI] [PubMed] [Google Scholar]
- 2.Ducharme NG. Delayed union, nonunion, and malunion. In: Nixon AJ, editor. Equine Fracture Repair. 1st ed. Philadelphia, Pennsylvania: Saunders; 1996. pp. 354–358. [Google Scholar]
- 3.Richards M, Huibregtse BA, Caplan AI, Goulet JA, Goldstein SA. Marrow-derived progenitor cell injections enhance new bone formation during distraction. J Orthop Res. 1999;17:900–908. doi: 10.1002/jor.1100170615. [DOI] [PubMed] [Google Scholar]
- 4.Stewart AA, Barrett JG, Byron CR, et al. Comparison of equine tendon-, muscle-, and bone marrow-derived cells cultured on tendon matrix. Am J Vet Res. 2009;70:750–757. doi: 10.2460/ajvr.70.6.750. [DOI] [PubMed] [Google Scholar]
- 5.Vidal MA, Kilroy GE, Lopez MJ, Johnson JR, Moore RM, Gimble JM. Characterization of equine adipose tissue-derived stromal cells: Adipogenic and osteogenic capacity and comparison with bone marrow-derived mesenchymal stromal cells. Vet Surg. 2007;36:613–622. doi: 10.1111/j.1532-950X.2007.00313.x. [DOI] [PubMed] [Google Scholar]
- 6.Ema H, Morita Y, Suda T. Heterogeneity and hierarchy of hematopoietic stem cells. Exp Hematol. 2014;42:74–82. doi: 10.1016/j.exphem.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 7.Phinney DG. Functional heterogeneity of mesenchymal stem cells: Implications for cell therapy. J Cell Biochem. 2012;113:2806–2812. doi: 10.1002/jcb.24166. [DOI] [PubMed] [Google Scholar]
- 8.Triffit JT. The stem cell of the osteoblast. In: Bilizekian J, Raisz L, Rodan G, editors. Principles of Bone Biology. San Diego, California: Academic Press; 1996. pp. 39–50. [Google Scholar]
- 9.Rostovskaya M, Anastassiadis K. Differential expression of surface markers in mouse bone marrow mesenchymal stromal cell subpopulations with distinct lineage commitment. PLoS One. 2012;7:e51221. doi: 10.1371/journal.pone.0051221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caballero S, Hazra S, Bhatwadekar A, et al. Circulating mononuclear progenitor cells: Differential roles for subpopulations in repair of retinal vascular injury. Invest Ophthalmol Vis Sci. 2013;54:3000–3009. doi: 10.1167/iovs.12-10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun JS, Wu SY, Lin FH. The role of muscle-derived stem cells in bone tissue engineering. Biomaterials. 2005;26:3953–3960. doi: 10.1016/j.biomaterials.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 12.Zhang L, Chan C. Isolation and enrichment of rat mesenchymal stem cells (MSCs) and separation of single-colony derived MSCs. J Vis Exp. 2010;22:1852. doi: 10.3791/1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zohar R, McCulloch CA, Sampath K, Sodek J. Flow cytometric analysis of recombinant human osteogenic protein-1 (BMP-7) responsive subpopulations from fetal rat calvaria based on intracellular osteopontin content. Matrix Biol. 1998;16:295–306. doi: 10.1016/s0945-053x(98)90001-4. [DOI] [PubMed] [Google Scholar]
- 14.Li TS, Cheng K, Malliaras K, et al. Direct comparison of different stem cell types and subpopulations reveals superior paracrine potency and myocardial repair efficacy with cardiosphere-derived cells. J Am Coll Cardiol. 2012;59:942–953. doi: 10.1016/j.jacc.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zimmerlin L, Donnenberg VS, Pfeifer ME, et al. Stromal vascular progenitors in adult human adipose tissue. Cytometry A. 2010;77:22–30. doi: 10.1002/cyto.a.20813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pascucci L, Curina G, Mercati F, et al. Flow cytometric characterization of culture expanded multipotent mesenchymal stromal cells (MSCs) from horse adipose tissue: Towards the definition of minimal stemness criteria. Vet Immunol Immunopathol. 2011;144:499–506. doi: 10.1016/j.vetimm.2011.07.017. [DOI] [PubMed] [Google Scholar]
- 17.Corradetti B, Lange-Consiglio A, Barucca M, Cremonesi F, Bizzaro D. Size-sieved subpopulations of mesenchymal stem cells from intervascular and perivascular equine umbilical cord matrix. Cell Prolif. 2011;44:330–342. doi: 10.1111/j.1365-2184.2011.00759.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Radtke CL, Nino-Fong R, Esparza Gonzalez BP, McDuffee LA. Application of a novel sorting system for equine mesenchymal stem cells (MSCs) Can J Vet Res. 2014;78:290–6. [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang Y, Jahagirdar BN, Reinhardt RL, et al. Pluripotency of mesenchymal stem cells derived from adult marrow. Nature. 2002;418:41–49. doi: 10.1038/nature00870. [DOI] [PubMed] [Google Scholar]
- 20.Toupadakis CA, Wong A, Genetos DC, et al. Comparison of the osteogenic potential of equine mesenchymal stem cells from bone marrow, adipose tissue, umbilical cord blood, and umbilical cord tissue. Am J Vet Res. 2010;71:1237–1245. doi: 10.2460/ajvr.71.10.1237. [DOI] [PubMed] [Google Scholar]
- 21.Roda B, Lanzoni G, Alviano F, et al. A novel stem cell tag-less sorting method. Stem Cell Rev. 2009;5:420–427. doi: 10.1007/s12015-009-9088-7. [DOI] [PubMed] [Google Scholar]
- 22.De Schauwer C, Meyer E, Van de Walle GR, Van Soom A. Markers of stemness in equine mesenchymal stem cells: A plea for uniformity. Theriogenology. 2011;75:1431–1443. doi: 10.1016/j.theriogenology.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 23.Roda B, Zattoni A, Reschiglian P, et al. Field-flow fractionation in bioanalysis: A review of recent trends. Anal Chim Acta. 2009;635:132–143. doi: 10.1016/j.aca.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 24.Roda B, Reschiglian P, Zattoni A, et al. A tag-less method of sorting stem cells from clinical specimens and separating mesenchymal from epithelial progenitor cells. Cytometry B Clin Cytom. 2009;76:285–290. doi: 10.1002/cyto.b.20472. [DOI] [PubMed] [Google Scholar]
- 25.Reschiglian P, Zattoni A, Roda B, Michelini E, Roda A. Field-flow fractionation and biotechnology. Trends Biotechnol. 2005;23:475–483. doi: 10.1016/j.tibtech.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 26.Roda B, Reschiglian P, Alviano F, et al. Gravitational field-flow fractionation of human hemopoietic stem cells. J Chromatogr A. 2009;1216:9081–9087. doi: 10.1016/j.chroma.2009.07.024. [DOI] [PubMed] [Google Scholar]
- 27.Radtke CL, Nino-Fong R, Esparza Gonzalez BP, Stryhn H, McDuffee LA. Characterization and osteogenic potential of equine muscle tissue- and periosteal tissue-derived mesenchymal stem cells in comparison with bone marrow- and adipose tissue-derived mesenchymal stem cells. Am J Vet Res. 2013;74:790–800. doi: 10.2460/ajvr.74.5.790. [DOI] [PubMed] [Google Scholar]
- 28.Nino-Fong R, McDuffee LA, Esparza Gonzalez BP, Kumar MR, Merschrod SEF, Poduska KM. Scaffold effects on osteogenic differentiation of equine mesenchymal stem cells: An in vitro comparative study. Macromol Biosci. 2013;13:348–355. doi: 10.1002/mabi.201200355. [DOI] [PubMed] [Google Scholar]
- 29.Kisiel AH, McDuffee LA, Masaoud E, Bailey TR, Esparza Gonzalez BP, Nino-Fong R. Isolation, characterization, and in vitro proliferation of canine mesenchymal stem cells derived from bone marrow, adipose tissue, muscle, and periosteum. Am J Vet Res. 2012;73:1305–1317. doi: 10.2460/ajvr.73.8.1305. [DOI] [PubMed] [Google Scholar]
- 30.Bhargava U, Bar-Lev M, Bellows CG, Aubin JE. Ultrastructural analysis of bone nodules formed in vitro by isolated fetal rat calvaria cells. Bone. 1988;9:155–163. doi: 10.1016/8756-3282(88)90005-1. [DOI] [PubMed] [Google Scholar]
- 31.Burstone MS. Histochemical observations on enzymatic processes in bones and teeth. Ann N Y Acad Sci. 1960;85:431–444. doi: 10.1111/j.1749-6632.1960.tb49972.x. [DOI] [PubMed] [Google Scholar]
- 32.De Schauwer C, Piepers S, Van de Walle GR, et al. In search for cross-reactivity to immunophenotype equine mesenchymal stromal cells by multicolor flow cytometry. Cytometry A. 2012;81:312–323. doi: 10.1002/cyto.a.22026. [DOI] [PubMed] [Google Scholar]
- 33.Guest DJ, Ousey JC, Smith MRW. Defining the expression of marker genes in equine mesenchymal stromal cells. Stem Cells Cloning. 2008;1:1–9. doi: 10.2147/sccaa.s3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Braun J, Hack A, Weis-Klemm M, et al. Evaluation of the osteogenic and chondrogenic differentiation capacities of equine adipose tissue-derived mesenchymal stem cells. Am J Vet Res. 2010;71:1228–1236. doi: 10.2460/ajvr.71.10.1228. [DOI] [PubMed] [Google Scholar]
- 35.Parker RA, Clegg PD, Taylor SE. The in vitro effects of antibiotics on cell viability and gene expression of equine bone marrow-derived mesenchymal stromal cells. Equine Vet J. 2012;44:355–360. doi: 10.1111/j.2042-3306.2011.00437.x. [DOI] [PubMed] [Google Scholar]
- 36.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 37.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 38.Weiss L, Sakai H. The hematopoietic stroma. Am J Anat. 1984;170:447–463. doi: 10.1002/aja.1001700317. [DOI] [PubMed] [Google Scholar]
- 39.Whitlock CA, Tidmarsh GF, Muller-Sieburg C, Weissman IL. Bone marrow stromal cell lines with lymphopoietic activity express high levels of a pre-B neoplasia-associated molecule. Cell. 1987;48:1009–1021. doi: 10.1016/0092-8674(87)90709-4. [DOI] [PubMed] [Google Scholar]
- 40.Zhang J, Li L. Stem cell niche: Microenvironment and beyond. J Biol Chem. 2008;283:9499–9503. doi: 10.1074/jbc.R700043200. [DOI] [PubMed] [Google Scholar]
- 41.Kolf CM, Cho E, Tuan RS. Mesenchymal stromal cells. Biology of adult mesenchymal stem cells: Regulation of niche, self-renewal and differentiation. Arthritis Res Ther. 2007;9:204. doi: 10.1186/ar2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fortier LA, Nixon AJ, Williams J, Cable CS. Isolation and chondrocytic differentiation of equine bone marrow-derived mesenchymal stem cells. Am J Vet Res. 1998;59:1182–1187. [PubMed] [Google Scholar]
- 43.Roda B, Cioffi N, Ditaranto N, et al. Biocompatible channels for field-flow fractionation of biological samples: Correlation between surface composition and operating performance. Anal Bioanal Chem. 2005;381:639–646. doi: 10.1007/s00216-004-2860-2. [DOI] [PubMed] [Google Scholar]
- 44.McDuffee LA, Anderson GI. In vitro comparison of equine cancellous bone graft donor sites and tibial periosteum as sources of viable osteoprogenitors. Vet Surg. 2003;32:455–463. doi: 10.1053/jvet.2003.50060. [DOI] [PubMed] [Google Scholar]
- 45.Glynn ER, Londono AS, Zinn SA, Hoagland TA, Govoni KE. Culture conditions for equine bone marrow mesenchymal stem cells and expression of key transcription factors during their differentiation into osteoblasts. J Anim Sci Biotechnol. 2013;4:40. doi: 10.1186/2049-1891-4-40. [DOI] [PMC free article] [PubMed] [Google Scholar]