Abstract
Pododermatitis is a disease of concern for mink breeders in Canada and worldwide, as it causes discomfort and lowers the breeding rates on farms affected by the disease. Unfortunately, the etiology and pathogenesis of pododermatitis are still unknown. In this study, we compared Staphylococcus spp. and Streptococcus canis isolates from healthy mink with isolates from animals with pododermatitis on 2 farms in Ontario. Almost all hemolytic Staphylococcus spp. isolated were shown to be Staphylococcus delphini Group A by 16S ribosomal ribonucleic acid (rRNA) sequence analysis and polymerase chain reaction (PCR). Pulsed-field gel electrophoresis (PFGE) did not reveal any S. delphini or S. canis clonal lineages specifically associated with pododermatitis, which suggests that these bacteria do not act as primary pathogens, but does not dismiss their potential roles as opportunistic pathogens. While S. delphini and S. canis were the most prevalent bacterial pathogens in mink pododermatitis, they were also present in samples from healthy mink. Arcanobacterium phocae is occasionally isolated from pododermatitis cases, but is difficult to recover with conventional culture methods due to its slow growth. A quantitative real-time PCR was developed for the detection of A. phocae and was tested on 138 samples of footpad tissues from 14 farms. The bacterium was detected only in pododermatitis-endemic farms in Canada and was at higher concentrations in tissues from infected footpads than in healthy tissues. This finding suggests that A. phocae is involved in the pathogenesis of pododermatitis.
Résumé
La pododermatite est une maladie qui préoccupe les éleveurs de visons au Canada et ailleurs dans le monde, étant donné qu’elle cause un inconfort et diminue les taux de reproduction dans les fermes où la maladie est retrouvée. Malheureusement, l’étiologie et la pathogénie de la pododermatite sont encore inconnues. Dans la présente étude nous avons comparé des isolats de Staphylococcus spp. et Streptococcus canis provenant de visons en santé à ceux provenant d’animaux avec pododermatite de deux fermes en Ontario. Presque tous les isolats hémolytiques de Staphylococcus spp. ont été identifiés comme étant des S. delphini Groupe A par analyse des séquences de l’ARN ribosomal (ARNr) 16S et par réaction d’amplification en chaîne par la polymérase (PCR). L’analyse par électrophorèse en gel à champs pulsés (PFGE) n’a pas permis de mettre en évidence pour S. delphini ou S. canis de lignées clonales associées spécifiquement à la pododermatite, ce qui suggère que ces bactéries n’agissent pas en tant qu’agents pathogènes primaires, mais ne diminue pas pour autant leur rôle potentiel comme agents pathogènes opportunistes. Malgré le fait que S. delphini et S. canis étaient les bactéries pathogènes les plus prévalentes dans les cas de pododermatite chez le vison, elles étaient également présentes dans des échantillons provenant de visons en santé. Arcanobacterium phocae est isolé occasionnellement de cas de pododermatite, mais il est difficile de l’isoler en utilisant des méthodes conventionnelles de culture étant donné sa croissance lente. Une épreuve PCR quantitative en temps réel fut développée pour la détection d’A. phocae et fut testée sur 138 échantillon de tissus des coussinets plantaires provenant de 14 fermes. La bactérie fut détectée seulement de fermes canadienne où la pododermatite était endémique et se retrouvait en plus grande concentrations dans les tissus de coussinets plantaires infectés comparativement à des tissus sains. Ces résultats suggèrent qu’A. phocae est impliqué dans la pathogénie de la pododermatite.
(Traduit par Docteur Serge Messier)
Introduction
Pododermatitis is a significant problem in farmed mink (Neovison vison) and may impact the welfare and reproductive success of these animals (1,2). It can appear as acute, severe ulceration on footpads with occasional further infection of nail beds (1) and also as more chronic hyperkeratotic purulent plantar dermatitis (Figure 1). The disease appears to become endemic in affected farms and often recurs during successive breeding seasons. The more acute form of pododermatitis, which is also reported in other countries (3), has historically been associated with feeding seal meat, primarily derived from east coast harp seal (Phoca groenlandica), to mink (1,2). Although a primary mechanical etiology associated with increasing weight of breeding animals can be suspected for the chronic form of the disease, no specific microbial pathogen has been identified to date in association with pododermatitis lesions.
Figure 1.
Plantar hyperkeratotic and suppurative pododermatitis on the hind (left) and front feet (right) representative of lesions submitted by Canadian mink farmers for A. phocae PCR testing.
Multiple bacterial pathogens have been isolated from footpad lesions, with Staphylococcus spp. being the most frequent (D. Bruce Hunter, personal communication). Staphylococcal species are commonly associated with opportunistic infections of the skin. The most common species are Staphylococcus aureus, Staphylococcus intermedius, Staphylococcus pseudintermedius, and Staphylococcus delphini (4,5). The latter 3 coagulase-positive species make up the S. intermedius group (SIG) and are difficult to differentiate using phenotypic methods (6,7). In canine and feline infections, S. pseudintermedius is the most prevalent SIG species (4). Staphylococcus intermedius has been the most frequently identified species in cases of mink pododermatitis in Canada (4), but recent findings from Denmark suggest that S. delphini is indeed the major coagulase-positive species found in Mustelidae and mink in particular (8).
Much like the SIG, β-hemolytic Streptococcus spp. are normal residents of the skin, respiratory, genitourinary, and intestinal tracts of many animals, including dogs, horses, cats, and mink (9,10). They are also common opportunistic pathogens, causing a wide range of diseases, including mild to severe skin infections (9–11). Streptococcus canis is a common cause of infections in dogs and many other mammals (12,13) and has been regularly isolated from cases of pododermatitis in Canadian mink (14). The role of S. canis in footpad lesions is presently not known, as it is a common member of the natural microflora of healthy animals. Arcanobacterium spp. are commonly isolated from infected wounds of animals and humans (15–18), but can also be found in specimens from healthy individuals.
Although it is recovered much less frequently than streptococci and staphylococci, Arcanobacterium phocae is occasionally isolated from pododermatitis lesions. Its relatively slow growth and pinpoint sized colonies suggest that A. phocae may be commonly overgrown by other bacteria in culture and thus overlooked as a potential pathogen in pododermatitis. Arcanobacterium phocae is closely related to the animal and human pathogens Arcanobacterium pyogenes [now Trueperella pyogenes (19)], and Arcanobacterium haemolyticum, respectively. This suggests that it may share similar pathogenic properties with these species (20). Interestingly, A. phocae was first isolated from harbor seals (Phoca vitulina) and grey seals (Halichoerus grypus) in 1997 (21). A subsequent study (22) suggested that in the past, A. phocae may have been frequently misidentified as Listeria ivanovii, and has been underreported. It may be more significant as a pathogen than previously thought.
The objectives of this study were to: i) identify the major staphylococcal species from pododermatitis cases in Canadian mink using reliable molecular methods; ii) assess whether particular strains of Staphylococcus spp. and S. canis are associated with pododermatitis using pulsed-field gel electrophoresis (PFGE); and iii) develop and apply a quantitative real-time polymerase chain reaction (PCR) assay for the detection of A. phocae in order to compare tissues from feet of healthy mink and mink with pododermatitis lesions.
Materials and methods
Bacterial isolates and growth conditions
Samples collected from 60 mink, both healthy and with macroscopic evidence of pododermatitis at postmortem examination, were received for diagnostic testing by the Animal Health Laboratory at the University of Guelph from 2 separate farms in Canada. Swabs were taken of the oral cavity, feet, and rectum and were used for bacterial culture and identification. When footpad lesions were observed, swabs were taken of these areas specifically. Hemolytic Staphylococcus spp. (n = 56), S. canis (n = 13), and A. phocae (n = 2) isolates were grown at 35°C with 5% carbon dioxide (CO2) on blood agar plates. No systematic search for and characterization of anaerobes was carried out during this study. An additional A. phocae from older frozen samples of mink footpads and a Trueperella pyogenes isolate from a clinical infection in cattle were obtained from the Animal Health Laboratory at the University of Guelph, Ontario. A fourth A. phocae isolate, a human clinical A. haemolyticum isolate, and the Trueperella bernardiae ATCC 51728 strain were obtained from the National Microbiology Laboratory in Winnipeg, Manitoba. No mink were sacrificed for the purpose of the study and only animals killed by the farmers for reasons independent of the study were used.
Bacterial species identification
Fifty-six hemolytic Staphylococcus spp. isolates were recovered from the mink samples through the Animal Health Laboratory, of which 36 were confirmed as coagulase-positive. A fragment covering 98% of the 16S ribosomal ribonucleic acid (rRNA) gene was amplified from these 36 isolates using primers BSF8-20 (AGAGTTTGATCCTGGCTCAG) and 16S-1522R (AAGGAGGTGATCCARCCGCA), as described in a previous study (23). The resulting PCR products were sequenced and identified using nucleotide BLAST (24). A previously described PCR for identification of Group A and B S. delphini was further carried out on these 36 isolates, using the primers dea-R and dea-F, and deb-R4 and deb-F, respectively (25).
Streptococcus spp. and Arcanobacterium spp. were identified to the species level using standard culture techniques and MALDI-TOF analysis (Bruker Daltonics, Billerica, Massachusetts, USA). The identity of A. phocae was further confirmed using the 16S rRNA sequencing protocol previously described to eliminate the possibility of misidentification, especially in light of the recent discovery of Arcanobacterium canis (26).
Pulsed-field gel electrophoresis of S. delphini and S. canis
One isolate of S. delphini and S. canis was used per individual mink for further investigation (Table I). Four additional S. canis isolates from unrelated dogs were also compared to the original set of mink isolates. Staphylococcus delphini genomic deoxyribonucleic acid (DNA) was prepared for PFGE following standard protocols (27), with the addition of 4 U/mL lysostaphin (Cedarlane Labs, Burlington, Ontario) during bacterial lysis. Streptococcus canis DNA was prepared using the protocol described by DeWinter and Prescott (11). Genomic DNA from both species was digested with SmaI (New England BioLabs, Ipswich, Massachusetts, USA) before electrophoresis in a 1.0% agarose gel, using a CHEF-III electrophoresis unit (Bio-Rad Laboratories, Hercules, California, USA) in 0.5× Trisborate-ethylenediamine tetra-acetic acid (EDTA) buffer containing 200 μM thiourea (Fisher Scientific, Fair Lawn, New Jersey, USA). For S. delphini, gels were run at 14°C at 6V/cm with an angle of 120° and a linear ramping from 4 s to 10 s for 12 h, followed by 13 s to 20 s for 6 h. For S. canis, gels were run under the same conditions, but with linear ramping from 1 s to 20 s over 16 h. Gel images were analyzed using BioNumerics software v5.1 (Applied Maths, Austin, Texas, USA). Band matching was done using a 0.5% position tolerance for S. delphini and 1.0% for S. canis, with 0% optimization, and cluster analysis was carried out using the Dice similarity coefficient and unweighted pair group method with arithmetic mean (UPGMA).
Table I.
Number and source of S. delphini and S. canis isolates used for pulsed-field gel electrophoresis (PFGE) in this study
| Farm | Number of diseased mink/total mink | Number of swabs (oral, footpad, rectal) | Bacterial species | Number of pododermatitis-associated isolates/total isolates |
|---|---|---|---|---|
| A | 9/30 | 39 (10,15,14) | S. delphini | 5/15 |
| S. canis | 5/5 | |||
| B | 15/30 | 46 (15,17,14) | S. delphini | 1/6 |
| S. canis | 2/8 |
Real-time PCR design for the detection of A. phocae
Primers c (TTGTACACACCGCCCGTCA) and b (GGTACCTTAGATGTTTCAGTTC) were designed as a variation of the protocol described by Hassan et al (20) to amplify a 620 bp region of the 16S–23S intergenic spacer region (ISR) of 1 A. phocae isolate. Primers specific to A. phocae (Aphocae_F; CAAAAAACCAAAGATGCTCGCG and Aphocae_R; TTTGTTWTTTKKGGGTGTGGCTG) were subsequently designed to amplify a 250 bp internal fragment of the ISR region by comparing the sequences of these 620 bp products with sequences of the closely related species A. haemolyticum [GenBank: FN551181], T. pyogenes [GenBank: EU194563], and Trueperella bernardiae [GenBank: EU194562]. These primers were used in an amplification mix for real-time PCR (20 μL per reaction) consisting of 1X SYBR Green I Master (Roche Applied Science, Mannheim, Germany), 250 nM each primer, and 10 ng template DNA. To assess the specificity of the resulting real-time PCR protocol, genomic DNA was extracted from broth cultures of all 4 A. phocae isolates, as well as of E. coli ATCC 25922, and of the A. haemolyticum and T. pyogenes isolates previously mentioned. The Agencourt Genfind v2 kit (Beckman Coulter, Beverly, Massachusetts, USA) was used for this purpose, following manufacturer’s instructions, with the addition of 3 μL of 150 U/mL achromopeptidase enzyme (Sigma-Aldrich, St. Louis, Missouri, USA) for the lysis step. All samples were run in technical triplicate, including no template controls. Plates were run on a LightCycler 480 II (Roche Applied Science) with an initial cycle of 95°C for 10 min, followed by 45 cycles of 95°C for 15 s, 67°C for 15 s, and 72°C for 15 s. Melting peaks were determined by ramping at 0.2°C/s from 65°C to 99°C.
Ten-fold serial dilutions of broth cultures were used in order to determine the detection limit of the real-time PCR assay. Two A. phocae isolates, as well as A. haemolyticum and T. pyogenes isolates, were used in a dilution range between ~1 × 109 and 1 colony-forming unit (CFU)/mL. The 2 latter species were used as they had the highest ISR sequence identity with A. phocae. Genomic preparations were done for each bacterial serial dilution as previously described and 1 μL was used as template for real-time PCR.
Real-time PCR detection of A. phocae in footpad tissue samples
Additional mink feet were collected from 14 mink farms in central and eastern Canada for DNA extraction from footpad tissue. One hundred and thirty-eight feet were collected from 75 individual mink at time of pelting and from natural mortality during the summer/fall season. No animals were sacrificed in order to collect footpad samples for this study. Feet were collected by the farmers. Each foot was identified and packed separately in a sealed plastic bag and shipped on ice to the laboratory by express courier. Whenever possible, only the rear feet were used in the study, as they are most frequently affected by pododermatitis. Three farms with no history of the disease and 11 farms with ongoing occurrences of pododermatitis were sampled. From each of 9 of the 11 pododermatitis-associated farms, 3 animals showing no macroscopic lesions and 3 animals showing clear chronic pododermatitis lesions (Figure 1) were used. In the 2 remaining pododermatitis-associated farms, only feet with lesions could be obtained.
Twenty-five mg of healthy footpad tissue or tissue from pododermatitis lesions from each foot was used to purify DNA using the QIAamp DNA Mini Kit (Qiagen, Valencia, California, USA) according to the manufacturer’s instructions for tissue extraction and the DNA was eluted in a final volume of 200 μL AE buffer. Quantitative real-time PCR was carried out as previously described using 4 μL of template DNA. Each sample was run in technical triplicate. Using known concentrations of A. phocae DNA to create a standard curve between 10 ng/μL and 0.01 pg/μL, target DNA concentration in sample footpad tissues could then be calculated.
Statistical analysis
Stata/IC v13.0 software (StataCorp, College Station, Texas, USA) was used for statistical analysis of PCR results. A multi-level logistic regression model with random intercepts for animal and farm was used to assess the association between disease status (independent variable) on endemic farms and the detection of A. phocae (dependent variable); samples for pododermatitis from non-endemic farms were not included in this analysis. Additionally, a multi-level linear regression model, fitted with restricted maximum likelihood and random intercepts for animal and farm, was used to compare A. phocae concentration values in tissues from endemic and non-endemic farm-related mink. Disease status was again used as the independent variable. A log10 transformation was used on these values and 0.03 was added to each original value to assure homogeneity of variance and to account for zero values in the transformation, respectively. Foot status (left versus right) was excluded from these analyses as it had no significant effect on the outcomes. Pearson residuals and standardized residuals were assessed graphically to identify outliers in the multi-level logistic and multi-level linear models, respectively. In the multi-level linear regression model, the standardized residuals and best linear unbiased predictors (BLUPS) were graphically assessed to determine whether they met the assumptions of homogeneity of variance and normality. In the multi-level logistic regression, only the BLUPS needed to be assessed for these distributional assumptions.
Results
16S rRNA gene sequencing of Staphylococcus spp
Thirty-two of the 36 coagulase-positive staphylococci isolates investigated had 16S rRNA gene sequences 99.85% identical to that of S. delphini ATCC 49171 and 99.70% identical to S. intermedius ATCC 29663T. All 32 isolates were shown to be Group A S. delphini by PCR. Of the remaining 4 isolates, 2 were identified as S. cohnii and 2 as S. schleiferi subsp. coagulans.
Pulsed-field gel electrophoresis of S. delphini and S. canis
Pulsed-field gel electrophoresis (PFGE) was carried out for a subset of 21 of the 32 S. delphini isolates (Table I). Each isolate had a unique SmaI banding pattern. No clustering was visible for pododermatitis-derived S. delphini or for those from healthy mink (Figure 2). In 4 instances, pairs of isolates that shared more than 80% pattern similarity were obtained from 1 sick and 1 healthy mink. In addition, no clear clustering by farm was visible (Figure 2).
Figure 2.
Pulsed-field gel electrophoresis (PFGE)-based dendrogram of 21 SmaI digested S. delphini genomic DNA from healthy and diseased mink from 2 unrelated farms in Canada. Scale bar represents percent similarity based on Dice coefficients and UPGMA clustering. Asterisk (*) indicates 2 isolates taken from the same individual mink.
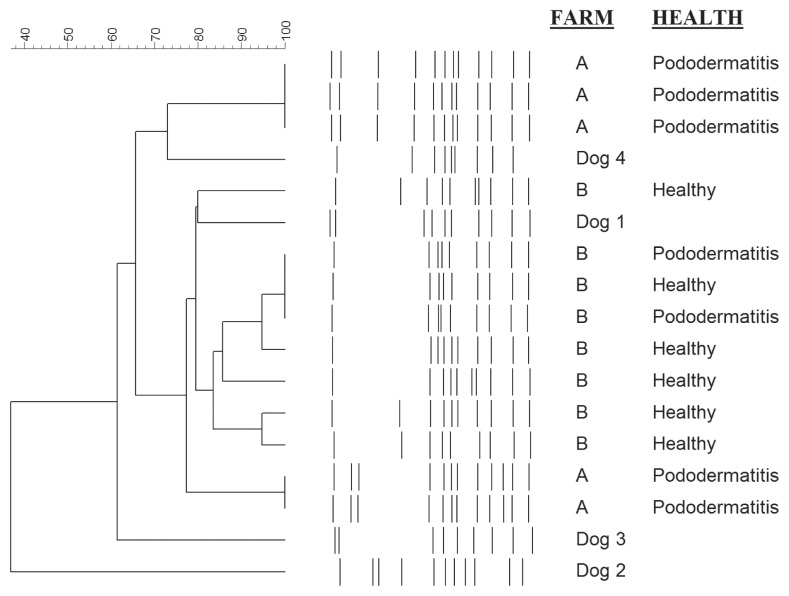
Based on PFGE results, all but 3 of the 13 S. canis isolates shared greater than 77% pattern similarity (Figure 3). Seven of the 13 isolates shared more than 83% pattern similarity and 3 isolates from individual infected mink on the same farm had identical PFGE patterns. The same result was observed on the second farm for 3 isolates from infected individuals. Although the canine isolates had unique PFGE patterns, they were closely related to those from mink (Figure 3). The 2 most closely related isolates from dogs showed 80% and 73% pattern similarity to S. canis isolates from mink. No clear-cut clustering of isolates by animal species, i.e., mink or dog, or by farm was visible.
Figure 3.
Pulsed-field gel electrophoresis (PFGE)-based dendrogram of 13 SmaI digested S. canis genomic DNA from healthy and diseased mink from 2 unrelated farms in Canada and clinical infections in 4 unrelated dogs. Scale bar represents percent similarity based on Dice coefficients and UPGMA clustering.
A. phocae intergenic spacer and real-time PCR for detecting A. phocae
The 620 bp ISR fragment from the A. phocae isolate from a mink with pododermatitis showed 98.8% identity with the published A. phocae ISR sequence (20). The next most closely related species was A. haemolyticum with only 82.9% identity.
Real-time PCR resulted in a product with a unique melting peak for A. phocae. Using 10 ng of purified genomic DNA, the 4 isolates of A. phocae were detected with crossing points (Cp) between 12.9 and 14.8 cycles. No PCR amplification signal was detectable for T. bernardiae or Escherichia coli. Non-specific signals were detected for A. haemolyticum and T. pyogenes only after Cp 32.4. Melting peak temperatures were also clearly distinct, with A. phocae at 84.5°C, A. haemolyticum at 85.5°C, and T. pyogenes at 91.3°C.
The detection limit for the real-time PCR was of ~5 CFUs/mL of broth culture for A. phocae and non-specific DNA amplification ceased completely below concentrations of 2.6 × 106 CFU/mL for A. haemolyticum and below 2.4 × 107 CFU/mL for T. pyogenes.
Detection of A. phocae in tissue samples
Figure 1 shows a pododermatitis representative of most lesions observed in the current study. Real-time PCR was run for 45 cycles and the average concentration was determined using Cp from each footpad sample based on technical triplicate results. No amplification was observed in any healthy control farm samples (n = 0/24). Of the footpads collected from endemic farms, 39% (n = 21/54) of healthy tissues and 62% (n = 37/60) of diseased tissues tested positive for A. phocae. The average concentrations from endemic farm samples were 11.9 pg/mg tissue (median = 0.00) from healthy tissues and 340.2 pg/mg (median = 1.34) from diseased tissues.
Association between disease status and the detection/concentration of A. phocae
Logistic regression analysis (Table IIa) showed that the odds of detecting A. phocae on endemic farms was significantly higher among foot samples with lesions than in samples without lesions, with most of the variance among farms being explained at the farm-level.
Table IIa.
Results of a multi-level logistic regression model used to evaluate the association between disease status on endemic farms and the detection of A. phocae using a multi-level mixed-effects logistic regression model
| Variable | OR | 95% CI | P-value |
|---|---|---|---|
| Health status | |||
| Healthy-endemic | Referent | ||
| Pododermatitis | 7.15 | 1.34 to 38.03 | 0.021 |
| Variance component | Variancea (95% CI) | VPC | |
| Farm-level | 6.22 (1.28 to 30.10) | 52.1% | |
| Animal-level | 2.43 (0.32 to 18.20) | 20.3% | |
Total variance = 11.9 using latent-variable technique for sample-level variance, i.e., π2/3 = 3.29).
OR — odds ratio; CI — confidence interval; VPC — Variance partition coefficient.
Multi-level linear regression analysis (Table IIb) showed that the concentration of A. phocae was not significantly different between samples without lesions from endemic and non-endemic farms. The concentration of this bacterium was greater in samples from animals with lesions, however, than in samples from animals without lesions. The greatest proportion of the variance was again explained at the farm-level. Based on visual inspection of the residuals and BLUPS, both multi-level models fit the data.
Table IIb.
Results of the multi-level mixed-effects linear regression model used to compare A. phocae concentration values in tissues from mink with and without pododermatitis
| Variable | 10βa | 95% CI | P-value |
|---|---|---|---|
| Health status | |||
| Healthy-endemic | 7.76 | 0.43 to 144.54 | 0.167 |
| Pododermatitis | 35.5 | 2.00 to 645.65 | 0.015 |
| Variance component | Variance (95% CI) | VPC | |
| Farm-level | 0.77 (0.29 to 2.06) | 41.9% | |
| Animal-level | 0.43 (0.21 to 0.88) | 23.4% | |
| Residual | 0.64 (0.45 to 0.91) | 35.7% | |
Back-transformation (from log10) for each coefficient.
CI — confidence interval; VPC — Variance partition coefficient.
Discussion
Although it was previously believed that most of the coagulase-positive staphylococcal isolates from mink pododermatitis are S. intermedius, 16S rRNA gene sequencing and PCR typing showed that Group A S. delphini is the major coagulase-positive staphylococcal species found in footpad lesions from mink in Canada. The S. intermedius group (SIG) bacteria can be difficult to identify and have been widely misidentified in the past (28,29). Our results correlate with recent reports of S. delphini in mink from other countries and confirm its strong association with this Mustelidae species on a broader geographical basis (8).
In many bacterial species, specific clones are associated with disease (30) and we hypothesized that particular S. canis and S. delphini strains may be associated with the development of pododermatitis in mink. Our results demonstrated a high diversity in PFGE patterns for S. delphini from both healthy and diseased mink. Isolates from lesions did not show any signs of clustering, however, and therefore did not support the hypothesis that a distinct S. delphini clonal lineage is associated with pododermatitis. Staphylococcus delphini was originally isolated from skin lesions of sea mammals (31), which supports the suspected association of pododermatitis with feeding seal meat to mink, although there are no reports available in the literature on its isolation from seals specifically. Our PFGE results also showed that no specific S. canis clonal lineage could be associated with pododermatitis, and in some cases, isolates from healthy mink footpads and from pododermatitis lesions had indistinguishable banding patterns. Thus, our results provide no evidence to support the hypothesis that specific S. delphini or S. canis clones are associated with pododermatitis in mink. In an attempt to further investigate the relationships between S. canis from mink and dogs, a few isolates from canine samples were also subjected to PFGE. Two of these isolates were genetically closely related to those from mink and no clear clustering of either mink or canine isolates was apparent. Despite the small number of isolates tested, our results therefore strongly suggest that no specific lineages of S. canis have adapted to either species.
Although other bacteria are recovered far more frequently from mink, A. phocae may appear to play a role in pododermatitis. As A. phocae grows much slower than other bacteria and forms very small colonies, it is likely often missed or overgrown by other bacteria during routine culture from tissue samples/swabs containing mixed bacterial populations such as is the case with mink feet samples. Thus, it may be present in more pododermatitis lesions than traditional bacterial culture methods would indicate. The development of a real-time PCR assay for detecting A. phocae allows detection of the bacterium directly from tissue samples without the interference caused by other bacterial species. By comparing tissue samples from pododermatitis-free and endemic farms, we have observed a clear gradient in the occurrence of A. phocae: the bacterium was only detected in samples from endemic farms, with significantly larger amounts found in diseased tissues than in tissues from mink showing no macroscopic signs of pododermatitis. Most importantly, the observation that A. phocae was never found in farms with no previous history of the disease suggests a clear association between the bacterium and the potential for development of pododermatitis. These findings demonstrate that, although A. phocae could not be detected in every case of pododermatitis, it is strongly associated with the lesions and farms where the problem occurs. The distribution of the variance components from the multi-level models also strongly suggests that farm-level interventions would have the greatest impact on the prevalence/concentration of this bacterium. Thus, A. phocae may play a role as an opportunistic pathogen in mink pododermatitis and further investigations on this organism are warranted.
In conclusion, the present study confirmed that, contrary to previous reports, Group A S. delphini is the major coagulase-positive staphylococcus species found in pododermatitis lesions from mink and that no specific restricted clone of this species or of S. canis is associated with this disease. In addition, our results demonstrated a significant association of A. phocae with pododermatitis and corroborated the previously described link between feeding seal meat and the emergence of pododermatitis in mink. This raises the hypothesis that temporary changes in feeding practices may have introduced a new pathogen in farmed mink, which has persisted after the practice was discontinued. Finally, a new quantitative PCR was developed during this study that will form a solid basis for further epidemiological investigations on the role of A. phocae in mink.
Acknowledgments
The authors acknowledge Dr. Bruce Hunter’s lifetime of hard work and dedication to veterinary research and animal welfare and his role in spearheading this research project. He is missed by all who knew him. The authors thank the collaborators of the Animal Health Laboratory at the University of Guelph for culture and identification of bacterial isolates. We also thank Kathryn Bernard from the National Microbiology Laboratory in Winnipeg for providing control isolates of T. bernardiae, A. haemolyticum, and A. phocae. This project was funded by the Canada Mink Breeders Association.
References
- 1.Bröjer C. MSc dissertation. Guelph, Ontario: University of Guelph; 2000. Pododermatitis in farmed mink in Canada. [Google Scholar]
- 2.Ontario Aquaculture Research and Services Coordinating Committee. 2004 Strategic Report (For the Period 2005 — 2009). Presented to the Ontario Animal Research and Services Committee; 2004. p. 106. [Google Scholar]
- 3.Fernandez-Antonio R, Frailde LD, Losada AP, et al. Pododermatitis in farmed mink (Neovison vison) in Spain. Proceedings of the IX International Scientific Congress in Fur Animal Production; 2008. [Google Scholar]
- 4.Devriese LA, Hermans K, Baele M, Haesebrouck F. Staphylococcus pseudintermedius versus Staphylococcus intermedius. Vet Microbiol. 2009;133:206–207. doi: 10.1016/j.vetmic.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 5.Ruscher C, Lubke-Becker A, Wleklinski CG, Soba A, Wieler LH, Walther B. Prevalence of Methicillin-resistant Staphylococcus pseudintermedius isolated from clinical samples of companion animals and equidaes. Vet Microbiol. 2009;136:197–201. doi: 10.1016/j.vetmic.2008.10.023. [DOI] [PubMed] [Google Scholar]
- 6.Devriese LA, Vancanneyt M, Baele M, et al. Staphylococcus pseudintermedius sp. nov., a coagulase-positive species from animals. Int J Syst Evol Microbiol. 2005;55:1569–1573. doi: 10.1099/ijs.0.63413-0. [DOI] [PubMed] [Google Scholar]
- 7.Decristophoris P, Fasola A, Benagli C, Tonolla M, Petrini O. Identification of Staphylococcus intermedius Group by MALDI-TOF MS. Syst Appl Microbiol. 2011;34:45–51. doi: 10.1016/j.syapm.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 8.Guardabassi L, Schmidt KR, Petersen TS, et al. Mustelidae are natural hosts of Staphylococcus delphini group A. Vet Microbiol. 2012;159:351–353. doi: 10.1016/j.vetmic.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 9.DeWinter LM, Low DE, Prescott JF. Virulence of Streptococcus canis from canine streptococcal toxic shock syndrome and necrotizing fasciitis. Vet Microbiol. 1999;70:95–110. doi: 10.1016/s0378-1135(99)00128-5. [DOI] [PubMed] [Google Scholar]
- 10.Kruger EF, Byrne BA, Pesavento P, Hurley KF, Lindsay LL, Sykes JE. Relationship between clinical manifestations and pulsed-field gel profiles of Streptococcus canis isolates from dogs and cats. Vet Microbiol. 2010;146:167–171. doi: 10.1016/j.vetmic.2010.04.026. [DOI] [PubMed] [Google Scholar]
- 11.DeWinter LM, Prescott JF. Relatedness of Streptococcus canis from canine streptococcal toxic shock syndrome and necrotizing fasciitis. Can J Vet Res. 1999;63:90–95. [PMC free article] [PubMed] [Google Scholar]
- 12.Hassan AA, Akineden O, Usleber E. Identification of Streptococcus canis isolated from milk of dairy cows with subclinical mastitis. J Clin Microbiol. 2005;43:1234–1238. doi: 10.1128/JCM.43.3.1234-1238.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galperine T, Cazorla C, Blanchard E, Boineau F, Ragnaud JM, Neau D. Streptococcus canis infections in humans: Retrospective study of 54 patients. J Infect. 2007;55:23–26. doi: 10.1016/j.jinf.2006.12.013. [DOI] [PubMed] [Google Scholar]
- 14.Martínez J, Vidaña B, Cruz-Arambulo R, Slavic D, Tapscott B, Brash ML. Bacterial diskospondylitis in juvenile mink from 2 Ontario mink farms. Can Vet J. 2013;54:859–863. [PMC free article] [PubMed] [Google Scholar]
- 15.Jost BH, Billington SJ. Arcanobacterium pyogenes: Molecular pathogenesis of an animal opportunist. Antonie Van Leeuwenhoek. 2005;88:87–102. doi: 10.1007/s10482-005-2316-5. [DOI] [PubMed] [Google Scholar]
- 16.Hijazin M, Ulbegi-Mohyla H, Alber J, et al. Molecular identification and further characterization of Arcanobacterium pyogenes isolated from bovine mastitis and from various other origins. J Dairy Sci. 2011;94:1813–1819. doi: 10.3168/jds.2010-3678. [DOI] [PubMed] [Google Scholar]
- 17.Clarke TM, Citron DM, Towfigh S. The conundrum of the gram-positive rod: Are we missing important pathogens in complicated skin and soft-tissue infections? A case report and review of the literature. Surg Infect (Larchmt) 2010;11:65–72. doi: 10.1089/sur.2008.085. [DOI] [PubMed] [Google Scholar]
- 18.Foster G, Hunt B. Distribution of Arcanobacterium pluranimalium in animals examined in veterinary laboratories in the United Kingdom. J Vet Diagn Invest. 2011;23:962–964. doi: 10.1177/1040638711416632. [DOI] [PubMed] [Google Scholar]
- 19.Yassin AF, Hupfer H, Siering C, Schumann P. Comparative chemotaxonomic and phylogenetic studies on the genus Arcanobacterium Collins et al. 1982 emend. Lehnen et al. 2006: Proposal for Trueperella gen. nov. and emended description of the genus Arcanobacterium. Int J Syst Evol Microbiol. 2011;61:1265–1274. doi: 10.1099/ijs.0.020032-0. [DOI] [PubMed] [Google Scholar]
- 20.Hassan AA, Mohyla H, Kanbar T, et al. Molecular identification of Arcanobacterium bialowiezense and Arcanobacterium bonasi based on 16S–23S rRNA intergenic spacer region sequences. Vet Microbiol. 2008;130:410–414. doi: 10.1016/j.vetmic.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 21.Ramos CP, Foster G, Collins MD. Phylogenetic analysis of the genus Actinomyces based on 16S rRNA gene sequences: Description of Arcanobacterium phocae sp. nov., Arcanobacterium bernardiae comb. nov., and Arcanobacterium pyogenes comb. nov. Int J Syst Bacteriol. 1997;47:46–53. doi: 10.1099/00207713-47-1-46. [DOI] [PubMed] [Google Scholar]
- 22.Johnson SP, Jang S, Gulland FM, et al. Characterization and clinical manifestations of Arcanobacterium phocae infections in marine mammals stranded along the central California coast. J Wildl Dis. 2003;39:136–144. doi: 10.7589/0090-3558-39.1.136. [DOI] [PubMed] [Google Scholar]
- 23.Cai H, Archambault M, Prescott JF. 16S ribosomal RNA sequence-based identification of veterinary clinical bacteria. J Vet Diagn Invest. 2003;15:465–469. doi: 10.1177/104063870301500511. [DOI] [PubMed] [Google Scholar]
- 24.Altschul SF, Madden TL, Schaffer AA, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sasaki T, Tsubakishita S, Tanaka Y, et al. Multiplex-PCR method for species identification of coagulase-positive staphylococci. J Clin Microbiol. 2010;48:765–769. doi: 10.1128/JCM.01232-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hijazin M, Prenger-Bernighoff E, Sammra O, et al. Arcanobacterium canis sp. nov., isolated from an otitis externa of a dog and emended description of the genus Arcanobacterium Collins et al. 1983 emend. Yassin et al. 2011. Int J Syst Evol Microbiol. 2012;62:2201–2205. doi: 10.1099/ijs.0.037150-0. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd ed. Vol. 1. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2001. Pulsed-field gel electrophoresis; pp. 5.55–5.90. [Google Scholar]
- 28.Bannoehr J, Franco A, Iurescia M, Battisti A, Fitzgerald JR. Molecular diagnostic identification of Staphylococcus pseudintermedius. J Clin Microbiol. 2009;47:469–471. doi: 10.1128/JCM.01915-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sasaki T, Kikuchi K, Tanaka Y, Takahashi N, Kamata S, Hiramatsu K. Reclassification of phenotypically identified Staphylococcus intermedius strains. J Clin Microbiol. 2007;45:2770–2778. doi: 10.1128/JCM.00360-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Musser JM. Molecular population genetic analysis of emerged bacterial pathogens: Selected insights. Emerg Infect Dis. 1996;2:1–17. doi: 10.3201/eid0201.960101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Varaldo PE, Kilpper-Balz R, Biavasco F, Satta G, Schleifer KH. Staphylococcus delphini sp. nov., a coagulase-positive species isolated from dolphins. Int J Sys Bact. 1988;38:436–439. [Google Scholar]