Abstract
Cocaine esterase (CocE) has been known as the most efficient native enzyme for metabolizing the naturally occurring cocaine. A major obstacle to the clinical application of CocE is the thermoinstability of native CocE with a half-life of only ~11 min at physiological temperature (37°C). It is highly desirable to develop a thermostable mutant of CocE for therapeutic treatment of cocaine overdose and addiction. To establish a structure-thermostability relationship, we carried out molecular dynamics (MD) simulations at 400 K on wild-type CocE and previously known thermostable mutants, demonstrating that the thermostability of the active form of the enzyme correlates with the fluctuation (characterized as the RMSD and RMSF of atomic positions) of the catalytic residues (Y44, S117, Y118, H287, and D259) in the simulated enzyme. In light of the structure-thermostability correlation, further computational modeling including MD simulations at 400 K predicted that the active site structure of the L169K mutant should be more thermostable. The prediction has been confirmed by wet experimental tests showing that the active form of the L169K mutant had a half-life of 570 min at 37°C, which is significantly longer than those of the wild-type and previously known thermostable mutants. The encouraging outcome suggests that the high-temperature MD simulations and the structure-thermostability may be considered as a valuable tool for computational design of thermostable mutants of an enzyme.
Introduction
Cocaine is considered to be the most addictive substance abused by millions of people worldwide.1,2,3,4 The devastating medical and social consequences of cocaine addiction have made the development of an effective pharmacological treatment a high priority.3,5 It has been found that cocaine exerts its effects on the central nervous system (CNS) through its blocking the re-uptake of neurotransmitter dopamine, thus potentiating the effects of dopamine in the synapse.6,7,8 The classic pharmacodynamic approach has failed to produce a feasible small-molecule antagonist of dopamine transporter (DAT) because of the difficulties inherent in blocking a blocker like cocaine without affecting the normal function of DAT.2,4,6 Alternatively, pharmacokinetic approach has become the most promising strategy, because this strategy aims at accelerating the hydrolysis of cocaine and, therefore, eliminating cocaine quickly from the peripheral circulation. 9,10,11,12,13,14 For this purpose, the principal plasma butyrylcholinesterase (BChE), 11,12,13,15 the bacterial cocaine esterase (CocE),10,14,16,17,18,19,20,21,22,23,24 and anti-cocaine catalytic antibody9,25,26,27 are the promising choices to be developed as a potential anti-cocaine agent for therapeutic treatment of cocaine overdose and abuse. Among them, CocE22,24 is the most efficient natural enzyme against the naturally occurring cocaine, i.e. (−)-cocaine. Native CocE is capable of protecting against cocaine-induced lethality, but its duration of effectiveness is very short, with a half-life (τ1/2) of only ~11 minutes at physiological temperature (37 °C).17,22 It is highly desirable to develop thermostable mutants of CocE for the treatment of cocaine toxicity.
In general, protein thermostabilization has become a hot topic in the field of protein engineering. During the last decade, extensive experimental and computational studies have been performed to search for possible structural determinants of stability. 28,29,30,31 Several rational design methods have been developed, and successfully applied to design thermostable proteins and mostly single domain enzymes.32,33,34,35,36,37,38,39,40,41,42,43,44,45 Among these various rational design methods, high-temperature molecular dynamics (MD) simulations have been shown to be a robust protocol to explore the structure-thermostability-activity relationship and the effects of environmental factors like pH and solvent on the stability.46,47,48,49,50 Compared with rational design of non-catalytic proteins, the design of a thermostable enzyme is more challenging, because the active site structure of an enzyme and its dynamic behavior during the catalytic reaction are finely tuned for the optimum catalytic efficiency.40,51,52 To thermostabilize an enzyme without losing its catalytic efficiency, one needs to understand the structure-thermostability-activity relationship of the particular enzyme, and then develop a reliable computational strategy and protocol to predict thermostable mutations. The predicted mutants are expected to have minimal shift in the backbone structure of the enzyme, and not to disrupt the active site structure or quench its flexibility. When an enzyme becomes inactive, it does not necessarily mean that the enzyme is unfolded or partially unstructured, because the inactivation could be merely due to some minor structural changes on the active site of the enzyme.32,50,51 There is no direct correlation relationship between the free energy of folding and half-life of the catalytic activity. A mutation lowering the folding free energy does not necessarily result in a longer half-life of the catalytic activity. For these reasons, it is very important to understand the inactivation mechanism when designing thermostable mutants of an enzyme.
CocE consists of three distinct domains with totally 574 amino acids.10 The fold of domain I (amino acid residues from #1 to #144 and from #241 to #354) is similar to the fold of canonical α/β-hydrolase. The seven α-helices of domain II (amino acid residues from #145 to #240) is inserted between strands β6 and β7 of domain I, and domain III (amino acid residues from #355 to #574) takes a jelly roll-like topology. The most particular feature of CocE structure is that the active site is located at the interface of these three domains, i.e. all three domains contribute to the active site pocket. Such particular location of the active site and the large size of CocE have made unprecedented challenges to our rational design of thermostable mutants. In our previous studies,22,23 we designed and discovered several thermostable mutants of CocE through computational design, followed by in vitro and in vivo studies. The designed CocE mutants, i.e. T172R, G173Q, and T172RG173Q, have significantly increased the thermostability of the enzyme in vitro and in vivo. Results obtained from our molecular modeling and molecular dynamics (MD) simulations revealed that the enzyme was stabilized by enhanced intra-molecular interactions resulting from these specific mutations. For example, the G173Q mutation brings in additional hydrogen bonding between the side chain of Q173 and the backbone of P43. However, it is theoretically uncertain whether or not these enhanced intramolecular interactions also help to stabilize the active site of CocE. If so, how are the effects of the enhanced intramolecular interactions relayed to the active site of CocE? Answers to these questions are very important for us to better understand the thermostabilization mechanism of CocE, and such understanding will also help us to rationally design more thermostable mutants of CocE with a longer in vivo half-life in the treatment of cocaine overdose.
In the present study, we first performed high-temperature MD simulations on our previously designed T172R, G173Q, and T172R/G173Q mutants of CocE in order to find structural determinants for thermostability of CocE. The results indicated that these CocE mutants had much smaller structural fluctuations for the catalytic residues (including the catalytic triad S117-H287-D259 and oxyanion hole consisting of Y44 and Y118) during the MD simulation, compared to wild-type CocE. In light of the structure-thermostability correlation for these CocE mutants, further high-temperature MD simulation and analysis revealed that the L169K mutant had even smaller structural fluctuations for the same catalytic residues, predicting that the L169K mutant should be more thermostable. The computational prediction has been confirmed by wet experimental tests in vitro and in vivo studies in the labs of our collaborators. Here we report the computational studies, whereas the corresponding experimental studies have been reported elsewhere (the computational studies described in this report were actually completed before the experimental studies described in ref. 24, but the submission of this report for publication was delayed considerably due to some unexpected reasons). The agreement between our computational results and experimental data suggests that the high-temperature MD simulation is a valuable approach for computational design of novel, thermostable mutants of proteins as the therapeutic candidates.
Computational Methods
High-temperature MD simulations were performed on wild-type CocE and its mutants involved in this study by using the Sander module of the Amber 8 program package.53 All MD simulations started from the X-ray crystal structures (PDB entry codes 3I2J for wild-type CocE, 3I2I for T172R mutant, 3I2G for G173Q mutant, and 3I2F for T172R/G173Q mutant).24 For all of the high-temperature MD simulations, the lengths of covalent bonds involving hydrogen atoms were constrained with the SHAKE algorithm.54 The particle mesh Ewald (PME) method 55,56 was used to treat long-range electrostatic interactions. A 12 Å cutoff for non-bonded interactions was used, and the non-bonded list was updated every 25 steps. The motion of the center of mass of the system was removed every 1,000 steps. The Berendsen temperature coupling method57 was used to couple the system to a thermal bath of a target temperature. Periodic boundary conditions with isotropic molecule-based scaling were applied. All the high-temperature MD simulations at T = 400 K were carried out at constant temperature and volume (NTV ensemble), and the time step was 1 fs.
Each system (wild-type CocE or its mutant) was solvated in a rectangular box of TIP3P water molecules58 with a minimum solute-wall distance of 10 Å, and the original water molecules and chloride ions (Cl−) from the X-ray crystal structure24 were kept. Sodium counter ions (Na+) were added to neutralize the solvated system. Each of the solvated systems was energy-minimized by using the Sander module of Amber8 with a nonbonded cutoff of 12 Å and a conjugate gradient energy-minimization method.53 After 20,000 steps of energy minimization for all atoms in the system, 80 ps MD simulations were performed for water molecules and ions (Na+ and Cl−) with constant temperature and constant volume (NTV ensemble) at T = 298.15 K. After another 5,000 steps of energy minimization, the whole system was gradually heated from 10.15 K to 298.15 K by weak-coupling method,57 and equilibrated for 500 ps at constant temperature and constant pressure (NTP ensemble). After that, the system was heated to 400 K and equilibrated for 80 ps at NTV ensemble. Finally the MD simulation at 400 K was kept running to the targeted length of production.
Results and Discussion
It is extremely challenging to study the thermostability of CocE by carrying out MD simulations. This is because the half-life of wild-type CocE at body temperature (~11 min) is too short for use as a drug, but too long for performing a MD simulation to simulate the inactivation process. A practical, fully relaxed MD simulation (with a required time step of 1 or 2 fs) on a protein system like CocE can be performed for as long as nanoseconds using currently available supercomputers. Instead, we carried out high-temperature MD simulations on the protein structures based on an analysis of the kinetic relationship between the inactivation rate constant (kina) and the temperature (T) for a given inactivation free energy barrier (ΔGina) of the inactivation process,22 i.e.
| (1) |
where kB is the Boltzmann’s constant, h is Planck’s constant, and R is the gas constant. As discussed in our previous report,22 Eq. (1) is based on an extended use of the well-known vibrational transition state theory. It is also based on an implicit assumption that the rate-determining step of the inactivation process is vibration-like. According to Eq. (1), the rate constant kina of the enzyme inactivation is dependent on the temperature for a given inactivation free energy barrier. The higher the temperature, the larger the rate constant and, therefore, the half-life of the active enzyme is shorter. This kinetic understanding enabled us to uncover the inactivation pathway of CocE through performing MD simulation at an appropriately high temperature. The high temperature used in the MD simulations is physiologically irrelevant, but can considerably shorten the time of the protein inactivation process and, thus, allows us to observe the protein inactivation process within a much shorter period of time. An implicit hypothesis used in this approach is that the relative stability of various proteins (wild-type CocE and its mutants) at the high temperature correlates with the relative stability of the same proteins at the body or room temperature.
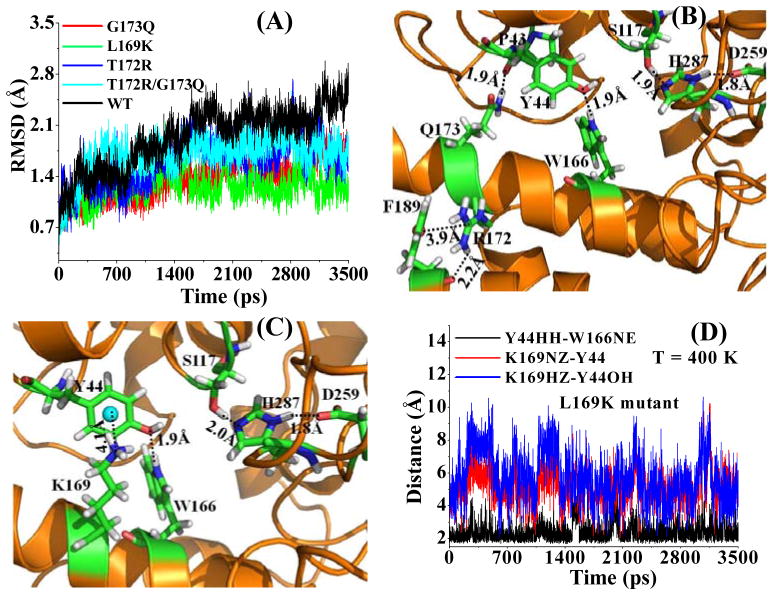
Our previously designed T172R, G173Q, and T172R/G173Q mutants22 of CocE were expected to be more thermostable than the wild-type without losing the catalytic activity. By performing high-temperature MD simulations, we were able to see whether these designed CocE mutants are more structurally stabilized. Our previous MD simulation and computational analysis22 focused on the changes of the overall protein structure and intramolecular interactions involving the mutated amino acid residues, but did not account for the effects of the structural fluctuations on the protein thermostability. In the present study, we focused on the effects of amino acid mutations on the structural fluctuations of the active site and computationally correlated the structural fluctuations with the protein thermostability for the first time. For this purpose, we first performed MD simulations on the wild-type CocE and three known thermostable mutants (i.e. T172R, G173Q, and T172R/G173Q) at 400 K and tracked the changes of root-mean square deviation (RMSD) of atomic positions from the corresponding initial positions for catalytic residues including the catalytic triad (consisting of S117, H287, and D259) and oxyanion hole (consisting of Y44 and Y118) during the MD simulations. We are particularly interested in the fluctuations of the catalytic residues, because their overall fluctuation is expected to correlate with the thermostability of the active form of the enzyme. Depicted in Fig. 1A are the results of tracked RMSD curves for the five catalytic residues during the MD simulations. As shown in Fig. 1A, these catalytic residues of wild-type CocE have the largest (average) RMSD value during the MD simulations after ~1.5 ns. The average RMSD values for the CocE mutants simulated are all smaller than that for the wild-type after ~1.5 ns, suggesting that all of the simulated mutants should have a more stable active site structure.
Fig. 1.
(A) Time-dependence of the root-mean square deviation (RMSD) of the high-temperature (T = 400 K) MD-simulated atomic positions of all heavy atoms of the catalytic residues (Y44, S117, Y118, D259, and H287) from those in the starting structure. (B) Energy-minimized structure of the T172R/G173Q mutant. The enzyme is represented as colored ribbon. Amino acid residues including the catalytic triad (S117-H287-D259) are shown as stick and colored by atom types. The hydrogen bonding and cation-π interactions are represented as dashed lines with distances labeled. (C) Energy-minimized structure of the L169K mutant, which is shown in a similar way as that in (B). (D) Time-dependence of important internuclear distances from the high-temperature (T = 400 K) MD-simulated L169K mutant. Y44HH-W166NE represents the distance between the hydroxyl hydrogen (HH) of Y44 side chain and the nitrogen (NE) atom of W166 side chain, K169NZ-Y44 stands for the cation-π distance between the nitrogen (NZ) atom on the cationic head of K169 side chain and the center of the phenyl ring of Y44 side chain, and K169HZ-Y44OH refers to the shortest distance between the hydroxyl oxygen of Y44 side chain and the protons on the cationic head of K169 side chain. All of the data are based on the production MD.
To understand why the T172R and G173Q mutations can help to stabilize the active site structure of the enzyme, we analyzed the modeled structure of the T172R/G173Q mutant. Depicted in Fig. 1B is the active site structure of the T172R/G173Q mutant. Domain II (residues #145 to #240) of CocE is the least stable part of the protein. As depicted in Fig. 1B, the side chain of Q173 is hydrogen bonded with the backbone of P43, whereas the side chain of R172 interacts with the side chain of F189 through typical cation-π interactions. All of these enhanced local intramolecular interactions stabilize the secondary structure of domain II of CocE. The effect of the local stabilization is relayed to the active site through the neighboring residues and/or simply through the secondary structure of domain II. For example, as both P43 and Y44 are located at the same loop, the catalytic residue Y44 becomes more stable when P43 is stabilized through the hydrogen bonding with Q173. The relay of stabilization effect makes the active site structure of CocE more persistent during the MD simulation at 400 K compared to the wild-type enzyme and, therefore, more stable at physiological temperature (37°C).
Compared to the single mutants T172R and G173Q, the local intramolecular interactions (Fig. 1B) in the T172R/G173Q mutant should be stronger. However, the stronger local intramolecular interactions are not reflected in the RMSD values depicted in Fig. 1A because the overall RMSD for the T172R/G173Q mutant is actually similar to those for the T172R and G173Q mutants. It should be pointed out that the RMSD depicted in Fig. 1A are all relative to the deviations of the atomic positions of the catalytic residues from those in the corresponding starting structures. Whereas the RMSD from the starting structure is the most important indicator (primary factor) of the active site stability of the enzyme, another significant indicator (secondary factor) of the active site stability might be the root-mean square fluctuation (RMSF) of the atomic positions of the catalytic residues relative to the MD-simulated average structure during the high-temperature MD simulations. The secondary factor associated with RMSF favors the T172R/G173Q mutant in comparison with the T172R and G173Q mutants. The average RMSF value during the high-temperature MD simulation is 1.12 Å for the T172R mutant, 1.02 Å for the G173Q mutant, and 0.93 Å for the T172R/G173Q mutant. The relative RMSF values help us to better understand why the T172R/G173Q mutant should be more thermostable than the T172R and G173Q mutants when their RMSD values depicted in Fig. 1A are comparable.
To summarize the computational results, the RMSD values depicted in Fig. 1A indicate that, compared to the active form of wild-type CocE, the active forms of the T172R, G173Q, and T172R/G173Q mutants all should be more thermostable than the wild-type. The calculated average RMSF values further suggest that the T172R/G173Q mutant should be more thermostable than the corresponding single mutants. The computational results are qualitatively consistent with the experimental data at 37°C in both the in vitro and in vivo experimental activity tests.22 For example, the in vitro half-lives of the active enzymes at 37°C were ~11 min (wild-type), ~78 min (T172R), ~75 min (G173Q), and ~305 min (T172R/G173Q). The half-life mentioned here refers to the time required to decrease the enzyme activity by 50%. The qualitative agreement between the computational and experimental data suggests that the high-temperature MD simulations may be used as a valuable tool to predict new mutants of CocE with an improved thermostability.
Beyond the reported T172R, G173Q, and T172R/G173Q mutants, we carried out further computational design of thermostable mutants of CocE, leading to the prediction that the L169K mutant of CocE should be more thermostable. Our computational modeling for the mutant design was a two-step approach. The first step is the initial screening, in which the energetic effects of possible mutations on amino acid residues were estimated by using our previously reported fast computational approach.22 This fast approach22 calculates the shift in the interaction energy between the mutated residue and the remaining part of the enzyme, through the combined application of the Amber53 and Delphi59 programs, without performing any MD simulations. The structures of both native CocE and mutants were optimized (through energy minimization) prior to the interaction energy calculations. The second step was based on the MD simulations at 400 K on the mutants selected in the first step. The initial screening with the fast computational approach revealed that the L169K mutant should be more stable compared to the wild-type and the above-mentioned mutants. We further examined the L169K mutant by performing the MD simulation at 400 K in the same way as we did for the wild-type and the T172R, G173Q, and T172R/G173Q mutants. The simulated results are also depicted in Fig. 1A, C, and D.
As shown in Fig. 1A, the L169K mutant is associated with the smallest (average) RMSD value for the five catalytic residues during the MD simulation at 400 K after ~1.5 ns. The average RMSF value (0.92 Å) is also the smallest within the proteins simulated. As seen in Fig. 1C, the cationic head of K169 side chain has the favorable cation-π interaction with the phenyl ring of catalytic residue Y44 and, therefore, directly stabilizes the active site structure. Associated with the cation-π interaction, the other interactions with Y44 also become more stable in the L169K mutant. As shown in Fig. 1D, the hydrogen bond between the hydroxyl group of Y44 side chain and the NE atom of W166 side chain was persistent during the MD simulation even at 400 K.
The data from the MD simulations at 400 K further suggested that the L169K mutant should have a higher thermostability compared to wild-type CocE and its T172R, G173Q, and T172R/G173Q mutants. The high thermostability of the active form of the L169K mutant has been confirmed by wet experimental tests in vitro and in vivo based on our computational prediction. The detailed experimental procedure and data have been described in a separate report.24 In particular, the activity data24 revealed that the active form of the L169K mutant had a half-life of 570 min at 37°C. The half-life of 570 min observed for the L169K mutant is significantly longer than those observed for the T172R, G173Q, and T172R/G173Q mutants.
Conclusion
The molecular dynamics (MD) simulations on wild-type cocaine esterase (CocE) and our previously reported thermostable mutants (i.e. T172R, G173Q, and T172RG173Q) of CocE at 400 K revealed that the thermostability of the active form of the enzyme correlates with the fluctuation of the active site structure in the simulated enzyme. The fluctuation of the active site structure is represented by the RMSD and RMSF of atomic positions in all of the catalytic residues that form the catalytic triad (S117, H287, and D259) and oxyanion hole (Y44 and Y118). All of the thermostable mutants examined have significantly smaller RMSD values compared to the wild-type. Based on the simulated structures, the thermostable mutations enhanced the local intramolecular interactions that stabilize the secondary structure of domain II of CocE. The effect of the local stabilization is relayed to the active site through the neighboring residues and/or simply through the secondary structure of domain II. The relay of stabilization effect makes the active site structure more persistent during the MD simulation at 400 K compared to the wild-type and, therefore, more stable at physiological temperature (37°C).
In light of the correlation between the RMSD/RMSF of the simulated atomic positions in the catalytic residues and the thermostability of the active form of the enzyme, further computational modeling including MD simulations at 400 K predicted that the active site structure of the L169K mutant should be more stable. In the simulated structure, the cationic head of K169 side chain had the favorable cation-π interaction with the phenyl ring of catalytic residue Y44 and, therefore, directly stabilizes the active site structure. The computational prediction has been confirmed by wet experimental tests showing that the active form of the L169K mutant had a half-life of 570 min at 37°C, which is significantly longer than those of wild-type CocE and the previously known thermostable mutants. The encouraging outcome suggests that the high-temperature MD simulations and the correlation between the RMSD/RMSF of the simulated atomic positions in the catalytic residues and the thermostability of the active form of the enzyme may be considered as a valuable tool for computational design of thermostable mutants of an enzyme.
Acknowledgments
This work was supported by NIH (grants R01 DA025100, R01 DA021416, and R01 DA013930). The authors also acknowledge the Center for Computational Sciences (CCS) at the University of Kentucky for supercomputing time on an IBM X-series Supercomputer Cluster consisting of 340 nodes or 1,360 processors and on a Dell Supercomputer Cluster consisting of 388 nodes or 4,816 processors.
References
- 1.Mendelson JH, Mello NK. New Eng J Med. 1996;334:965. doi: 10.1056/NEJM199604113341507. [DOI] [PubMed] [Google Scholar]
- 2.Sparenborg S, Vocci F, Zukin S. Drug Alcohol Depend. 1997;48:149. doi: 10.1016/s0376-8716(97)00072-0. [DOI] [PubMed] [Google Scholar]
- 3.Gorelick DA. Drug Alcohol Depend. 1997;48:159. doi: 10.1016/s0376-8716(97)00119-1. [DOI] [PubMed] [Google Scholar]
- 4.Singh S. Chem Rev. 2000;100:925. doi: 10.1021/cr9700538. [DOI] [PubMed] [Google Scholar]
- 5.Paula S, Tabet MR, Farr CD, Norman AB, Jr, Ball WJ. J Med Chem. 2004;47:133. doi: 10.1021/jm030351z. [DOI] [PubMed] [Google Scholar]
- 6.Gaintdinov RR, Sotmikova TD, Caron MG. Trends Pharmacol Sci. 2002;231:367. doi: 10.1016/s0165-6147(02)02044-8. [DOI] [PubMed] [Google Scholar]
- 7.Torres GE, Gainetdinov RR, Caron MG. Nat Rev Neurosci. 2003;4:13. doi: 10.1038/nrn1008. [DOI] [PubMed] [Google Scholar]
- 8.Chen R, Tilley MR, Wei H, Zhou F, Zhou FM, Ching S, Quan N, Stephens RL, Hill ER, Nottoli T, Han DD, Gu HH. Proc Natl Acad Sci USA. 2006;103:9333. doi: 10.1073/pnas.0600905103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Landry DW, Zhao K, Yang GXQ, Glickman M, Georgiadis TM. Science. 1993;259:1899. doi: 10.1126/science.8456315. [DOI] [PubMed] [Google Scholar]
- 10.Larsen NA, Turner JM, Stevens J, Rosser SJ, Basran A, Lerner RA, Bruce NC, Wilson IA. Nature Struct Biol. 2002;9:17. doi: 10.1038/nsb742. [DOI] [PubMed] [Google Scholar]
- 11.Zhan CG, Zheng F, Landry DW. J Am Chem Soc. 2003;125:2462. doi: 10.1021/ja020850+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng F, Yang W, Ko MC, Liu J, Cho H, Gao D, Tong M, Tai HH, Woods JH, Zhan CG. J Am Chem Soc. 2008;130:12148. doi: 10.1021/ja803646t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang W, Pan Y, Zheng F, Cho H, Tai HH, Zhan CG. Biophys J. 2009;96:1931. doi: 10.1016/j.bpj.2008.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jutkiewicz EM, Baladi MG, Cooper ZD, Narasimhan D, Sunahara RK, Woods JH. Ann Emerg Med. 2009;54:409. doi: 10.1016/j.annemergmed.2008.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pan Y, Gao G, Yang W, Cho H, Yang G, Tai HH, Zhan CG. Proc Natl Acad Sci USA. 2005;102(46):16656. doi: 10.1073/pnas.0507332102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turner JM, Larsen NA, Basran A, Barbas CF, III, Bruce NC, Wilson IA, Lerner RA. Biochem. 2002;41:12297. doi: 10.1021/bi026131p. [DOI] [PubMed] [Google Scholar]
- 17.Cooper ZD, Narasimhan D, Sunahara RK, Mierzejewski P, Jutkiewicz EM, Larsen NA, Wilson IA, Landry DW, Woods JH. Mol Pharmacol. 2006;70:1885. doi: 10.1124/mol.106.025999. [DOI] [PubMed] [Google Scholar]
- 18.Ko MC, Bowen LD, Narasimhan D, Berlin AA, Lukacs NW, Sunahara RK, Cooper ZD, Woods JH. J Pharmacol Exp Ther. 2007;320:926. doi: 10.1124/jpet.106.114223. [DOI] [PubMed] [Google Scholar]
- 19.Ko MC, Narasimhan D, Berlin AA, Lukacs NW, Sunahara RK, Woods JH. Drug Alcohol Depend. 2009;101:202. doi: 10.1016/j.drugalcdep.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wood S, Narasimhan D, Cooper Z, Sunahara R, Woods JH. Drug Alcohol Depend. 2009;106(2–3):219. doi: 10.1016/j.drugalcdep.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collines GT, Brim RL, Narasimhan D, Ko MC, Sunahara RK, Zhan CG, Woods JH. J Pharmacol Exp Ther. 2009;331(2):445. doi: 10.1124/jpet.108.150029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao D, Narasimhan D, Macdonald J, Brim R, Ko MC, Landry DW, Woods JH, Sunahara RK, Zhan CG. Mol Pharmacol. 2009;75:318. doi: 10.1124/mol.108.049486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J, Hamza A, Zhan CG. J Am Chem Soc. 2009;131:11964. doi: 10.1021/ja903990p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Narasimhan D, Nance M, Gao D, Ko MC, Macdonald J, Yoon D, Landry DW, Woods JH, Zhan CG, Tesmer J, Sunahara RK. Protein Eng Des Sel. 2010;23(7):537. doi: 10.1093/protein/gzq025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mets B, Winger G, Cabrera C, Seo S, Jamdar S, Yang G, Zhao K, Briscoe RJ, Almonte R, Woods JH, Landry DW. Proc Natl Acad Sci U S A. 1998;95(17):10176. doi: 10.1073/pnas.95.17.10176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larsen NA, de Prada P, Deng SX, Mittal A, Braskett M, Zhu X, Wilson IA, Landry DW. Biochem. 2004;43(25):8067. doi: 10.1021/bi049495l. [DOI] [PubMed] [Google Scholar]
- 27.Pan Y, Gao D, Zhan CG. J Am Chem Soc. 2008;130:5140. doi: 10.1021/ja077972s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dahiyat BI. Curr Opin Biotechnol. 1999;10:387. doi: 10.1016/S0958-1669(99)80070-6. [DOI] [PubMed] [Google Scholar]
- 29.Lin YS. Proteins. 2008;73:53. doi: 10.1002/prot.22049. [DOI] [PubMed] [Google Scholar]
- 30.Bendová-biedermannová L, Hobza H, Vondrášek J. Proteins. 2008;72:402. doi: 10.1002/prot.21938. [DOI] [PubMed] [Google Scholar]
- 31.Olufsen M, Papaleo E, Smalås AO, Brandsdal BO. Proteins. 2008;71:1219. doi: 10.1002/prot.21815. [DOI] [PubMed] [Google Scholar]
- 32.Schweiker KL, MakhatadzeI GL. Methods Enzymol. 2009;454:175. doi: 10.1016/S0076-6879(08)03807-X. [DOI] [PubMed] [Google Scholar]
- 33.Malakauskas SM, Mayo SL. Nature Struct Biol. 1998;5:470. doi: 10.1038/nsb0698-470. [DOI] [PubMed] [Google Scholar]
- 34.Lehmann M, Wyss M. Curr Opin Biotechnol. 2001;12:371. doi: 10.1016/s0958-1669(00)00229-9. [DOI] [PubMed] [Google Scholar]
- 35.Dantas G, Kuhlman B, Callender D, Wong M, Baker D. J Mol Biol. 2003;332:449. doi: 10.1016/s0022-2836(03)00888-x. [DOI] [PubMed] [Google Scholar]
- 36.Kuhlman B, Dantas G, Ireton GC, Varani G, Stoddard BL, Baker D. Science. 2003;302:1364. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- 37.Dwyer MA, Looger LL, Hellinga HW. Science. 2004;304:1967. doi: 10.1126/science.1098432. [DOI] [PubMed] [Google Scholar]
- 38.Schweiker K, Zarrine-Afsar A, Davindson AR, Makhatadze GI. Protein Sci. 2007;16:2694. doi: 10.1110/ps.073091607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yosef E, Politi R, Choi MH, Shifman JM. J Mol Boil. 2009;385:1470. doi: 10.1016/j.jmb.2008.09.053. [DOI] [PubMed] [Google Scholar]
- 40.Korkegian A, Black ME, Baker d, Stoddard BL. Science. 2005;308:857. doi: 10.1126/science.1107387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Das R, Baker D. Annu Rev Biochem. 2008;77:363. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- 42.Xiao Z, Bergeron H, Grosse S, Beauchemin M, Garron ML, Shaya D, Sulea T, Cygler M, Lau PCK. Appl Environ Microbiol. 2008;74:1183. doi: 10.1128/AEM.02220-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gribenko AV, Patel MM, Liu J, McCallum SA, Wang C, Makhatadze GI. Proc Natl Acad Sci USA. 2009;106:2601. doi: 10.1073/pnas.0808220106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruiu L, Roque ACA, Taipa MA, Lowe CR. J Mol Recognit. 2006;19:372. doi: 10.1002/jmr.782. [DOI] [PubMed] [Google Scholar]
- 45.Sousa IT, Ruiu L, Lowe CR, Taipa MA. J Mol Recognit. 2009;22:83. doi: 10.1002/jmr.900. [DOI] [PubMed] [Google Scholar]
- 46.Mazumder-Shivakumar D, Bruice TC. Proc Natl Acad Soc USA. 2004;101:14379. doi: 10.1073/pnas.0406002101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pedone E, Saviano M, Bartolucci S, Rossi M, Ausili A, Scirè A, Bertoli E, Tanfani MF. J Proteome Res. 2005;4:1972. doi: 10.1021/pr050152z. [DOI] [PubMed] [Google Scholar]
- 48.Chiu WC, You JY, Wang WC. J Mol Biol. 2006;359:741. doi: 10.1016/j.jmb.2006.03.063. [DOI] [PubMed] [Google Scholar]
- 49.Skopalík J, Anzenbacher P, Otyepka M. J Phys Chem B. 2008;112:8165. doi: 10.1021/jp800311c. [DOI] [PubMed] [Google Scholar]
- 50.Aurilia V, Rioux-Dubé JF, Marabotti A, Pézolet M, D’Auria S. J Phys Chem B. 2009;113:7753. doi: 10.1021/jp901921r. [DOI] [PubMed] [Google Scholar]
- 51.Rutherford K, Daggett V. Biochem. 2009;48:6450. doi: 10.1021/bi900174v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xie B-B, Bian F, Chen X-L, He H-L, Guo J, Gao X, Zheng Y-X, Chen b, Zhou B-C, Zhang Y-Z. J Biol Chem. 2009;284:9257. doi: 10.1074/jbc.M808421200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Case DA, Darden TA, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Wang B, Pearlman DA, Crowley M, Brozell S, Tsui V, Gohlke H, Mongan J, Hornak V, Cui G, Beroza P, Schafmeister C, Caldwell JW, Ross WS, Kollman PA. AMBER 8. University of California; San Francisco: 2004. [Google Scholar]
- 54.Ryckaert J, Ciccotti PG, Berendsen HJC. J Comput Phys. 1977;23:327. [Google Scholar]
- 55.Darden T, York D, Pedersen L. J Chem Phys. 1993;98:10089. [Google Scholar]
- 56.Toukmaji A, Sagui C, Board J, Darden T. J Chem Phys. 2000;113:10913. [Google Scholar]
- 57.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. J Chem Phys. 1984;81:3684. [Google Scholar]
- 58.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW. J Chem Phys. 1983;79:926. [Google Scholar]
- 59.Gilson MK, Sharp KA, Honig BH. J Comput Chem. 1988;9:327. [Google Scholar]