Abstract
Objective
Hemophagocytes (HPCs) are activated macrophages that have engulfed other hematopoietic cells. Rarely identified in normal spleen and bone marrow, their excess characterizes many cytokine storm syndromes, particularly Macrophage Activation Syndrome (MAS) and Hemophagocytic Lymphohistiocytosis (HLH). The functions of HPCs, and thus their significance in acute inflammatory conditions, remain unclear.
Methods
HPCs were generated in wild-type mice using repeated Toll-like Receptor 9 stimulation and Interkeukin-10 receptor blockade. RNA was extracted from HPCs that were isolated by laser capture microdissection. Transcriptional profiles of HPCs were then compared to those of resting splenic macrophages. Additionally, a diverse cohort of patients with excess hemophagocytosis on clinical bone marrow evaluation was identified. Immunohistochemistry of these patients’ bone marrow samples was performed for markers of classical (CD64) or alternative (CD163 and CD206) macrophage activation.
Results
Differential gene expression and Gene Set Enrichment Analyses identified upregulation of genes and gene sets associated with alternative-activation in HPCs. Immunohistochemistry of HPCs in human bone marrow samples showed universal staining of HPCs for CD163, but rarely for CD206 or CD64.
Conclusion
Laser-captured murine TLR9-induced HPCs had a transcriptional profile similar to alternatively activated macrophages. Additionally, HPC expression CD163 was confirmed in a uniquely diverse cohort of patients. Collectively, these data support the hypothesis that HPCs have immunoregulatory or clean-up functions.
INTRODUCTION
Macrophages reside in organs throughout the body where they are involved in diverse functions including pathogen sensing, pro- and anti-inflammatory immune responses, and wound healing. Macrophage activation has been described in a continuum from classically- (M1) to alternatively-activated (broadly categorized as M2) (1). M1 macrophages have pro-inflammatory functions that often result in tissue damage, while M2 macrophages participate in immunoregulation and tissue remodeling. Thus, macrophage functions can be fluid and varied, proinflammatory or anti-inflammatory, depending on the mix of signals they receive.
Morphologically, hemophagocytes (HPCs) are macrophages that have engulfed other hematopoietic cells. The evaluation for hemophagocytosis is an important aspect of the diagnosis and management of Macrophage Activation Syndrome (MAS) and Hemophagocytic Lymphohistiocytosis (HLH). MAS and HLH are complex cytokine storm disorders that can complicate various infectious, rheumatic, or malignant diseases (2). HLH can also be caused by primary genetic defects in cytotoxicity.
Work in animals and humans has suggested both pro- and anti-inflammatory roles for HPCs. Evidence for a pro-inflammatory role derives from the importance of interferon (IFN)-γ acting on macrophages to drive animal models of MAS/HLH (3), and from the localization of pro-inflammatory cytokines in MAS patients’ liver biopsies (4). The evidence for alternative activation includes the expression of the scavenger receptor CD163 on HPCs (2) as well as the detection of anti-inflammatory functions in murine erythrophagocytes identified by flow cytometry (3, 5).
In this report, we show that the transcriptional program of morphologically-identified murine HPCs is consistent with alternative activation. We then confirm expression of CD163 on bone marrow HPCs from a uniquely broad human cohort. While the roles of macrophages in hemophagocytic syndromes remain imprecise, these results suggest that murine TLR9-induced HPCs are alternatively activated and that their presence may be beneficial to the control or clearance of inflammation.
PATIENTS, MATERIALS, AND METHODS
Isolation of Murine HPCs
Fulminant MAS was induced with repeated Toll-like Receptor (TLR9)-stimulation (via CpG administration) and IL-10 receptor (IL10R) blockade as previously described (6). Splenic touch preps were made on nuclease free polyethylene naphthalate membrane-coated slides (Zeiss) and immediately Wright-Giemsa stained. A pediatric hematopathologist with expertise in hemophagocytic syndromes (MP) morphologically identified HPCs from TLR9-stimulated, IL10R-blocked mice or resting macrophages from saline-treated mice. From each mouse, twenty cells were captured, using the Zeiss/P.A.L.M. laser microdissection microscope (Figure 1A & B), isolated, pooled, and processed in aggregate.
Figure 1.
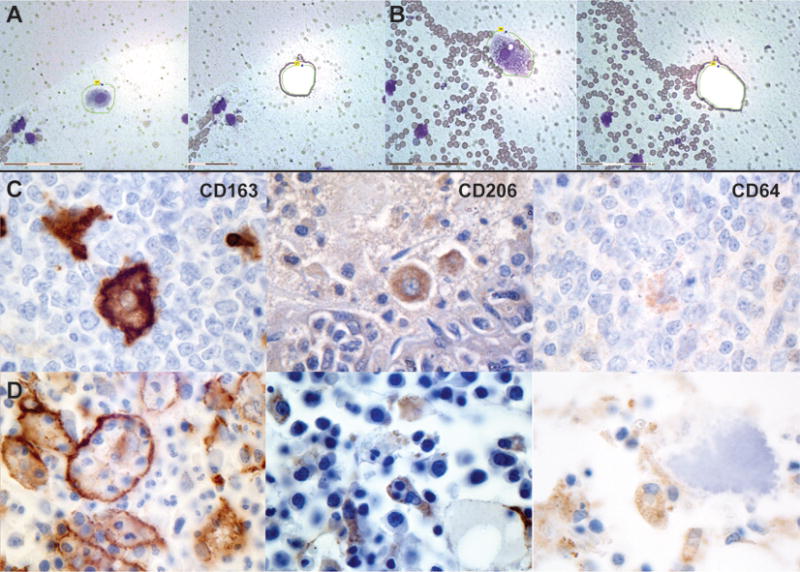
Murine and human hemophagocytes. (A) Laser capture microdissection of a resting murine splenic macrophage and (B) a murine HPC. (C & D) HPCs from a heterogeneous cohort express CD163, and rarely CD206 or CD64. (C) Positive controls for immunohistochemical stains in tonsil tissue from patients without hemophagocytic disease. (D) Bone marrow biopsies from representative patients with hemophagocytic disease immunostained for CD163, CD206, or CD64 as detailed in Materials and Methods. All images are 100X magnification and slides are counterstained with hematoxylin.
RNA isolation and microarray
cDNA libraries were generated from the RNA of pooled, microdissected cells using the WT-Ovation One-Direct Amplification System (NuGen Technologies) according to manufacturer’s instructions. Fragmented cDNA was then hybridized to Affymetrix Mouse Gene ST 1.0 microarrays, washed, stained and scanned with the Affymetrix Scanner 3000 7G.
Transcriptional Analysis
Affymetrix Expression Console software was used to perform quality control, excluding two chips from each group and leaving four biological replicates per group for analysis. Microarrays were preprocessed using robust multiarray analysis (7) (Gene Expression Omnibus accession number GSE47430). Probe sets lacking gene symbol annotation or with a mean log2 intensity less than five among HPC samples were filtered out. Log2 transformed expression data were analyzed using the R statistical computing language. Differentially expressed genes (DEGs) were defined as those with at least 1.5-fold difference in expression in HPCs versus resting macrophages. DEGs were tested for statistical significance using Student’s t-test with multiple testing correction using false discovery rate (FDR). Functional enrichment analysis among DEGs was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.7 (8). Gene ontology terms for biological pathways, molecular function and cellular components were tested, as were Kyoto Encyclopedia of Genes and Genomes (KEGG) biological pathways. Genes passing filtering criteria were used as the background gene list in the enrichment analysis.
Gene Set Enrichment Analysis (GSEA)
Gene sets were identified by searching the Molecular Signatures Database (MSigDB) for the term “inflammation” (9). Gene sets with substantial overlap were excluded. Initially, 17 gene sets were included in this analysis, but the “Phagocytosis” gene set was excluded as too few probes were included in our dataset (Table II). MOgene expression data were translated into Human Genome Organization (HUGO) gene symbols before comparison with selected gene sets, as per GSEA protocol. Gene sets were analyzed using gene set permutation, and prior to analysis we chose the following threshold for significant enrichment: nominal p-value ≤0.05 and FDR ≤5% (9).
Table II.
Gene Set Enrichment Analysis of TLR9-induced HPCs versus Resting Macrophages.
| Gene Set | Size | N-ES | Nom. P-val | FDR |
|---|---|---|---|---|
| Proteasome* | 43 | 2.42 | < 0.001 | < 0.001 |
| Upregulated in M2 vs M1* | 73 | 1.85 | < 0.001 | 0.009 |
| NOD-like Receptor Signaling* | 54 | 1.74 | 0.002 | 0.022 |
| Innate Immunity Signaling | 97 | 1.49 | 0.015 | 0.133 |
| Actin Regulation | 195 | 1.48 | 0.007 | 0.116 |
| Cytosolic DNA Sensing | 46 | 1.48 | 0.058 | 0.101 |
| TLR Signaling | 90 | 1.41 | 0.044 | 0.127 |
| Upregulated in M1 vs M2 | 76 | 1.32 | 0.080 | 0.188 |
| Endocytosis | 160 | 1.23 | 0.12 | 0.283 |
| TLR3 Cascade | 56 | 1.04 | 0.391 | 0.545 |
| TLR4 Cascade | 25 | 0.93 | 0.556 | 0.717 |
| IL-10 Pathway | 17 | 0.84 | 0.663 | 0.839 |
| Integrin Pathway | 37 | 0.61 | 0.963 | 1 |
| TLR9 Cascade | 20 | 0.54 | 0.969 | 0.989 |
| Inflammatory Pathway | 25 | −0.95 | 0.547 | 1 |
| IL-1R Pathway | 30 | −0.67 | 0.921 | 0.946 |
N-ES=Normalized Enrichment Score, FDR=False Discovery Rate,
=Significantly enriched Gene Set per criteria in Materials & Methods.
Patient Selection and Immunohistochemistry
We performed an unbiased, retrospective search of the pathology database of The Children’s Hospital of Philadelphia for bone marrow specimens performed between 1998 and 2011 whose clinical report documented excessive hemophagocytosis. Thirty-seven samples from 34 patients were identified and verified as having excess hemophagocytosis. These samples had been decalcified, fixed in acetic acid-zinc-formalin (AZF), and paraffin-embedded per institutional protocol.
Immunohistochemistry was performed for CD64 (1:25; clone 10.1, Abcam), CD163 (1:200; clone 10D6; Vector Laboratories) and CD206 (1:500; clone 5C11, Abnova/Novus Biologicals). Heat antigen retrieval was utilized for CD64, at pH 6, and CD163, at pH 9. Avidin-biotin complex signal amplification was used for CD206 and a polymeric–based method for CD64 and CD163. Detection was performed with horseradish peroxidase and 3,3′ Diaminobenzidine (DAB) chromogen, and the sections were counterstained with hematoxylin. Adequate positive and negative staining controls were performed on AZF fixed tonsils (Figure 1C). Patient sections were evaluated for the presence or absence of immunostained HPCs by a blinded hematopathologist with expertise in hemophagocytic diseases (MP).
Chart review identified clinical information (available upon request) related to each sample. This study was approved by the institutional review board of The Children’s Hospital of Philadelphia.
RESULTS
Genes associated with macrophage activation and regulation of inflammation are among the most upregulated in murine TLR9-induced HPCs
Forty-five genes met our criteria for differential expression (Table I and data not shown). No genes with decreased expression in HPCs were found to be statistically significant as defined in the Methods. Genes encoding the ribosomal subunits Rps20 and Rpl35a were among the most highly upregulated, suggesting regulation of global protein synthesis by HPCs. Consistent with this, functional enrichment analysis of all DEGs suggested that “translation” was a significant biological function of HPCs (Gene Ontology biological process annotation, Benjamini-corrected p-value=0.002). We further found the KEGG “Ribosome” pathway was significantly enriched (Benjamini-corrected p-value=1.6×10−7). Another group of upregulated genes were related to cellular energetics. Two genes encoding subunits of the mitochondrial respiratory protein complex cytochrome c oxidase (COX6C, COX6A1) were upregulated, as well as the glycolytic enzyme GAPDH. Accordingly, the KEGG “Oxidative Phosphorylation” pathway was significantly enriched in genes differentially expressed by HPCs (Benjamini-corrected p-value=4.2×10−4). Thus, these data suggest TLR9-induced HPCs are acting to meet demands for energy and protein synthesis.
Table I.
Highly Differentially Expressed Genes in TLR9-induced HPCs versus Resting Macrophages.
| Gene | Protein Product | Fold Change | P-value | FDR |
|---|---|---|---|---|
| Rps20 | Ribosomal Protein S20 | 22.6 | 0.0063 | 0.146 |
| β2m | β-2 microglobulin | 12.5 | 0.0292 | 0.206 |
| Rpl35a* | Ribosomal Protein L35A | 8.5 | 4.29e-4 | 0.055 |
| Saa3 | Serum Amyloid A 3 | 7.9 | 0.0014 | 0.055 |
| Ifitm2 | Interferon-inducible transmembrane protein 3 | 6.7 | 0.0445 | 0.210 |
| Cox6c | Cytochrome c oxidase, subunit Vic | 6.2 | 0.0238 | 0.174 |
| Ftl* | Ferritin light chain | 5.8 | 0.0028 | 0.055 |
| Tmsb10 | Thymosin β10 | 4.9 | 0.0430 | 0.210 |
| Gapdh* | Glyceraldehyde 3 phosphate dehydrogenase | 4.9 | 0.0019 | 0.055 |
| Cox6a1 | Cytochrome c oxidase subunit VIa, polypeptide 1 | 4.2 | 0.0388 | 0.210 |
| Usmg5 | Upregulated during skeletal muscle growth 5 | 4.2 | 0.0412 | 0.210 |
| Tmsb4x | Thymosin β4 | 4.1 | 0.0427 | 0.210 |
| S100a6 | S100 calcium binding protein A6 (calcyclin) | 4.1 | 6.27e-4 | 0.055 |
Genes associated with ≥ 4-fold induction in HPCs versus resting macrophages.
=Target identified by more than one probe. FDR=False Discovery Rate
Genes associated with macrophage stimulation and activation were also among the most differentially expressed. β2m and IFITM2 are upregulated with anti-viral responses, while SAA3 and S100A6 are parts of the acute phase response. Upregulation of the ferritin light chain (FTL) gene by HPCs is consistent with the hyperferritinemia characteristic of MAS/HLH. Finally, the homologues Thymosin β10 and β4 may play regenerative and anti-inflammatory roles in macrophages.
Gene Set Enrichment Analysis Suggests Alternative Polarization of murine TLR9-induced HPCs
To refine our analysis of differential gene expression, we tested for upregulation of gene sets associated with a variety of relevant transcriptional programs in HPCs. Only three gene sets fulfilled our criteria for significance: proteasomal degradation, nod-like receptor signaling, and the set of genes upregulated by M2 versus M1 polarized macrophages (Table II; Supplemental Figure I) (10). Notably, the set of genes upregulated in M1 versus M2 macrophages was not significantly enriched in HPCs.
Hemophagocytes from a diverse human cohort uniformly express CD163, and rarely express CD206 or CD64
We identified a longitudinal cohort of human bone marrow samples that on biopsy or aspirate showed excess hemophagocytosis regardless of diagnosis. These samples represented hematologic malignancies (lymphoma-1, Langerhans-cell histiocytosis-3, and post-transplant lymphoproliferative disease-3), infections (Epstein Barr Virus-6, bacterial-2, parvoviral-1, and fungal-1), immunodeficiencies (paroxysmal nocturnal hemoglobinuria-1 and Munc13-4 deficiency-1), rheumatic diseases (systemic and other forms of Juvenile Idiopathic Arthritis-9 (JIA) and dermatomyositis-1) and idiopathic causes of hemophagocytosis-9).
Regardless of disease of origin or treatment stage, bone marrow samples demonstrated positive staining of HPCs for CD163 in all tested samples (Figure 1C). CD163 is a scavenger receptor that binds and internalizes hemoglobin–haptoglobin complexes, which then activate heme-oxygenase 1 and induce the synthesis of ferritin (11). Five samples, representing diverse diseases, stained positively for CD206, a mannose-receptor and marker of alternative or M2-macrophage activation (Figure 1C) (1). Two samples, both from patients with systemic infections, showed positive staining of HPCs for CD64, the Fcγ-receptor 1 and a marker of classical/M1-macrophage activation (Figure 1C) (1). No samples demonstrated positive staining of HPCs for both CD64 and CD206.
DISCUSSION
Hemophagocytes are enigmatic cells appearing in a variety of inflammatory contexts. Their presence as part of diagnostic criteria for MAS and HLH often influences important medical decisions regarding evaluation and treatment. However, our poor understanding of the function of these cells complicates interpretation of their presence or absence. Our analysis of the transcriptional program of morphologically-identified murine TLR9-induced HPCs suggests that they are alternatively activated.
The “fulminant TLR9-MAS” model, including IL-10 receptor blockade, was chosen due to the homogeneity of hemophagocytosis, the absence of confounding by infection or genetic alteration, and the fact that mice treated with TLR9 stimulation or IL10R blockade alone fail to develop HPCs (6). Examination of the genes most upregulated by HPCs showed, among others, induction of Ftl (ferritin light-chain) and Saa3. Astoundingly high ferritin levels are characteristic of MAS/HLH, and ferritin has been associated with anti-inflammatory transcripts in sJIA (12). Both ferritin and SAA3 production have been associated with anti-inflammatory activity in macrophages (11, 13).
We also found that cytochrome oxidase genes, oxidative phosphorylation pathways, and the canonically glycolytic enzyme GAPDH were upregulated in HPCs. While this might suggest induction of both glycolysis and oxidative phosphorylation, GAPDH is increasingly associated with a variety of non-glycolytic functions including inhibition of cytokine translation (14).
The M2 versus M1 gene set was generated by evaluating genes upregulated by in vivo skewed M2 macrophages compared to M1 skewed macrophages (10). We found that this M2-associated gene set, but not the M1 gene set, was strongly associated with genes differentially upregulated by HPCs (Table II), supporting alternative activation. The other enriched gene sets, nod-like receptor signaling and proteasome activity, may be associated with scavenger receptor activity (15) or the degrading requirements of hemophagocytosis, respectively.
RNA from laser-captured splenic macrophages served as the basis for our microarrays. Notably, macrophages derived from splenic touch-preps may not be entirely comparable to those found in the bone marrow. Additionally, our gene expression analyses were limited by the necessary pooling of individual cells’ RNA from any single mouse, and by small input amounts of RNA. Thus, the potential for M1 versus M2 heterogeneity exists within mice and even within individual cells. Additionally, these results have not been validated by quantitation of specific targets. The limited starting material made protein quantitation impossible. Additionally, the process of amplifying small input RNA has limited our ability to quantitate at the RNA level: the RNA fragments that result from this process, while of ideal size for microarray, were of insufficient length for conventional quantitative PCR. However, our analyses were able to detect changes associated with macrophage activation. Specifically, our transcriptomic data suggest that the dominant program of murine HPCs induced by TLR9 stimulation and IL10R blockade is skewed towards alternative activation. Accordingly, recent data support the notion that TLR9-driven HPCs arise independently of a number of pro-inflammatory cytokines including IFN-γ (6) and may exert anti-inflammatory effects (5). Given the prominent role of IL-10 in alternative macrophage activation (1), future investigations may help illuminate the specific role of IL-10 in the development of HPCs.
Immunohistochemistry for selected markers in human bone marrow samples supported CD163 expression by HPCs in a cohort that, to our knowledge, is unique in terms of its breadth of diagnoses. The association of CD163 with alternative activation is well established (11). Future studies should attempt to validate further the findings of TLR9-stimulated murine HPCs in human samples.
Beginning with morphologically identified HPCs, we have shown transcriptional evidence for alternative activation of murine HPCs from an infection-free model, as well as immunohistochemical evidence that human HPCs from a diversity of sources express CD163. These data support the body of literature that HPCs are alternatively-activated macrophages occurring as a common response to systemic inflammation. In MAS/HLH there is likely heterogeneity in macrophage function, and therapeutically depleting all macrophages could actually be detrimental. Further study is warranted to better understand the induction of hemophagocytosis, the precise functions of HPCs, and the roles these cells play in regulating immunopathology.
Acknowledgments
The authors are extremely grateful to Daniel Martinez, Joanne Mauger, and Eric Rappaport for technical assistance.
This project was supported by a grant from the Histiocytosis Association. SC was supported by NIH F32-AI-98337 and a Rheumatology Research Foundation Scientist Development Award. PCR was supported by Fundação Calouste de Gulbenkian and Fundação para a Ciência e a Tecnologia. EB was supported by NIH K08-AI-79396, an HHMI Early Career Physician Scientist Award, and an Arthritis Foundation Innovative Research Grant.
ABBREVIATIONS
- DEG
Differentially Expressed Gene
- FDR
False Discovery Rate
- GSEA
Gene Set Enrichment Analysis
- HLH
Hemophagocytic Lymphohistiocytosis
- HO1
Heme-oxygenase 1
- HPC
Hemophagocyte
- IFN
Interferon
- IL
Interleukin
- MAS
Macrophage Activation Syndrome
- sJIA
Systemic Juvenile Idiopathic Arthritis
- TLR
Toll-like Receptor.
References
- 1.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8(12):958–69. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Canna SW, Behrens EM. Not all hemophagocytes are created equally: appreciating the heterogeneity of the hemophagocytic syndromes. Curr Opin Rheumatol. 2012;24(1):113–8. doi: 10.1097/BOR.0b013e32834dd37e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zoller EE, Lykens JE, Terrell CE, Aliberti J, Filipovich AH, Henson PM, et al. Hemophagocytosis causes a consumptive anemia of inflammation. J Exp Med. 2011;208(6):1203–14. doi: 10.1084/jem.20102538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Billiau AD, Roskams T, Van Damme-Lombaerts R, Matthys P, Wouters C. Macrophage activation syndrome: characteristic findings on liver biopsy illustrating the key role of activated, IFN-gamma-producing lymphocytes and IL-6- and TNF-alpha-producing macrophages. Blood. 2005;105(4):1648–51. doi: 10.1182/blood-2004-08-2997. [DOI] [PubMed] [Google Scholar]
- 5.Ohyagi H, Onai N, Sato T, Yotsumoto S, Liu J, Akiba H, et al. Monocyte-derived dendritic cells perform hemophagocytosis to fine-tune excessive immune responses. Immunity. 2013;39(3):584–98. doi: 10.1016/j.immuni.2013.06.019. [DOI] [PubMed] [Google Scholar]
- 6.Canna SW, Wrobel J, Chu N, Kreiger PA, Paessler M, Behrens EM. Interferon-gamma mediates anemia but is dispensable for fulminant toll-like receptor 9-induced macrophage activation syndrome and hemophagocytosis in mice. Arthritis Rheum. 2013;65(7):1764–75. doi: 10.1002/art.37958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 8.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 9.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coates PJ, Rundle JK, Lorimore SA, Wright EG. Indirect macrophage responses to ionizing radiation: implications for genotype-dependent bystander signaling. Cancer Res. 2008;68(2):450–6. doi: 10.1158/0008-5472.CAN-07-3050. [DOI] [PubMed] [Google Scholar]
- 11.Schaer CA, Schoedon G, Imhof A, Kurrer MO, Schaer DJ. Constitutive endocytosis of CD163 mediates hemoglobin-heme uptake and determines the noninflammatory and protective transcriptional response of macrophages to hemoglobin. Circ Res. 2006;99(9):943–50. doi: 10.1161/01.RES.0000247067.34173.1b. [DOI] [PubMed] [Google Scholar]
- 12.Fall N, Barnes M, Thornton S, Luyrink L, Olson J, Ilowite NT, et al. Gene expression profiling of peripheral blood from patients with untreated new-onset systemic juvenile idiopathic arthritis reveals molecular heterogeneity that may predict macrophage activation syndrome. Arthritis Rheum. 2007;56(11):3793–804. doi: 10.1002/art.22981. [DOI] [PubMed] [Google Scholar]
- 13.Nguyen KD, Macaubas C, Nadeau KC, Truong P, Yoon T, Lee T, et al. Serum amyloid A overrides Treg anergy via monocyte-dependent and Treg-intrinsic, SOCS3-associated pathways. Blood. 2011;117(14):3793–8. doi: 10.1182/blood-2010-11-318832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang CH, Curtis JD, Maggi LB, Jr, Faubert B, Villarino AV, O’Sullivan D, et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell. 2013;153(6):1239–51. doi: 10.1016/j.cell.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mukhopadhyay S, Varin A, Chen Y, Liu B, Tryggvason K, Gordon S. SR-A/MARCO-mediated ligand delivery enhances intracellular TLR and NLR function, but ligand scavenging from cell surface limits TLR4 response to pathogens. Blood. 2011;117(4):1319–28. doi: 10.1182/blood-2010-03-276733. [DOI] [PubMed] [Google Scholar]


