Abstract
A three step protocol for protein S-nitrosothiol conversion to fluorescent mixed disulfides with purified proteins, referred to as the thiosulfonate switch, is explored which involves: 1) thiol blocking at pH 4.0 using S-phenylsulfonylcysteine (SPSC); 2) trapping of protein S-nitrosothiols as their S-phenylsulfonylcysteines employing sodium benzenesulfinate; and 3) tagging the protein thiosulfonate with a fluorescent rhodamine based probe bearing a reactive thiol (Rhod-SH), which forms a mixed disulfide between the probe and the formerly S-nitrosated cysteine residue. S-nitrosated bovine serum albumin and the S-nitrosated C-terminally truncated form of AdhR-SH (alcohol dehydrogenase regulator) designated as AdhR*-SNO were selectively labelled by the thiosulfonate switch both individually and in protein mixtures containing free thiols. This protocol features the facile reaction of thiols with S-phenylsulfonylcysteines forming mixed disulfides at mild acidic pH (pH = 4.0) in both the initial blocking step as well as in the conversion of protein-S-sulfonylcysteines to form stable fluorescent disulfides. Labelling was monitored by TOF-MS and gel electrophoresis. Proteolysis and peptide analysis of the resulting digest identified the cysteine residues containing mixed disulfides bearing the fluorescent probe, Rhod-SH.
Introduction
Protein S-nitrosothiols, defined as a covalent bond between a cysteine thiol and an NO equivalent, have been identified as important posttranslational modifications in cell biology.1-3 S-nitrosothiols function as a means toward nitric oxide cellular signal transduction4,5 and play a role in both physiological as well as pathophysiological processes.6,7 A wide array of biological processes have been found to be effected by protein S-nitrosothiols including transcriptional regulation,8,9 blood flow,10 neuronal function,11,12 and phosphorylation.13 Aberrant S-nitrosation has been implicated in various cancers,14 cardiovascular disease,15 and neurodegenerative diseases16,17 and may well contribute to the resistance of bacterial pathogens to both host-generated and endogenous sources of nitric oxide.18,19
The biological relevance of protein S-nitrosothiols has led to intense interest in variations of the biotin switch technique,20,21 which switches the S-nitrosothiol group into a thiol by reduction with ascorbate. The resulting thiol is labelled, provided that native free thiols are blocked before reduction. This method has facilitated extensive proteomic studies by use of gel electrophoresis,22-24 resin capture,25 microarray based analysis,26 and proteolysis followed by amino acid sequencing.27,28 Phenyl mercury compounds have also been employed on resins and with biotin tags for enrichment and comprehensive detection of protein S-nitrosothiols.29,30
Due to the intricate chemistry associated with S-nitrosothiol detection, there is the likelihood that the field will require a variety of complementary approaches. Switchless protocols have been developed that utilize S-nitrosothiol ligation to triarylphosphine derivatives31 to form sulfenamides,32 which can be further manipulated to form disulfides,33 thioethers,34 or dehydroalanines.35 These reactions are promising but are limited by aqueous solubility issues and compatibility with amines and carboxylates. Water soluble triphenylphosphine probes have been developed that convert protein-S-nitrosothiols into S-alkylphosphonium salts detectable by mass spectrometry.36 While these reactions exhibit advantages by reacting directly with S-nitrosothiols, the resulting S-alkylphosphonium salts are not stable to gel electrophoresis or tryptic digest. This work has been expanded by incorporating triarylphosphines with ester and thioester functionalities along with water solubilizing groups.37 While low molecular weight S-nitrosothiols such as S-nitrosoglutathione (GSNO) have been detected through mass fragmentation, this method has not been applied to protein-S-nitrosothiols.
We have recently published38 a detailed study of the reaction between S-nitrosated bovine serum albumin (BSA-SNO) and benzenesulfinic acid sodium salt (PhSO2Na). PhSO2Na was shown to react with BSA-SNO forming the corresponding S-phenylthiosulfonate, BSA-SSO2Ph, and N-hydroxybenzene-sulfonamide in a 1:1 ratio (Scheme 1) within minutes. N-hydroxybenzenesulfonamide, also known as Piloty’s acid,39 is not an NO donor at neutral and acidic pH. It should be noted that in the biotin switch technique, ascorbate reacts with S-nitrosothiols giving rise to O-nitrosoascorbate, which releases NO within minutes at neutral pH.40 PhSO2Na was shown to be unreactive to the protein thiol and disulfides in bovine serum albumin.
Scheme 1.

BSA-SNO is converted into BSA-SSO2Ph and PhSO2NHOH upon reaction with PhSO2Na at pH 4.0.
While PhSO2Na has been employed as an S-nitrosothiol probe,38 the reporter group linkage, i.e. the S-phenylsufonylcysteine functional group, is susceptible to hydrolysis at neutral pH resulting in diminished signal. An expanded methodology that displaces S-phenylthiosulfonates with a thiol containing fluorescent probe resulting in a stable disulfide linkage has the potential of addressing this shortcoming and is the focus of the work presented here.
Herein, we explore a new protocol, denoted as the thiosulfonate switch, that traps protein S-nitrosothiols as mixed disulfides bearing a fluorescent probe at pH 4.0. This protocol involves initial blocking of protein thiols by S-phenylsulfonylcysteine (SPSC),41 which forms cysteine bearing mixed disulfides. It should be noted that the byproduct of the initial blocking step is benzenesulfinate itself. Subsequent addition of PhSO2Na converts protein S-nitrosothiols into protein S-phenylthiosulfonates. Addition of a highly water soluble zwitterionic rhodamine based fluorophore incorporating a reactive thiol, denoted Rhod-SH (1), reacts with protein S-phenylthiosulfonates, giving rise to a mixed disulfide between the probe and the formerly S-nitrosated cysteine residue. Since PhSO2Na does not react with thiols,38 the sequential addition of a thiol derivatized fluorophore is performed without purification.
Results and Discussion
Control Reactions
(i) Stability of GSNO at different pH’s with and without DTT
The thiosulfonate switch protocol employs a mild acidic pH in order to convert S-nitrosothiols into their corresponding S-phenylthiosulfonates.38 In order to assess the stability of S-nitrosothiols at pH 4.0, the absorbance at 545 nm of a 10 mM GSNO solution (ε = 15 M−1cm−1, 545 nm, nN→π*)41 was monitored (Fig. 1) over 50 min in pH 4.0 buffer. This was repeated at pH 7.0 (100 mM HEPES, 1 mM EDTA) and minimal degradation of GSNO was observed at either pH. Denitrosation reactions were also monitored between GSNO and dithiothreitol (DTT) (10 mM each) at varying pHs. When DTT undergoes transnitrosation, an unstable S-nitrosothiol is formed which is converted to a disulfide along with concomitant release of nitroxyl (HNO).42-44 A 1% decrease in absorbance is observed when DTT is added to GSNO at pH 4.0 over 50 min. This is in stark contrast to the results obtained at pH 4.8 (100 mM ammonium formate, 1 mM EDTA), 5.5 (100 mM MES, 1 mM EDTA), and 7.0 (100 mM HEPES, 1 mM EDTA), where 17%, 47%, and 75% (respectively) of the absorbance is lost. This is consistent with previously reported findings where the rate of transnitrosation increases with increasing pH.45 This stability trend of S-nitrosothiols demonstrated here (Fig. 1) suggests that there are clear advantages to designing a proteomics-based detection strategy at mild acidic pHs provided that protein solubility issues can be overcome.
Fig. 1.
% GSNO over time (min) taken from absorbance readings at 545 nm at different pH’s with and without equimolar DTT.
(ii) Reactions of PhSO2Na and Rhod-SH (1) with GSNO, GSH and GSSG
First, GSNO was employed as the substrate to undergo two of the essential steps in the thiosulfonate switch: S-nitrosothiol conversion to an S-phenylthiosulfonate and displacement with a thiol probe forming a mixed disulfide. Conversion of GSNO to GSSO2Ph was performed by adding excess benzenesulfinic acid sodium salt (250 eq) at pH 4.0 (100 mM ammonium formate, 1 mM EDTA) (Scheme 2). After 30 min, the fluorescent rhodamine based probe bearing a reactive thiol, Rhod-SH (1) (see Experimental for synthesis and optical characterization), was added at 1 eq and after 10 min, the product Rhod-SSG, was observed at 95% conversion based on the reverse phase HPLC trace (See Fig. S1) with detection at λmax of Rhod-SH (1) (562 nm). When this experiment was repeated replacing GSNO with either GSSG or GSH, only Rhod-SH (1) was observed, confirming the selectivity of the reaction towards S-nitrosothiols. These reactions also confirm that the thiol probe, Rhod-SH (1), does not readily undergo disulfide exchange at pH 4.0 but does exchange with S-phenylsulfonylglutathione in the presence of excess PhSO2Na.
Scheme 2.
Reactions of PhSO2Na and Rhod-SH (1) on GSNO, GSH and GSSG.
(iii) Rhod-SH (1) control reactions with thiol blockers and disulfides
The reactivity of SPSC was compared to well-known commercially available thiol blockers such as MMTS, NEM, and IAM by reacting each with Rhod-SH (1) at pH 4.0 (Scheme 3). While SPSC has been shown to react with thiols at basic pH (0.1 M sodium bicarbonate),46 the reaction at mild acidic pH’s is unreported to our knowledge. In addition, reactions with disulfides GSSG and (Gly-Cys)2 were also observed in order to assess the reactivity of Rhod-SH (1) with disulfides over extended time. For both disulfides, no product formation was observed after 1 h. For thiol blocking reactions, SPSC reacted with Rhod-SH (1) the fastest with approximately full conversion in 1 min. MMTS reacted the second fastest with 22% conversion over 1 min and 97% conversion after 30 min. Therefore, both SPSC and MMTS are efficient thiol blockers at pH 4.0. The reaction with NEM was slow at pH 4.0 with only 48% conversion over 1 h while IAM showed no product formation in 1 h. All products were confirmed by collecting the corresponding HPLC elution peak followed by HRMS analysis (see Fig. S2 for HPLC traces).
Scheme 3.
Reactions of Rhod-SH (1) with GSSG, (GlyCys)2, SPSC, MMTS, NEM, and IAM at pH 4.0.
(iv) SPSC control reactions
While SPSC reacts readily with Rhod-SH (1), a series of thiols with varying pKa’s were blocked with SPSC in order to validate that SPSC reacts with thiols and not thiolates (Scheme 4). A solution of thiol (2 mM) and SPSC (1.8 mM) in pH 4.0 buffer was analyzed by reverse phase HPLC in tandem with UV-vis detection. Reactions were monitored by measuring absorbance at 265 nm (λmax of SPSC and PhSO2Na)38 at Rt= 10 min (PhSO2Na) and Rt= 12 min (SPSC) (retention times were determined by running authentic samples). Conversion of SPSC to PhSO2Na went to completion after 2 min for penicillamine and glutathione and 87% completion for captopril. Therefore, SPSC undergoes facile reactions with thiols at pH 4.0 and does not require the deprotonation of thiols to form thiolates for conversion.
Scheme 4.
Reactions of SPSC with a series of thiols of varying pKa.
The hydrolysis of a 2 mM solution of SPSC at different pH’s was also measured using the same HPLC method as above over 1 h (Fig. 2). Hydrolysis at pH 4.0, 4.8, 5.5, and 6.0 was detected at 2%, 5%, 9%, and 31%, respectively. At pH 7.0 the hydrolysis was almost complete (96%). Minimal detection of benzenesulfonic acid (Rt= 5.5 min) was observed. This confirms that S-phenylsulfonylcysteines are unstable at neutral pH. Since common proteomic methodology requires neutral to slightly basic pH, such as polyacrylamide-SDS gel electrophoresis and trypsinolysis, S-phenylsulfonylcysteines are an unlikely choice as a reporter adduct for protein-S-nitrosothiols. Conversion with a thiol probe readily generates mixed disulfides, which are the linkage utilized in the biotin switch technique used in protein-S-nitrosothiol proteomics.20
Fig. 2.
% Conversion of SPSC to PhSO2Na over 1 h as measured from absorbance detection at 265 nm from RP HPLC analysis.
Labeling of Individual Protein-S-nitrosothiols with Rhod-SH (1)
(i) TOF analysis of Rhod-SH (1) labelled BSA-SNO
S-nitrosated bovine serum albumin (BSA-SNO) was used to probe the reaction between an S-nitrosated protein and PhSO2Na and Rhod-SH (1). Serum albumin has been implicated as an NO reservoir in plasma47 and exerts NO bioactivity by transnitrosation to other proteins such as cell surface Protein Disulfide Isomerase (csPDI).48 The detection of BSA-SNO by the biotin switch assay is hindered by a slow reduction rate with ascorbate.49 BSA contains a single reactive thiol (Cys34) and 17 protein disulfides which allows for evaluation of selectivity of S-nitrosothiol labeling employing PhSO2Na and Rhod-SH (1) in the presence of disulfides.
Initial TOF-MS analysis of commercial BSA (sigma-Aldrich) shows that in addition to the free thiol (66,430 amu), covalent adducts were detected including the mixed disulfide BSA-SSCys (66,549 amu), and glycosylation adduct BSA(Glc) (66,592 amu) which have been observed previously.38,50 The free thiol content of BSA was found to be 49% employing the DTDP method.38,51 S-nitrosation of BSA with 5 eq Cys-SNO in HEN buffer (100 mM HEPES, 1 mM EDTA, 0.1 mM neocuproine, pH 8.0) for 1 h in the dark was 90% complete (44% overall SNO) based on a Griess/Saville assay as modified by Basu et. al.52 BSA-SNO was purified and the buffer was exchanged to pH 4.0 (100 mM ammonium formate, 1 mM EDTA) by centrifugation through a Sephadex G25 column.
When PhSO2Na (250 eq) followed by Rhod-SH (1) (2 eq) was added to BSA-SNO (44% SNO content, 100 μM total protein, 100 μL) at pH 4.0, the mixed disulfide BSA-SS-Rhod (67,376 amu) was detected as the major product within 30 min (Fig. 3, blue, middle). The glycosylated adduct, BSA(Glc)-SS-Rhod (67,538 amu), was also detected as a minor product. In a control reaction with commercially available BSA (Sigma) employing PhSO2Na and Rhod-SH (1), no fluorescent adducts were detected (Fig. 3, red, top). Other peaks were observed corresponding to unidentified BSA variants. After labelling of BSA-SNO, these variants show a corresponding mixed disulfide peak (+946 amu, +SRhod).
Fig. 3.
Deconvoluted mass spectra of either (red, top): BSA (100 μM) or (blue, middle): BSA-SNO (44% SNO content, 100 μM total protein) after addition of PhSO2Na and Rhod-SH (1) for 30 min. (green, bottom): BSA-SNO (22% SNO content, 28% free thiol, 100 μM total protein) after sequential addition of SPSC (1 h), PhSO2Na (30 min), and Rhod-SH (1) (10 min).
As shown in Fig. 3, (red, top) commercial BSA and the contaminant BSA-SSCys upon exposure to PhSO2Na and Rhod-SH (1) does not give rise to BSA-SS-Rhod. However, protein thiols can react with protein S-phenylsulfonylcysteines resulting in diminished signal. A thiol blocking step before SNO labelling addresses this concern. To demonstrate the blocking of free thiols at pH 4.0, BSA was partially nitrosated (22% SNO, 28% free thiol) by addition of GSNO (5 eq, 1 h, HEN pH 7.0) in the dark followed by buffer exchange to pH 4.0. The addition of S-phenylsulfonylcysteine (SPSC, 1 eq, 1 h) to the mixture of BSA-SNO and BSA (100 μM, 10 nmol total protein) resulted in blocking of the free thiol group (Fig. 3, green, bottom) as seen by the increase in the BSA-SSCys peak (66,549 amu) as well as a new peak corresponding to BSA(Glc)SSCys (66,711 amu). Sequential addition of PhSO2Na (250 eq, 30 min) and Rhod-SH (1) (3 eq, 10 min) formed BSA-SS-Rhod (67,376 amu).
(ii) Trypsinolysis of BSA-SS-Rhod
To demonstrate the compatibility of the mixed disulfide reporter group with trypsinolysis and confirm the location of the tagged cysteine residue, BSA-SS-Rhod was subjected to trypsin digestion. Toward this end, BSA-SNO (44% SNO, 100 μM total protein) was exposed to PhSO2Na (250 eq) and Rhod-SH (1) (2 eq) at pH 4.0 for 30 min. MMTS was added to trap the excess dye and safeguard against disulfide scrambling between residual Rhod-SH (1) and protein disulfides after buffer exchange to pH 7.5. Trypsinolysis was performed (10 h, 37 °C) on BSA-SS-Rhod and the peptides obtained were separated by reverse phase HPLC and analyzed by UV-vis in tandem with mass spectrometry. As seen in Fig. 4 (blue), the BSA-SS-Rhod proteolysis produced two major peptides that exhibited strong absorbance at 565 nm. The isotopic pattern (Table S1) of the major peak (3.85 min) corresponds to the fifth fully digested tryptic peptide from the C-terminus [GLVLIAFSQYLQQC(SSRhod)PFDEHVK)] wherein Cys34 is a mixed disulfide derived from Rhod-SH (1) (T5SSRhod) while the first peak (3.7 min) in the HPLC trace corresponds to a tryptic peptide with two miscleavages [FKDLGEEHFKG LVLIAFSQYLQQC(SSRhod)PFDEHVK] wherein Cys34 is a mixed disulfide derived from Rhod-SH (1) (T3-5SSRhod) (Fig. S4, Table S1). The S-methyl capped dye species RhodSSMe (4.35 min), as well as the dimer RhodSSRhod (4.68 min), were also observed. The small peak at 4.5 min was also identified as a derivative of RhodSSRhod lacking an ethyl group (Table S1).
Fig. 4.
Blue: Chromatogram for a tryptic digest of BSA-SS-Rhod (λ= 565 nm). Red: Chromatogram for a tryptic digest of BSA subjected to the same reaction and digestion conditions.
The same protocol was used on BSA as a control (Fig. 4, red), which exhibited low absorbance overall, demonstrating the selectivity of the technique. The major peaks in the control experiment correspond to RhodSSMe and RhodSSRhod (Fig. 4, red).
(iii) 1D Gel Electrophoresis on BSA-SS-Rhod
One dimensional SDS-PAGE nonreducing gels were run on the samples of BSA-SS-Rhod prepared from a BSA-SNO dilution series containing the same amount of total protein (6.6 μg) with varying amounts of BSA-SNO (1.8, 1.2, 0.6, 0.3, 0.15, and 0.0 μg). In each dilution series, BSA-SNO was subjected to the sequential addition of SPSC (0.8 nmol, 1 h), PhSO2Na (2.5 μmol, 30 min) and Rhod-SH (1) (2 nmol, 10 min) at pH 4.0. Before addition of non-reducing sample buffer and gel electrophoresis (pH 8.0), an excess of MMTS (80 nmol) was added to block free Rhod-SH (1). After electrophoresis, the gels were scanned at 532 nm with a 30 nm band pass filter at 580 nm. A linear response was observed between spot volume and μg BSA-SNO over the dilution range as seen in Fig. 5.
Fig. 5.
Spot volume vs. μg BSA-SS-Rhod from 1D SDS-PAGE nonreducing gels. BSA-SNO (1.8, 1.2, 0.6, 0.3, 0.15, and 0.0 μg) was subjected to the sequential addition of SPSC (1 h), PhSO2Na (30 min), Rhod-SH (1) (10 min), and MMTS (5 min). The error bars represent the standard deviation. The image above is the fluorescent image of the gel and coomassie stain.
(iv) AdhR*SNO synthesis and labelling by the thiosulfonate switch
In order to evaluate the general utility of the thiosulfonate switch to react with biologically important S-nitrosothiols, we investigated the ability of two homologous (46% identity) bacterial transcriptional activators, NmlR (novel MerR-like regulator) from Streptococcus pneumoniae and AdhR (alcohol dehydrogenase regulator) from Bacillus subtilis, to undergo S-nitrosation. NmlR protects S. pneumoniae from nitric oxide stress required for survival of this human pathogen in the bloodstream,53 while AdhR protects B. subtilis from naturally occurring carbon electrophiles such as methylglyoxal and formaldehyde. AdhR activates transcription of Fdh and cysteine proteinase YraA, both of which protect against aldehyde toxicity.54 Recent studies also suggest that NmlR may be capable of sensing carbon electrophiles in the cell.55 Both NmlR and AdhR possess a single cysteine residue, Cys52, derivatization of which is linked to transcriptional activation via an unknown mechanism.
Purified recombinant and reduced NmlR (13,463 amu) and AdhR*-SH (14,438 amu), a C-terminally truncated and functional (Fig. S5) variant of AdhR-SH (see Experimental section for expression and purification details), each readily react with GNSO to form NmlR-SNO and AdhR*-SNO, unambiguously established by their characteristic absorption spectra (Fig. S6). We reasoned that AdhR*-SNO may be difficult to reduce with ascorbate since S-nitrosation of Cys52 is regulatory, converting AdhR*-SH from a weak repressor to a potent activator of transcription.54 To test this, AdhR*-SH was nitrosated with GSNO (10 eq, 25 mM HEPES, 400 mM NaCl, 1 mM EDTA, 0.1 mM bathocuproine disulfonate, pH=7.0, 1 h) in the dark and the SNO content was determined to be 53% by UV-Vis (AdhR*-SH: ε280nm = 12,950 cm−1M−1 and AdhR*-SNO: ε340nm =1000 cm−1M −1, Fig. S6). Fig. 6 (dotted line) shows the deconvoluted mass spectrum of AdhR*-SNO (14,467 amu) and AdhR*-SH (14,438 amu) mixture. When exposed to ascorbate (20 mM) at pH 7.5 (HEN buffer) under ambient conditions for 1 h, we observed only a modest 10% increase in free thiol relative to the starting mixture (Fig. 6, solid line). While denaturing agents, heat, and increased ascorbate concentration would likely increase the conversion of S-nitrosothiol to thiol as employed in the biotin switch technique,20 this initial experiment demonstrates a slow reaction between AdhR*-SNO and ascorbate much like the reaction between BSA-SNO and ascorbate.49
Fig. 6.
Deconvoluted mass spectra of (dotted line): AdhR*-SNO; (solid line): AdhR*-SNO+ ascorbate (20 mM), 1 h. Buffer= 100 mM HEPES, 1 mM EDTA, 0.1 mM neocuproine (HEN), pH 7.5. AdhR*-SNO = 53% SNO, 47% free thiol, 25 μM total protein.
A series of mixing experiments were next carried out in order to establish the specificity of the thiosulfonate switch to uniquely detect S-nitrosated cysteines in the presence of underivatized cysteines. Initially, the thiosulfonate switch was carried out on a mixture of AdhR*-SNO (53%) and AdhR*-SH (47%) (25 μM total protein concentration, 0.2 nmol). SPSC was used to block the free thiol over 1 h to form AdhR*-SSCys (14,557 amu) while subsequent addition of PhSO2Na (30 min) and Rhod-SH (1) (10 min) gave rise to AdhR*-SS-Rhod (15,384 amu) (Fig. 7, red, top). When subjecting underivatized AdhR*-SH (25 μM) to the identical protocol, only the blocked AdhR*-SSCys was observed (Fig. 7, blue, bottom).
Fig. 7.
Deconvoluted mass spectra after labelling by the thiosulfonate switch of (red, top): AdhR*-SNO (53% SNO, 47% free thiol, 25 μM total protein); (blue, bottom): AdhR*-SH (25 μM). thiosulfonate switch = SPSC (1 h) + PhSO2Na (30 min) + Rhod-SH (1) (10 min).
Protein-S-nitrosothiol labelling of mixtures of BSA-SNO/AdhR*-SH and BSA/AdhR*-SNO
(i) Thiosulfonate switch labelling of BSA-SNO/AdhR*-SH
To demonstrate the selectivity of the thiosulfonate switch protocol toward S-nitrosothiols in the presence of a second protein bearing a free thiol, a mixture of BSA-SNO (52% SNO, negligible free thiol content) and underivatized AdhR*-SH (50 μM, 5 nmol total protein each) at pH 4.0 was subjected to the thiosulfonate switch. SPSC was added to block the free thiol of AdhR*-SH for 1 h followed by the sequential addition of PhSO2Na (30 min) and Rhod-SH (1) (10 min). Fig. 8A shows the BSA-SS-Rhod adduct (67,376 amu) is dominant 10 min after addition of Rhod-SH (1) while AdhR*-SH was completely converted to AdhR*-SSCys (14,557 amu). Scrambling of the reporter group was not observed, as noted by the arrow pointing to the mass of the undetected Rhod-SH (1) adduct, AdhR*-SS-Rhod. The labelled protein mixture was precipitated in acetone, dissolved in 20 mM Tris, 1% deoxycholate (pH 7.7) and proteolyzed with trypsin for 24 h at 37 °C. The peptides were analysed by reverse phase HPLC/MS with absorbance detection at 565 nm. Two absorbance peaks were observed corresponding to labelled peptides QQC(SSRhod)PF and LQQC(SSRhod)PF from the BSA sequence (Fig. S8, Table S1). 1D SDS-PAGE nonreducing gel electrophoresis was performed on BSA-SNO (52% SNO, negligible free thiol content) and underivatized AdhR*-SH (5 μM, 0.25 nmol total protein each) after thiosulfonate switch labelling. MMTS (4 μL, 20 mM,) was added to quench the excess fluorophore. As shown in Fig. 8B, labelling was selective for the BSA-SNO with the background labelling of AdhR*-SH minimal (<1%). The image shows the actual gel spots with their corresponding molecular weights (Fig. 8B). Coomassie staining of the gel is shown in Fig. S9.
Fig. 8.
Deconvoluted spectrum of thiosulfonate switch labelled (A) BSA-SNO# and AdhR*-SH (50 μM each) and (B) Spot volume of BSA-SNO# and AdhR*-SH (5 μM each) labelled by the thiosulfonate switch and run on a 1D SDS-PAGE nonreducing gel. The image shows the actual gel spots with their corresponding molecular weights. BSA-SNO#= 52% SNO, 0% free thiol.
(ii) Thiosulfonate switch labelling of BSA/AdhR*-SNO
In the reciprocal experiment, a mixture of AdhR*-SNO (53% SNO content, 47% free thiol) and underivatized BSA (50 μM, 5 nmol total protein each) was subjected to the thiosulfonate switch and the deconvoluted spectra are shown in Fig. 9A. Both AdhR*-SSCys (14,557 amu) and AdhR*-SS-Rhod (15,384 amu) were detected as anticipated. BSA-SSCys was also formed and BSA was not labelled by Rhod-SH (1) as evidenced by the absence of BSA-SS-Rhod. The mixture was precipitated in acetone, dissolved in 20 mM Tris, 1 % deoxycholate (pH 7.7) and proteolyzed with trypsin for 24 h at 37 °C. The absorbance peaks observed correspond to the labelled tryptic peptide C(SSRhod)MR and overdigested peptide C(SSRhod)M from the AdhR sequence (Fig S8, Table S1). 1D SDS-PAGE nonreducing gel electrophoresis of a BSA and AdhR*-SNO (53% SNO content, 47% free thiol) mixture (5 μM, 0.25 nmol total protein each) labeled by the thiosulfonate switch shows that labeling was selective for AdhR*-SNO and background labeling of BSA was minimal (~1%) (Fig. 9B). The image shows the actual gel spots with their corresponding molecular weights (Fig. 9B). Coomassie staining of the gel is shown in Fig. S9.
Fig. 9.
(A) Deconvoluted spectrum of thiosulfonate switch labeled BSA and AdhR*-SNO (50 μM each) (B) Spot volume of BSA and AdhR*-SNO (5 μM each) labelled by TST and run on a 1D SDS-PAGE nonreducing gel. The image shows the actual gel spots with their corresponding molecular weights. Buffer = 100 mM ammonium formate, 1 mM EDTA, pH 4.0. AdhR*-SNO = 53% SNO, 47% free thiol.
To further confirm the identity of the AdhR*-SNO adduct, AdhR*-SS-Rhod was proteolyzed by Glu-C, which allows the detection of a larger labelled peptide. AdhR*-SNO (53% SNO, 100 uM) was subjected to labelling by the thiosulfonate switch and the protein was precipitated in acetone and dissolved in 100 mM ammonium bicarbonate, (pH 8.0), deoxycholate (1%), and Glu-C (8 μg/mL) and heated to 37 °C for 16 h. The peptide mixture was analyzed by reverse phase HPLC and detected the labeled peptide FIKC(SSRhod)MRNAGLSIE from the AdhR*-SH sequence (Table S1).
Conclusions
A new method designated as the thiosulfonate switch for protein-S-nitrosothiol conversion to mixed disulfides in mild acidic aqueous solution has been developed. This method utilizes the conversion of S-nitrosothiols to S-phenylthiosulfonates mediated by benzenesulfinate. Both BSA-SNO and AdhR*-SNO are readily converted within minutes into their respective S-phenylsulfonylcysteines through the addition of benzenesulfinic acid sodium salt. The thiol-modified rhodamine fluorescent probe, Rhod-SH (1), upon addition to the protein S-phenylthiosulfonates, provides the corresponding fluorescent mixed disulfides. This robust labelling protocol is compatible with 1D non-reducing SDS PAGE and tryptic proteolysis followed by peptide analysis. The thiosulfonate switch is unique to other S-nitrosothiol detection strategies such as the biotin-switch technique in that it is not reliant upon the reduction of the S-nitrosothiol functionality to a thiol. Therefore, incomplete thiol blocking does not generate false positives, which is a major obstacle to overcome using reductive techniques. Thiols have been shown, however, to react readily with S-phenylsulfonylcysteines and therefore have the potential of diminishing signal. This discovery has been exploited as both a thiol blocking strategy and as a means to convert the thiosulfonate to a mixed disulfide, a more robust reporter group for proteolysis/peptide analysis and gel electrophoresis. The thiol blocking reagent, S-phenyl sulfonylcysteine (SPSC), that efficiently blocks free thiols at pH 4.0 in the presence of S-nitrosothiols, is used to block thiols that possess reactivity and proximity to displace protein-S-phenylsulfonylcysteines. Residual SPSC does not interfere with the subsequent reactions and therefore purification steps are not required. While this study has examined labelling of purified proteins BSA-SNO and AdhR*-SNO, future work will explore the labelling of the S-nitroso proteome of biological systems, including human and bacterial cells, in lysate form. Also, cross reactivity with other cysteine posttranslational modifications such as sulfenic acids and persulfides will be evaluated by global modifications of lysates. On the whole, this study has allowed for new observations in thiol and S-nitrosothiol chemistry, which will aid in developing prospective S-nitrosothiol detection strategies.
Experimental
Synthesis and characterization
(i) Materials
Boc-Cys(Trt)-OSu was purchased from Advanced Chemtech. Adipic anhydride was purchased from Wako Chemicals USA. All other reagents were purchased from Aldrich. Methylene chloride, triethylamine and N-methylmorpholine were distilled over CaH2. Reversed phase HPLC purification was performed with a Waters 2487 Dual l Absorbance detector, 600 controller and pump, and a Phenomenex Synergi 4μ Polar RP 80A HPLC column (250 × 21.2 mm,) using Waters Empower 3 software. Compound 3 was purified by reverse phase HPLC using a Shimadzu SPD-10AV UV-vis detector, a Shimadzu LC-8A pump, a Phenomenex Synergi 4μ Polar RP 80A HPLC column (250 × 30 mm), a Shimadzu SIL-10AP autosampler, and a Shimadzu FRC-10A fraction collector using Shimadzu Lab Solutions software. Compound 9 was purified and control reactions were monitored by reversed phase HPLC using a Shimadzu SPD-M20A prominence diode array detector, an LC-20AB prominence liquid chromotography pump, and a Phenomenex Synergi 4μ Polar RP 80A HPLC column (250 × 10.00 mm) using Shimadzu Lab Solutions software. A gradient of A (0.1 % TFA, H2O) and B (0.1 % TFA, 20:1 CH3CN, H2O) was used in all cases. NMR spectroscopy was performed on Bruker Avance DPX 300 or Bruker Avance DRX 500 NMR spectrometer.
(ii) Synthetic Procedures
S-phenylsulfonylcysteine (SPSC)
The synthesis of SPSC was performed by modifying a literature procedure.41 To a stirred solution of L-cysteine (240 mg, 2.0 mmol) in 2 N HCl (2 mL) was slowly added a solution of NaNO2 (138 mg, 2.0 mmol) in water (0.5 mL). After 40 min at 5 °C the deep red solution was treated with a solution of PhSO2Na (650 mg, 4.0 mmol) in water (4 mL) and stirred for 1 h at 5 °C and 4 h at rt. The precipitate was filtered and washed with water (2 × 4 mL), acetone (2 × 3 mL) and ether (2 × 3 mL). Drying by lyophilization provided 332 mg of SPSC as a white solid (66% yield): 1H NMR (300 MHz, d-DMSO): δ 3.30-3.15 (m, 2H), 3.35-3.41 (m, 1H), 7.66-7.80 (m, 3H), 7.91 (d, 2H, J= 7.7 Hz); 13C NMR (125 MHz, CD3OD): δ 36.36, 53.91, 128.50, 131.08, 135.90, 169.79; HRMS-EI (m/z): calcd for C9H11NO4S2 [M + H]+ 262.0202, found 262.0227.
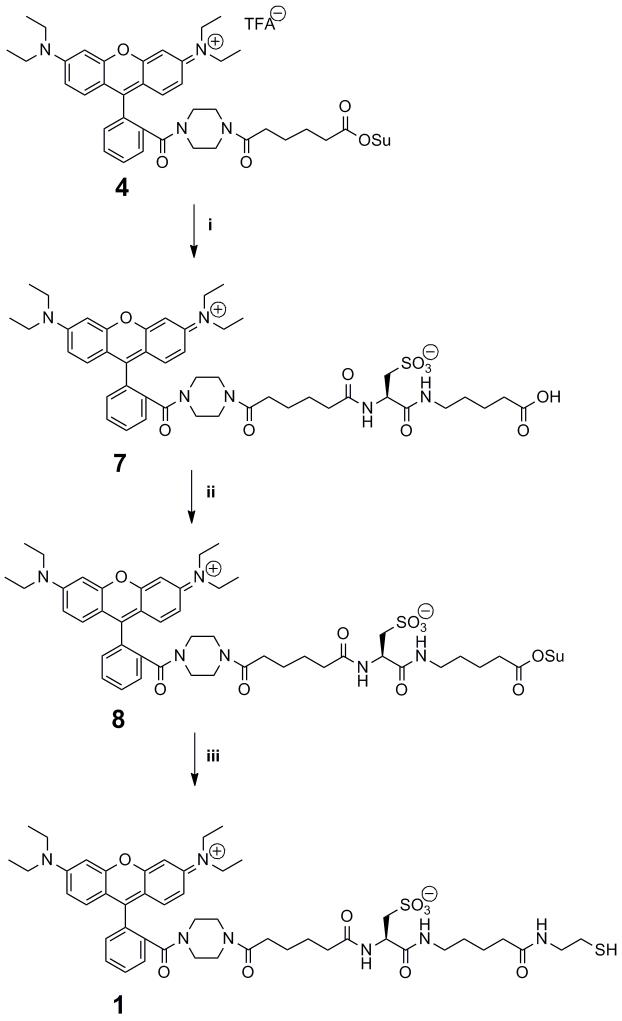
N-(9-(2-(4-(5-carboxypentanoyl)piperazine-1-carbonyl)phenyl)-6-(diethylamino)-3H-xanthen-3-ylidene)-N-ethylethanaminium 2,2,2-trifluoroacetate (3)
The synthesis of N-(6-(diethylamino)-9-(2-(piperazine-1-carbonyl) phenyl)-3H-xanthen-3-ylidene)-N-ethylethanaminium 2,2,2-trifluoroacetate (2) was performed by following a literature procedure.56 To a solution of 2 (2.39 g, 4.37 mmol), adipic anhydride (700.5 mg, 5.47 mmol), and 4,4-dimethylaminopyridine (659 mg, 5.39 mmol) in dry methylene chloride (15 mL) was added dropwise freshly distilled triethylamine (0.65 mL, 4.66 mmol). The solution was stirred for 2 h under Argon at which time the reaction was complete by TLC analysis (Rf= 0.8, 2:1 CH2Cl2/MeOH). The resulting solution was partitioned between 100 mL EtOAc and 150 mL 1 M K2CO3 and washed with 100 mL × 3 EtOAc to remove starting material. NaCl (15 g) was added to the aqueous layer which was then extracted with 2:1 isopropanol / methylene chloride until clear. The organic layer was dried with Na2SO4 and the solvent was removed in vacuo. The resulting red solid was dissolved in 50 mL of chloroform and filtered through a 0.45 mm filter (Whatman, Spartan 13). The solvent was removed in vacuo and the resulting red solid was purified by reverse phase HPLC (B 40% to 65%, 0-12.5 min, 65% to 95%, 12.5-25 min, 95% to 50%, 25-30 min, flow= 25 mL/min, λ= 525 nm, Rt = 14 min, multiple runs) and after lyophilization provided 2.48 g of 3 as a red powder (84 % yield): 1H NMR (300 MHz, CD3OD): δ 1.31 (t, 12H, J = 7.0), 1.59 (m, 4H), 2.30 (m, 2H), 2.37 (m, 2H), 3.39 (m, 8H), 3.69 (q, 8H, J = 7.1 Hz), 6.97 (m, 2H), 7.08 (m, 2H), 7.28 (m, 2H), 7.52 (m, 1H), 7.74 (m, 3H); 13C NMR (125 MHz, CD3OD): δ 12.97, 25.71, 25.87, 33.68, 34.66, 42.88, 47.04, 97.50, 115.00, 115.54, 129.06, 131.44, 131.91, 132.43, 133.31, 136.65, 157.15, 157.35, 159.41, 161.41, 169.69, 174.01, 177.22; HRMS-EI (m/z): calcd for C38H47N4O5+ [M]+ 639.3541, found 639.3528.
N-(6-(diethylamino)-9-(2-(4-(6-((2,5-dioxopyrrolidin-1-yl)oxy)-6-oxohexanoyl)piperazine-1-carbonyl)phenyl)-3H-xanthen-3-ylidene)-N-ethylethanaminium 2,2,2-trifluoroacetate (4)
To an Argon flushed flask cooled to 0 °C containing 3 (141 mg, 0.19 mm) and N-hydroxysuccinimide (430 mg, 3.74 mmol, 20 eq) was added dry DCM (12 mL) followed by dicyclohexylcarbodiamide (386 mg, 1.87 mm, 10 eq). The reaction was complete by TLC analysis (Rf(product) = 0.7, Rf(3)= 0.4, 9:1 CH2Cl2/MeOH) in 2.5 hr at which time the solvent was removed in vacuo. The resulting red solid was purified by reversed phase HPLC (B 40% to 70% over 30 min, flow= 20 mL/min, λ= 530 nm, Rt = 24 min) and after lyophilization provided 134 mg of 4 as a red powder (84%): 1H NMR (500 MHz, CD3OD): δ 1.31 (t, 12H, J = 7.0), 1.67 (m, 2H), 1.74 (m, 2H), 2.40 (t, 2H, J= 7.1 Hz), 2.65 (m, 2H), 2.82 (s, 4H), 3.39 (m, 7H), 3.49 (m, 1H), 3.69 (q, 8H, J = 7.0 Hz), 6.97 (m, 2H), 7.08 (m, 2H), 7.29 (m, 2H), 7.53 (m, 1H), 7.74 (m, 1H), 7.78 (m, 2H); 13C NMR (125 MHz, CD3OD): δ 12.96, 25.37, 25.47, 26.19, 26.63, 31.40, 33.49, 34.85, 42.89, 46.12, 47.04, 97.50, 115.00, 115.38, 115.54, 119.20, 129.06, 131.43, 131.91, 132.44, 133.31, 136.65, 157.17, 157.35, 159.42, 160.27, 160.77, 161.28, 169.72, 170.30, 171.93, 173.79; HRMS-EI (m/z): calcd for C42H50N5O7+ [M]+ 736.3705, found 736.3719.
5-((R)-2-((N-Boc)-amino)-(3-tritylthio)-propionamido)-pentanoic acid (5)
A mixture of 5-aminovaleric acid hydrochloride (1.40 g, 9.11 mmol), Boc-Cys(Trt)-OSu (5.0 g, 8.92 mmol) and Na2CO3 (1.89 g, 17.8 mmol) in H2O (52.5 mL) and 1,4-Dioxane (52.5 mL) was stirred at ambient temperature for 2 d before the solvents were removed in vacuo. The residue was dissolved in H2O (1 L) and acidified to pH 2 using 1 M HCl, at which point a white precipitate fell out of solution. The precipitate was filtered and washed with H2O, followed by drying under vacuum providing 4.40 g of a white solid (88%): FTIR (CH2Cl2): 700 (s), 742 (s), 1166 (s), 1490 (s), 1527 (s), 1708 (s), 2341 (s), 2359 (s), 2931 (s), 2975 (s) 3304 (br); 1H NMR (500 MHz, D2O): δ 1.40 (s, 9H), 1.49-1.62 (m, 4H), 2.28 (t, 2H, J = 7.0 Hz), 2.51-2.59 (m, 2H), 3.15-3.22 (m, 2H), 4.08 (m, 1H), 6.08 (d, 1H, J= 8.0 Hz), 7.23 (t, 3H, J = 7.2 Hz), 7.35 (t, 6H, J = 7.5 Hz,), 7.40 (d, 6H, J = 7.5 Hz,); 13C NMR (125 MHz, CDCl3): δ 22.05, 28.44, 28.86, 33.59, 34.01, 39.25, 53.82, 67.36, 80.67, 127.06, 128.23, 129.75, 144.59, 155.74, 170.87, 177.82; HRMS-EI (m/z): calcd for C32 H38N2O5S [M+Na]+ 585.2399, found 585.2393.
5-((R)-2-amino-sulfono-propionamido)-pentanoic acid (6)
Triethylsilane (4.58 mL, 28.6 mmol) and TFA (40.4 mL) were added to a solution of acid 5 (8.075 g, 14.3 mmol) in anhydrous DCM (40.4 mL) stirred under Argon at ambient temperature for 1.25 h before the solvents were removed in vacuo. The white residue was partitioned between Et2O and H2O. The aqueous phase was removed in vacuo providing a yellow oil. A solution of 30% H2O2 (22.6 mL) and formic acid (203.4 mL) were allowed to stand together at ambient temperature for 1 h. The solution was used to dissolve the yellow oil, at which point the mixture was stirred at ambient temperature for 1.5 h. The solvent was removed in vacuo, and the white solid was dissolved in water. The water was removed in vacuo and the white solid was dried by lyophilization providing 3.38 g of 6 (81%): 1H NMR (500 MHz, D2O): δ 1.49-1.58 (m, 4H), 2.35 (t, 2H, J = 7.0 Hz), 3.21 (m, 2H), 3.31 (m, 1H), 3.39 (m, 1H), 4.30 (dd, 1H. J= 8.7, 4.1 Hz); 13C NMR (125 MHz, D2O): δ 21.48, 27.42, 33.18, 39.29, 49.95, 50.26, 167.15, 178.57; HRMS-EI (m/z): calcd for C8H16N2O6S [M+Na]+ 291.0627, found 291.0609.
(R)-3-((4-carboxybutyl)amino)-2-(6-(4-(2-(6-(diethylamino)-3-(diethyliminio)-3H-xanthen-9-yl)benzoyl)piperazin-1-yl)-6-oxohexanamido)-3-oxopropane-1-sulfonate (7)
To an Argon flushed flask containing 6 (424 mg, 1.58 mm) was added dry DMF (15 mL) followed by NMM (0.34 mL, 3.16 mmol). A solution of 4 (134 mg, 0.158 mmol) in dry DCM (5 mL) was added dropwise to the solution and stirred. The reaction was complete by TLC analysis (Rf(product) = 0.0, Rf(4) = 0.7, 9:1 CH2Cl2/MeOH) after 23 h. Removal of the solvent by lyophilization gave rise to a red solid which was purified by reverse phase HPLC (B 40% to 70% over 30 min, flow= 20 mL/min, λ= 530 nm, Rt= 10 min) and after lyophilization afforded 87 mg of 7 as a red powder (62% yield): 1H NMR (300 MHz, CD3OD): δ 1.31 (t, 12H, J = 7.0 Hz), 1.58 (m, 8H), 2.31 (m, 6H), 3.17 (m, 4H), 3.38 (m, 7H), 3.54 (m, 1H), 3.69 (q, 8H, J = 7.0 Hz), 4.60 (m, 1H), 6.97 (s, 2H), 7.09 (m, 2H), 7.30 (d, 2H, J = 9.5 Hz), 7.51 (m, 1H), 7.76 (m, 3H); 13C NMR (125 MHz, CD3OD): δ 13.02, 23.40, 25.95, 26.11, 29.84, 33.60, 33.90, 34.60, 36.51, 40.27, 42.86, 43.33, 47.06, 52.74, 52.92, 97.47, 115.00, 115.71, 129.15, 131.39, 131.47, 131.87, 132.34, 133.35, 136.72, 157.34, 159.40, 169.67, 172.84, 174.24, 175.78, 177.26; HRMS-EI (m/z): calcd for C46H60N6O10S [M+Na]+ 911.3984, found 911.3972.
(R)-2-(6-(4-(2-(6-(diethylamino)-3-(diethyliminio)-3H-xanthen-9-yl)benzoyl)piperazin-1-yl)-6-oxohexanamido)-3-((5-((2,5-dioxopyrrolidin-1-yl)oxy)-5-oxopentyl)amino)-3-oxopropane-1-sulfonate (8)
To an Argon flushed flask containing 7 (38 mg, 43 mmol) and N-hydroxysuccinimide (104 mg, 0.9 mmol) was added dry DMF (2 mL) followed by DCC (93 mg, 0.45 mmol, 10 eq) and stirred. The reaction was complete by TLC analysis (Rf(product) = 0.7, Rf(8) = 0.4, 3:1 CH2Cl2/MeOH) after 17 h at which time the solvent was removed by lyophilization. The resulting red solid was purified by reverse phase HPLC (B 40% to 70% over 30 min, flow= 4 mL/min, λ= 530 nm, Rt= 12 min) and after lyophilization afforded 38 mg of 9 as a red powder (91% yield): 1H-NMR (300 MHz, CD3OD) δ 1.31 (t, 12H, J = 7.0 Hz), 1.60 (m, 6H), 1.71 (m, 2H), 2.27 (m, 2H), 2.35 (m, 2H), 2.64 (t, 2H, J = 7.0 Hz), 2.81 (s, 4H), 3.19 (m, 4H), 3.39 (m, 7H), 3.54 (m, 1H), 3.69 (m, 8H), 4.60 (m, 1H), 6.96 (s, 2H), 7.13 (m, 2H), 7.30 (d, 2H, J= 9.4 Hz), 7.51 (m, 1H), 7.77 (m, 3H); 13C-NMR (125 MHz, CD3OD): δ 13.01, 23.09, 25.90, 26.12, 26.65, 29.46, 31.33, 33.64, 33.90, 34.85, 36.47, 39.93, 42.21, 42.91, 43.31, 46.22, 47.06, 52.69, 52.87, 97.46, 115.01, 115.72, 129.16, 131.48, 131.88, 132.34, 132.56, 133.36, 136.74, 157.35, 159.42, 160.20, 169.66, 170.33, 171.93, 172.93, 174.25, 175.78; HRMS-EI (m/z): calcd C50H63N7O12S [M+2Na]+2 515.7020, found 515.7041.
(R)-2-(6-(4-(2-(6-(diethylamino)-3-(diethyliminio)-3H-xanthen-9-yl)benzoyl)piperazin-1-yl)-6-oxohexanamido)-3-((5-((2-mercaptoethyl)amino)-5-oxopentyl)amino)-3-oxopropane-1-sulfonate (Rhod-SH, 1)
Argon was bubbled through dry DMF (1 mL) for 20 min at which time NHS ester 8 (19 mg, 0.019 mmol) was added followed by cysteamine hydrochloride (3.2 mg, 0.028 mmol). Then NMM (6 μL, 0.127 mmol) was added and the solution was stirred for 23 h under Argon at which time the solvent was removed by lyophilization and the resulting red solid was purified via reverse phase HPLC (30% B to 60% B over 30 min, 20 mL/min flow, λ = 530 nm, product Rt = 19 min) and after lyophilization provided 11 mg of 1 (60% yield) as a red powder: 1H NMR (500 MHz, CD3OD): δ 1.31 (t, 12H, J = 6.8 Hz), 1.51 (m, 2H), 1.61 (m, 6H), 2.19 (m, 2H), 2.27 (m, 2H), 2.37 (m, 2H), 2.58 (m, 2H), 3.19 (m, 4H), 3.40 (m, 8H), 3.53 (m, 2H), 3.69 (m, 8H), 4.61 (m, 1H), 6.96 (s, 2H), 7.08 (m, 1H), 7.13 (m, 1H), 7.29 (d, 2H, J = 9.4 Hz), 7.52 (m, 1H), 7.73 (m, 1H), 7.78 (m, 2H); 13C NMR (125 MHz, CD3OD): δ 13.02, 24.35, 24.62, 25.93, 26.14, 29.79, 33.64, 33.90, 36.57, 36.72, 40.18, 42.23, 42.92, 43.35, 44.04, 46.22, 47.07, 52.78, 97.49, 115.03, 115.62, 115.73, 129.17, 131.39, 131.49, 131.88, 132.55, 133.37, 136.76, 157.39, 159.43, 169.71, 172.85, 174.26, 175.76, 176.24; HRMS-EI (m/z): calcd C48H65N7O9S2 [M + H]+ 948.4358, found 948.4363.
(iii) Optical Characterization of Rhod-SH, (1)
Absorbance and emission profiles for Rhod-SH (1), were determined using both a Varian Cary 6000i UV-VIS NIR Spectrophotometer and a Varian Cary Eclipse Fluorescence Spectrophotometer. Ethanol was used as the spectroscopic solvent, utilizing 1 cm cuvettes for both absorbance and emission readings. Molecular extinction coefficient values were determined by finding the slope of the concentration versus absorbance at λmax (562 nm, 86,000 M−1cm−1). The quantum yield was determined in triplicate by comparing 1 to Rhodamine B (510 nm excitation, QY= 70%).57 Dilution series’ were made with a maximum absorbance of 0.1 at 510 nm. The quantum yield was determined to be 48±0.1%. Fig. S10 shows the absorbance and emission profiles for 1.
Control reactions
(i) Reactions of PhSO2Na and Rhod-SH (1) on GSNO, GSH and GSSG
GSNO was synthesized and characterized using a previously developed method.38,41 All solutions were in pH 4.0 buffer (100 mM ammonium formate, 1 mM EDTA). GSNO (20 μL, 1 mM) was added to PhSO2Na (50 μL, 0.1 M) and incubated for 30 min at which time Rhod-SH (1) (4 μL, 5 mM) was added. After 10 min, an aliquot was taken and analyzed by reversed phase HPLC with absorbance detection at λmax of Rhod-SH (1) (B 5% to 70% 0-20 min, 70% to 100%, 20-30 min, flow= 4 mL/min, λ= 562 nm). Rhod-SH (1) exhibited a retention time of 18.9 min. The reaction with GSNO results in a new peak with a retention time of 17.3 min (95%) while the peak at 18.9 min is drastically diminished. HRMS analysis of the 17.3 min peak confirmed the product to be the mixed disulfide Rhod-SSG (HRMS-EI (m/z): calcd C58H81N10O15S3+ [M]+ 1254.5040 found 1254.5031.
(ii) Rhod-SH (1) control reactions with thiol blockers and disulfides
Thiol blocking reactions were performed at pH 4.0 (100 mM HCO2NH4, 1 mM EDTA) between 0.27 mM Rhod-SH (1) and 0.27 mM thiol blocker (SPTC, MMTS, NEM, IAM, GSSG, or (Gly-Cys)2). The reactions were monitored by taking aliquots at given time points and analysing by reverse phase HPLC (B 5% to 70% 0-20 min, 70% to 100%, 20-30 min, flow= 4 mL/min, λ= 562 nm). The products were further confirmed by HRMS: Rhod-SSCys: Rt= 17.7 min; HRMS (m/z): calc for C51H70N8O11S3 [M+H]+ 1067.4399, found 1067.4375; Rhod-SSMe: Rt= 20.2 min; HRMS (m/z): calc for C49H67N7O9S3 [M+H]+ 994.4235, found 994.4200; Rhod-S-NEM: Rt= 19.5 min; HRMS (m/z): calc for C54H72N8O11S2 [M+H]+ 1073.4835, found 1073.4808.
(iii) SPSC control reactions
SPSC (2mM) in the following buffers were analyzed by reverse phase HPLC (B 5% to 70% 0-20 min, 70% to 100%, 20-30 min, flow= 4 mL/min, λ= 265 nm) after 1 h: pH 4.0 and 4.8: 100 mM ammonum formate, 1 mM EDTA; pH 5.5, 6.0: 100 mM MES, 1 mM EDTA; pH 7.0: 100 mM HEPES, 1 mM EDTA.
Protein Analysis
(i) Reactions of BSA-SNO and PhSO2Na and Rhod-SH (1)
To an amber vial containing a solution of BSA-SNO (44% SNO content, 100 μM total protein, 100 μL, 10 nmol, pH 4.0 buffer) was added PhSO2Na (5 μL, 0.5 M, 2.5 μmol, pH 4.0 buffer, 250 eq) followed by Rhod-SH (1) (2 μL, 10 mM, pH 4.0 buffer, 20 nmol, 2 eq). After 30 min, TOF MS analysis was performed (Fig. 3, blue, middle). A control reaction was performed under identical conditions employing commercial BSA and analyzed by TOF MS (Fig. 3, red, top). The BSA-SS-Rhod solution was subjected to trypsinolysis (see below).
(ii) Thiosulfonate switch labeling of BSA-SNO
To an amber vial containing a solution of BSA-SNO (22% SNO content, 28% free thiol, 100 μM total protein, pH 4.0 buffer, 100 μL, 10 nmol) was added S-phenylthiosulfonylcysteine (SPSC) (10 μL, 1 mM, pH 4.0 buffer, 10 nmol). After 1 h, PhSO2Na (5 μL, 0.5 M, pH 4.0 buffer, 2.5 μmol, 250 eq) was added. After 30 min, Rhod-SH (1) (3 μL, 10 mM, pH 4.0 buffer, 30 nmol, 3 eq) was added and after 10 min, TOF MS analysis was performed (Fig. 3, green, bottom).
(iii) TOF MS analysis
Intact-protein mass spectra were collected via HPLC-ESI on a Bruker microTOF. A Phenomenex size-exclusion column (Phenomenex S2000, 300x4.6 mm) was used in denaturing conditions to separate the proteins from buffer and small molecules. The solvent consisted of 50% water, 50% acetonitrile, with 0.1% formic acid, at a flow rate of 1.0 mL/min: the protein eluted in a wide band at approximately 5 minutes.
(iv) Trypsinolysis and peptide analysis on BSA-SS-Rhod
The BSA-SS-Rhod solution prepared and analyzed in Fig. 3 (blue, middle) was buffer exchanged with Sephadex G25 to HEN (pH 7.5). To 80 μL of BSA-SS-Rhod (pH 7.5) in an amber vial was added MMTS (2 μL, 20 mM in DMF, 40 nmol). Trypsin (Trypsin Gold, Promega, 2 μL, 0.1 mg/mL in water) was added and the solution was heated to 37 °C for 10 h in the dark. The solution was centrifuged and the supernatant was analyzed by reverse phase HPLC in tandem with MS and UV/vis detection (Fig. 4). Before peptide analysis, TOF MS analysis was performed using a Biosep column to ensure protein digestion. Digested samples were eluted through an Agilent Zorbax reverse phase column with 1.8 μm particles and dimensions of 50 mm length and 2.1 mm diameter with an Agilent 1290 HPLC system and detected by UV-Vis and mass spectrometry. The column was maintained at 50 °C with a flow rate of 500 μL/min. Chromatography was as follows: solvent consisted of H2O with 0.1% (v/v) formic acid for channel “A” and acetonitrile with 0.1% formic acid for channel “B”. Following column equilibration at 5% B, the sample was injected via autosampler, and the column was flushed for 1.0 min to waste. From 1.0 min to the end of the run, the column eluant was directed to the MS source. From 1.0 min to 7.0 min, the gradient was linearly ramped from 5% to 95% B. From 7.0 to 9.0 min, the gradient was held at 95%, and from 9.0 to 10.0 minutes the gradient was held at 5% for re-equilibration. A UV detector was used in-line following the column and prior to the mass spectrometer. The mass spectrometer used was a Bruker microTOF with ESI source: resolution is approximately 10,000 and accuracy is 10 ppm (Table S1). Source parameters were as follows: drying gas 5.5 L/min, nebulizer 4.0 bar, capillary voltage 4500 V, capillary exit 100 V. Spectra were collected in positive mode from 50 to 1600 m/z at a rate of 1 Hz. The compounds of interest were detected using a combination of UV detection at 565 nm and MS ion intensities. Compound identification was confirmed via accurate-mass isotope distribution matches compared to calculated values based upon theoretical formulas (Table S1).
(v) 1D SDS-PAGE nonreducing gels on BSA-SS-Rhod
A solution of BSA-SNO (36% SNO content, pH 4.0 buffer) was diluted with a solution of BSA to generate a dilution series with identical amounts of total protein. To each vial was added SPSC (8 μL, 100 μM) and after 1 h, PhSO2Na (5 μL, 0.5 M, pH 4.0 buffer, 2.5 μmol) was added. After 30 min, Rhod-SH (1) (4 μL, 0.5 mM, pH 4.0 buffer, 2 nmol) was added and after 10 min, MMTS (4 μL, 20 mM, pH 4.0 buffer, 80 nmol) was added. 15 μL of the final reaction solutions were added to 5 μL non-reducing sample buffer. For each lane, 15 μL were added and the gel was run with Tris-HEPES-SDS buffer pH 8.0 at 100 V through a 4-20% Precise Protein Gel from Thermo Scientific using a Biorad Power Pac HC and mini protean tetracell. Precision plus all blue standards from Biorad were used as molecular weight markers. Gels were washed for 1 h in distilled water and fluorescence images were acquired on a Typhoon 9210 (GE Healthcare) scanning at 400 PMT, 100 micron, with excitation and emission filters at 532/580 nm. Image J was used to determine relative fluorescence intensities. After imaging, the gel was stained with coomassie.
(vi) Bacterial expression and purification of S. pneumoniae D39 NmlR53 and B. subtilis AdhR54
The nmlR gene was amplified from S. pneumoniae D39 genomic DNA (locus tag SPD_1637) using primers 5′AGGTCATATGAATATTAAATCTGCCAGTG-3′ and 5′-CCGGATCCCATTTAAAATTTTCCTTC-3′ containing NdeI and BamHI restriction sites (underlined). The adhR gene was amplified from B. subtilis str. 168 genomic DNA using primers 5′GGGCATATGAATATTGCTCAGGTGGCAAAG-3′ and GGGGGGGATCCCTACCTTCT-TTGCTCCACTGATCC also containing NdeI and BamHI restriction sites. Following double digestion and ligation into pET-3a, all plasmids were confirmed to contain the desired gene by DNA sequencing. Bacterial expression of NmlR and AdhR proteins was carried out in E. coli BL21(DE3)/pLysS essentially as described previously.58 Briefly, cell pellets were resuspended in a lysis buffer containing 25 mM Tris, 1 M NaCl, 2 mM DTT, 2 mM EDTA, pH 8.0, lysed via sonication, nucleic acids precipitated by polyethyleneimine (PEI) addition, followed by protein precipitation of the PEI supernatant by ammonium sulfate essentially as described previously.59 Following ammonium sulfate precipitation, the pellet was resuspended and dialyzed into Buffer A (25 mM Tris-HCl, 2 mM EDTA, 1 mM DTT, pH 8.0) supplemented with 50 mM NaCl, loaded onto an in-house packed Q (quaternary ethylamine)-column equilibrated with the same buffer and eluted by running a linear gradient in Buffer A plus 750 mM NaCl (4 mL fractions were collected). The highest purity fractions as assessed by SDS-PAGE were pooled, concentrated, and dialyzed again into Buffer A/50 mM NaCl, loaded onto a MonoQ column equilibrated with the same buffer, and eluted with Buffer A/750 mM NaCl over a 25-30 column volume gradient, collecting 2 mL fractions. Highest purity fractions were again pooled, concentrated, and further purified by gel filtration chromatography in either a G75 or G200 column 16/60 (GE Life Sciences), equilibrated with Buffer G (25 mM Hepes, 200 mM NaCl, 2 mM EDTA, 1 mM DTT, pH 7.0), collecting 1 mL fractions. Fractions exhibiting >95% purity as judged by SDS-PAGE were combined, concentrated, and stored in Buffer G at −80 °C. Molecular masses determined by ESI MS or MALDI MS for NmlR and AdhR were 13,462 D (13,463.4 D expected; residues 1-117) and 14,438 D (16,415.8 D expected), respectively. Tandem MS/MS of purified AdhR reveals a C-terminal tryptic peptide R|ENEAKL123 giving an expected molecular mass of 14,438.6 D (14,438 D observed; residues 1-123). We designate this C-terminally truncated form of AdhR as AdhR*-SH. This C-terminal tail is not found in Spn NmlR (117 residues) and both reduced AdhR*-SH and NmlR bind to a 30-base pair nml operator promoter-containing DNA (NmlO) with high affinity (Fig. S5). NmlR and AdhR*-SH are 46% identical over 117 residues and both contain a single Cys, Cys52. Following gel filtration, NmlR and AdhR*-SH stocks were incubated with 5 mM TCEP, and then dialyzed anaerobically into a degassed buffer containing 10 mM Hepes, 200 mM NaCl, pH 7.0. The reduced thiol content (1 expected) was verified using a DTNB assay carried out as previously described60 and was found to be greater than 80% in all preparations.
(vii) (ix) NmlR and AdhR*-SH C52 reactivity experiments
GSNO was either purchased (Axxora) or synthesized using a procedure also used to prepare S-nitrosylated cysteine (CysNO). Briefly, a 1.1:1 ratio of sodium nitrite to cysteine or glutathione was mixed in 1 M HCl in darkness and under argon.41,61 Following a 15 min reaction, a 10-fold volumetric excess of ice cold acetone was added to the solution to precipitate CysNO or GSNO. The precipitate was filtered and then washed with water, acetone, and finally anhydrous diethyl ether. The resulting pink powder was allowed to dry overnight in darkness. The powder was then resuspended in either 20 % DMSO or reaction buffer and stored at −80 °C until use. GSNO and CysNO stock concentrations were estimated using the ε340 nm value for GSNO of 1000 cm−1 M−1.61 GSNO and CysNO stocks were aliquoted into 100-200 μL aliquots into either 20% DMSO or reaction buffer and stored at −80 °C until used. C52 reactivity experiments were conducted by adding a 10-fold (GSNO) or 15-fold (CysSNO) molar excess of NO donor to 1 mM (AdhR*-SH) or 0.1 mM (NmlR) in 0.4 mL (AdhR*-SH) or 4 mL (NmlR), respectively, in Buffer R (25 mM Hepes, 400 mM NaCl, 1 mM EDTA) containing 0.1 mM BCS, the latter used to chelate any Cu(I) formed during the reaction.61 Reactions were allowed to proceed for 1 h at room temperature anaerobically with rotational mixing on a LabQuake. A HiPrep 26/10 desalting column was run in Buffer R and the absorbance at 280 nm, 338 nm, and 575 nm monitored in darkness (Fig. S6A). At the conclusion of the run, NmlR-SNO and AdhR*-SNO fractions were concentrated, and the UV-Vis absorbance spectrum measured (Fig. S6B). The percentage of S-NO reacted thiol was estimated by the 280 nm/340 nm ratio (see main text). An NmlR prep reacted with GSNO was additionally injected onto a μRPC C2/C18 column (GE Life Sciences) reverse phase column with buffers of 0.1% formic acid (Buffer A) and 0.1% formic acid, 60% acetonitrile (Buffer B), with reduced, S-glutathionylated and S-nitrosylated NmlRs (NmlR-SNO) eluted with a very shallow gradient (Fig. S7). All C52 adducts of NmlR and AdhR*-SH were routinely analyzed by ESI/MS equipped with either a C4 or C8 column with buffers of 0.1% formic acid (Buffer A) and 90% acetonitrile, 0.1% formic acid (Buffer B), running either a 7 minute or 20 minute gradient from 10% to 90% Buffer B.
(viii) AdhR*-SNO Reduction with Ascorbate
To an amber vial containing AdhR*-SNO (53% SNO, 47% free thiol, 25 μM total protein, HEN, pH 7.5, 100 μL, 3 nmol) was added ascorbate (1 M, 2 μL, 2 μmol). After 1 h, the solution was analyzed by TOF MS (Fig. 6).
(ix) Thiosulfonate switch labelling of AdhR*-SNO
To an amber vial containing AdhR*-SNO (53% SNO, 47% free thiol, 25 μM total protein, pH 4.0 buffer, 8 μL, 0.2 nmol) was added SPSC (4 μL, 130 μM, pH 4.0 buffer, 0.5 nmol). After 1 h, PhSO2Na (5 μL, 0.25 M, pH 4.0 buffer, 1.25 μmol) was added and after 30 min, Rhod-SH (1) (4 μL, 0.5 mM, pH 4.0 buffer, 2 nmol) was added. After 10 min, the solution was analyzed by TOF MS (Fig. 7, red, top). AdhR*-SH was subjected to the same procedure and analysis (Fig. 7, blue, bottom).
(x) Thiosulfonate switch labelling of protein mixtures
BSA-SNO (52% SNO, 0% free thiol) and AdhR*-SH were mixed in an amber vial to 50 μM total protein each (100 μL total, pH 4.0 buffer, 5 nmol each). To the protein mixture was added SPSC (10 μL, 1 mM, pH 4.0 buffer, 10 nmol). After 1 h, PhSO2Na (5 μL, 0.5 M, pH 4.0 buffer, 2.5 μmol) was added and after 30 min, Rhod-SH (1) (2 μL, 10 mM, pH 4.0 buffer, 20 nmol) was added. After 10 min, the solution was then analyzed by TOF MS (Fig. 8A). 80 μL of the protein mixture was precipitated in cold acetone (240 μL) and kept at −20 °C for 1 h. After centrifugation, the acetone was removed and the resulting pellet was dissolved in 20 mM Tris (1% deoxycholate, pH 7.7, 80 μL). Trypsin (5 μL, 1 mg/mL, sigma-Aldrich) was added and the vial was heated to 37 °C for 24 h. After cooling to room temperature, formic acid (1 M) was added until a pH of 3.0 was reached and the mixture was centrifuged to separate the precipitated deoxycholate. The supernatant was analyzed by reverse phase HPLC in tandem with MS and UV/vis detection. Detection of peptide products for the mixing experiments (Fig. S8) was performed in the same manner as the BSA-SS-Rhod digest, except with longer column and chromatography to ensure accurate detection of multiple peptides. An Agilent Zorbax C18 column with 1.8 μm particles and dimensions of 100 mm length and 2.1 mm diameter was used for separation, with an Agilent 1290 HPLC system. The column was maintained at 50 °C with a flow rate of 600 μL/min. Chromatography was as follows: solvent consisted of H2O with 0.1% (v/v) formic acid for channel “A” and acetonitrile with 0.1% formic acid for channel “B”. Following column equilibration at 20% B, the sample was injected via autosampler, and the column was flushed for 1.0 min to waste. From 1.0 min to the end of the run, the column eluant was directed to the MS source. From 1.0 min to 26.0 min, the gradient was linearly ramped from 20% to 45% B. From 26.0 to 28.0 min, the gradient was held at 95%, and from 28.0 to 30.0 minutes the gradient was held at 20% for re-equilibration. The higher percentage of “B” in this experiment was tailored for the specific retention time of dye-labeled peptides. A UV detector was used in-line following the column and prior to the mass spectrometer. The mass spectrometer used was an Agilent 6538 QToF with ESI source: resolution is approximately 20,000 and accuracy is 1 ppm. Source parameters were as follows: drying gas 8.0 L/min, nebulizer 55 psi, capillary voltage 3500 V, and capillary exit 120 V. Spectra were collected in positive mode from 50 to 1700 m/z at a rate of 1 Hz. The compounds of interest were detected using a combination of UV detection at 565 nm and MS ion intensities. Compound identification was confirmed via accurate-mass isotope distribution matches compared to calculated values based upon theoretical formulas (Table S1). In a parallel experiment, a mixture of AdhR*-SNO (47% SNO, 53% SH) and commercial BSA (50 μM total protein each, 100 μL total, pH 4.0 buffer) was subjected to the entire protocol and analysis.
(xiii) 1D SDS-PAGE nonreducing gels on protein mixtures subjected to labeling by the thiosulfonate switch
To an amber vial was added BSA-SNO (52% SNO, 25 μM total protein, pH 4.0 buffer, 10 μL, 0.25 nmol) and AdhR*-SH (25 μM total protein, 10 μL, 0.25 nmol) followed by 15 μL pH 4.0 buffer. SPSC (4 μL, 0.1 mM, pH 4.0 buffer, 0.4 nmol) was added. After 1 h, PhSO2Na (5 μL, 0.5 M, pH 4.0 buffer, 2.5 μmol) was added and after 30 min, Rhod-SH (1) (2 μL, 0.5 mM, pH 4.0 buffer, 1 nmol) was added. After 10 min, MMTS (4 μL, 20 mM, pH 4.0 buffer, 80 nmol) was added followed by non-reducing sample buffer. Image J was used to determine relative fluorescence intensities (Fig. 8B). In a parallel experiment, AdhR*-SNO (47% SNO, 53% SH) and commercial BSA was subjected to the entire protocol (Fig. 9B).
Supplementary Material
Scheme 5.

Synthesis of S-phenylsulfonylcysteine (SPSC).
Scheme 6.
Synthesis of compounds 2-4.
Scheme 7.
Synthesis of cysteic acid side chain 6. i. Boc-Cys(Trt)-OSu, Na2CO3, aq. dioxane, rt, 2 d, 88%, ii. Et3SiH, TFA, CH2Cl2, rt, 1 h, iii. HCO2H, H2O2, °C, 1 h, 81%, 2 steps.
Scheme 8.
Synthesis of Rhod-SH (1). i. 6, NMM, DMF, 23 h, 62%, ii. DCC, NHS, DMF, 17 h, 91%, iii. cysteamine hydrochloride, NMM, DMF, 22 h, 61%.
Acknowledgements
P.A.G., D.J.S., And E.A.D. Receive Support From The National Institutes Of Health (Cobre 5P20Rr024237-04 And P20Gm104935-05). D.P.G. Receives Support From Nih Grants Gm042569 And Gm097225. We Would Like To Acknowledge David Fouchard For Synthetic Support. The Mass Spectrometry, Proteomics And Metabolomics Core Facility Receives Support From The Murdock Charitable Trust And Inbre Mt Grant No. P20 Rr-16455-08.
Footnotes
Electronic Supplementary Information (ESI) available: [Fig. S1-Fig. S10 and 1H and 13C NMRs of new compounds]. See DOI: 10.1039/c000000x/
references
- 1.Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Nat. Rev. Mol .Cell. Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 2.Stamler JS, Simon DI, Osborne JA, Mullins ME, Jaraki O, Michel T, Singel DJ, Loscalzo J. Proc. Natl. Acad. Sci. USA. 1992;89:444–448. doi: 10.1073/pnas.89.1.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Broniowska KA, Hogg N. Antioxid. Redox. Sign. 2012;17:969. doi: 10.1089/ars.2012.4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miersch S, Mutus B. Clin. Biochem. 2005;38:777–791. doi: 10.1016/j.clinbiochem.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 5.Marozkina NV, Gaston B. BBA – Gen. Subjects. 2012;6:722–729. doi: 10.1016/j.bbagen.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foster MW, Hess DT, Stamler JS. Trends. Mol. Med. 2009;15:391–404. doi: 10.1016/j.molmed.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anand P, Stamler JS. J. Mol. Med. 2012;90:233–244. doi: 10.1007/s00109-012-0878-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marshall HE, Merchant K, Stamler JS. FASEB J. 2000;14:1889–1900. doi: 10.1096/fj.00.011rev. [DOI] [PubMed] [Google Scholar]
- 9.Sha Y, Marshall HE. BBA-Gen. Subjects. 2012;6:701–711. doi: 10.1016/j.bbagen.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singel DJ, Stamler JS. Annu. Rev. Physiol. 2005;67:99–145. doi: 10.1146/annurev.physiol.67.060603.090918. [DOI] [PubMed] [Google Scholar]
- 11.Lipton SA, Choi Y-B, Pan Z-H, Lei SZ, Chem H-SV, Sucher NJ, Loscalzo JL, Singel DJ, Stamler JS. Nature. 1993;364:626–632. doi: 10.1038/364626a0. [DOI] [PubMed] [Google Scholar]
- 12.Shahani N, Sawa A. BBA-Gen. Subjects. 2012;6:736–742. doi: 10.1016/j.bbagen.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hess DT, Stamler JS. J. Biol. Chem. 2012;287:4411–4418. doi: 10.1074/jbc.R111.285742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Z. Cancer Lett. 2012;320:123–129. doi: 10.1016/j.canlet.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 15.Maron BA, Tang S-S, Loscalzo JL. Antioxid. Redox. Sign. 2012;18:270. doi: 10.1089/ars.2012.4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chung KKK, David KK. Nitric Oxide. 2010;22:290–295. doi: 10.1016/j.niox.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 17.Nakamura T, Lipton SA. Antioxid. Redox. Sign. 2008;10:87. doi: 10.1089/ars.2007.1858. [DOI] [PubMed] [Google Scholar]
- 18.Seth D, Hausladen A, Wang YJ, Stamler JS. Science. 2012;336:470–3. doi: 10.1126/science.1215643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gusarov I, Shatalin K, Starodubtseva M, Nudler E. Science. 2009;325:1380–4. doi: 10.1126/science.1175439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, H S. Snyder, Nat. Cell Biol. 2001;3:193–197. doi: 10.1038/35055104. [DOI] [PubMed] [Google Scholar]
- 21.Forrester MT, Foster MW, Benhar M, Stamler JS. Free Radical Bio. Med. 2009;46:119–126. doi: 10.1016/j.freeradbiomed.2008.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chouchani ET, Hurd TR, Nadtochiy SM, Brookes PS, Fearnley IM, Lilley KS, Smith RAJ, Murphy MP. Biochem. J. 2010;430:49–59. doi: 10.1042/BJ20100633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Santhanam L, Gucek M, Brown TR, Mansharamani M, Ryoo S, Lemmon CA, Romer L, Shoukas AA, Berkowitz DE, Cole RN. Nitric Oxide. 2008;19:295–302. doi: 10.1016/j.niox.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han P, Zhou X, Huang B, Zhang X, Chen C. Anal. Chem. 2008;377:150–155. doi: 10.1016/j.ab.2008.03.023. [DOI] [PubMed] [Google Scholar]
- 25.Forrester MT, Thompson JW, Foster MW, Nogueira L, Moseley MA, Stamler JS. Nature Biotechnol. 2009;27:557–559. doi: 10.1038/nbt.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Foster MW, Forrester MT, Stamler JS. Proc. Natl. Acad. Sci. USA. 2009;45:18948. doi: 10.1073/pnas.0900729106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hao G, Derakhshan B, Shi L, Campagne F, Gross SS. Proc. Natl. Acad. Sci. USA. 2006;103:1012–1017. doi: 10.1073/pnas.0508412103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Derakhshan B, Wille PC, Gross SS. Nature Protoc. 2007;2:1685–1691. doi: 10.1038/nprot.2007.210. [DOI] [PubMed] [Google Scholar]
- 29.Doulias P-T, Greene JL, Greco TM, Tenopoulou M, Seeholzer SH, Dunbrack RL, Ischiropoulos H. Proc. Natl. Acad. Sci. USA. 2010;107:16958. doi: 10.1073/pnas.1008036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doulias P-T, Tenopoulou M, Greene JL, Raju K, Ischiropoulos H. Sci. Signal. 2013;6:rs1. doi: 10.1126/scisignal.2003252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang H, Xian M. Angew. Chem. Int. Ed. 2008;47:6598–6601. doi: 10.1002/anie.200801654. [DOI] [PubMed] [Google Scholar]
- 32.Pan J, Xian M. Chem. Commun. 2011;47:352–354. doi: 10.1039/c0cc02076a. [DOI] [PubMed] [Google Scholar]
- 33.Zhang J, Li S, Zhang D, Wang H, Whorton AR, Xian M. Org. Lett. 2010;12:4208–4211. doi: 10.1021/ol101863s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang D, Devarie-Baez O, Pan J, Wang H, Xian M. Org. Lett. 2010;12:5674. doi: 10.1021/ol102491n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang H, Zhang J, Xian M. J. Am. Chem. Soc. 2009;131:13238–13239. doi: 10.1021/ja905558w. [DOI] [PubMed] [Google Scholar]
- 36.Bechtold E, Reisz JA, Klomsiri C, Tsang AW, Wright MW, Poole LB, Furdui CM, King SB. ACS Chem. Biol. 2010;5:405–414. doi: 10.1021/cb900302u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seneviratne U, Godoy LC, Wishnok JS, Wogan GN, Tannenbaum SR. J. Am. Chem. Soc. 2013;135:7693–7704. doi: 10.1021/ja401565w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reeves BD, Hilmer JK, Mellmann L, Hartzheim M, Poffenberger K, Johnson K, Joshi N, Singel DJ, Grieco PA. Tet. Lett. 2013;54:5707–5710. doi: 10.1016/j.tetlet.2013.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DuMond JF, King SB. Antioxid. Redox. Sign. 2011;14:1637–1648. doi: 10.1089/ars.2010.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kirsch M, Buscher A-M, Aker S, Schulz R, de Groot H. Org. Biomol. Chem. 2009;7:1954–1962. doi: 10.1039/b901046g. [DOI] [PubMed] [Google Scholar]
- 41.Hart TW. Tet. Lett. 1985;26:2013–2016. [Google Scholar]
- 42.Arnelle DR, Stamler JS. Arch. Biochem. Biophys. 1995;318:279–285. doi: 10.1006/abbi.1995.1231. [DOI] [PubMed] [Google Scholar]
- 43.Stoyanovsky DA, Tyurina YY, Tyurin VA, Anand D, Mandavia DN, Gius D, Ivanova J, Pitt B, Billiar TR, Kagan VE. J. Am. Chem. Soc. 2005;127:15815–15823. doi: 10.1021/ja0529135. [DOI] [PubMed] [Google Scholar]
- 44.Liebeskind S, Korth H-G, de Groot H, Kirsch M. Org. Biomol. Chem. 2008;6:2560–2573. doi: 10.1039/b801583j. [DOI] [PubMed] [Google Scholar]
- 45.Liu ZG, Rudd MA, Freedman JE, Loscalzo J. Journal Pharmacol. Exp. Ther. 1998;284:526–534. [PubMed] [Google Scholar]
- 46.Hart TW, Vine MB, Walden NR. Tet. Lett. 1985;26:3879–3882. [Google Scholar]
- 47.Stamler JS, Jaraki O, Osborne J, Simon DI, Keaney J, Vita J, Singel DJ, Valeri CR, Loscalzo J. Proc. Natl. Acad. Sci. USA. 1992;89:7674–7677. doi: 10.1073/pnas.89.16.7674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang L-M, St. Croix C, Cao R, Wasserloos K, Watkins SC, Stevens T, Li S, Tyurin V, Kagan VE, Pitt BR. Exp. Biol. and Med. 2006;231:1507–1515. doi: 10.1177/153537020623100909. [DOI] [PubMed] [Google Scholar]
- 49.Zhang Y, Keszler A, Broniowska KA, Hogg N. Free Radical Biol. Med. 2005;38:874–881. doi: 10.1016/j.freeradbiomed.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 50.Beck JL, Ambahera S, Yong SR, Sheil MM, de Jersey J, Ralph SF. Anal. Biochem. 2004;325:326–336. doi: 10.1016/j.ab.2003.10.041. [DOI] [PubMed] [Google Scholar]
- 51.Riener CK, Kada G, Gruber HJ. Anal. Bioanal. Chem. 2002;373:266–276 C. doi: 10.1007/s00216-002-1347-2. [DOI] [PubMed] [Google Scholar]
- 52.Basu S, Hill JD, Shields H, Huang J, King SB, Kim-Shapiro DB. Nitric Oxide. 2006;15:1–4. doi: 10.1016/j.niox.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 53.Stroeher UH, Kidd SP, Stafford SL, Jennings MP, Paton JC, McEwan AG. J. Infect. Dis. 2007;196:1820–6. doi: 10.1086/523107. [DOI] [PubMed] [Google Scholar]
- 54.Huyen NTT, Eiamphungporn W, Mader U, Liebeke M, Lalk M, Hecker M, Helmann JD, Antelmann H. Mol. Microbiol. 2009;71:876–894. doi: 10.1111/j.1365-2958.2008.06568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McEwan AG, Djoko KY, Chen NH, Counago RL, Kidd SP, Potter AJ, Jennings MP. Adv. Microb. Physiol. 2011;58:1–22. doi: 10.1016/B978-0-12-381043-4.00001-5. [DOI] [PubMed] [Google Scholar]
- 56.Nguyen T, Francis MB. Org Lett, 2003;5:3245–3248. doi: 10.1021/ol035135z. [DOI] [PubMed] [Google Scholar]
- 57.Arbeloa L, Ojeda FPR, Arbeloa IL. J. Luminesc. 1989;44:105–112. [Google Scholar]
- 58.Arunkumar AI, Campanello GC, Giedroc DP. Proc. Natl. Acad. Sci. USA. 2009;106:18177–18182. doi: 10.1073/pnas.0905558106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Campanello GC, Ma Z, Grossoehme NE, Guerra AJ, Ward BP, Dimarchi RD, Ye Y, Dann CE, Giedroc DP. J. Mol. Biol. 2013;425:1143–57. doi: 10.1016/j.jmb.2013.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Busenlehner LS, Giedroc DP. J. Inorg. Biochem. 2006;100:1024. doi: 10.1016/j.jinorgbio.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 61.Williams DL. Acc. Chem. Res. 1999;32:869–876. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.