Abstract
Background
Urochloa humidicola is a warm-season grass commonly used as forage in the tropics and is recognized for its tolerance to seasonal flooding. This grass is an important forage species for the Cerrado and Amazon regions of Brazil. U. humidicola is a polyploid species with variable ploidy (6X–9X) and facultative apomixis with high phenotypic plasticity. However, this apomixis and ploidy, as well as the limited knowledge of the genetic basis of the germplasm collection, have constrained genetic breeding activities, yet microsatellite markers may enable a better understanding of the species’ genetic composition. This study aimed to develop and characterize new polymorphic microsatellite molecular markers in U. humidicola and to evaluate their transferability to other Urochloa species.
Findings
A set of microsatellite markers for U. humidicola was identified from two new enriched genomic DNA libraries: the first library was constructed from a single sexual genotype and the second from a pool of eight apomictic genotypes selected on the basis of previous results. Of the 114 loci developed, 72 primer pairs presented a good amplification product, and 64 were polymorphic among the 34 genotypes tested. The number of bands per simple sequence repeat (SSR) locus ranged from 1 to 29, with a mean of 9.6 bands per locus. The mean polymorphism information content (PIC) of all loci was 0.77, and the mean discrimination power (DP) was 0.87. STRUCTURE analysis revealed differences among U. humidicola accessions, hybrids, and other Urochloa accessions. The transferability of these microsatellites was evaluated in four species of the genus, U. brizantha, U. decumbens, U. ruziziensis, and U. dictyoneura, and the percentage of transferability ranged from 58.33% to 69.44% depending on the species.
Conclusions
This work reports new polymorphic microsatellite markers for U. humidicola that can be used for breeding programs of this and other Urochloa species, including genetic linkage mapping, quantitative trait loci identification, and marker-assisted selection.
Keywords: Microsatellite, Genomic library, SSR transferability, Forage, Grass
Findings
Background
Urochloa humidicola (Rendle) Morrone & Zuloaga (syn. Brachiaria humidicola (Rendle) Schweick.), commonly known as koronivia grass, is a perennial tropical grass native to eastern Africa that was introduced to Brazil in the 1950s [1,2]. U. humidicola is an apomictic polyploid species with variable levels of ploidy (6X–9X) [3-7].
In Brazil, the grasses of the genus Urochloa occupy 85% of the cultivated pasture areas [8]. U. humidicola is cultivated as forage in several tropical regions worldwide and is particularly recognized for its tolerance to poorly draining soils, seasonal flooding, and infertile acidic soils [9]. For this reason, this species has been largely exploited in the tropics as a forage option over other Urochloa grasses, mostly in the African savannas and similar environments, such as the Brazilian Cerrado [7].
The development and adoption of new U. humidicola cultivars with a broad genetic base are crucial for the diversification of forage pastures in the tropics, primarily because there are few cultivars of this species in Brazil (Tully, Llanero, and BRS Tupi). However, the development of new cultivars must be a dynamic process, providing cultivars with high nutritional value, increased biotic and abiotic resistance, and economic competitiveness.
Molecular markers are important tools to the progress of breeding programs, and their utilization would favor a more dynamic development of new cultivars of this species. However, there is a lack of information about the U. humidicola genome. Indeed, little or nothing is known about the number of genes, distribution of gene families, abundance and diversity of retro-elements, QTL localization of traits of economic importance, genome colinearity with model species, or abundance of repetitive sequences. Molecular markers are widely used in the fingerprinting of cultivars, the detection of genetic diversity in evaluating population structure in the mapping genes of interest, and in the selection of elite genotypes in breeding programs. SSR markers, in particular, are often used due to their codominant and multi-allelic characteristics [10]; moreover, they are highly site specific and transferable to related species [11].
Some microsatellite markers have already been developed for U. humidicola [12,13] and have been used for germplasm diversity studies [7,13], with all of them from the same microsatellite-enriched library constructed from genotype H016. Moreover, our research group identified four different gene pools among U. humidicola accessions; genotype H031 was found to be completely different from all other accessions, which was verified by a population structure analysis and by the fact that 18.5% of the tested markers did not amplify in this accession [7]. As a large number of markers are necessary for molecular breeding programs, our goal was to isolate and characterize new polymorphic microsatellite markers for U. humidicola genotype H031 (accession 12) to ensure that its genome was well represented by the new set of markers and also different accessions that belong to different gene pools and to test the transferability of these markers to four other Urochloa species (U. brizantha, U. decumbens, U. ruziziensis, and U. dictyoneura). The results were compared with previously reported data [12,13].
Methods
The plant material for library construction and marker validation was obtained from young leaves from several Urochloa genotypes. For the first library (Lb-1) construction, a single sexual genotype (H031) was used. For the second library (Lb-2) construction, a pool of eight apomictic genotypes (H010, H013, H015, H034, H041, H043, H101, and H108) was used. For marker validation, 34 genotypes were selected, consisting of 20 U. humidicola germplasm accessions, six intra-specific hybrids, and eight Urochloa accessions, as represented by two different accessions from each of the following species: U. brizantha, U. decumbens, U. ruziziensis, and U. dictyoneura. These genotypes were selected based on the four gene pools found by a previous study [7], from which two genotypes were selected from each gene pool. All of the accessions used are from the Urochloa germplasm collection maintained at Embrapa Beef Cattle, Campo Grande, MS, Brazil. They have been personally identified by S. A. Renvoize, from the Royal Botanic Gardens, Kew, UK and their identity have been confirmed by C. B. do Valle when transferred to Brazil [9]. The annotation numbers, accession numbers (as recorded in Embrapa Beef Cattle (EBC) and Center for Tropical Agriculture (CIAT)), genotypes, and species identifications are shown in Table 1. Genomic DNA was extracted from freeze-dried leaf samples using the CTAB method [14]. The DNA samples were evaluated on a 1% agarose gel and quantified by comparison to known quantities of uncut λ phage DNA (Invitrogen, Carlsbad, CA, USA).
Table 1.
Genotypes of U. humidicola and four species of the genus Urochloa used for the characterization and transferability analyses of new microsatellite markers
| AN | CIAT | BRA | EBC | Genotype | Species |
|---|---|---|---|---|---|
| 1 | 16181 | 4821 | H004 | germplasm accession | U. humidicola |
| 2 | 16182 | 4839 | H005 | germplasm accession | U. humidicola |
| 3 | 16867 | 4863 | H006 | germplasm accession | U. humidicola |
| 4 | 16871 | 4901 | H008 | germplasm accession | U. humidicola |
| 5 | 16880 | 4952 | H010 | germplasm accession | U. humidicola |
| 6 | 16882 | 4979 | H012 | germplasm accession | U. humidicola |
| 7 | 16886 | 5011 | H013 | germplasm accession | U. humidicola |
| 8 | 26141 | 5088 | H015 | germplasm accession | U. humidicola |
| 9 | 26149 | 5118 | H016 | germplasm accession | U. humidicola |
| 10 | 16877 | 4928 | H023 | germplasm accession | U. humidicola |
| 11 | 16894 | 5070 | H030 | germplasm accession | U. humidicola |
| 12 | 26146 | 5100 | H031 | germplasm accession | U. humidicola |
| 13 | 26413 | 6131 | H035 | germplasm accession | U. humidicola |
| 14 | 26432 | 6203 | H041 | germplasm accession | U. humidicola |
| 15 | 16884 | 4995 | H044 | germplasm accession | U. humidicola |
| 16 | NA | NA | H048 | germplasm accession | U. humidicola |
| 17 | NA | 1929 | H107 | germplasm accession | U. humidicola |
| 18 | 6705 | 2208 | H112 | germplasm accession | U. humidicola |
| 19 | 6133 | 1449 | H125 | germplasm accession | U. humidicola |
| 20 | 6369 | 0370 | H126 | germplasm accession | U. humidicola |
| 21 | - | - | 20 | hybrid | U. humidicola |
| 22 | - | - | 45 | hybrid | U. humidicola |
| 23 | - | - | 184 | hybrid | U. humidicola |
| 24 | - | - | 215 | hybrid | U. humidicola |
| 25 | - | - | 264 | hybrid | U. humidicola |
| 26 | - | - | 320 | hybrid | U. humidicola |
| 27 | 16162 | - | B057 | germplasm accession | U. brizantha |
| 28 | 16467 | - | B166 | germplasm accession | U. brizantha |
| 29 | 16499 | 004481 | D009 | germplasm accession | U. decumbens |
| 30 | 26300 | 004707 | D028 | germplasm accession | U. decumbens |
| 31 | 26163 | 005568 | R102 | germplasm accession | U. ruziziensis |
| 32 | 26174 | 005614 | R104 | germplasm accession | U. ruziziensis |
| 33 | 16186 | 007889 | DT157 | germplasm accession | U. dictyoneura |
| 34 | 16188 | 007901 | DT159 | germplasm accession | U. dictyoneura |
NA: not available, AN: annotation number, CIAT: Center for Tropical Agriculture, BRA: codes from EMBRAPA, EBC: codes from EMBRAPA Beef Cattle.
Genomic DNA was restriction digested with Afa I (Invitrogen), enriched in microsatellite fragments using (CT)8 and (GT)8 probes, and then used to construct a microsatellite-enriched library following the protocol of Billotte et al. [15]. The enriched microsatellite fragments were cloned into pGEM-T (Promega, Madison, WI), and the ligation products were used to transform Escherichia coli XL1-Blue competent cells. All 94 clones from both libraries were sequenced with an ABI 377 automated sequencer (Applied Biosystems, Foster City, CA) using the BigDye terminator cycle sequencing kit (Applied Biosystems, Foster City, CA).
The microsatellites were identified using MISA software [16]. Only mono-nucleotides with twelve or more repeats, di-nucleotides with six or more repeats, tri-nucleotides with four or more repeats, and tetra-, penta-, and hexa-nucleotides with three or more repeats were considered. Primer pairs were designed using the Primer Select 5.01 software (DNASTAR Inc.) and the Primer3Plus software [17]. Polymerase chain reactions (PCRs) were carried out as previously described [12]. The amplification products were resolved by electrophoresis through 3% agarose gels prior to vertical electrophoresis through 6% denaturing polyacrylamide gels. The gels were then silver stained [18], and the product sizes were determined by comparison to a 10-bp DNA ladder (Invitrogen, Carlsbad, CA).
Polyploid microsatellite genotyping is difficult due to the closeness of fragment sizes, stutter peaks observed and allele overlap due to multiple alleles of the same size. Few methods have been developed to overcome allele overlapping and estimate the allele frequencies, such as the estimation of alleles based on the electropherogram peak ratios [19] or the statistical estimation of allele frequencies [20]. However, for the present study work, we restricted the project to describe the new SSR markers, which were visually scored based on the presence (1) or absence (0) of a band in the polyacrilamide gels for each of the Urochloa genotypes. PIC (Polymorphic Information Content) [21] and DP (Discriminatory Power) [22] values were calculated to estimate polymorphisms at each locus.
The microsatellite scores for the 34 individuals were evaluated using a model-based method with Bayesian clustering approach in STRUCTURE software version 2.2 [23-25]. The admixture model was tested with 200,000 replicates for burn-in and 100,000 replicates for Markov Chain Monte Carlo (MCMC) processes through ten iterations (runs). The numbers of clusters (K) were tested from 2 to 20. The optimal number of clusters was estimated using the ΔK value, as previously described [26], and the final graphs were visualized using the STRUCTURE HARVESTER software [27]. The individuals were grouped into clusters according to the association coefficient (Q) proportion of each allelic pool in an individual.
A joint analysis (Lb-c) was performed with the data from the polymorphic loci derived from the new libraries Lb-1 and Lb-2. Data from a previous study [12] that used SSRs developed from accession 9 (H016) were used to compare the three libraries. The data were reanalyzed under the same parameters as those used for the new libraries, resulting in Lb-3. Another joint analysis (Lb-ct) was performed with data from the three libraries together (Lb-1, Lb-2, and Lb-3). The results obtained by STRUCTURE software were permuted by CLUMPP software [28], and the figures were generated using DISTRUCT software [29].
Results
Microsatellite enrichment success for the U. humidicola DNA libraries was 79.0% for Lb-1 and 61.2% for Lb-2. From all of the sequenced clones, 183 microsatellites were identified. Di-nucleotide repeats were the most abundant class of microsatellites detected, representing 76.4% and 72.7% of the loci for Lb-1 and Lb-2, respectively, followed by mono-nucleotide and tetra-nucleotide repeats. Perfect microsatellites were the most abundant (Table 2).
Table 2.
Characterization of new microsatellite-enriched libraries from U. humidicola
| Library | Lb-1 | Lb-2 | |
|---|---|---|---|
| Total clones sequenced | 86.0 | 80.0 | |
| Sequences containing microsatellites (%) | 79.0 | 61.2 | |
| Total number of SSRs identified | 106.0 | 77.0 | |
| Type of repeat (%) | |||
| By nucleotide string | Mono-nucleotides | 12.7 | 6.5 |
| Di-nucleotides | 76.4 | 72.7 | |
| Tri-nucleotides | 1.9 | 5.2 | |
| Tetra-nucleotides | 5.6 | 11.6 | |
| Penta-nucleotides | 2.8 | 3.9 | |
| Hexa-nucleotides | 0.9 | 0.0 | |
| By form | Perfect | 79.1 | 80.6 |
| Imperfect | 9.3 | 1.6 | |
| Perfect Compound | 5.8 | 9.7 | |
| Imperfect Compound | 5.8 | 8.1 | |
Of the 114 SSR primer pairs designed and tested, 72 were successfully amplified in U. humidicola genotypes, and 64 SSRs were polymorphic. A description of the number of alleles per locus and PIC and DP values for both the U. humidicola accessions and Urochloa accessions is presented in Table 3. The loci BhUNICAMP68 to BhUNICAMP108 are derived from Lb-1, and the loci BhUNICAMP109 to BhUNICAMP139 are derived from Lb-2. Based on the allelic frequencies estimated by STRUCTURE software, 36.43% of the alleles are rare (frequency < 0.05), 60.06% are intermediate alleles (0.05 < frequency < 0.30), and 3.50% are abundant alleles (frequency > 0.30).
Table 3.
Characterization of the 72 polymorphic SSR markers developed for U. humidicola
| SSR locus | GenBank accession number | Repeat motif | Ta (°C) a | Primer sequences (5′-3′) | Urochloa species accessions* | U. humidicola accessions** | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Size range (bp) | A b | PIC c | A b | PIC c | DP d | |||||
| BhUNICAMP068 | KM068303 | (CACACC)4(CA)17 | 58.5 | F_CCACAAACGTGAACACATACA R_AGGGACGGAAACACCCTTAG | 226-261 | 10 | 0.87 | 10 | 0.87 | 0.95 |
| BhUNICAMP069 | KM068304 | (TC)25 | 64.5 | F_GAGGAACTCCTTTGGGTAGA R_TTCAGAGAGAGGATGGTATAGAG | 285-300 | 2 | 0.36 | 2 | 0.36 | 0.58 |
| BhUNICAMP070 | KM068305 | (GT)9 | 65 | F_CCCCGGTCTCGACCTATC R_GAGGCTGCCCCCTTACTC | 174-214 | 12 | 0.84 | 6 | 0.78 | 0.54 |
| BhUNICAMP071 | KM068306 | (AC)11 | 65 | F_CGCAACGAAGCTCCAATAG R_CGATCGCAAGCGTGTATCTA | 160-228 | 11 | 0.86 | 11 | 0.86 | 0.94 |
| BhUNICAMP072 | KM068307 | (GT)7 | 56.5 | F_CCCCATGTAAACAACCGTAGA R_CCATGGTTGACCGCTAGAA | 174-186 | 3 | 0.56 | 3 | 0.56 | 0.85 |
| BhUNICAMP073 | KM068308 | (TG)10 | 60 | F_TGAACATGTGAATGCCCACT R_ATTGCAGGATGCGGACTCTA | 240-304 | 10 | 0.85 | 10 | 0.85 | 0.94 |
| BhUNICAMP074 | KM068309 | (CT)6 | 58.5 | F_ACGAACGATCCGACCAACTA R_TGCTTACGAGACGGCATAGA | 231-255 | 7 | 0.81 | 7 | 0.81 | 0.92 |
| BhUNICAMP075 | KM068310 | (TC)22 | 50 | F_TGAATGCTTTTGTCCTGGTATC R_ACGTGCAGCAGCAACAGTA | 148-236 | 28 | 0.95 | 24 | 0.95 | 0.98 |
| BhUNICAMP076 | KM068311 | (AC)18 | 51.5 | F_CCGATGGTCAAAGGTCAGTT R_GGTGGGCATATACCATGTTT | 206-234 | 10 | 0.84 | 10 | 0.84 | 0.66 |
| BhUNICAMP077 | KM068312 | (AC)7 | 65 | F_CGGGAAGTCCTACTCCGTAA R_GGAGCTCAAGGTAGGGATTG | 212-230 | 8 | 0.83 | 8 | 0.83 | 0.93 |
| BhUNICAMP078 | KM068313 | (GT)7 | 58.5 | F_ACCAGTGCACGTCTGAAAGA R_CGATCACTGCTGCGTCATA | 216-218 | 2 | 0.35 | 2 | 0.35 | 0.52 |
| BhUNICAMP079 | KM068314 | (AG)12G(GA)17 | 62.5 | F_GGATTGAAAGTTGGAGCACA R_GCATGCTGTGAAGGAGGTTA | 180-222 | 17 | 0.92 | 17 | 0.92 | 0.96 |
| BhUNICAMP080 | KM068315 | (GA)26 | 50 | F_CAAGCCTCTTCATGCAAGTAAC R_TGTCATACCCCCATGATTAAGA | 176-230 | 22 | 0.93 | 21 | 0.93 | 0.93 |
| BhUNICAMP081 | KM068316 | (AGC)5ACAAT(CA)11 | 55 | F_CTGGCATGGGTCCCTTTAC R_TCTTCTTCCTCCAGCCACAT | 160-179 | 5 | 0.75 | 5 | 0.75 | 0.95 |
| BhUNICAMP082 | KM068317 | (CA)23 | 60 | F_TTGCCGGGAACAGTTATACA R_GAAGCTCTATCAAACAGCCCT | 157-192 | 9 | 0.82 | 9 | 0.82 | 0.92 |
| BhUNICAMP083 | KM068318 | (AG)22 | 56.5 | F_AAACATGCACCGTCATAACT R_GGGCTTGATTCATTTGTTA | 152-190 | 6 | 0.68 | 4 | 0.68 | 0.77 |
| BhUNICAMP084 | KM068319 | (TG)15 | 65 | F_GGCGAAGACCATACCCTGTA R_TGCTGGTGGAAGAAGATGAA | 159-182 | 9 | 0.80 | 9 | 0.80 | 0.96 |
| BhUNICAMP085 | KM068320 | (GT)9 | 60 | F_CGATTTATCGACGACCGAGT R_CCTTACTCGCAGGTCTGTCC | 158-171 | 5 | 0.76 | 5 | 0.76 | 0.64 |
| BhUNICAMP086 | KM068321 | (TC)19 | 65 | F_AGTTGAATGGGCTGAACCAT R_TGCACTTCCAGGATCAGACA | 238-326 | 10 | 0.82 | 10 | 0.82 | 0.93 |
| BhUNICAMP087 | KM068322 | (GT)10 | 50 | F_GGCCATTTCTAGCCAAACAA R_CCTTACTCGCAGGTCTGTCC | 240 | 1 | 0.00 | 1 | 0.00 | 0.00 |
| BhUNICAMP088 | KM068323 | (TG)12 | 65 | F_AGAGGTTCCATGGACATTGC R_CTCATCAACAGACGCCTGAA | 178 | 1 | 0.00 | 1 | 0.00 | 0.00 |
| BhUNICAMP089 | KM068324 | (AC)7 | 65 | F_CCGGATAGAAGGTCTGAACG R_AGTCGTCGAAGCGAGCTCTA | 175 | 1 | 0.00 | 1 | 0.00 | 0.00 |
| BhUNICAMP090 | KM068325 | (CA)10 | 65 | F_CAGAGTAAGCTTCCGGGACA R_CGATTTATCGACGACCGAGT | 200-300 | 12 | 0.85 | 11 | 0.85 | 0.91 |
| BhUNICAMP091 | KM068326 | (AC)8 | 65 | F_CTTGTGCCACTTCCACCTTT R_TCGTGTGGACACTTCCTCTG | 120-150 | 9 | 0.83 | 9 | 0.83 | 0.95 |
| BhUNICAMP092 | KM068327 | (TG)6 | 65 | F_ATGCCTTGCTCCCACTAACA R_TAAATGCTCCAGCGACCTTC | 135-168 | 11 | 0.85 | 11 | 0.85 | 0.91 |
| BhUNICAMP093 | KM068328 | (AAG)4 | 65 | F_GGAGCGCTAATTTCGTTCAG R_CCTCCGTTCTCGCTAATGAC | 230 | 1 | 0.00 | 1 | 0.00 | 0.00 |
| BhUNICAMP094 | KM068329 | (TG)7 | 65 | F_TTGGAGCTTTCCCTAGCTCA R_GAACAAGAAGGGAGGAAGCA | 272-290 | 4 | 0.31 | 4 | 0.31 | 0.39 |
| BhUNICAMP095 | KM068330 | (TC)16(TG)14 | 65 | F_GGGTTGGCCTACACACTGAT R_CGCACGACATTGATACCTTG | 268-320 | 6 | 0.75 | 6 | 0.75 | 0.92 |
| BhUNICAMP096 | KM068331 | (TC)8TT(TC)40 | 65 | F_TGTTCTGCTCACTGGTTTGG R_TCAGCTCTCTACGGCTGGAT | 157-255 | 11 | 0.87 | 11 | 0.87 | 0.95 |
| BhUNICAMP097 | KM068332 | (GT)6 | 65 | F_GCGAGCTACCGAGGTATTTG R_ACGTCAATGTCGAGCTTCCT | 129-148 | 5 | 0.69 | 5 | 0.69 | 0.80 |
| BhUNICAMP098 | KM068333 | (GT)10(G)18 | 65 | F_GGACTGGTCGTCTTTCCATC R_GCTTTCTGCAAGCGGTAGAT | 250-312 | 9 | 0.85 | 9 | 0.85 | 0.95 |
| BhUNICAMP099 | KM068334 | (CA)10TG(GA)10 | 65 | F_TTTGTGGCACCTGCAGAATA R_CGCTTCGTGCTGACAGATTA | 124-174 | 16 | 0.91 | 16 | 0.91 | 0.99 |
| BhUNICAMP100 | KM068335 | (TG)12 | 65 | F_GCGCCATGGTTTCATCTATT R_GGTGGTTCCTCGTGTGAGAT | 178-219 | 7 | 0.79 | 7 | 0.79 | 0.98 |
| BhUNICAMP101 | KM068336 | (TG)28 | 65 | F_GGTAAAGAAGGGCCGGACT R_GCATGGCATGTTCCTACTGA | 128-184 | 14 | 0.89 | 12 | 0.89 | 0.97 |
| BhUNICAMP102 | KM068337 | (GCGA)4 | 65 | F_TGGTGGGCTCCACTATCTCT R_TCCGCCATCTCTCCTCTCT | 224-260 | 12 | 0.89 | 12 | 0.89 | 0.94 |
| BhUNICAMP103 | KM068338 | (CT)22 | 65 | F_AGCTCTCCCGCCTCTCTCT R_CATCCACACCGTCTCTCTCA | 100-156 | 14 | 0.91 | 14 | 0.91 | 0.96 |
| BhUNICAMP104 | KM068339 | (TG)26 | 60 | F_ACGACGACCTAATGGGTGAA R_ACCCAGCAACAAATCTCGTC | 190-274 | 15 | 0.87 | 13 | 0.87 | 0.96 |
| BhUNICAMP105 | KM068340 | (AC)10ATACACACACAC(AG)53 | 50 | F_CTCCATCACGTGCTTGCTAA R_GTGTGATCGGCTGGAGATTT | 100-176 | 30 | 0.93 | 29 | 0.93 | 0.98 |
| BhUNICAMP106 | KM068341 | (TTTGT)3 | 50 | F_GCTGTTCGGAGAGGAATCTG R_ATGAGAGGAGGGAAGGAAGG | 135-155 | 8 | 0.79 | 7 | 0.79 | 0.91 |
| BhUNICAMP107 | KM068342 | (GA)18 | 50 | F_GGGTCAGTGTCGTCTCAGTTT R_CAGATTCCTCTCCGAACAGC | 118-190 | 26 | 0.94 | 26 | 0.94 | 0.98 |
| BhUNICAMP108 | KM068343 | (CT)16 | 65 | F_TTGCCATTACTGGATCTGGA R_GCGCCACCCATAACTTAAA | 112-160 | 14 | 0.85 | 13 | 0.85 | 0.94 |
| BhUNICAMP109 | KM068344 | (GT)9 | 60 | F_AGCGAGTCAAGCACAAGGAT R_GGGTCCAATCTCCCTCTCTC | 186-226 | 9 | 0.82 | 9 | 0.82 | 0.93 |
| BhUNICAMP110 | KM068345 | (TG)8 | 65 | F_TCTGCATCCACTAGGCTCAG R_TCCTCCACCTTCTTTCCGTA | 148-164 | 4 | 0.39 | 4 | 0.39 | 0.46 |
| BhUNICAMP111 | KM068346 | (TG)27 | 65 | F_AACTCCGACTATCTTCCAGTTGA R_AATGCATGGGTAGGATCTGC | 250-330 | 15 | 0.89 | 15 | 0.89 | 0.96 |
| BhUNICAMP112 | KM068347 | (AC)26 | 65 | F_GACCAAACCCTCCGAAGTTA R_GGTTGCAACTACACGACCAG | 246-300 | 10 | 0.81 | 10 | 0.81 | 0.94 |
| BhUNICAMP113 | KM068348 | (CGTG)3 | 63 | F_AACTTCGAGAGGTTCGTCCA R_ACCGGCAATCTATCCGTGT | 144-179 | 3 | 0.45 | 3 | 0.45 | 0.51 |
| BhUNICAMP114 | KM068349 | (CT)21 | 63 | F_TATACAAGGCGCATCCACAA R_GCTCTTTCCTCACGCTGTTC | 200-266 | 15 | 0.89 | 15 | 0.89 | 0.96 |
| BhUNICAMP115 | KM068350 | (AC)27(AT)7 | 60 | F_CTTCCTGCCAATAAGCGAAG R_CGAGCTTCCAGATTCTTTGG | 240 | 1 | 0.00 | 1 | 0.00 | 0.00 |
| BhUNICAMP116 | KM068351 | (TG)8 | 65 | F_CTCCGCACCGCTTAAATTAG R_GTTGGAAATGGTGCTTCCAC | 288-306 | 3 | 0.52 | 3 | 0.52 | 0.62 |
| BhUNICAMP117 | KM068352 | (TGA)7 | 65 | F_CCAACTGAACGGCCATACTT R_CCCACAAAGGAACCCTGAT | 290-300 | 4 | 0.61 | 4 | 0.61 | 0.77 |
| BhUNICAMP118 | KM068353 | (AG)9 | 50 | F_CTGCATAACTTTCAGCCATCTC R_TTGGCACAACTGGAACGTAG | 149 | 1 | 0.00 | 1 | 0.00 | 0.00 |
| BhUNICAMP119 | KM068354 | (AAG)7 | 65 | F_AAGGGCGTGATGTTCTGAAG R_AGGCCAAACGAATTTCTCAA | 189-204 | 4 | 0.66 | 4 | 0.66 | 0.82 |
| BhUNICAMP120 | KM068355 | (AT)8ACACACACACG(CA)9 | 65 | F_TCCAGCAGTGTGTTCCTCAG R_ACCAGGAGTGCATAGCCAAG | 190-200 | 6 | 0.71 | 6 | 0.71 | 0.75 |
| BhUNICAMP121 | KM068356 | (TC)12 | 65 | F_CGCTACTGCTGCACACAAAT R_CTGAGTGCGCCGTATGTTTA | 170-195 | 6 | 0.71 | 6 | 0.71 | 0.92 |
| BhUNICAMP122 | KM068357 | (GT)15 | 65 | F_AGGAAGGCTCGCACTCACTA R_CCAAAGGCGGTGGTTAGATA | 200-315 | 14 | 0.90 | 14 | 0.90 | 0.95 |
| BhUNICAMP123 | KM068358 | (TTA)4 | 65 | F_CCAAACTCTAGCTTTCACAGCA R_TTGGATCCACGTCAAACAAG | 280 | 1 | 0.00 | 1 | 0.00 | 0.00 |
| BhUNICAMP124 | KM068359 | (AG)23 | 65 | F_TTGGAGTTGCTGGGCTATTT R_GAACCAAGCATAAGGCAACA | 218-320 | 12 | 0.85 | 10 | 0.85 | 0.95 |
| BhUNICAMP125 | KM068360 | (GT)8GAATGTGTGT(GA)7 | 65 | F_TGTTATCAGTGCAGGTGTTGG R_GAGGCTGACGAAAGCTCAAC | 258-280 | 7 | 0.81 | 7 | 0.81 | 0.93 |
| BhUNICAMP126 | KM068361 | (AC)10 | 65 | F_GGGAACCCAGGGTATCGTAT R_CTCTCCCAGCGTCTTTCCTT | 210 | 1 | 0.00 | 1 | 0.00 | 0.00 |
| BhUNICAMP127 | KM068362 | (GT)6 | 65 | F_CCACCATTGCTTCCAGAGTAA R_ATTCGCCTCTCCTAGCACAA | 272-320 | 7 | 0.69 | 7 | 0.69 | 0.91 |
| BhUNICAMP128 | KM068363 | (GA)37 | 65 | F_TGCCTGGAGACTGAGAAAGG R_CCTGCAGCAGACCTTCACAT | 150-240 | 17 | 0.91 | 17 | 0.91 | 0.98 |
| BhUNICAMP129 | KM068364 | (AC)7ATGAA(CATG)3(CA)22 | 63 | F_TGTGTTTAGACCGCCAACAA R_TTATCGGCTCCCATTCACTC | 207-310 | 11 | 0.84 | 10 | 0.84 | 0.95 |
| BhUNICAMP130 | KM068365 | (AC)7 | 63 | F_ACGCAGGAGAACTGCGTATC R_ATGGGATCCAACCGAACATA | 236-300 | 12 | 0.79 | 11 | 0.79 | 0.87 |
| BhUNICAMP131 | KM068366 | (AC)7(A)16 | 60 | F_CATCAGATGCCTCAAACAGC R_GCAGGTGTGCAGCAAATAGA | 184-238 | 14 | 0.87 | 14 | 0.87 | 0.93 |
| BhUNICAMP132 | KM068367 | (TG)7(T)29 | 50 | F_TCACTAGTGCGTCTGCTGCT R_GCACTCCATTGCAGACCTAAG | 184-196 | 4 | 0.53 | 3 | 0.53 | 0.63 |
| BhUNICAMP133 | KM068368 | (TG)10 | 50 | F_CATGACTTATGTCCTTGGTGGA R_TCGACAGTGGAGCCACAA | 114-162 | 19 | 0.89 | 16 | 0.89 | 0.97 |
| BhUNICAMP134 | KM068369 | (CCGG)3 | 60 | F_CAAACGGAGGAAGAGAGACG R_GGTGTCAATGCAGCCAAGTA | 114-135 | 9 | 0.75 | 5 | 0.75 | 0.83 |
| BhUNICAMP135 | KM068370 | (AG)27 | 65 | F_CATGAGCCATCTCGTTGTTG R_TGCATTGACTTGACGTCTCC | 176-260 | 14 | 0.90 | 9 | 0.90 | 0.91 |
| BhUNICAMP136 | KM068371 | (AC)9(ACAA)3 | 50 | F_TCCTGGTAAAGTTCCTCGTCA R_ACAACAATGCACGTCGAGAA | 225-290 | 7 | 0.75 | 6 | 0.75 | 0.93 |
| BhUNICAMP137 | KM068372 | (GA)23 | 65 | F_TAGGTTTGGGTGGCACTAGG R_CTCCATGCTGCGTTGCTAT | 258-320 | 11 | 0.85 | 9 | 0.85 | 0.91 |
| BhUNICAMP138 | KM068373 | (T)12 | 60 | F_TGCTCATGTGGGTCACATTT R_TGTGTGCCTGTGTGATGCTA | 270-288 | 5 | 0.70 | 5 | 0.70 | 0.95 |
| BhUNICAMP139 | KM068374 | (AAAAG)3 | 65 | F_TCCTTTCTTTGAGCCGAGAG R_GCTGATGCTGACATCAAGGA | 248-294 | 6 | 0.67 | 5 | 0.67 | 0.97 |
| Total average | 10.26 | 0.77 | 9.60 | 0.77 | 0.87 | |||||
| Lb-1 average | 11.05 | 0.79 | 10.48 | 0.79 | 0.87 | |||||
| Lb-2 average | 9.18 | 0.75 | 8.40 | 0.75 | 0.86 | |||||
*Species evaluated: Urochloa humidicola (Rendle) Morrone & Zuloaga, Urochloa brizantha (Hochst. ex A. Rich.) R.D. Webster, Urochloa decumbens (Stapf) R.D. Webster, Urochloa dictyoneura (Figure & De Not.) Veldkamp, Urochloa ruziziensis (R. Germ. & C.M. Evrard) Crins.
**Hybrids included.
aAmplification temperature (°C).
bMaximum number of alleles observed.
cPolymorphism Information Content.
dDiscrimination Power.
A survey of the potential transferability of the microsatellite markers from U. humidicola to other Urochloa species identified that 61.11% of the 72 markers resulted in amplified PCR products in at least one U. brizantha genotype, 58.33% were amplified in U. decumbens, 59.72% were amplified in U. ruziziensis, and 69.44% were amplified in U. dictyoneura. The number of successfully amplified genotypes per number of genotypes tested per species is shown in Table 4.
Table 4.
Cross-amplification of the 72 SSR markers among other Urochloa species
| Transferability a,b | ||||
|---|---|---|---|---|
| SSR locus | U. brizantha | U. decumben | U. ruziziensis | U. dictyoneura |
| BhUNICAMP068 | 0/2 | 0/2 | 0/2 | 0/2 |
| BhUNICAMP069 | 0/2 | 0/2 | 0/2 | 0/2 |
| BhUNICAMP070 | 2/2 | 2/2 | 2/2 | 2/2 |
| BhUNICAMP071 | 0/2 | 0/2 | 0/2 | 2/2 |
| BhUNICAMP072 | 1/2 | 1/2 | 0/2 | 1/2 |
| BhUNICAMP073 | 0/2 | 0/2 | 0/2 | 2/2 |
| BhUNICAMP074 | 0/2 | 0/2 | 0/2 | 0/2 |
| BhUNICAMP075 | 2/2 | 2/2 | 2/2 | 2/2 |
| BhUNICAMP076 | 2/2 | 2/2 | 2/2 | 1/2 |
| BhUNICAMP077 | 2/2 | 2/2 | 2/2 | 2/2 |
| BhUNICAMP078 | 1/2 | 0/2 | 1/2 | 1/2 |
| BhUNICAMP079 | 2/2 | 2/2 | 2/2 | 1/2 |
| BhUNICAMP080 | 2/2 | 1/2 | 2/2 | 1/2 |
| BhUNICAMP081 | 0/2 | 0/2 | 0/2 | 0/2 |
| BhUNICAMP082 | 2/2 | 0/2 | 1/2 | 1/2 |
| BhUNICAMP083 | 1/2 | 1/2 | 1/2 | 2/2 |
| BhUNICAMP084 | 2/2 | 2/2 | 2/2 | 2/2 |
| BhUNICAMP085 | 2/2 | 2/2 | 2/2 | 2/2 |
| BhUNICAMP086 | 2/2 | 2/2 | 2/2 | 2/2 |
| BhUNICAMP087 | 2/2 | 2/2 | 2/2 | 2/2 |
| BhUNICAMP088 | 2/2 | 2/2 | 2/2 | 2/2 |
| BhUNICAMP089 | 0/2 | 0/2 | 0/2 | 0/2 |
| BhUNICAMP090 | 2/2 | 2/2 | 2/2 | 2/2 |
| BhUNICAMP091 | 0/2 | 0/2 | 0/2 | 2/2 |
| BhUNICAMP092 | 0/2 | 0/2 | 0/2 | 2/2 |
| BhUNICAMP093 | 0/2 | 0/2 | 0/2 | 0/2 |
| BhUNICAMP094 | 2/2 | 2/2 | 1/2 | 2/2 |
| BhUNICAMP095 | 0/2 | 0/2 | 0/2 | 0/2 |
| BhUNICAMP096 | 2/2 | 2/2 | 1/2 | 2/2 |
| BhUNICAMP097 | 2/2 | 2/2 | 2/2 | 2/2 |
| BhUNICAMP098 | 0/2 | 0/2 | 0/2 | 0/2 |
| BhUNICAMP099 | 0/2 | 0/2 | 0/2 | 0/2 |
| BhUNICAMP100 | 0/2 | 0/2 | 0/2 | 0/2 |
| BhUNICAMP101 | 2/2 | 2/2 | 2/2 | 2/2 |
| BhUNICAMP102 | 0/2 | 0/2 | 0/2 | 0/2 |
| BhUNICAMP103 | 2/2 | 1/2 | 2/2 | 2/2 |
| BhUNICAMP104 | 2/2 | 2/2 | 2/2 | 2/2 |
| BhUNICAMP105 | 2/2 | 2/2 | 2/2 | 2/2 |
| BhUNICAMP106 | 2/2 | 2/2 | 2/2 | 2/2 |
| BhUNICAMP107 | 2/2 | 2/2 | 2/2 | 1/2 |
| BhUNICAMP108 | 2/2 | 2/2 | 2/2 | 1/2 |
| BhUNICAMP109 | 2/2 | 2/2 | 2/2 | 2/2 |
| BhUNICAMP110 | 2/2 | 2/2 | 2/2 | 2/2 |
| BhUNICAMP111 | 2/2 | 2/2 | 2/2 | 2/2 |
| BhUNICAMP112 | 2/2 | 2/2 | 2/2 | 2/2 |
| BhUNICAMP113 | 0/2 | 0/2 | 0/2 | 0/2 |
| BhUNICAMP114 | 0/2 | 0/2 | 0/2 | 0/2 |
| BhUNICAMP115 | 0/2 | 0/2 | 0/2 | 0/2 |
| BhUNICAMP116 | 2/2 | 2/2 | 2/2 | 2/2 |
| BhUNICAMP117 | 0/2 | 0/2 | 0/2 | 2/2 |
| BhUNICAMP118 | 0/2 | 0/2 | 0/2 | 0/2 |
| BhUNICAMP119 | 0/2 | 0/2 | 0/2 | 0/2 |
| BhUNICAMP120 | 2/2 | 2/2 | 2/2 | 2/2 |
| BhUNICAMP121 | 2/2 | 2/2 | 2/2 | 2/2 |
| BhUNICAMP122 | 0/2 | 0/2 | 0/2 | 0/2 |
| BhUNICAMP123 | 0/2 | 0/2 | 0/2 | 2/2 |
| BhUNICAMP124 | 2/2 | 2/2 | 2/2 | 0/2 |
| BhUNICAMP125 | 0/2 | 0/2 | 0/2 | 0/2 |
| BhUNICAMP126 | 2/2 | 2/2 | 2/2 | 2/2 |
| BhUNICAMP127 | 0/2 | 0/2 | 0/2 | 0/2 |
| BhUNICAMP128 | 0/2 | 0/2 | 0/2 | 0/2 |
| BhUNICAMP129 | 2/2 | 2/2 | 2/2 | 0/2 |
| BhUNICAMP130 | 2/2 | 2/2 | 2/2 | 2/2 |
| BhUNICAMP131 | 2/2 | 2/2 | 2/2 | 2/2 |
| BhUNICAMP132 | 0/2 | 0/2 | 0/2 | 2/2 |
| BhUNICAMP133 | 2/2 | 2/2 | 2/2 | 2/2 |
| BhUNICAMP134 | 2/2 | 2/2 | 2/2 | 2/2 |
| BhUNICAMP135 | 2/2 | 2/2 | 2/2 | 2/2 |
| BhUNICAMP136 | 2/2 | 2/2 | 1/2 | 2/2 |
| BhUNICAMP137 | 2/2 | 2/2 | 2/2 | 2/2 |
| BhUNICAMP138 | 0/2 | 0/2 | 0/2 | 2/2 |
| BhUNICAMP139 | 2/2 | 2/2 | 2/2 | 2/2 |
| Total | 44 | 42 | 43 | 50 |
| Amplification % | 61,11 | 58,33 | 59,72 | 69,44 |
aNumber of successfully amplified genotypes/Number of tested genotypes.
bNomenclatural classification: Urochloa humidicola (Rendle) Morrone & Zuloaga, Urochloa brizantha (Hochst. ex A. Rich.) R.D. Webster, Urochloa decumbens (Stapf) R.D. Webster, Urochloa dictyoneura (Figure & De Not.) Veldkamp, Urochloa ruziziensis (R. Germ. & C.M. Evrard) Crins.
The population structure analysis based on SSR allelic data showed differentiation among the U. humidicola accessions, hybrids, and other Urochloa species. The STRUCTURE analysis for Lb-1 and Lb-2 and the joint analysis of data from both libraries (Lb-c) showed K = 18, K = 17, and K = 17 allelic pools, respectively, with each one represented by a different color in Figure 1. Clusters I to V were composed of U. humidicola accessions. Cluster VI was composed of two U. humidicola accessions (accessions 9 and 12) and six hybrids derived from a controlled cross between these two accessions. The other Urochloa species were grouped into Clusters VII and VIII for Lb-1 and Lb-c and in Cluster VII for Lb-2.
Figure 1.
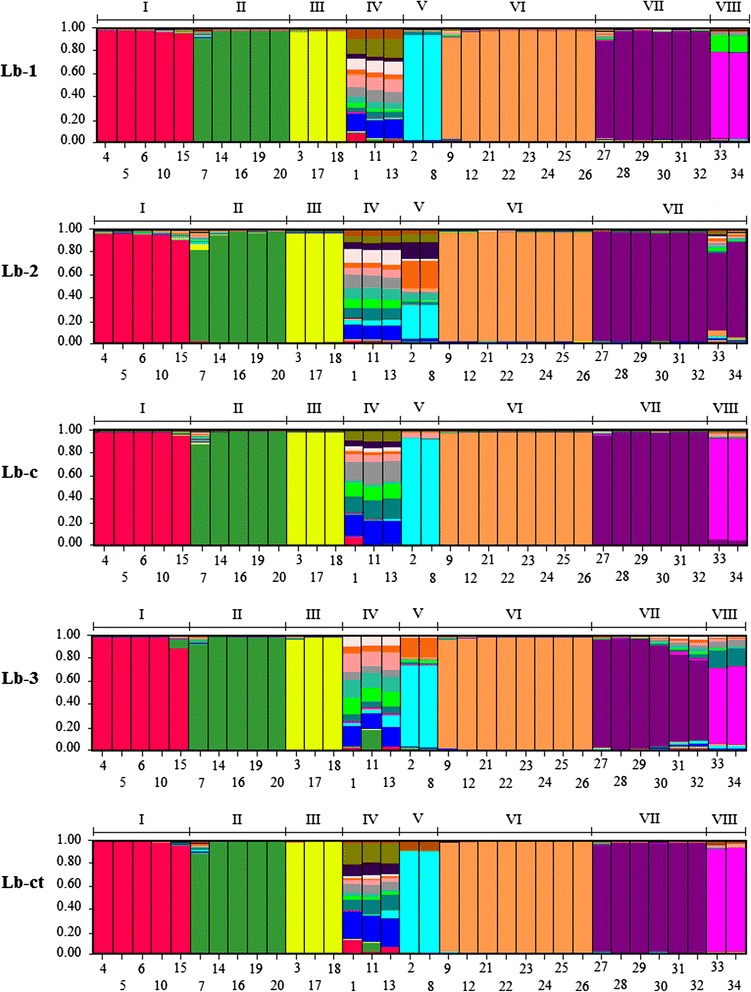
Analysis performed with STRUCTURE software. Lb-1: Library constructed from a sexual accession (H031), Lb-2: Library constructed from a pool of eight apomictic accessions, Lb-3: Library constructed from an apomictic accession (H016) [12], Lb-c: Joint analysis of Lb-1 and Lb-2, Lb-ct: Joint analysis of Lb-1, Lb-2, and Lb-3. Each of the 34 genotypes is represented by a single column divided into colored segments with lengths proportional to each of the allelic pools inferred by K through Evanno method [24]. Each K is represented by a different color and Lb-1 presented K = 18, Lb-2 K = 17, Lb-c K = 17, Lb-3 K = 15, and Lb-ct K = 18. The individuals were grouped into clusters according to the Q proportion of each allelic pool in an individual. Eight clusters were identified for Lb-1, Lb-c, Lb-3, and Lb-ct (I to VIII) and seven clusters for Lb-2 (I to VII). The left scale indicates the association coefficient (Q) for the assignment of genotypes into groups. The genotypes are named according to the annotated numbers listed in Table 1.
The STRUCTURE analysis for Lb-3 and Lb-ct showed K = 15 and K = 18 allelic pools, respectively (Figure 1), classified in the same clusters as for Lb-1 and Lb-c.
Discussion
In the present study, we described 72 new SSRs for U. humidicola, 64 of which are polymorphic. Along with the 67 previous developed SSRs [12,13], these markers contribute to the genetic breeding of the species and other species of the genus Urochloa in efforts to obtain new cultivars and better understanding of the species genetic, through genetic mapping, marker-assisted selection, genome sequencing and synteny.
The increased occurrence of di-nucleotide motifs for Lb-1 and Lb-2 is in accordance with the enrichment of both libraries with (CT)8 and (GT)8 probes. In addition, Morgante et al. [30] reported a higher occurrence of microsatellites with di-nucleotide motifs in plants, which may have been a contributing factor in our observation.
Among the microsatellites analyzed, 88% had a polymorphism among the evaluated genotypes. The most informative loci in this panel of SSRs were those with the highest PIC and DP values (BhUNICAMP075 and BhUNICAMP107). Locus BhUNICAMP094 showed the lowest values for PIC and DP, at 0.3165 and 0.3969, respectively, even though it was amplified in all the Urochloa species evaluated. This also occurred with the BhUNICAMP030 locus [12]. Both loci may be useful markers for studies in Urochloa because it may be the result of a conserved region among the species studied herein. Monomorphic loci may be useful in other studies.
The transferability rates of the loci from U. humidicola to four other species were very similar. Although these results were not highly variable, U. dictyoneura presented the highest transferability, corroborating the genetic closeness between U. dictyoneura and U. humidicola, as has been previously described [2,31] and the results obtained in another study [13].
For the population structure analysis, different numbers of allelic pools [K] were observed for all analyses. However, the individual composition presented in each cluster was maintained into Lb-1, Lb-c, Lb-3, and Lb-ct analyses, but for Lb-2 analysis, the Clusters VII and VIII were grouped into Cluster VII.
The genotypes of the species U. brizantha, U. decumbens, and U. ruziziensis were grouped into the same cluster in all the analyses. However, the U. dictyoneura genotypes were grouped separately from the other species for all the analyses, except for Lb-2, with the four species grouping into Cluster VII.
In all analyses, Cluster VI included accessions 9 and 12, and six hybrids derived from crosses between these two accessions grouped together. However, in a previous study, the progenitors did not group together with the hybrids [13], as only runs from K = 1 to K = 10 were performed. These hybrids are part of an F1 population that is being mapped with the SSRs described in this study and previously published [12,13].
Although some discrepancies were found among the three libraries (Lb-1, Lb-2, and Lb-3), the set of loci belonging to each was able to satisfactorily differentiate the accessions. Comparing the three libraries developed, Lb-1 presented the highest number of allelic pools, which may be correlated to the usage of the accession H031, a highly diverse genotype, as described by [7]. The genotype used for the enriched library construction directly influences the results. The joint analysis of the three libraries (Lb-ct) would be the most recommended way to differentiate among accessions, because it uses loci derived from many different genotypes, conferring a greater reliability of the observed results.
These markers are immediately useful for U. humidicola breeding programs, aiding in areas such as the construction of linkage and QTL maps, gene flow and mating system evaluation, and marker-assisted selection.
Availability of supporting data
The datasets supporting the results of this article are included in the article.
Acknowledgements
The authors thank Dr. Cacilda Borges do Valle, Dr. Leticia Jungmann and the Brazilian Agricultural Research Corporation (EMBRAPA Beef Cattle) for providing the Urochloa accessions used. This work was supported by grants from Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP, 2008/52197-4) and a scholarship to FAO (2013/14903-2). JCSS is a recipient of a scholarship from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES-EMBRAPA Program), and MAB is a recipient of a scholarship from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). APS is a recipient of a research fellowship from CNPq.
Abbreviations
- A
Maximum number of alleles observed
- AN
Annotation number
- BRA
Codes of the accessions from EMBRAPA
- CAPES
Coordination of Improvement of Higher Education Personnel
- CIAT
Center for Tropical Agriculture
- CTAB
Cetyltrimethyl ammonium bromide
- DNA
Deoxyribonucleic acid
- DP
Discrimination power
- EBC
Embrapa beef cattle
- EMBRAPA
Brazilian Agricultural Research Corporation
- FAPESP
Foundation for Research Support of the State of Sao Paulo
- K
Number of clusters
- Lb-1
Library construction from a sexual accession (H031)
- Lb-2
Library construction from a pool of eight apomictic accessions
- Lb-3
Library construction from an apomictic accession (H016) [12]
- Lb-c
Joint analysis of Lb-1 and Lb-2
- Lb-ct
Joint analysis of Lb-1, Lb-2, and Lb-3
- MCMC
Markov Chain Monte Carlo
- NA
Not available
- bp
Base pairs
- PCR
Polymerase chain reaction
- PIC
Polymorphism information content
- Q
Association coefficient from STRUCTURE analysis
- QTL
Quantitative trait locus
- SSR
Simple sequence repeat
- Ta (°C)
Annealing temperature
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JCSS and MAB developed the microsatellite-enriched libraries, participated in the microsatellite marker validation, performed the statistical analysis, and drafted the manuscript. JCSS and FAO carried out computational searches for microsatellite identification and drafted the manuscript. BBZV participated in the design and implementation of the study and helped to draft the manuscript. APS conceived of and supervised the study and helped to draft the manuscript. All authors read and approved the final manuscript.
Contributor Information
Jean CS Santos, Email: jsantos.agro@gmail.com.
Mariana A Barreto, Email: maribiologista@gmail.com.
Fernanda A Oliveira, Email: f.ancelmo.o@gmail.com.
Bianca BZ Vigna, Email: bianca.vigna@embrapa.br.
Anete P Souza, Email: anete@unicamp.br.
References
- 1.Sendulsky T. Brachiaria: taxonomy of cultivated and native species in Brazil. Hoehnea. 1978;7:99–139. [Google Scholar]
- 2.Renvoize SA, Clayton WD, Kabuye CHS. Morphology, Taxonomy and Natural Distribution of Brachiaria (Trin.) Griseb. In: Miles JW, Maass BL, Valle CB, editors. Brachiaria: Biology, agronomy, and improvement. Embrapa/CIAT; 1996. p.1-15.
- 3.Adamowski EV, Boldrini KR, Pagliarini MS, Valle CB. Abnormal cytokinesis in microsporogenesis of Brachiaria humidicola (Poaceae: Paniceae) Genet Mol Res. 2007;6:616–21. [PubMed] [Google Scholar]
- 4.Boldrini KR, Pagliarini MS, do Valle CB. Meiotic behavior of a nonaploid accession endorses x = 6 for Brachiaria humidicola (Poaceae) Genet Mol Res. 2009;8:1444–50. doi: 10.4238/vol8-4gmr679. [DOI] [PubMed] [Google Scholar]
- 5.Boldrini KR, Micheletti PL, Gallo PH, Mendes-Bonato AB, Pagliarini MS, do Valle CB. Origin of a polyploid accession of Brachiaria humidicola (Poaceae: Panicoideae: Paniceae) Genet Mol Res. 2009;8:888–95. doi: 10.4238/vol8-3gmr617. [DOI] [PubMed] [Google Scholar]
- 6.Boldrini KR, Pagliarini MS, do Valle CB. Evidence of natural hybridization in Brachiaria humidicola (Rendle) Schweick. (Poaceae: Panicoideae: Paniceae) J Genet. 2010;89:91–4. doi: 10.1007/s12041-010-0016-z. [DOI] [PubMed] [Google Scholar]
- 7.Jungmann L, Vigna BBZ, Boldrini KR, Sousa ACB, do Valle CB, Resende RMS, et al. Genetic diversity and population structure analysis of the tropical pasture grass Brachiaria humidicola based on microsatellites, cytogenetics, morphological traits, and geographical origin. Genome. 2010;53:698–709. doi: 10.1139/G10-055. [DOI] [PubMed] [Google Scholar]
- 8.Moreira LM, Martuscello JA, Fonseca DM, Mistura C, Morais RV, Júnior JIR. Perfilhamento, acúmulo de forragem e composição bromatológica do capim-braquiária adubado com nitrogênio. In: Revista Brasileira de Zootecnia.2009. p. 1675–1684. http://www.scielo.br/pdf/rbz/v38n9/06.pdf. Accessed: 02 April 2014.
- 9.Keller-Grein G, Maass BL, Hanson J. Natural variation in Brachiaria and existing germplasm collections. In: Miles JW, Maass BL, Valle CB, editors. Brachiaria: biology, agronomy and improvement. Embrapa/CIAT; 1996. p.16-42.
- 10.Rauscher G, Simko I. Development of genomic SSR markers for fingerprinting lettuce (Lactuca sativa L.) cultivars and mapping genes. BMC Plant Biol. 2013;13:11. doi: 10.1186/1471-2229-13-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhat PR, Krishnakumar V, Hendre PS, Rajendrakumar P, Varshney RK, Aggarwal RK. Identification and characterization of expressed sequence tags-derived simple sequence repeats markers from robusta coffee variety ‘CxR’ (an interspecific hybrid of Coffea canephora x Coffea acongensis) Mol Ecol Notes. 2005;5:80–3. doi: 10.1111/j.1471-8286.2004.00839.x. [DOI] [Google Scholar]
- 12.Jungmann L, Vigna BBZ, Paiva J, Sousa ACB, do Valle CB, Laborda PR, et al. Development of microsatellite markers for Brachiaria humidicola (Rendle) Schweick. Conserv Genet Resour. 2009;01:475–9. doi: 10.1007/s12686-009-9111-y. [DOI] [Google Scholar]
- 13.Vigna BBZ, Alleoni GC, Jungmann L, do Valle CB, Souza AP. New microsatellite markers developed from Urochloa humidicola (Poaceae) and cross amplification in different Urochloa species. BMC Res Notes. 2011;4:523. doi: 10.1186/1756-0500-4-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987;19:11–5. [Google Scholar]
- 15.Billotte N, Lagoda PJR, Risterucci AM, Baurens FC. Microsatellite-enriched libraries: applied methodology for the development of SSR markers in tropical crops. Fruits. 1999;54:277–88. [Google Scholar]
- 16.Thiel T. MISA — MIcroSAtellite identification tool, Version 1.0. In: MISA — MIcroSAtellite identification tool. Leibniz Institute of Plant Genetics and Crop Plant Research. 2001. http://pgrc.ipk-gatersleben.de/misa/misa.html. Accessed 21 August 2012.
- 17.Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JA. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007;35:W71–4. doi: 10.1093/nar/gkm306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Creste S, Tulmann Neto A, Figueira A. Detection of single sequence repeat polymorphisms in denature polyacrylamide sequencing gels by silver staining. Plant Mol Bio Rep. 2001;19:299–306. doi: 10.1007/BF02772828. [DOI] [Google Scholar]
- 19.Esselink GD, Nybom H, Vosman B. Assignment of allelic configuration in polyploids using the MAC-PR (microsatellite DNA allele counting—peak ratios) method. Theor Appl Genet. 2004;109:402–8. doi: 10.1007/s00122-004-1645-5. [DOI] [PubMed] [Google Scholar]
- 20.Clark LV, Jasieniuk M. Polysat: an R package for polyploid microsatellite analysis. Mol Ecol Resour. 2011;11:562–6. doi: 10.1111/j.1755-0998.2011.02985.x. [DOI] [PubMed] [Google Scholar]
- 21.Mateescu RG, Zhang Z, Tsai K, Phavaphutanon J, Burton Wursten NI, Lust G, et al. Analysis of allele fidelity, polymorphic information content, and density of microsatellites in a genome-wide screening for hip dysplasia in a crossbreed pedigree. J Hered. 2005;96:847–53. doi: 10.1093/jhered/esi109. [DOI] [PubMed] [Google Scholar]
- 22.Tessier C, David J, This P, Boursiquot JM, Charrier A. Optimization of the choice of molecular markers for varietal identification in Vitis vinifera L. Theor Appl Genet. 1999;98:171–7. doi: 10.1007/s001220051054. [DOI] [Google Scholar]
- 23.Pritchard J, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–59. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–87. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes. 2007;7:574–8. doi: 10.1111/j.1471-8286.2007.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–20. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- 27.Earl DA, von Holdt BM. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Res. 2012;4:359–61. doi: 10.1007/s12686-011-9548-7. [DOI] [Google Scholar]
- 28.Jakobsson M, Rosenberg NA. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–6. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- 29.Rosenberg NA. DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes. 2004;4:137–8. doi: 10.1046/j.1471-8286.2003.00566.x. [DOI] [Google Scholar]
- 30.Morgante M, Hanafey M, Powell W. Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat Genet. 2002;30:194–200. doi: 10.1038/ng822. [DOI] [PubMed] [Google Scholar]
- 31.Gonzalez AMT, Morton CM. Molecular and morphological phylogenetic analysis of Brachiaria and Urochloa (Poaceae) Mol Phylogenet Evol. 2005;37:36–44. doi: 10.1016/j.ympev.2005.06.003. [DOI] [PubMed] [Google Scholar]


