Abstract
In dividing animal cells, a microtubule (MT)-based bipolar spindle governs chromosome movement. Current models propose that the spindle facilitates and/or generates translocating forces by regionally depolymerizing the kinetochore fibers (k-fibers) that bind each chromosome. It is unclear how conserved these sites and the resultant chromosome-moving mechanisms are between different dividing cell types because of the technical challenges of quantitatively studying MTs in many specimens. In particular, our knowledge of MT kinetics during the sperm-producing male meiotic divisions remains in its infancy. In this study, I use an easy-to-implement photobleaching-based assay for measuring spindle MT dynamics in primary cultures of meiotic spermatocytes isolated from the fruit fly Drosophila melanogaster. By use of standard scanning confocal microscopy features, fiducial marks were photobleached on fluorescent protein (FP)-tagged MTs. These were followed by time-lapse imaging during different division stages, and their displacement rates were calculated using public domain software. I find that k-fibers continually shorten at their poles during metaphase and anaphase A through the process of MT flux. Anaphase chromosome movement is complemented by Pac-Man, the shortening of the k-fiber at its chromosomal interface. Thus, Drosophila spermatocytes share the sites of spindle dynamism and mechanisms of chromosome movement with mitotic cells. The data reveal the applicability of the photobleaching assay for measuring MT dynamics in primary cultures. This approach can be readily applied to other systems.
Keywords: meiosis, microtubule flux, time-lapse imaging, scanning confocal microscopy, Pac-Man
INTRODUCTION
Light microscopy has been an invaluable tool in dissecting the mechanisms by which animal cells divide and propagate. It has been more than century since Flemming identified the fundamental structures of cell division, the chromosomes, and bipolar spindle.1 We now know that the spindle is a tubulin polymer-based scaffold that is essential for chromosome alignment at metaphase and subsequent anaphase segregation. Movement requires kinetochore fibers (k-fiber), specialized bundles of microtubules (MTs) that link the kinetochore of each chromosome’s centromere to the spindle’s pole. Errors in these interactions and the regulated events responsible for chromosome partitioning can lead to aneuploidy and a host of pathologies, including cancer, sterility, and developmental defects.2,3 Although the dynamic events of cell division can be observed to differing extents in living cells using various transmitted light imaging techniques (e.g., differential interference contrast, phase-contrast, polarization), the optical properties of the cellular milieu makes their investigation most amenable to study by epifluorescence microscopy.
Seminal early attempts to visualize spindle MTs in living specimens used mitotic tissue culture cells and the pressure-mediated microinjection of tubulin subunits conjugated to a fluorescently labeled probe. The progressive incorporation of these molecules into the dynamically exchanging tubulin polymer allowed for direct MT observation. By monitoring the integration of the fluorescent signal or following the displacement of marks generated by photobleaching labeled MTs or photoactivating and imaging previously nonradiating tags, the sites and rates of tubulin incorporation, MT elongation, and depolymerization were determined.4–7 The spatial resolution of MT dynamics studies was improved with the introduction of fluorescence speckle microscopy.8 This method is based on the integration of substoichiometric amounts of fluorescently tagged tubulin into the polymer. As the MT stochastically incorporates subunits from labeled and unlabeled pools, “speckles” appear along the length of MTs. These can be visualized by epifluorescence microscopy without adapting the imaging setup or using additional illumination sources as may be necessary for photobleaching or excitation manipulations. By quantitatively tracking the punctae, the kinetics of k-fiber MT elongation and shortening at the chromosome proximal MT plus ends or the spindle pole associated minus ends can be determined. When combined with functional assays, they reveal that regional k-fiber dynamics are key determinants of chromosome movement.9–12
Not all specimens are amenable to microinjection of recombinant proteins. The technical constraints of the microinjection approach have hindered the analysis of MT dynamics in different cell types. Some cells may not bind securely enough to the substrate for needle insertion. They may also be too fragile to recover from the mechanical insult inherent in the procedure. Alternatively, samples may be maintained in cultures that preclude the use of conventional apparatus.
Gene fusions between genetically encoded fluorescent protein (FP) tags, such as enhanced green fluorescent protein (EGFP) or any of a spectrum of other colored variants, and MT-associated proteins provide an alternative to microinjection for visualizing MTs in living cells. Tagged transgenes can be rapidly engineered and introduced into virtually all model systems. In addition, depending on the regulatory elements used, the protein fusion can be selectively induced offering cell type- or stage-specific expression. Because FPs can be fused with MT-associated proteins that exclusively target either the plus or minus ends,13–16 they can be used as polarity markers in vivo. However, this may be disadvantageous in some contexts as such proteins do not allow the visualization of the entire MT polymer. Whole MT imaging is needed for accurately documenting changes in net MT length and position. In addition, FP-tagged MT-associated proteins may not bind along the entire length of MT’s lattice, or they may associate preferentially with different spindle MT subpopulations. This can lead to a misrepresentation of MT size or distribution. These shortcomings can be averted by expressing tubulin that has been labeled with a FP. As the position of the FP tag may alter protein behavior, protein fusion functionality must be assessed before use. Once validated, FP-tubulin represents a convenient and reliable method for studying the spindle in living material that is analogous to the microinjection studies described above.
The ability to readily quantify and compare spindle MT dynamics in different experimental systems is critical to understanding the processes responsible for chromosome movement and maintaining genetic integrity. Studies in mitotic models, as well as male meiotic spermatocytes isolated from the crane fly Nephrotoma suturalis, suggest that the sites and rates of MT growth and shrinkage can vary over the course of division and between dividing cell types. In vertebrate systems, metaphase chromosomes continue to oscillate after reaching the metaphase plate at the spindle equator, a phenomenon that does not appear to occur in insect mitotic or meiotic cells.17–21 Although the data are incomplete, even in cases where no chromosome displacement or k-fiber elongation is observed, tubulin appears to be incorporated continually at the plus ends of k-fiber MTs.22 The absence of k-fiber extension is attributed to the process of MT flux, the shortening and translocation of MTs at their minus ends.6,23 Because the rates of growth and shrinkage are balanced, the k-fiber undergoes treadmilling and maintains a steady-state length as subunits are exchanged with the cellular pool.4–7,21–26 The function of MT flux during metaphase is controversial, and it has been implicated in roles ranging from chromosome alignment to increasing the fidelity of anaphase chromosome partitioning.12,22,24,27
By contrast, MT flux has an established and conserved role in the chromosome-to-pole movements of anaphase A. In mitosis, the depolymerization of k-fiber minus ends pulls disjoining chromosomes away from the equator.11,22,24 Absolute flux velocities can vary between experimental systems, but most studies report that it remains constant or slows relative to metaphase and accounts for a minority of polewards chromosome travel. The remaining segregation movement is attributed to Pac-Man activity. This mechanism uses kinetochore associated motor proteins and other local force producers to move the chromosome polewards as the k-fiber shortens at its plus ends.4,7,11,24,28 It is noteworthy that experiments in meiotic crane fly cells indicate that k-fibers do not undergo anaphase MT plus-end depolymerization. Rather, k-fiber MTs exhibit a continuous anti-Pac-Man behavior marked by plus-end elongation. The chromosomes ultimately reach opposing poles through the elevation of the flux rate.21 The evolutionary advantage of this variation is unclear. Given the paucity of data on MT dynamics in male meiotic systems, it also remains unknown if such a mechanistic divergence between mitosis and male meiosis is conserved or occurs uniquely when examining crane flies.
The primary spermatocyte of the fruit fly Drosophila melanogaster is a well-established system for studying the meiotic divisions of the male germline. Drosophila has exquisite genetics, and spermatocytes are amenable to high-resolution live-cell light microscopy investigations. The specialized and differentiated nature of these cells prevents their in vitro propagation. Therefore, different primary culturing methods have been developed for their study. These vary according to the duration of the experiment. All require the dissection and removal of the testes from the organism and the extraction of the dividing cells from within. Short-term observations (several hours) commonly rely on covering individual isolated cells with inert and gas permeable oils, which has an additional cell-flattening effect.17,19,29 Longer investigations (12 h or more) use bathing of cells, isolated as intact multicellular cysts, in tissue culture media supplemented with serum.30 Neither culturing approach is compatible with pressure-based microinjection. Spermatocytes only partially adhere to the coverslip, and the longer term cultures use enclosed chambers. The properties of the cortex and membrane system further make the cells resistant to conventional approaches (unpublished observations). Insect spermatocytes from other organisms are similarly difficult to microinject.31 In several instances, this has been overcome through the implementation of specialized or purpose-engineered instrumentation,21,32 which may not be available to most laboratories.
FP technology allows for the live-cell imaging of MTs in primary cultures of Drosophila male meiotic cells. The continuous MT labeling observed when overexpressing FP-tagged tubulin in spermatocytes makes it a valuable tool for studying MT distribution. Such fusions have shown that the meiotic spindle assembles and matures through novel and complex pathways that differ from mitotic tissues.33,34 In contrast, these FP-tubulin fusions appear poorly suited for investigating MT dynamics. Spindle MTs are labeled homogeneously along their lengths. The absence of any fluorescence heterogeneities to serve as internal references or speckles12 precludes the identification of sites of growth or shrinkage. Thus, despite its recognized utility as a male meiotic model system and amenability to high-resolution live-cell imaging, the dynamic properties of the Drosophila spermatocyte spindle remain undefined.
To elucidate how the spindle directs chromosome movement in male meiosis, I used a photobleaching-based assay to characterize MT dynamics in Drosophila spermatocytes during metaphase and anaphase A. A scanning confocal microscope was used to photobleach fiducial marks onto the k-fibers of cells expressing EGFP-tagged tubulin. The displacement of these marks along fluorescing MTs was quantified relative to their plus and minus ends using the public domain program ImageJ (National Institutes of Health, Bethesda, MD, USA).35 This revealed the sites and rates of k-fiber MT growth and shrinkage. The data indicate that flux occurs continuously during metaphase and anaphase. Its rate increases during the latter where, along with Pac-Man, flux mediates chromosome segregation. Thus, as with mitotic systems, both flux and Pac-Man are the determinants of chromosome movement in Drosophila spermatocytes. These results demonstrate the utility of combining photobleaching with FP fusions as a tool for studying MT dynamics in spermatocyte primary cultures. Because the assay is based on standard scanning confocal microscope features and public domain software, it can be applied to other specimens with FP-tagged MTs without additional equipment investment or modification of established live-cell imaging protocols.
Materials and Methods
Drosophila Husbandry
All experiments used flies ubiquitously expressing β-tubulin56D::EGFP as described previously.36 Flies were reared at 25°C according to standard procedures.
Drosophila primary culturing
Experiments were performed using newly eclosed adults. Testes were dissected from individuals in phosphate-buffered saline (pH 7.4) and cleaned of unwanted adherent tissues. These organs were transferred subsequently to an upright microscope imaging chamber.19 This was constructed by placing a drop of molten VALAP (1:1:1 Vaseline petroleum jelly/lanolin/paraffin) on each corner of a clean number 1 thickness coverslip that had been coated previously with 0.1 mg/ml concanavalin A (Sigma-Aldrich, St. Louis, MO, USA). Two concentric circles were drawn in the center of the coverslip using Vaseline petroleum jelly extruded from a syringe. The innermost circle was filled with Voltalef 10s oil (VWR International, Radnor, PA, USA). After immersion of the testes in the oil, additional aqueous contaminants were blotted away with strips of number 1 Whatman filter paper. The testes then were ruptured, and the cells were spread using super fine forceps (ProSciTech, Kirwan, Australia). The coverslip with cells was inverted, positioned centrally on a glass slide, and gently squashed to seal the chamber. Under these conditions, cells remained viable for several hours.19
Scanning confocal microscopy and photobleaching
Culture chambers were placed on a Leica SP5 upright scanning confocal microscope system and maintained at ambient room temperature (24 ± 1°C). Cells were imaged using a Leica DM6000B microscope outfitted with a 63× apochromatic numerical aperture 1.4 lens. Image acquisition was controlled by the Leica LAS AF software (version 2.7.3.9723; Leica Microsystems, Buffalo Grove, IL, USA). Every 2 seconds, the 488 nm laser line (20% laser power) from an argon laser (operating at 20% of maximum output) was used to illuminate a single optical plane with the EGFP (detection window: 493–571 nm; pinhole 1.2 airy units), and differential interference contrast (DIC) images simultaneously captured by independent detectors. To decrease acquisition time, the scan area was set to 512 × 512 pixels (zoom factor of 6; final pixel size ∼80 nm), and no image averaging was performed.
Photobleaching was carried out with “FRAP Wizard” mode of the LAS AF software. This entailed defining regions of interest (ROIs) between the k-fiber’s chromosome proximal plus ends and the minus ends found at the spindle poles. The fluorescence signal was bleached by increasing the 488 nm laser line power to 100% and sequentially scanning each ROI at least 6 times.
Flux and Pac-Man Velocity Determination
All image processing and analyses were performed using the public domain software program ImageJ (version 1.49) (http://www.imagej.nih.gov/ij/).35 This consisted of importing the Leica data sets into ImageJ using the Bio-Formats Importer plugin (http://www.openmicroscopy.org/site/support/bio-formats5/). The fluorescence and transmitted light channels were separated and viewed as independent 8-bit sequences. All analyses were performed on fluorescence channel data with the transmitted light images only serving for reference or being used for illustration purposes. The background fluorescence was removed by subtracting the mean intensity value measured from the area occupied by the chromosomes. No discrimination was made between autosome and sex chromosome k-fibers. Only those k-fibers that could be traced clearly from their plus ends to the spindle pole for 20 or more frames were selected for analysis. These were isolated by rotating the sequence first using bicubic interpolation, such that the k-fiber was parallel to the x or y axis. The rotated data then were copied within an ROI that excluded all other spindle features, with the exception of the k-fiber and spindle pole/centrosome. To correct any registration issues within the sequence, the “StackReg” plugin37 (http://bigwww.epfl.ch/thevenaz/stackreg/) was applied using a “Rigid Body” transformation. A maximum z-projection was made of the aligned stack to confirm that the spindle pole and centrosome remained stationary and to reveal any change in k-fiber position over time. A multisegmented line was drawn atop the projected k-fiber path. This was increased to a 4 pixel width and duplicated onto the aligned stack. To generate a kymograph along this line, the “kbi Kymograph” plugin from the KbiPlugins collection (http://hasezawa.ib.k.u-tokyo.ac.jp/zp/Kbi/ImageJKbiPlugins) was used. After calibration, the slopes of the k-fiber’s plus end or the bleached region relative to the poles and centrosome (as an indicator of the minus end) were determined with the “kbi KymoMeasure” plugin from the same collection and converted into a velocity. Pac-Man rates were determined by subtracting the anaphase flux rate from the rate of plus end shortening. All calculations and distribution determinations were performed in Microsoft (MS) Excel.
Figures were prepared by capturing selected frames from time-lapse microscopy sequences and saving them as 8-bit grayscale or 24-bit color tag image file format (.tiff) images in ImageJ. These images along with graphs generated in MS Excel were then imported into Adobe Photoshop CC for final figure generation.
RESULTS AND DISCUSSION
Toward defining the dynamic properties of the male meiotic spindle and its contribution to chromosome movement, I used primary spermatocytes isolated from the fruit fly D. melanogaster that stably express β-tubulin56D::EGFP. These cells are well suited to meiotic studies by live-cell light microscopy. The metaphase spindle is relatively large (∼25 µm) allowing for the easy discrimination of the 4 chromosomes and their individual k-fibers by DIC and simultaneous scanning confocal microscopy of the EGFP signal using the 488 nm laser line from an argon laser (Fig. 1A). Although the cells became flattened by the Voltalef oil and concanavalin A,34 their spindles commonly rotated in the z axis. Attempts to restrict rotation with higher concentrations of concanavalin A were unsuccessful with cells becoming damaged mechanically during culturing or forming aberrant spindles as a result of being overly spread. Therefore, all imaging was done with the pinhole expanded from the default 1.0 airy units to 1.2 airy units increasing the optical section thickness to ∼0.9 µm.
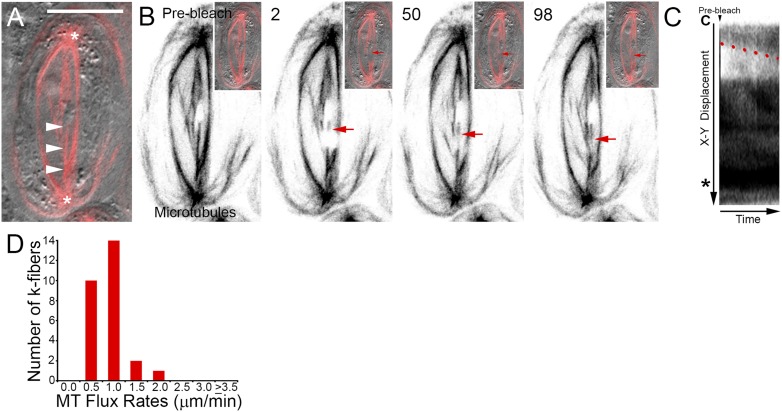
Figure 1.
Drosophila primary spermatocyte kinetochore fibers undergo microtubule flux during metaphase. A) Dual channel scanning confocal image showing spindle morphology and chromosome position. MTs labeled with EGFP are shown in red, and the DIC transmitted light image is in gray. Arrowheads denote the k-fiber of interest, (*) indicates centrosomes. Scale bar is 10 µm. B) Selected frames taken from a time-lapse sequence of the cell shown in A. At the onset of the experiment, the k-fiber is homogeneously fluorescent. After photobleaching, a nonfluorescent bar is visible (arrow) that can be tracked as it moves progressively polewards (2–98). Insets show corresponding composite EGFP and DIC images. Time is in seconds relative to the photobleaching event. C) Kymograph of the photobleached k-fiber. Over time, the bleached region travels away from the chromosome (C) and toward the centrosome (*). No change in k-fiber length occurs as the mark moves. The rate of flux can be calculated from the slope of the line (red) delineating the displacement of the bleached zone. D) Distribution of metaphase flux rates.
Studies in mitotic and male meiotic models suggest that the fluxing of k-fibers is a conserved process among animal spindles. To date, there is only indirect evidence of MT flux in Drosophila primary spermatocytes. This comes from transmitted light observations of metaphase spindle shortening following MT plus end stabilization with the agent Taxol,19 an event correlated with flux in other systems.26,38,39 To determine the dynamic status of metaphase k-fibers, time-lapse recordings were made of engineered fiducial marks positioned midway between the kinetochores and the spindle poles. This was done by iteratively scanning a user-defined rectangular ROI with the 488 nm laser line at increased laser power until the fluorescence signal intensity was reduced to levels approaching the non-MT containing background. Adjacent MT polymerization events sometimes diminished the distinctness of the bleached zone. Nevertheless, the absence of fluorescence within this region provided a marker whose displacement over time could be tracked along the otherwise fluorescent k-fiber length and relative to the chromosome-associated plus end and spindle pole and centrosome proximal minus end. The bleached regions appeared to consistently travel toward the spindle poles (Fig. 1B, arrows) without any notable change in k-fiber or spindle length. This was confirmed by generating kymograph plots using the public domain program ImageJ. These graphical summaries show the displacement of the mark over time as represented in Fig. 1C. Neither the chromosome proximal plus end (C) nor the centrosome (*) and spindle pole adjacent minus end signals exhibit any positional change during the course of the experiment as would be expected from alterations in k-fiber length. Therefore, the poleward movement of the bleached mark (red line) is attributed to the fluxing of MTs and the associated shortening and translocation of k-fiber minus ends. Quantification of the slopes delineated by the bleached region revealed the average MT flux rate to be 0.6 ± 0.1 µm/min (N = 27). Most k-fibers fluxed between 0.5 and 1.0 µm/min (Fig. 1D, 24/27). These rates are similar to those reported for crane-fly spermatocytes (∼0.7 µm/min21,24). It is noteworthy that, although Drosophila spermatocyte chromosomes exhibit little movement following metaphase alignment,19,40,41 opposing k-fibers within a homologous pair could flux at rates varying by up to 2-fold. This is unexpected as equatorial chromosome positioning in spermatocytes is sensitive to the number of MTs in each k-fiber.42 The photobleaching assay does not quantify kinetochore MT numbers. However, here it reveals an imbalance in MT dynamics and presumptive polewards acting forces. Understanding how chromosomes remain aligned stably under these conditions will require further investigation.
Scanning confocal microscopy was used next to study spindle MT dynamics during anaphase A chromosome segregation. Unlike metaphase, k-fibers did not always remain straight during this time and sometimes appeared bent or to have undergone slight rotations near the spindle poles as the chromosomes traveled polewards. The rate of chromosome segregation as indicated by decreasing k-fiber length was 2.2 ± 0.2 µm/min (N = 30). Further inspection revealed this to be biphasic: a slow chromosome disjunction phase (average rate: 1.1 ± 0.2 µm/min; N = 8) and a more rapid shortening that occurred after the chromosomes had separated (average rate: 2.6 ± 0.2 µm/min; N = 22).
To determine whether chromosome segregation is governed by k-fiber MT depolymerization at the minus end (flux) or the plus end (Pac-Man), the photobleaching assay was applied further. Attempts to follow photobleached marks generated in late metaphase and before chromosome segregation were unsuccessful because of the spindle rocking out of the plane of focus. Therefore, cells were selected in which anaphase A had been initiated. In 100% of the k-fibers followed (N = 30), the photobleached mark, which was generated between the pole and the chromosome, moved polewards consistent with MT flux. Concurrent with this, the distance between the chromosome at the k-fiber’s plus end and the photobleached mark also decreased. Therefore, as with mitotic systems, Drosophila male meiotic cells engage both flux and Pac-Man activities (Fig. 2).
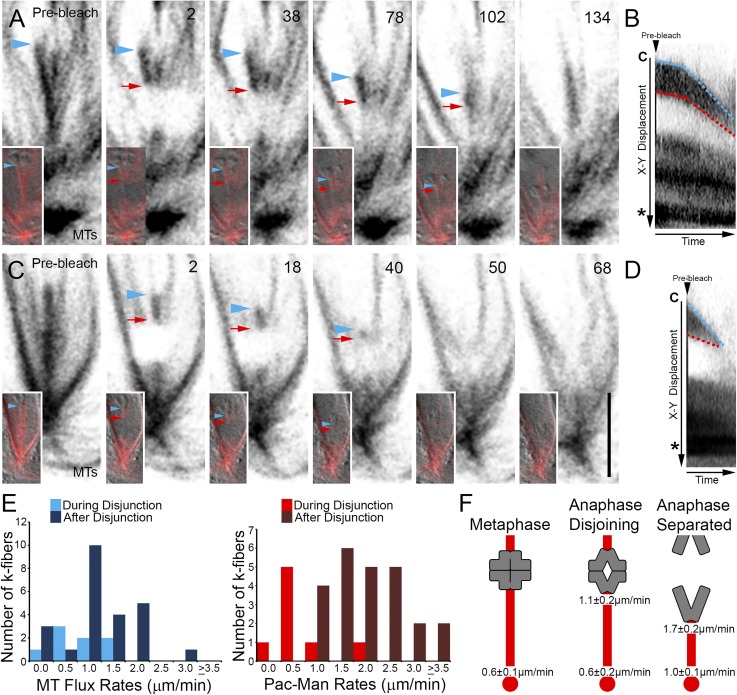
Figure 2.
Drosophila primary spermatocyte chromosomes segregate by Pac-Man and flux mechanisms. A) Selected frames taken from a time-lapse sequence of a cell entering and progressing through anaphase A. Insets show MTs overlaid on the DIC image. The k-fiber plus end and chromosome position is denoted by blue arrowheads, the boundary of the bleach region is indicated by red arrows. After the photobleaching event, the chromosome and bleached mark move polewards (2–134). The chromosome approaches the bleached mark before the k-fiber signal is lost. Time is in seconds relative to photobleaching. B) Kymograph of the photobleached k-fiber in A. The separation distance between the chromosome (C) and bleached mark decreases as both move toward the pole and centrosome (*) revealing k-fiber plus end depolymerization (Pac-Man) and minus end MT flux. Demarcation of the k-fiber’s plus end (blue lines) and that of the bleached region (red lines) indicate distinct slow and fast k-fiber dynamic phases. C) Selected frames from a time-lapse series of an anaphase cell following chromosome disjunction. Similar to A, as the chromosome (blue arrowheads) segregates, it overtakes the polewards moving bleached mark (red arrows), demonstrating Pac-Man and flux activities. Time is in seconds relative to the photobleaching event. Scale bar is 5 µm. D) Kymograph of the photobleached k-fiber in C confirms the rapid shortening of the k-fiber plus end adjacent to the chromosome (C; blue line) by Pac-Man while the entire k-fiber shortens at the minus end drawing the bleached fiducial mark toward the centrosome (*) as it undergoes MT flux. E) Distribution of MT flux and Pac-Man rates during chromosome disjunction and after their separation. F) Graphical summary of the kinetic findings of this study. K-fibers are shown in red and chromosomes in gray. Numbers are the average local depolymerization rates for k-fiber MT plus or minus ends during the indicated period.
The observation of Pac-Man was unexpected given findings made in crane-fly spermatocytes. To identify the extent that both this and flux contribute to chromosome movement in Drosophila, kymographs were generated and the rates at which k-fiber plus ends and the photobleached regions moved polewards were determined. As shown in Fig. 2, the two segregation mechanisms changed velocity throughout the course of anaphase. During disjunction, the k-fibers fluxed on average at 0.6 ± 0.2 µm/min (N = 8), a value virtually indistinguishable from metaphase. This increased to 1.0 ± 0.1 µm/min (N = 22) after the chromosomes had separated. Analysis of Pac-Man rates within the same periods revealed that, whereas k-fiber MT plus ends shortened at 1.1 ± 0.2 µm/min (N = 8) at anaphase onset, this increased by ∼55% to 1.7 ± 0.2 µm/min (N = 22) once the homologs were no longer attached to one another (Fig. 2E). The sites and rates of MT dynamism found in this study are summarized in Fig. 2F.
The pathways regulating k-fiber MT dynamics are not fully understood. It has been proposed that the application of mechanical resistance against the kinetochore promotes k-fiber plus-end elongation.10 If correct, the loss of the antagonizing force that occurs following chromosome disjunction should shift the k-fiber plus-end dynamic state from polymerization to a neutral or depolymerization one and induce the transition from metaphase treadmilling to anaphase Pac-Man. This prediction has been observed in insect spermatocytes. In anaphase crane flies, the premature dissolution of the homologs by laser microbeam paused the k-fiber plus end polymerization characteristic of these cells.43 Moreover, under some conditions, Pac-Man was induced leading to plus-end shortening.44 Moreover, the data reported here for Drosophila male meiotic cells directly correlate the up-regulation of plus end depolymerization with the naturally occurring diminution of resistance that follows chromosome separation. The molecules that govern Drosophila spermatocyte MT dynamics for chromosome movement await identification. In mitosis, Pac-Man and MT flux are driven by members of the kinesin-13 family of MT depolymerases.11,25,45 The conservation of these meiotic processes with their mitotic counterparts suggests similar molecular players. However, recent reports from Drosophila indicate that spindle assembly pathways vary between the two dividing cell types.34,46–48 Therefore, it remains to be determined whether the plus and minus end effectors are shared or whether spermatocytes use alternative depolymerization mechanisms. Elucidating the molecules involved is an exciting future challenge that will require the application of the photobleaching assay to cells manipulated by genetic or pharmacological methods.
In this study, I applied a photobleaching and FP-based approach for studying MT dynamics in primary cultures of EGFP-tagged tubulin expressing Drosophila spermatocytes. The approach uses standard scanning confocal microscope features and public domain software. The photobleaching assay lends itself to the study of specimens that are amenable to FP expression and live-cell imaging but resistant to conventional microinjection of exogenous labeled proteins. The required photomanipulations take advantage of standard scanning confocal microscope functions. Image acquisition and FP photobleaching use the same laser line but with different power attenuations. Therefore, the assay can be applied to any FP fusion that can be visualized on the available scanning confocal microscope setup. No additional equipment, technical expertise, or expenditure is required. Moreover, the assay does not require specialized sample preparation or culturing chambers beyond those normally used for live-cell microscopy, facilitating its application to tissue and primary cell cultures of divergent origins and which require different maintenance conditions. By defining multiple ROIs that are iteratively irradiated until photobleached, the user can simultaneously demarcate different MT subpopulations within in the same experiment, allowing for direct behavioral comparison. Experimental data are imported into the computer program ImageJ (National Institutes of Health) for processing and analysis. ImageJ is a highly adaptable public domain software package that performs functions commonly associated with commercial products without any associated cost. It has an interactive global user community that continually develops additional software “plug-ins” that perform a vast array of data import/export, processing, and analysis operations.35 The combination of photobleaching with ImageJ-based data manipulation offers an easy to use system for quantifying MT dynamics that can be implemented in most laboratory settings. The experimental flexibility in FP probe usage and specimen preparation makes it a powerful system for studying MT dynamics in different sample types. Information gained through performing such comparative studies is fundamental to developing a comprehensive understanding of MTs dynamics for cell division and physiology.
Acknowledgments
All imaging was performed at Massey University’s Manawatu Microscopy and Imaging Centre. This work supported by a grant from Massey University Research Fund to M.S.S. The author states there are no competing interests, financial or otherwise.
REFERENCES
- 1.Wilson EB. The Cell in Development and Heredity. 3 ed NY: MacMillan Co; 1925. [Google Scholar]
- 2.Compton DA. Mechanisms of aneuploidy. Curr Opin Cell Biol 2011;23:109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel JJ, Amon A. New insights into the troubles of aneuploidy. Annu Rev Cell Dev Biol 2012;28:189–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gorbsky GJ, Sammak PJ, Borisy GG. Microtubule dynamics and chromosome motion visualized in living anaphase cells. J Cell Biol 1988;106:1185–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gorbsky GJ, Borisy GG. Microtubules of the kinetochore fiber turn over in metaphase but not in anaphase. J Cell Biol 1989;109:653–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitchison TJ. Polewards microtubule flux in the mitotic spindle: evidence from photoactivation of fluorescence. J.Cell Biol. 1989;109:637–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhai Y, Kronebusch PJ, Borisy GG. Kinetochore microtubule dynamics and the metaphase-anaphase transition. J Cell Biol 1995;131:721–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Waterman-Storer CM, Desai A, Bulinski JC, Salmon ED. Fluorescent speckle microscopy, a method to visualize the dynamics of protein assemblies in living cells. Curr Biol 1998;8:1227–1230. [DOI] [PubMed] [Google Scholar]
- 9.Brust-Mascher I, Scholey JM. Microtubule flux and sliding in mitotic spindles of Drosophila embryos. Mol. Biol. Cell 2002;13:3967–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maddox P, Straight A, Coughlin P, Mitchison TJ, Salmon ED. Direct observation of microtubule dynamics at kinetochores in Xenopus extract spindles: implications for spindle mechanics. J. Cell Biol. 2003;162:377–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rogers GC, Rogers SL, Schwimmer TA, et al. . Two mitotic kinesins cooperate to drive sister chromatid separation during anaphase. Nature 2004;427:364–370. [DOI] [PubMed] [Google Scholar]
- 12.Matos I, Pereira AJ, Lince-Faria M, Cameron LA, Salmon ED, Maiato H. Synchronizing chromosome segregation by flux-dependent force equalization at kinetochores. J. Cell Biol. 2009;186:11–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perez F, Diamantopoulos GS, Stalder R, Kreis TE. CLIP-170 highlights growing microtubule ends in vivo. Cell 1999;96:517–527. [DOI] [PubMed] [Google Scholar]
- 14.Galjart N. Plus-end-tracking proteins and their interactions at microtubule ends. Curr. Biol. 2010;20:R528–R537. [DOI] [PubMed] [Google Scholar]
- 15.Goodwin SS, Vale RD. Patronin regulates the microtubule network by protecting microtubule minus ends. Cell 2010;143:263–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hendershott MC, Vale RD. Regulation of microtubule minus-end dynamics by CAMSAPs and Patronin. Proc Natl Acad Sci USA 2014;111:5860–5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Church K, Lin HP. Kinetochore microtubules and chromosome movement during prometaphase in Drosophila melanogaster spermatocytes studied in life and with the electron microscope. Chromosoma 1985;92:273–282. [DOI] [PubMed] [Google Scholar]
- 18.Skibbens RV, Skeen VP, Salmon ED. Directional instability of kinetochore motility during chromosome congression and segregation in mitotic newt lung cells: a push-pull mechanism. J Cell Biol 1993;122:859–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Savoian MS, Goldberg ML, Rieder CL. The rate of poleward chromosome motion is attenuated in Drosophila zw10 and rod mutants. Nat. Cell Biol. 2000;2:948–952. [DOI] [PubMed] [Google Scholar]
- 20.Savoian MS, Rieder CL. Mitosis in primary cultures of Drosophila melanogaster larval neuroblasts. J. Cell Sci. 2002;115:3061–3072. [DOI] [PubMed] [Google Scholar]
- 21.LaFountain JR Jr, Cohan CS, Siegel AJ, LaFountain DJ. Direct visualization of microtubule flux during metaphase and anaphase in crane-fly spermatocytes. Mol Biol Cell 2004;15:5724–5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buster DW, Zhang D, Sharp DJ. Poleward tubulin flux in spindles: regulation and function in mitotic cells. Mol. Biol. Cell 2007;18:3094–3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitchison T, Evans L, Schulze E, Kirschner M.. Sites of microtubule assembly and disassembly in the mitotic spindle. Cell 1986;45:515–527. [DOI] [PubMed] [Google Scholar]
- 24.Rogers GC, Rogers SL, Sharp DJ. Spindle microtubules in flux. J. Cell Sci. 2005;118:1105–1116. [DOI] [PubMed] [Google Scholar]
- 25.Ganem NJ, Upton K, Compton DA. Efficient mitosis in human cells lacking poleward microtubule flux. Curr. Biol. 2005;15:1827–1832. [DOI] [PubMed] [Google Scholar]
- 26.Wilson PJ, Forer A. Effects of nanomolar taxol on crane-fly spermatocyte spindles indicate that acetylation of kinetochore microtubules can be used as a marker of poleward tubulin flux. Cell Motil Cytoskeleton 1997;37:20–32. [DOI] [PubMed] [Google Scholar]
- 27.Ganem NJ, Compton DA. Functional roles of poleward microtubule flux during mitosis. Cell Cycle 2006;5:481–485. [DOI] [PubMed] [Google Scholar]
- 28.Sharp DJ, Rogers GC, Scholey JM. Cytoplasmic dynein is required for poleward chromosome movement during mitosis in Drosophila embryos. Nat. Cell Biol. 2000;2:922–930. [DOI] [PubMed] [Google Scholar]
- 29.Rebollo E, Gonzalez C.. Time-lapse imaging of male meiosis by phase-contrast and fluorescence microscopy. Methods Mol. Biol. 2004;247:77–87. [DOI] [PubMed] [Google Scholar]
- 30.Vazquez J, Belmont AS, Sedat JW. Multiple regimes of constrained chromosome motion are regulated in the interphase Drosophila nucleus. Curr. Biol. 2001;11:1227–1239. [DOI] [PubMed] [Google Scholar]
- 31.Czaban BB, Forer A, Wise DA. Microinjection into crane-fly spermatocytes. Biochem. Cell Biol. 1993;71:222–288. [DOI] [PubMed] [Google Scholar]
- 32.Alsop GB, Zhang D. Microtubules are the only structural constituent of the spindle apparatus required for induction of cell cleavage. J. Cell Biol. 2003;162:383–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rebollo E, Llamazares S, Reina J, Gonzalez C.. Contribution of noncentrosomal microtubules to spindle assembly in Drosophila spermatocytes. PLoS Biol. 2004;2:54–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Savoian MS, Glover DM. Differing requirements for Augmin in male meiotic and mitotic spindle formation in Drosophila. Open Biol 2014;4:140047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Collins TJ. ImageJ for microscopy. Biotechniques 2007; 43(1, Suppl)25–30. [DOI] [PubMed] [Google Scholar]
- 36.Inoue YH, Savoian MS, Suzuki T, Mathe E, Yamamoto MT, Glover DM. Mutations in orbit/mast reveal that the central spindle is comprised of two microtubule populations, those that initiate cleavage and those that propagate furrow ingression. J. Cell Biol. 2004;166:49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thévenaz P, Ruttimann UE, Unser M. A pyramid approach to subpixel registration based on intensity. IEEE Trans Image Process 1998;7:27–41. [DOI] [PubMed] [Google Scholar]
- 38.Wilson P, Forer A. Identifying the site of microtubule polymerization during regrowth of UV-sheared kinetochore fibres using antibodies against acetylated alpha-tubulin. Cell Biol Int Rep 1989;13:823–832. [DOI] [PubMed] [Google Scholar]
- 39.Waters JC, Mitchison TJ, Rieder CL, Salmon ED. The kinetochore microtubule minus-end disassembly associated with poleward flux produces a force that can do work. Mol. Biol. Cell. 1996;7:1547–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Savoian MS, Gatt MK, Riparbelli MG, Callaini G, Glover DM. Drosophila Klp67A is required for proper chromosome congression and segregation during meiosis I. J Cell Sci. 2004;117:3669–3677. [DOI] [PubMed] [Google Scholar]
- 41.Church K, Lin HP. Kinetochore microtubules and chromosome movement during prometaphase in Drosophila melanogaster spermatocytes studied in life and with the electron microscope. Chromosoma 1985;92:273–282. [DOI] [PubMed] [Google Scholar]
- 42.Hays TS, Salmon ED. Poleward force at the kinetochore in metaphase depends on the number of kinetochore microtubules. J. Cell Biol. 1990;110:391–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.LaFountain JR Jr, Cohan CS, Oldenbourg R. Functional states of kinetochores revealed by laser microsurgery and fluorescent speckle microscopy. Mol. Biol. Cell. 2011;22:4801–4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.LaFountain JR Jr, Cohan CS, Oldenbourg R. Pac-man motility of kinetochores unleashed by laser microsurgery. Mol. Biol. Cell. 2012;23:3133–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rath U, Sharp DJ. The molecular basis of anaphase A in animal cells. Chromosome Res 2011;19:423–432. [DOI] [PubMed] [Google Scholar]
- 46.Giansanti MG, Farkas RM, Bonaccorsi S, et al. . Genetic dissection of meiotic cytokinesis in Drosophila males. Mol. Biol. Cell 2004;15:2509–2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duncan T, Wakefield JG. 50 ways to build a spindle: the complexity of microtubule generation during mitosis. Chromosome Res. 2011;19:321–333. [DOI] [PubMed] [Google Scholar]
- 48.Bucciarelli E, Pellacani C, Naim V, Palena A, Gatti M, Somma MP. Drosophila Dgt6 interacts with Ndc80, Msps/XMAP215, and gamma-tubulin to promote kinetochore-driven MT formation. Curr. Biol. 2009;19:1–7. [DOI] [PubMed] [Google Scholar]