Abstract
Purpose.
Familial exudative vitreoretinopathy (FEVR) is a developmental disease that can cause visual impairment and retinal detachment at a young age. Four genes involved in the Wnt signaling pathway were previously linked to this disease: NDP, FDZ4, LRP5, and TSPAN12. Identification of novel disease-causing alleles allows for a deeper understanding of the disease, better molecular diagnosis, and improved treatment.
Methods.
Sequencing libraries from 92 FEVR patients were generated using a custom capture panel to enrich for 163 known retinal disease-causing genes in humans. Samples were processed using next generation sequencing (NGS) techniques followed by data analysis to identify and classify single nucleotide variants and small insertions and deletions. Sanger validation and segregation testing were used to verify suspected variants.
Results.
Of the cohort of 92, 45 patients were potentially solved (48.9%). Solved cases resulted from the determination of 49 unique mutations, 41 of which are novel. Of the novel variants discovered, 13 were highly likely to cause FEVR due to the nature of these variants (frameshifting indels, splicing mutations, and nonsense variants types). To our knowledge, this is the largest study of a FEVR cohort using NGS.
Conclusions.
We were able to determine probable disease-causing variants in a large number of FEVR patients, the majority of which were novel. Knowledge of these variants will help to further characterize and diagnose FEVR.
Keywords: next-generation sequencing, FEVR, novel alleles, familial segregation
We were able to determine probable disease-causing variants in 45 of 92 FEVR patients, the majority of which were novel. To our knowledge, this is the largest study of a FEVR cohort using NGS. Knowledge of these variants will help to further characterize and diagnose FEVR.
Introduction
Familial exudative vitreoretinopathy (FEVR) is a genetic disorder affecting retinal blood vessel development in young children.1 Although this disease is associated with a variety of symptomatic outcomes that can appear at any age, all cases of FEVR share a common initial observation of avascularity in the peripheral retina. This primary feature typically is followed by neovascularization of the retina. The overabundance and abnormal growth of blood vessels can lead to fibrovascular proliferation, vitreoretinal traction, retinal folds, retinal tears, and ultimately, retinal detachment and total vision loss.
Much of our understanding of the molecular mechanism of FEVR has come from genetic studies. Currently, a total of five genes have been associated with FEVR. Four of these genes are known to be involved in the Wnt/β-catenin signaling pathway, including frizzled-4 (FZD4), low-density lipoprotein receptor-related protein 5 (LRP5), Norrie disease protein (NDP), and tetraspanin-12 (TSPAN12). The FZD4 gene encodes a G-coupled receptor that forms a transmembrane receptor complex with binding partners LRP5 and TSPAN12. The NDP gene expression is restricted within the retina and specific neural tissue, and functions as a ligand to the receptor complex. Upon binding to the receptor complex, a cytoplasmic signal is propagated to activate β-catenin, which, in turn, regulates the transcription of many genes required for proper blood vessel formation in the retina. The fifth gene associated with FEVR, zinc finger protein 408 (ZNF408), was discovered recently and shown to be required for normal vascular development in zebrafish. Most pedigrees displaying FEVR follow an autosomal dominant inheritance model due to haploinsufficiency of one of the FEVR-associated proteins; however, there are exceptions. These include the possible recessive inheritance pattern seen with mutations in LRP5 and TSPAN12, as well as the X-linked recessive inheritance pattern seen with NDP.
To date, the contribution of mutations in each known FEVR disease gene to the disease is not well documented due the limited number of studies for systematic molecular characterization of the patient cohorts. Several studies have been reported where exons of known FEVR-associated genes were PCR amplified and Sanger sequenced to identify potential mutations in a cohort of FEVR patients.2–14 The vast majority of these studies have focused on screening mutations in one gene. There has been only a single instance in which all four FEVR genes have been screened within the same cohort over the course of three reports.15–17 However, the size of the cohort screened in these studies is small at only 53 patients. Mutation frequency in each gene varies among these studies. In general, disease-associated variants in FZD4 are the most abundant, attributing to approximately 18.6% of previously reported cases from 10 studies, in which the frequency ranged from 3% to 40%.2–13 The frequency of LRP5 mutation has been analyzed in four previous reports and displays a slightly lower prevalence with a mean rate of mutation of 15%, with a range from 10% to 25% across these studies.5,6,9,10 Familial exudative vitreoretinopathy–related NDP mutation rate is best described by a single previous study, in which a mutation rate of 6% was determined (4 of 62 FEVR patients).14 Similarly, only a single previous study provides an unbiased TSPAN12 mutation rate in a large cohort (90 participants) of 3%.18 The ZNF408 gene was reported recently and was found in only two FEVR families, including one Dutch family and a Japanese family.19 Based on these reported studies, we can roughly estimate that mutations in these five genes account for fewer than 50% of FEVR patients.
To gain a clearer picture of the mutation spectrum in FEVR patients, we performed a comprehensive molecular screen for all five genes associated with FEVR using next-generation sequencing (NGS) technology in a large cohort of 92 FEVR probands. Pathogenic mutations have been identified for 44 probands, approximately 48% of our collection. Strikingly, pathogenic mutations have been identified in all five known FEVR-associated genes where 87% of alleles were novel, demonstrating the heterogeneous nature of the disease at the gene and allele level. Frequency of mutation in each gene were calculated with LRP5 and FDZ4 being the most frequently mutated in our patient cohort, followed by TSPAN12, NDP, and ZNF408. Finally, mutations in the newly reported gene ZNF408 were found in one proband, strengthening its association with FEVR.
Materials and Methods
Study Subjects
This study was approved by the University of California, San Diego Institutional Review Board. We recruited 92 probands and their family members in this study based on the clinical diagnosis of FEVR. All participants were examined. The diagnosis of FEVR was established based on ophthalmic examination and fundus fluorescein angiography revealing at least one of the following classic findings: peripheral retinal avascularity, severe subretinal exudates, neovascularization, retinal fold or detachment, peripheral fibrovascular mass, macular ectopia, or vitreous hemorrhage.10 After a proband was seen, the rest of the family was enrolled in the study as affected or control subjects, depending on the results of the ophthalmic exam. Peripheral blood samples from family members who lived in the Southern California area in the United States were collected. All participants have signed an Institutional Review Board (IRB) approved Informed Consent Form, and all identifying information was removed before any research specific analysis.
Targeted DNA Capture and NGS
For each sample, paired-end libraries were created according to the Illumina standard protocol. Co-capture was performed on pool DNA libraries in groups of up to 48 samples. The capture probes were custom designed and produced by Roche Nimblegen, Inc. (Basel, Switzerland). Standard Roche Nimblegen methods were used to capture and wash hybridized DNA fragments. Captured sample DNA was sequenced on an Illumina HiSequation 2000 (Illumina, Inc., San Diego, CA, USA) according to the standard operating protocol.
Data Analysis
Sequenced reads were mapped to the hg19 human reference genome using the BWA alignment tool.20 Aligned reads were recalibrated and locally realigned using corresponding functions from the Genomic Analysis Tool Kit (GATK).21 Variants in each sample were determined using Atlas-SNP2 and Atlas-Indel to detect single nucleotide variants and indels, respectively.22 Variants were annotated using the following data sources: 1000 genome database,23 dbSNP,24 ESP5400,25 NIEHS95 exomes,26 RefSeq,27 dbNSFP,28 and internal data from previous studies. These sources (aside from RefSeq and dbNSFP) were especially used to filter improbable variants using frequency cutoffs of 0.5% and 0.1% for recessive and dominant variants, respectively. The RefSeq annotation was applied using the ANNOVAR suite,29 while other annotation was performed via custom perl scripts. The predicted functional effects of variants were determined using precomputed values of the PhyloP,30 SIFT,31 Polyphen2,32 LRT,33 and MutationTaster34 algorithms, collected within the dbNSFP database. Variants were annotated further based on population frequency information from 1000 genome project, ESP4500, and our internal database containing 11,000 controls. Only very rare variants were considered, with a dominant frequency cutoff of 0.05%. Each variant was searched further in dbSNP and HGMD to determine if it had been reported previously as pathogenic. Variants that were not found in these databases were considered novel. In silico functional predictive scores were calculated for all missense variants to evaluate their potential deleteriousness.
PCR and Sanger Sequencing Validation
Primer3 was used to design all PCR primers for Sanger sequence validation. Primers were designed to amplify approximately 500 bp region flanking each validated variant. After amplification, DNA was sequenced on an ABI 3730xI capillary sequencer and subsequently analyzed using Sequencher.
Results
A Large FEVR Patient Cohort
A total of 92 probands and their family members were recruited to participate in this study based on the diagnosis of FEVR in one or more member. This represents one of the largest FEVR patient cohorts. Of these families, 23 were large families of more than 10 known members included in the pedigree, 29 were families with fewer known members, and 40 were sporadic cases with no additional family information. Pedigree information is included for all large families in the Supplementary Material (Supplementary Figs. S1–S15).
Mutation Screen for Known FEVR-Related Genes
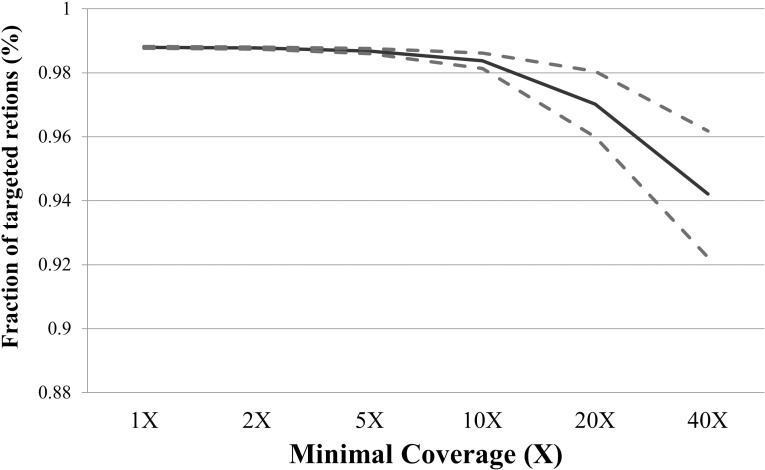
Target capture followed by NGS was performed on 92 probands of the patient cohort. Initial sequencing was performed using a custom retinal disease capture panel that included four FEVR disease-related genes, FDZ4, LRP5, NDP, and TSPAN12. More specifically, the custom retinal disease capture panel included probes for the coding region of 163 retinal disease genes (as selected by RetNet). Upon sequencing, reads then were analyzed via the bioinformatics pipeline as described in the Materials and Methods section. High overall sequence coverage was achieved for the target captured bases with a mean coverage of 174 reads per base, and a mean of 97.9% of all bases with greater than ×10 coverage. When only considering the coding regions of four known FEVR genes, we saw an average coverage of 184 reads per base, and a mean of 98.4% of all bases with greater than ×10 coverage (Fig. 1).
Figure 1.
Sequencing coverage achieved indicates high quality data. The solid line represents the percentage of targeted regions falling at or above the specified minimal coverage. The dashed lines represent the 95% confidence interval of this data.
As FEVR is a predominantly rare dominant inherited genetic disorder, variants were stringently filtered based on several databases of known variants. All but the variants found in less than 0.1% of normal controls were removed from our final set of variants (see Materials and Methods). This narrowed the average number of variants found in the four FEVR disease genes from 4.51 average variants per sample to an average of 0.59 variants per sample for further analysis.
Potentially Pathogenic Variants Identification
Following the method as described in the Materials and Methods section, reported and predicted pathogenic variants were found for a total of 45 probands (Tables 1, 2). Specifically, mutations in LRP5, FZD4, TSPN12, NDP, and ZNF408 account for 19.6%, 15.2%, 8.7%, 6.5%, and 1.1% of patients, respectively (Fig. 2). In addition, 48 cases remain unsolved, representing 52% of our patient cohort.
Table 1.
A Total of 37 Probands Carry Novel Mutations in Genes Associated With FEVR
|
Patient ID |
Gene |
Type |
Mutations |
SIFT |
Polyphen2 HDIV |
Polyphen2 Hvar |
LRT |
Mutation Taster |
PhyloP |
| 2082001 | LRP5 | Frameshift | c.2300delC, p.A767fs | N/A | N/A | N/A | N/A | N/A | N/A |
| 14023006 | LRP5 | Frameshift | c.2716_2719del, p.C906fs | N/A | N/A | N/A | N/A | N/A | N/A |
| 1007001 | LRP5 | Frameshift | c.2737dupT, p.Q912fs | N/A | N/A | N/A | N/A | N/A | N/A |
| 1377001 | LRP5 | Missense | c.4087G>A, p.D1363N | 0.08 | 1.0, D | 1.0, D | 0.000126, U | 0.999683, D | 2.453 |
| 1504001 | LRP5 | Missense | c.3310G>A, p.E1104K | 0.59 | 0.931, P | 0.336, B | 0.00016, U | 0.998586, D | 2.58 |
| 1548003 | LRP5 | Missense | c.2635C>T, p.R879C | 0 | 1.0, D | 0.997, D | 0.000391, U | 0.999954, D | 2.601 |
| 2978001 | LRP5 | Missense | c.4720C>G, p.P1574A | 0.07 | .0.897, P | 0.638, P | 0.000322, U | 0.999662, D | 2.367 |
| 14127002 | LRP5 | Missense | c.4517C>T, p.T1506M | 0 | 1.0, D | 0.999, D | 0.000255, U | 0.999743, D | 2.364 |
| 16311001 | LRP5 | Missense | c.1264G>A, p.A422T | 0 | 1.0, D | 0.997, D | 0.0003, U | 0.999782, D | 0.991 |
| 1079004 | LRP5 | Missense | c.1199C>A, p.A400E | 0 | 0.942, P | 0.852, P | 0.000302, U | 0.999921, D | 2.177 |
| 1346007 | LRP5 | Missense | c.4049T>G, p.L1350R | 0.53 | 0.464, P | 0.555, P | 0.024015, U | 0.997855, D | 0.843 |
| LRP5 | Missense | c.2543C>T, p.P848L | 0.01 | 1.0, D | 1.0, D | 0.000169, U | 0.999907, D | 2.554 | |
| 1348009 | LRP5 | Missense | c.1265C>T, p.A422V | 0 | 1.0, D | 0.997, D | 0.0003, U | 0.99984, D | 2.181 |
| LRP5 | Missense | c.1042C>T, p.R348W | 0 | 1.0, D | 0.998, D | 0.000754, U | 0.997426, D | 2.177 | |
| 3322001 | LRP5 | Missense | c.3782C>G, p.P1261R | 0.03 | 0.995, D | 0.981, D | 0.000173, U | 0.998741, D | 2.298 |
| LRP5 | Missense | c.1270G>A, p.D424N | 0 | 0.993, D | 0.85, P | 0.000145, U | 0.999756, D | 2.181 | |
| 7963001 | LRP5 | Missense | c.803G>A, p.G268E | 0.01 | 1, D | 1, D | 0.000224, U | 0.996877, D | 2.133 |
| LRP5 | Missense | c.1183C>T, p.R395W | 0.05 | 0.801, P | 0.327, B | 0.249244, U | 0.942468, D | 2.177 | |
| 1402003 | LRP5 | Splicing | c.2318G>A, p.G773D | N/A | N/A | N/A | N/A | N/A | N/A |
| LRP5 | Missense | c.119G>A, p.R40H | 0.11 | 1, D | 1, D | 0.000702, U | 0.999862, D | 1.148 | |
| 1027001 | FZD4 | Frameshift | c.662_663insA, p.I221fs | N/A | N/A | N/A | N/A | N/A | N/A |
| 1399004 | FZD4 | Frameshift | c.1507_1508del, p.G503fs | N/A | N/A | N/A | N/A | N/A | N/A |
| 1737001 | FZD4 | Stopgain | c.1488G>A, p.W496X | N/A | N/A | N/A | N/A | N/A | N/A |
| 2679002 | FZD4 | Stopgain | c.124G>T, p.E42X | N/A | N/A | N/A | N/A | N/A | N/A |
| 1076003 | FZD4 | Stopgain | c.G118T:p.E40X | N/A | N/A | N/A | N/A | N/A | N/A |
| 1328003 | FZD4 | Missense | c.134G>A, p.C45Y | – | 1, D | 0.995, D | 0, D | 0.999996, D | 2.387 |
| 1458001 | NDP | Missense | c.112C>T, p.R38C | 0 | 0.999, D | 0.82, P | 0.000009, D | 0.998712, D | 2.428 |
| 12876001 | NDP | Missense | c.314C>A, p.A105E | 0.07 | 0.999, D | 0.996, D | 0, D | 0.998867, D | 2.524 |
| 14500001 | NDP | Missense | c.362G>A, p.R121Q | 0.01 | 0.998, D | 0.988, D | 0, D | 0.99625, D | 2.524 |
| 7794001 | NDP | Stopgain | c.196G>T, p.E66X | N/A | N/A | N/A | N/A | N/A | N/A |
| 13788003 | NDP | Stopgain | c.196G>T, p.E66X | N/A | N/A | N/A | N/A | N/A | N/A |
| 14024003 | NDP | Stopgain | c.393C>A, p.C131X | N/A | N/A | N/A | N/A | N/A | N/A |
| 1400001 | TSPAN12 | Frameshift | c.581delA, p.H194fs | N/A | N/A | N/A | N/A | N/A | N/A |
| 7429001 | TSPAN12 | Frameshift | c.581delA, p.H194fs | N/A | N/A | N/A | N/A | N/A | N/A |
| 16300001 | TSPAN12 | Frameshift | c.601delC, p.L201fs | N/A | N/A | N/A | N/A | N/A | N/A |
| 810006 | TSPAN12 | Splicing | c.612+1G>A | N/A | N/A | N/A | N/A | N/A | N/A |
| 16305001 | TSPAN12 | Splicing | c.149+1G>A | N/A | N/A | N/A | N/A | N/A | N/A |
| 16307001 | TSPAN12 | Splicing | c.149+1G>A | N/A | N/A | N/A | N/A | N/A | N/A |
| 1491001 | TSPAN12 | Missense | c.308T>C, p.I103T | 0.06 | 0.996, D | 0.969, D | 0, D | 0.966537, D | 2.166 |
| 14022004 | ZNF408 | Missense | c.443G>A, p.R148Q | 0.3 | 0.101, B | 0.006, B | 0.521319, N | 0.01147, N | 1.487 |
| 13766001 | TSPAN12 | Missense | c.440C>A, p.T147N | 0.02 | 0.949, P | 0.884, P | 0, D | 0.965421, D | 2.783 |
| LRP5 | Missense | c.3304G>A, p.E1102K | 0.01 | 0.997, D | 0.793, P | 0.000159, U | 0.999856, D | 2.58 | |
| 16312001 | FZD4 | Missense | c.1589G>A, p.G530E | – | 0.59, P | 0.078, B | 0, D | 0.999742, D | 0.856 |
| LRP5 | Missense | c.1264G>A, p.A422T | 0 | 1.0, D | 0.997, D | 0.0003, U | 0.999782, D | 0.991 |
D, damaging; P, possibly damaging; B, benign; U, unknown; N, neutral; N/A, not applicable.
Table 2.
Nine Probands Carry Known Mutations in Two Genes Associated With FEVR
|
Patient ID |
Gene |
Type |
Mutation |
dbSNP ID |
PubMed ID |
| 1080001 | FZD4 | Frameshift | c.1501_1502del, p.S501fs | rs80358303 | 12172548 |
| 1347002 | FZD4 | Frameshift | c.1501_1502del, p.S501fs | rs80358303 | 12172548 |
| 14045002 | FZD4 | Frameshift | c.1282_1285del, p.N428fs | rs80358295 | N/A |
| 720001 | FZD4 | Missense | c.469A>G, p.M157V | rs80358286 | 15035989 |
| 874001 | FZD4 | Missense | c.313A>G, p.M105V | rs80358284 | 14507768 |
| 1344012 | FZD4 | Missense | c.610T>C, p.C204R | rs80358288 | 17093393 |
| 7067001 | FZD4 | Missense | c.313A>G, p.M105V | rs80358284 | 14507768 |
| 1079004 | LRP5 | Missense | c.1564G>A, p.A522T | rs80358309 | 15981244 |
| 14471001 | LRP5 | Missense | c.3403C>T, p.R1135C | rs143396225 | N/A |
Figure 2.
Solved cases based on associated disease genes. Cases with mutations in LRP5 have been split into separate categories for dominant and recessive cases. “Multiple” indicates that potentially causative variants were found in more than one disease-associated gene.
Mutations in LRP5.
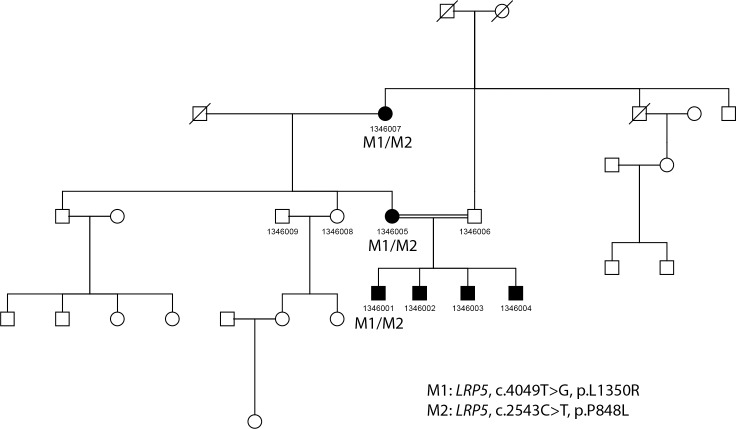
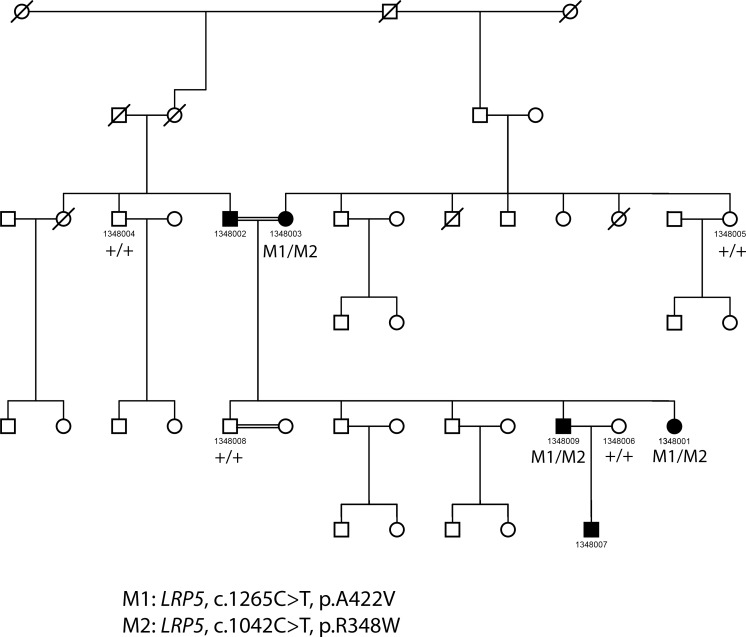
We found that LRP5 is the most frequently mutated gene in our FEVR patient cohort with putative mutations in LRP5 identified in 18 probands (19.6%). The LRP5 gene has been shown to exhibit dominant and recessive inheritance patterns. Recessive inheritance of LRP5 is most commonly due to compound heterozygous genotypes consisting of missense mutations. Recessive inheritance patterns have been observed in six probands who carry compound heterozygous mutations. In addition, putative loss-of-function (LOF) mutations were found in the other 12 probands with dominant inheritance patterns. In total, 23 distinct alleles have been observed. Strikingly, the vast majority of these alleles are novel with only two mutations, c.G1564A: p.A522T and c.C3403T:p.R1135C, having been reported previously. As detailed in Table 1, patients 2082001, 14023006, and 1007001 carried novel frameshift variants: c.2300delC:p.A767fs, c.2716_2719del:p.906_907del, and c.2737dupT:p.Q912fs, respectively. These novel frameshift variants are likely to be null alleles as they are located near the center of the protein sequence in the extracellular binding domain. As such, severe truncation in this location is likely to be pathogenic. Similarly, a novel splicing variant, c.G2318A:p.G773D, has been found in patient 1402003 and likely causes the exclusion of exon 7, a region containing an essential binding domain. In addition, a total of 19 novel missense variants were predicted to be pathogenic due to their rarity in the population, phylogenetic conservation, and in silico functional prediction. Segregation tests were conducted for families when additional members were available, including family 1346007, 1348009, 14023006, and 1402003, to further support the association of the variants with the disease (Table 3). Large pedigrees were available for family 1346007 and family 1348009. Figures 3 and 4 depict the segregation analysis of family 1346007 and 1348009, respectively, in which LRP5 mutations are found in each affected patient and only wild type variants are present in unaffected family members. In both cases, mutations are not seen individually in patients and, therefore, potential causation by a single variant could not be determined.
Table 3.
Segregation Testing was Used to Validate 14 Variants in 11 Families
|
Family ID |
Variant Tested |
Reported |
Patients With DNA Samples |
| 14023 | LRP5; c.2716_2719del, p.C906fs, frameshift | Novel | 14023001, 14023002, 14023003, 14023004, 14023005, 14023006* |
| 1346 | LRP5; c.4049T>G, p.L1350R, missense | Novel | 1346001, 1346002, 1346005, 1346007* |
| LRP5; c.2543C>T, p.P848L, missense | Novel | ||
| 1348 | LRP5; c.1265C>T, p.A422V, missense | Novel | 1348001, 1348003, 1348004, 1348005, 1348006, 1348008, 1348009* |
| LRP5; c.1042C>T, p.R348W, missense | Novel | ||
| 14020 | LRP5; c.2318G>A, p.G773D, splicing | Novel | 14020001, 14020002, 14020003* |
| LRP5; c.119G>A, p.R40H, missense | Novel | ||
| 1080 | FZD4; c.1501_1502del, p.S501fs, frameshift | Reported | 1080001,* 1080003 |
| 1404 | FZD4; c.1282_1285del, p.N428fs, frameshift | Reported | 14045001, 14045002* |
| 1344 | FZD4; c.610T>C, p.C204R, missense | Reported | 1344001, 1344002, 1344003, 1344004, 1344005, 1344006, 1344010, 1344012,* 1344017, 1344018 |
| 7794 | NDP; c.196G>T, p.E66X, stopgain | Novel | 7794001,* 7794002 |
| 13788 | NDP; c.196G>T, p.E66X, stopgain | Novel | 13788001, 13788003* |
| 14024 | NDP; c.393C>A, p.C131X, stopgain | Novel | 14024001, 14024003* |
| 810 | TSPAN12; c.612+1G>A, splicing | Novel | 810001, 810002, 810006,* 810007, 810008, 810020 |
| 14022 | ZNF408; c.443G>A, p.R148Q, missense | Novel | 14022001, 14022003,* 14022004 |
Patients who originally underwent NGS.
Figure 3.
Pedigree and mutation segregation of LRP5 mutation carried in family 1346.
Figure 4.
Pedigree and mutation segregation of LRP5 mutation carried in family 1348.
Mutations in FZD4.
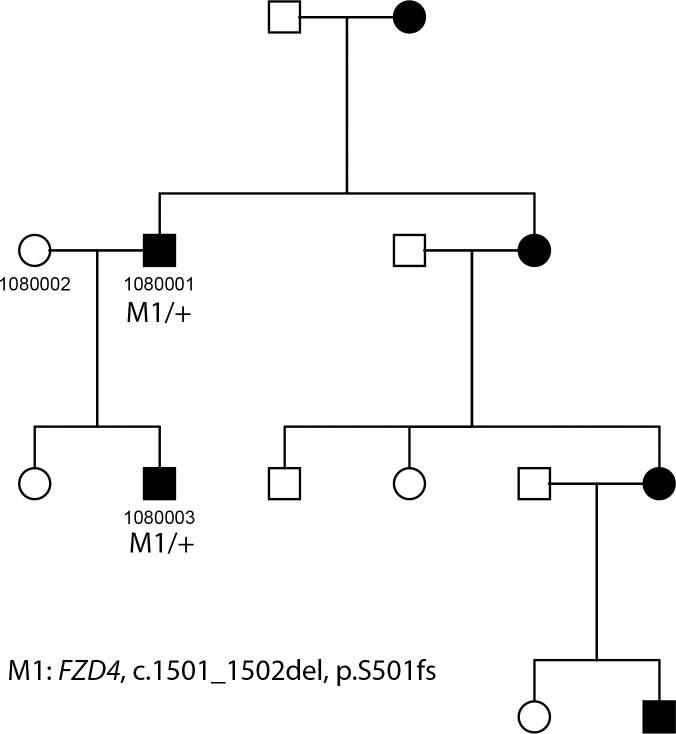
Mutations in FZD4 were found in 15% of our probands (14/92). A total of 13 FZD4 variants were identified, with one known variant, c.1501_1502del:p.501_501del, found in two patients. Of these 13 variants, six have been reported previously as causative of FEVR, while the other seven are novel (Tables 1, 2). Two novel frameshift mutations and three novel nonsense mutations have been identified. Three of these five variants disrupt the essential c-terminal PDZ domain.35 Specifically, c.662_663insA:p.I221fs was found in patient 1027001, which will lead to severe truncation of the protein, and c.1507_1508del:p.503_503del was found in patient 1399004 which will also lead to truncation of the PDZ domain that is essential for its function. Patient 1737001 displays nonsense variant c.G1488A:p.W496X at a similar location near the C-terminus, but before the PDZ-binding domain. Two severe nonsense mutations, c.G118T:p.E40X and c.G124T:p.E42X, located near the beginning of the protein, were found in patient 1076003 and 2679002, respectively. In addition, two novel missense variants were identified in FZD4. These missense variants are likely to be pathogenic due to the following reasons: first, both variants are rare in the population with allele frequency less than 0.1%; second, they are conserved across phylogeny; and third, their predicted detrimental function as calculated by four separate algorithms (Table 1). Familial segregation analysis was performed on families 1080, 1404, and 1344 (Table 3). Of these families, 1080 and 1344 were large pedigrees shown in Figures 5 and 6, respectively. Affected individuals in each case carried a single copy of the family specific FZD4 mutation, while there were not unaffected carriers in either case.
Figure 5.
Pedigree and mutation segregation of FZD4 mutation carried in family 1080.
Figure 6.
Pedigree and mutation segregation of FZD4 mutation carried in family 1344.
Mutations in NDP.
Six probands were discovered to harbor potentially pathogenic NDP alleles. Interestingly, all found alleles were novel, including two nonsense and three missense mutations. The nonsense variant, c.G196T:p.E66X, was found in two patients from independent families, 7794001 and 13788003. Examining the coding SNPs present in each patient reveals that the closest shared variant is more than 0.5 Mb upstream (671,532 bp). Additionally, there is an overall concordance of 33.3% when comparing the coding SNPs between these samples. This corresponds to the average pairwise concordance found in the cohort of 23.5%. These data show that these families were not closely related, and that these variants were not likely due to a distant founder effect. Three novel missense variants in NDP also were discovered, c.C112T:p.R38C, c.C314A:p.A105E, and c.G362A:p.R121Q from patients 1458001, 12876001, and 14500001, respectively. All three variants fit our criteria for rarity, conservation, and functional prediction (Table 1).
Mutations in TSPAN12.
Pathogenic variants in the TSPAN12 have been found in eight probands. Similarly to NDP, all six pathogenic variants of TSPAN12 discovered in this cohort are novel, including two frameshift, two splicing, and two missense variants. One frameshift variant (c.581delA:p.H194fs) is shared between two probands, 1400001 and 7429001. In examining SNPs found in probands, a small region from 18,026 bases upstream of this frameshift to 4103 bases downstream contains three shared SNPs and two unique SNPs. The closest shared SNP from the borders of this region is more than 5 Mb away. Although this may provide evidence of some sort of founder effect, it also is noted that the overall concordance of coding SNPs between these probands is approximately 21.5%, below the average pairwise concordance of the cohort. Haplotype analysis shows that these two probands are not closely related. This variant occurs in the sixth of seven exons, and likely causes truncation, leading to the loss of an extracellular, intramembranous, and cytoplasmic domain. The second frameshift variant, c.601delC:p.L201fs, occurs in proband 16300001 and likely causes a similar truncation. One splicing variant, (c.149+1G>A), was found in probands 16305001 and 16307001. This variant causes the exclusion of exon 2. The second splicing variant, c.612+1G>A, likely causes the exclusion of exon 6. Both missense variants, c.T308C:p.I103T and c.C440A:p.T147N, from probands 1491001 and 13766001, respectively, are likely to be pathogenic based on low population frequency, high conservation scores, and detrimental functional predictions. Neither variant appeared in any frequency control data sets. Both missense variants have high PhyloP scores of 2.166, and 2.783, respectively, indicating they are highly conserved across species. Finally, c.T308C:p.I103T was predicted to be functionally damaging by all four algorithms used, while c.C440A:p.T147N was predicted by all algorithms except polyphen2 (Table 1). Segregation testing was performed for variant c.612+1G>A originally found in proband 810006 with five additional family members, all but one of which were affected. The suspected variant was present in all affected family members, and absent in unaffected family member 810020 (Fig. 7).
Figure 7.
Pedigree and mutation segregation of TSPAN12 mutation carried in family 0810.
Mutations in ZNF408.
As the association between ZNF408 and FEVR has just been reported, it had not been included in our capture panel. To assess the frequency of ZNF408 mutation in our patient cohort, whole exome sequencing (WES) has been conducted for each proband who is negative for mutations in the other four FEVR-associated genes and variants in ZNF408 were assessed for their potential pathogenicity. A novel missense variant, c.G443A:p.R148Q, within the ZNF408 gene was identified in proband 14022004. This variant was not seen in any control database totaling approximately 20,000 individuals, indicating that it is very rare, and might be attributed to a dominant phenotype. Additionally, this variant is conserved in most vertebrates with a PhyloP score of 0.99 out of 1. The functional prediction algorithm Polyphen2 scored this variant as 0.33 which is “possibly damaging.” Finally, segregation testing has been conducted for this allele. As shown in Figure 8, two additional affected family members, 14022001 and 14022003, carry this variant, supporting the idea that this allele is pathogenic.
Figure 8.

Pedigree and mutation segregation of ZNF408 mutation carried by family 14022.
Discussion
In this study, we performed a comprehensive molecular screen for mutations in all five reported FEVR-associated genes in a large cohort of 92 unrelated FEVR patient families using a targeted NGS approach. As a result, we were able to obtain accurate overall solving rate as well as relative contribution of each known FEVR disease gene. In our report, we were able to identify putative pathogenic mutations for 48% of the patients, which is in line but slightly higher than the estimated 40% rate that we calculated based on published literature.
Our study allows us to obtain accurate estimation of the portion of patients carrying mutations in each known FEVR gene. In our patient cohort, LRP5 and FZD4 are most frequently mutated, each accounting for 19% and 15%, respectively. Mutation in NDP and TSPN12 are less frequent with each accounting for 7% and 6%. The ZNF408 gene is the most rare, with only one patient identified in our study. Variable frequency of mutation in these known FEVR disease genes have been reported by several previous studies. One main reason for the variation is due to smaller sample size. This is best demonstrated by a study by Boonstra et al.,5 in which eight of a cohort of 20 had FZD4 mutation, providing an outlier mutation frequency of 40% when compared to similar studies. In contrast, as a total of 92 probands were examined in our study, it allows better estimation of the mutation incidence, particularly for the ones at lower frequencies. Indeed, if we sum all previous studies together, the average frequency for mutation in each gene is similar to what is observed in our study. Another reason for the variation may come from differences in ethnicity. Examining other outliers in previous studies, we see a significantly lower than average number of cases attributed to FZD4 mutations in an Indian cohort examined by Nallathambi et al.,12 of only 3%. However, an additional outlier study by Qin et al.10 showed a similar frequency in a cohort of Chinese descent, while other cohorts were reported to have higher frequencies (Kondo et al.2 with 20%, and Jia et al.7 with 31%). As FEVR presents in the majority of cases as a dominant disease, it is expected that purifying selection limits ethnic or founder effect on associated mutation frequency. It is interesting to note that despite a few outliers, the overall mutation frequency is similar across all FEVR patients of many studied ethnicities. This observation can be explained by a model where the mutations for rare dominant disease are under negative selection pressure; therefore, is mainly due to sporadic mutation rather than founder effect. Indeed, in our study, no highly recurrent alleles have been identified. Further enforcing the notion, in respect to the mutation frequency of each FEVR disease gene, the frequency of mutations identified in this study strongly correlated with the length of each gene (e.g., 21 variants were found in LRP5, which has the largest coding region of the five genes). A Pearson correlation coefficient of 0.95 is observed when comparing the mutation load of FZD4, LRP5, TSPAN12, and LRP5 to the coding region size of each gene. It follows that the expectation of larger genetic regions being subject to increased frequency of mutation should apply to the four major FEVR genes.
Consistent with the idea that most of the alleles we identified in the FEVR patients are sporadic, newly occurring mutations, most of the alleles identified in our study are private and have never been reported before. Of the 48 alleles identified in this study, over 85% (41/48) are novel. The quantity of known variants discovered correlates well with the amount of previous work reported on each gene. The FZD4 was the first discovered, and is the most studied FEVR disease gene. In our patient cohort, four of six known variants were found in this gene. We expect that the portion of novel mutant alleles found in these genes will continue to be high. Therefore, to achieve a high detection rate when performing molecular diagnosis, it is essential to use sequencing based methods rather than hybridization based methods, where only reported alleles are interrogated. The NGS method used in this report allowed us to efficiently identify all of the most probable causative variants in known disease-associated genes within a large FEVR cohort at low cost.
In addition to known FEVR-associated disease genes, each proband was screened for potential mutations in all known retinal disease-related genes using a capture panel. Each proband that was negative for mutations in genes associated with FEVR was examined for other potential mutations in this collection of genes. One interesting feature of other types of retinal degeneration diseases is their extensive overlap in clinical symptoms and genetic abnormalities. For example, it has been observed that mutations in the same gene can lead to different clinical phenotypes. In contrast, no convincing pathogenic variants in other known retinal disease genes have been identified in our patient cohort. This is consistent with the idea that FEVR is primarily caused by vascular development defect in the retina, a mechanism that is different from many retinal diseases.
The discovery of FEVR-related disease genes has significant contribution to our understanding of the molecular mechanisms of angiogenesis in the retina, a key aspect of many common retinal diseases, such as age-related macular degeneration (AMD) and diabetic retinopathy. For example, the involvement of Wnt/β-catenin signaling pathway in vascular development in the retina was first supported in 2002 from a study by Robitaille et al.36 linking FZD4 mutations to FEVR.36 As early as 1996, Wnt/β-catenin signaling was being linked to changes in vascular development.37 Although FEVR was initially well characterized by its retinal vasculature centered phenotype, it wasn't until after FZD4 mutations were discovered in FEVR patients that the pathway and disease could be linked. Despite the comprehensive screen for all known FEVR and other retinal disease genes, the molecular cause in approximately 52% of our probands remain unsolved. Therefore, it represents an exciting opportunity for identification of novel FEVR disease genes, which might provide novel insights into angiogenesis in the retina.
In summary, a significant portion of FEVR patients (approximately 50%) can be diagnosed by mutations in one of the five genes. This molecular diagnosis, together with clinical phenotype, potentially provide useful information for disease diagnosis, prognosis, and genetic counseling. This is particularly valuable given the phenotypic variation found even within the same family. In the meantime, due to the sporadic nature of FEVR, the frequency of any specific variant in the affected population is small. As such, additional studies using high throughput methods, such as targeted capture NGS, are essential to obtain a complete list of potentially causative mutations and help to improve our ability to interpret the clinical significance of rare variants.
Acknowledgments
The authors thank all patients and families that participated in this study.
Supported by National Institutes of Health (NIH; Bethesda, MD, USA) shared Instrument Grant 1S10RR026550 (the NGS was conducted at the Functional Genomic Core [FGC] facility at Baylor College of Medicine); grants from Retinal Research Foundation, Foundation Fighting Blindness (BR-GE-0613-0618-BCM) and the National Eye Institute (NEI; Bethesda, MD, USA: R01EY022356, R01EY018571; RC); Research to Prevent Blindness (KZ); an NEI Center Core Grant for Vision Research (P30EY022589; CW); NEI Training Grant T32EY07102 (JS); and NIH Grant TL1TR00098 (VL). The authors alone are responsible for the content and writing of the paper.
Disclosure: J. Salvo, None; V. Lyubasyuk, None; M. Xu, None; H. Wang, None; F. Wang, None; D. Nguyen, None; K. Wang, None; H. Luo, None; C. Wen, None; C. Shi, None; D. Lin, None; K. Zhang, None; R. Chen, None
References
- 1. Shimouchi A, Takahashi A, Nagaoka T, Ishibazawa A, Yoshida A. Vitreomacular interface in patients with familial exudative vitreoretinopathy. Int Ophthalmol. 2013; 33: 711–715. [DOI] [PubMed] [Google Scholar]
- 2. Kondo H, Hayashi H, Oshima K, Tahira T, Hayashi K. Frizzled 4 gene (FZD4) mutations in patients with familial exudative vitreoretinopathy with variable expressivity. Br J Ophthalmol. 2003; 87: 1291–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Toomes C, Bottomley HM, Scott S, et al. Spectrum and frequency of FZD4 mutations in familial exudative vitreoretinopathy. Invest Ophthalmol Vis Sci. 2004; 45: 2083–2090. [DOI] [PubMed] [Google Scholar]
- 4. Drenser KA, Dailey W, Vinekar A, Dalal K, Capone A, Trese MT. Clinical presentation and genetic correlation of patients with mutations affecting the FZD4 gene. Arch Ophthalmol. 2009; 127: 1649–1654. [DOI] [PubMed] [Google Scholar]
- 5. Boonstra FN, van Nouhuys CE, Schuil J, et al. Clinical and molecular evaluation of probands and family members with familial exudative vitreoretinopathy. Invest Ophthalmol Vis Sci. 2009; 50: 4379–4385. [DOI] [PubMed] [Google Scholar]
- 6. Nikopoulos K, Venselaar H, Collin RW, et al. Overview of the mutation spectrum in familial exudative vitreoretinopathy and Norrie disease with identification of 21 novel variants in FZD4, LRP5, and NDP. Hum Mutat. 2010; 31: 656–666. [DOI] [PubMed] [Google Scholar]
- 7. Jia LY, Li XX, Yu WZ, Zeng WT, Liang C. Novel frizzled-4 gene mutations in Chinese patients with familial exudative vitreoretinopathy. Arch Ophthalmol. 2010; 128: 1341–1349. [DOI] [PubMed] [Google Scholar]
- 8. Robitaille JM, Zheng B, Wallace K, et al. The role of Frizzled-4 mutations in familial exudative vitreoretinopathy and Coats disease. Br J Ophthalmol. 2011; 95: 574–579. [DOI] [PubMed] [Google Scholar]
- 9. Toomes C, Bottomley HM, Jackson RM, et al. Mutations in LRP5 or FZD4 underlie the common familial exudative vitreoretinopathy locus on chromosome 11q. Am J Hum Genet. 2004; 74: 721–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Qin M, Hayashi H, Oshima K, Tahira T, Hayashi K, Kondo H. Complexity of the genotype-phenotype correlation in familial exudative vitreoretinopathy with mutations in the LRP5 and/or FZD4 genes. Hum Mutat. 2005; 26: 104–112. [DOI] [PubMed] [Google Scholar]
- 11. Kondo H, Kusaka S, Yoshinaga A, et al. Mutations in the TSPAN12 gene in Japanese patients with familial exudative vitreoretinopathy. Am J Ophthalmol. 2011; 151: 1095–1100. [DOI] [PubMed] [Google Scholar]
- 12. Nallathambi J, Shukla D, Rajendran A, Namperumalsamy P, Muthulakshmi R, Sundaresan P. Identification of novel FZD4 mutations in Indian patients with familial exudative vitreoretinopathy. Mol Vis. 2006; 12: 1086–1092. [PubMed] [Google Scholar]
- 13. MacDonald ML, Goldberg YP, Macfarlane J, Samuels ME, Trese MT, Shastry BS. Genetic variants of frizzled-4 gene in familial exudative vitreoretinopathy and advanced retinopathy of prematurity. Clin Genet. 2005; 67: 363–366. [DOI] [PubMed] [Google Scholar]
- 14. Kondo H, Qin M, Kusaka S, et al. Novel mutations in Norrie disease gene in Japanese patients with Norrie disease and familial exudative vitreoretinopathy. Invest Ophthalmol Vis Sci. 2007; 48: 1276–1282. [DOI] [PubMed] [Google Scholar]
- 15. Yang H, Xiao X, Li S, Mai G, Zhang Q. Novel TSPAN12 mutations in patients with familial exudative vitreoretinopathy and their associated phenotypes. Mol Vis. 2011; 17: 1128–1135. [PMC free article] [PubMed] [Google Scholar]
- 16. Yang H, Li S, Xiao X, Wang P, Guo X, Zhang Q. Identification of FZD4 and LRP5 mutations in 11 of 49 families with familial exudative vitreoretinopathy. Mol Vis. 2012; 18: 2438–2446. [PMC free article] [PubMed] [Google Scholar]
- 17. Yang H, Li S, Xiao X, Guo X, Zhang Q. Screening for NDP mutations in 44 unrelated patients with familial exudative vitreoretinopathy or Norrie disease. Curr Eye Res. 2012; 37: 726–729. [DOI] [PubMed] [Google Scholar]
- 18. Nikopoulos K, Gilissen C, Hoischen A, et al. Next-generation sequencing of a 40 Mb linkage interval reveals TSPAN12 mutations in patients with familial exudative vitreoretinopathy. Am J Hum Genet. 2010; 86: 240–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Collin RWJ, Nikopoulos K, Dona M, et al. ZNF408 is mutated in familial exudative vitreoretinopathy and is crucial for the development of zebrafish retinal vasculature. Proc Natl Acad Sci U S A. 2013; 110: 9856–9861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McKenna A, Hanna M, Banks E, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010; 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Challis D, Yu J, Evani US, et al. An integrative variant analysis suite for whole exome next-generation sequencing data. BMC Bioinformatics. 2012; 13: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010; 467: 1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sherry ST, Ward MH, Kholodov M, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001; 29: 308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Exome Variant Server, NHLBI GO Exome Sequencing Project (ESP) Seattle, WA, USA: Available at: http://evs.gs.washington.edu/EVS/. Accessed October 2013. [Google Scholar]
- 26. NIEHS Environmental Genome Project. Seattle, WA, USA: Available at: http://evs.gs.washington.edu/niehsExome. Accessed October 2013. [Google Scholar]
- 27. Pruitt KD, Tatusova T, Maglott DR. NCBI. Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005; 33: D501–D504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu X, Jian X. Boerwinkle E. dbNSFP: a lightweight database of human nonsynonymous SNPs and their functional predictions. Hum Mutat. 2011; 32: 894–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010; 38: e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2010; 4: 1073–1081. [DOI] [PubMed] [Google Scholar]
- 31. Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010; 20: 110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chun S, Fay JC. Identification of deleterious mutations within three human genomes. Genome Res. 2009; 19: 1553–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010; 7: 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010; 7: 575–576. [DOI] [PubMed] [Google Scholar]
- 35. Wheeler DS, Barrick SR, Grubisha MJ, Brufsky AM, Friedman PA, Romero G. Direct interaction between NHERF1 and Frizzled regulates β-catenin signaling. Oncogene. 2011; 30: 32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Robitaille J, MacDonald ML, Kaykas A, et al. Mutant frizzled-4 disrupts retinal angiogenesis in familial exudative vitreoretinopathy. Nat Genet. 2002; 32: 326–330. [DOI] [PubMed] [Google Scholar]
- 37. Monkley SJ, Delaney SJ, Pennisi DJ, Christiansen JH, Wainwright BJ. Targeted disruption of the Wnt2 gene results in placentation defects. Development. 1996; 122: 3343–3353. [DOI] [PubMed] [Google Scholar]