Abstract
The bacteriophage T4 head is an elongated icosahedron packed with 172 kb of linear double-stranded DNA and numerous proteins. The capsid is built from three essential proteins: gp23*, which forms the hexagonal capsid lattice; gp24*, which forms pentamers at 11 of the 12 vertices; and gp20, which forms the unique dodecameric portal vertex through which DNA enters during packaging and exits during infection.
Intensive work over more than half a century has led to a deep understanding of the phage T4 head. The atomic structure of gp24 has been determined. A structural model built for gp23 using its similarity to gp24 showed that the phage T4 major capsid protein has the same fold as numerous other icosahedral bacteriophages. However, phage T4 displays an unusual membrane and portal initiated assembly of a shape determining self-sufficient scaffolding core. Folding of gp23 requires the assistance of two chaperones, the Escherichia coli chaperone GroEL acting with the phage-coded gp23-specific cochaperone, gp31. The capsid also contains two nonessential outer capsid proteins, Hoc and Soc, which decorate the capsid surface. Through binding to adjacent gp23 subunits, Soc reinforces the capsid structure. Hoc and Soc have been used extensively in bipartite peptide display libraries and to display pathogen antigens, including those from human immunodeficiency virus (HIV), Neisseria meningitides, Bacillus anthracis, and foot and mouth disease virus. The structure of Ip1*, one of a number of multiple (>100) copy proteins packed and injected with DNA from the full head, shows it to be an inhibitor of one specific restriction endonuclease specifically targeting glycosylated hydroxymethyl cytosine DNA. Extensive mutagenesis, combined with atomic structures of the DNA packaging/terminase proteins gp16 and gp17, elucidated the ATPase and nuclease functional motifs involved in DNA translocation and headful DNA cutting. The cryoelectron microscopy structure of the T4 packaging machine showed a pentameric motor assembled with gp17 subunits on the portal vertex. Single molecule optical tweezers and fluorescence studies showed that the T4 motor packages DNA at the highest rate known and can package multiple segments. Förster resonance energy transfer–fluorescence correlation spectroscopy studies indicate that DNA gets compressed in the stalled motor and that the terminase-to-portal distance changes during translocation. Current evidence suggests a linear two-component (large terminase plus portal) translocation motor in which electrostatic forces generated by ATP hydrolysis drive DNA translocation by alternating the motor between tensed and relaxed states.
I. Introduction
The T4-type bacteriophages are ubiquitously distributed in nature and occupy environmental niches ranging from mammalian gut to soil, sewage, and oceans. More than 130 such viruses showing morphological features similar to phage T4 have been described; from the T4 superfamily, ∼1400 major capsid protein sequences have been correlated to its three-dimensional structure (Desplats and Krisch, 2003; Krisch and Comeau, 2008; Tetart et al., 1998). Features include a large elongated (prolate) head, contractile tail, and complex base plate with six long, kinked tail fibers emanating from it radially. Phage T4 continues to serve as an excellent model to elucidate the mechanisms of head structure, assembly, and DNA packaging not only of myoviridae but of other large icosahedral viruses such as herpes viruses, as well as providing a valuable biotechnology platform with diverse current applications. This chapter focuses on advances on the basic understanding of phage T4 head structure, assembly, and mechanism of DNA packaging. Application of some of this knowledge to develop phage T4 as a surface display and vaccine platform is also discussed. The reader is referred to the comprehensive review by Black et al. (1994) for earlier work (references through 1993) on T4 head assembly.
II. Structure and Assembly of Phage T4 Capsid
The overall architecture and dimensions of the phage T4 head (Fokine et al., 2004) are displayed in Fig. 1. The width and length of the elongated prolate icosahedron are Tend = 13 laevo and Tmid = 20 (86 nm wide and 120 nm long), and the copy numbers of gp23 (major capsid protein), gp24 (vertex protein), gp20 (portal protein), and nonessential display proteins Hoc and Soc are 930, 55, 12, 155, and 870, respectively (Fig. 1). Also, over 1000 internal proteins are packed together with the ∼170-kb linear double-stranded DNA (dsDNA) within the capsid.
Figure 1.
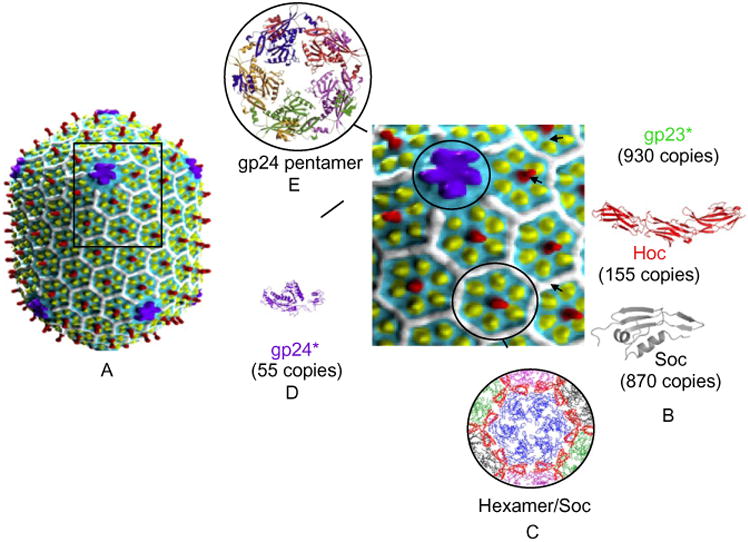
Structure of the bacteriophage T4 head. (A) CryoEM reconstruction of phage T4 capsid (Fokine et al., 2004); the square block shows enlarged view showing gp23 (yellow subunits), gp24 (purple subunits), Hoc (red subunits), and Soc (white subunits). (B) Structures of RB49 Hoc (red) and RB69Soc (gray). (C) Structural model showing one gp23 hexamer (blue) surrounded by six Soc trimers (red). Neighboring gp23 hexamers are shown in green, black, and magenta (Qin et al., 2009). (D) Structure of gp24 (Fokine et al., 2005). (E) Structural model of gp24 pentameric vertex.
Assembly of the phage T4 prohead (Fig. 2A) is remarkable in (i) being wholly dependent on a membrane-bound portal initiator; (ii) ability to form a correct capsid size scaffolding core in the absence of the major capsid protein; (iii) extensive processing of nearly all the capsid proteins (only the portal protein is not processed) at consensus IP2E- or LP2E-processing sites by the gp21 morphogenetic protease; cleavage distal to >5000 E- peptide bonds is followed by (iv) detachment of the fully processed prohead from the host membrane for DNA packaging in the cytoplasm. According to bioinformatics classification, the gp21 morphogenetic protease is thought to occupy an isolated niche among phage and viral prohead protease genes classified as serine proteases (Cheng et al., 2004; Liu and Mushegian, 2004). It should be noted that DNA packaging is initiated into the released processed procapsids already containing the internal proteins to be ejected (IPI, IPII, IPIII, and Alt) along with DNA upon infection. Following packaging, the head is “plugged” by attachment of the head completion (“neck”) proteins (gp13, gp14, and gp15) to the portal followed by assembly of the tail machine to form an infectious virus particle.
Figure 2.
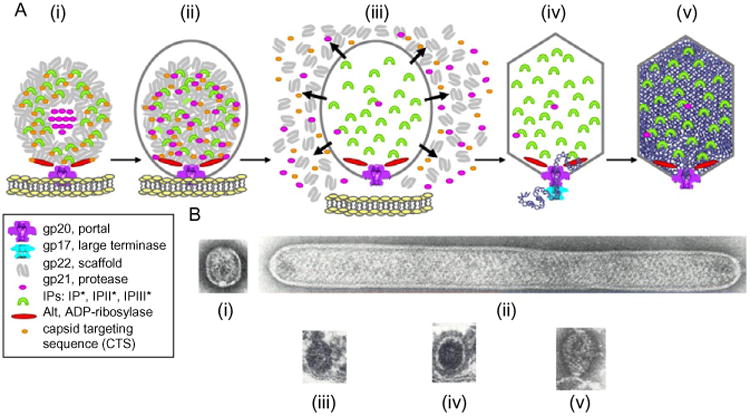
(A) Assembly of T4 prohead and head. (i) Membrane-bound portal initiates scaffolding core assembly; (ii) full prohead assembled on membrane; (iii) gp21 processing of core and procapsid components followed by detachment from membrane; (iv) DNA packaging into processed prohead; and (v) expanded fully packaged head. (B) Assembly intermediates from Black et al. (1994): (i) portal bound scaffolding core with procapsid; (ii) giant procapsid and core; (iii) in vivo-assembled naked core on membrane is precursor to (iv) full prohead; and (v) portal initiator assembles size-determining scaffolding core in vitro.
A gallery of unprocessed early prohead intermediates emphasizes a number of fundamental properties of T4 prohead assembly (Fig. 2B): (i) prominent and distinct internal prohead scaffolding core attached to the portal, (ii) aberrant giant proheads contain a giant core, (iii) “naked” cores without a procapsid shell are attached to the membrane-bound portal, (iv) can serve as prohead intermediates, and (v) core architecture is mainly imposed by assembly of the major scaffolding protein gp22, broken down to small peptides by the gp21 protease maturation. Mutations may also allow accumulation of other active intermediates such as vertex and distal cap-lacking proheads, showing that prohead assembly can diverge from a strictly linear pathway (Black et al., 1994).
Assembly of the membrane-bound portal initiator is dependent on a viral chaperone (gp40), and gp20 is found at the membrane and can be cross-linked to Tig, DnaK, and YidC proteins. It is likely that DnaK transports the protein to the membrane, while YidC may function as a membrane-associated chaperone binding gp20 to the surface of the lipid bilayer until prehead assembly is completed (Quinten and Kuhn, personal communication). Following processing, the “empty” prohead maintains its early small procapsid precursor structure (unexpanded pro-head, or empty small particle or esp) until irreversible expansion (∼50% volume increase) leads to the stable elp (expanded prohead or empty large particle) conformation; as discussed later, packaging can be initiated into either type particle in vitro.
A significant advance was the crystal structure of the vertex protein gp24 and, by inference, the structure of its close relative, the major capsid protein gp23 (Fokine et al., 2005). The gp24 and inferred gp23 structures are closely related to the structure of the major capsid protein of bacteriophage HK97, most probably also the same protein fold as the majority of tailed dsDNA bacteriophage major capsid proteins (Wikoff et al., 2000). This ∼0.3-nm resolution structure permits rationalization of head length mutations in the major capsid protein, as well as of mutations allowing bypass of the vertex protein. The former map to the periphery of the capsomer and the latter within the capsomer. It is likely that the special gp24 vertex protein of phage T4 is a relatively recent evolutionary addition as judged by the ease with which it can be bypassed. Cryoelectron (cryoEM) microscopy showed that in bypass mutants that substitute pentamers of the major capsid protein at the vertex, additional Soc decoration protein subunits surround these gp23* molecules, which does not occur in the gp23*–gp24* interfaces of the wild-type capsid (Fokine et al., 2006). Nevertheless, despite the rationalization of the major capsid protein affecting head size mutations, it should be noted that these divert only a relatively small fraction of the capsids to altered and variable sizes. The primary determinant of the normally invariant prohead shape is thought to be the internal gp22-based major scaffolding core, which normally grows concurrently with the shell (Black et al., 1994), although other core proteins (IPIII and Alt) also make a contribution to shape determination (Fig. 2). However, little progress has been made in establishing the exact mechanism of correct capsid size determination. Thus numerous “recent” T-even isolates are found to display increased and apparently invariant capsid prolate icosahedra; for example, KVP40, 254 kb, apparently has a single Tmid greater than the 170-kb T4 Tmid = 20)(Miller et al., 2003). However, despite providing interesting material bearing on the T-even head size determination mechanism, few if any in-depth studies have been carried out on these phages to determine whether the morphogenetic core, the major capsid protein, or other factors are responsible for the different and precisely determined volumes of their capsids.
A. Folding of the major capsid protein gp23
Folding and assembly of the phage T4 major capsid protein gp23 into the prohead require special utilization of the GroEL chaperonin system and an essential phage cochaperonin, gp31. gp31 replaces the GroES cochaperonin utilized for folding the 10–15% of E. coli proteins that require folding by the GroEL folding chamber. Although T4 gp31 and the closely related RB49 cochaperonin CocO have been demonstrated to replace the GroES function for all essential E. coli protein folding, the GroES-gp31 relationship is not reciprocal; that is, GroES cannot replace gp31 to fold gp23 because of special folding requirements of the latter protein (Andreadis and Black, 1998; Keppel et al., 2002). The N terminus of gp23 that is removed by protease processing appears to strongly target associated fusion proteins to the GroEL chaperonin (Bakkes et al., 2005; Clare et al., 2006; Snyder and Tarkowski, 2005). Especially perplexing, given this dedicated gp23 folding system, is the ability of mutant gp23 to fold without the gp31–GroEL system; four GroEL bypass gp23 amino acid missense mutations additively confer almost 50% folding efficiency and phage production without the GroEL chaperone. Binding of gp23 to the GroEL folding cage shows features distinct from those of most bound E. coli proteins. However, it has been shown that gp23 is able to interact with the GroEL–GroES complex, although not productively (Calmat et al., 2009). Unlike substrates such as RUBISCO, gp23 occupies both chambers of the GroEL folding cage, and only gp31 is able to promote efficient capped single “cis” chamber folding, apparently by creating a larger folding chamber (Clare et al., 2009). On the basis of the gp24 inferred structure of gp23 and structures of the GroES and gp31 complexed GroEL folding chambers, support for a critical increased chamber size to accommodate gp23 has been advanced as the explanation for the gp31 specificity (Clare et al., 2006). However, because comparable size T-even phage gp31 homologues display preference for folding their own gp23s, more subtle features of the various T-even phage structured folding cages may also determine specificity. Fluorescence experiments support earlier evidence that gp23 adds to the prohead as a hexamer rather than as a monomer; however, the structure of the early hexamer or its precursors is unknown but likely differs substantially from the mature capsid gp24-derived gp23 structure, as gp23 undergoes complex structural transformations from early to mature capsid (Black et al., 1994; Stortelder et al., 2006).
The novel additional phage T4 GroEL-related gene 39.2 has been discovered and shown to restore GroEL function in certain hosts and to certain GroEL mutations. This small protein (58 residues) is speculated to function to strengthen the GroEL interaction with various host cochaperones by favoring the open GroEL state over the closed state (D. Ang and C. Georgopoulos, personal communication).
III. Structure of the Phage T4 Head
A. Packaged DNA
Packaged phage T4 DNA shares a number of general features with other tailed dsDNA phages: 2.5-nm side-to-side packing of predominantly B-form duplex DNA condensed to ∼500 mg/ml. However, other features differ among phages; for example, T4 DNA is packed in an orientation parallel to the prolate and giant head tail axis together with ∼1000 molecules of imbedded and mobile internal proteins, unlike the DNA arrangement that traverses the head-tail axis and is arranged around an internal protein core as seen in phage T7 (Cerritelli et al., 1997). Use of the capsid-targeting sequence of internal proteins allows encapsidation of foreign proteins such as green fluorescent protein (GFP) and staphylococcal nuclease within the DNA of active virus (Mullaney and Black, 1998; Mullaney et al., 2000). Digestion by the latter nuclease upon addition of calcium yields a pattern of short DNA fragments, predominantly a 160-bp repeat (Mullaney and Black, 1998). This pattern supports a discontinuous pattern of DNA packing such as in the icosahedral-bend, liquid-crystal, or spiral-fold models among a number of proposed models for phage T4 (Fig. 3). Experimental evidence bearing on these models was summarized in Mullaney et al. (2000) and in a review of a large-tailed, dsDNA-containing bacteriophage-condensed genome structure that supports markedly different inner capsid structures among such phages, with the condensed genome structure of a single phage type unlikely to be precisely determined or to fit a single general structure (Black and Thomas, 2012).
Figure 3.
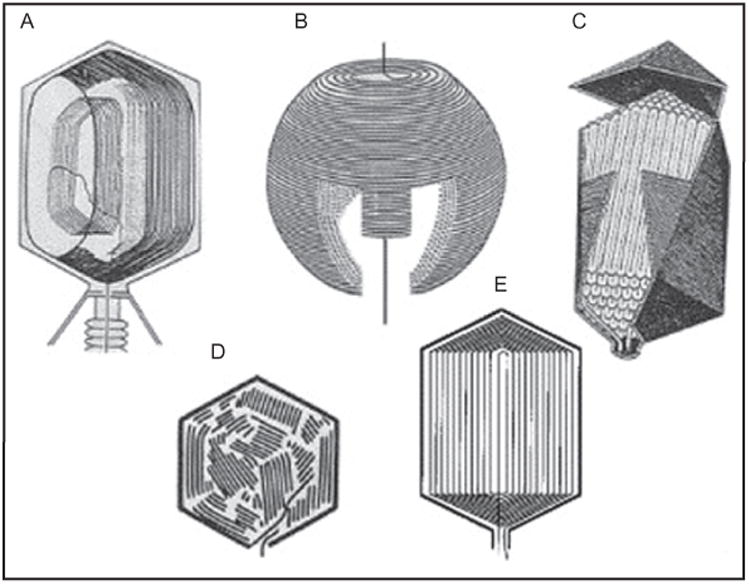
Models of packaged T4 DNA structure. (a) T4 DNA is packed longitudinally to the head–tail axis (Earnshaw et al., 1978), unlike the transverse packaging in T7 capsids (Cerritelli et al., 1997) (b). Other models shown include spiral fold (c), liquid crystal (d), and icosahedral bend (e). Both packaged T4 DNA ends are located in the portal (Ray et al., 2010a). For references and evidence bearing on packaged models see (Mullaney and Black, 1998).
B. Packaged proteins
In addition to the uncertain arrangement at the nucleotide level of packaged phage DNA, the structure of other internal components is poorly understood in comparison to surface capsid proteins. The internal protein I* (IPI*) of phage T4 is injected to protect the DNA from a two subunit GmrS+GmrD glucose modified restriction endonuclease of a pathogenic E. coli that digests glucosylated hydroxymethylcytosine DNA of T-even phages (Bair and Black, 2007; Bair et al., 2007). The 76 residue proteolyzed mature form of the protein has a novel compact protein fold consisting of two β sheets flanked with N- and C-terminal α helices, a structure that is required for its inhibitor activity that is apparently due to binding the GmrS/GmrD proteins (Fig. 4) (Rifat et al., 2008). A single chain GmrS/GmrD homologue enzyme with 90% identity in its sequence to the two subunit enzyme has evolved IPI* inhibitor immunity. It thus appears that the phage T-evens have coevolved with their hosts, a diverse and highly specific set of internal proteins to counter the hmC modification-dependent restriction endonucleases. Consequently, the internal protein components of the T-even phages are a highly diverse set of defense proteins against diverse attack enzymes with only a conserved capsid targeting sequence to encapsidate the proteins into the precursor scaffolding core (Repoila et al., 1994).
Figure 4.
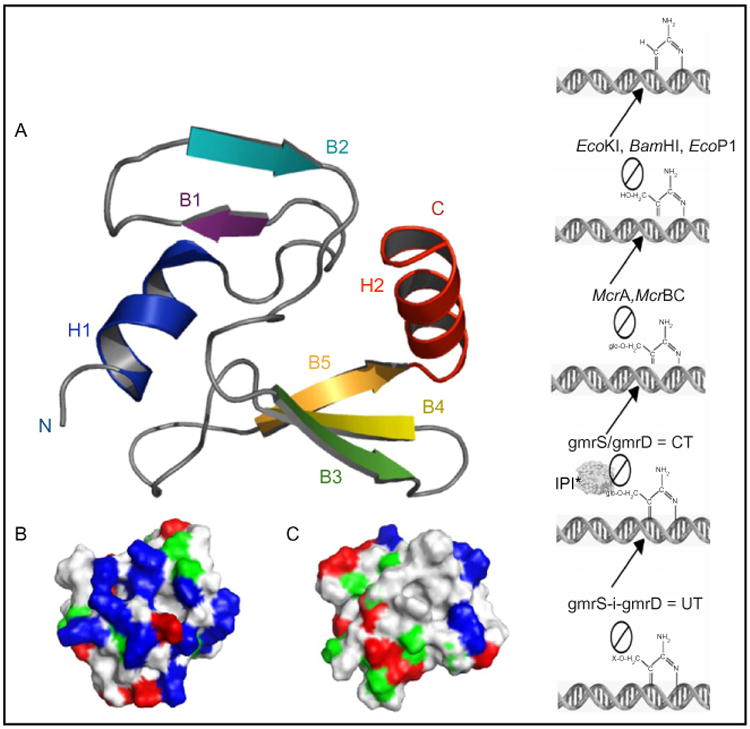
Structure and function of T4 internal protein I*. Nuclear magnetic resonance structure of IPI*, a highly specific inhibitor of the two-subunit CT (GmrS/GmrD) glucosyl-hmC DNA directed restriction endonuclease (right); DNA modifications blocking such enzymes are shown. The IPI* structure is compact with an asymmetric charge distribution on the faces (blue are basic residues) that may allow rapid DNA-bound ejection through the portal and tail without unfolding–refolding.
Genes 2 and 4 of phage T4 likely are associated in function, and gp2 was shown previously by Goldberg and co-workers to be able to protect the ends of mature T4 DNA from the recBCD exonuclease V, likely by binding to DNA termini (Lipińska et al., 1989). The gp2 protein has not been identified within the phage head because of its low abundance, but evidence for its presence in the head comes from the fact that gp2 can be added to gp2-deficient full heads to confer exonuclease V protection. Thus gp2 affects head–tail joining as well as protecting the DNA ends likely with as few as two copies per particle binding the two DNA ends (Wang et al., 2000). It is unknown whether following packaging a DNA end descends part way into the tail poised for ejection as in some other phages. Neck proteins complete the head and are part of an extended family of phage components (Cardarelli et al., 2010).
Solid-state nuclear magnetic resonance analysis of phage T4 particle shows that DNA is largely B form and allows its electrostatic interactions to be tabulated (Yu and Schaefer, 2008). This study reveals high-resolution interactions bearing on the internal structure of the phage T4 head. The DNA phosphate negative charge is balanced among lysyl amines, polyamines, and mono and divalent cations. Interestingly, among positively charged amino acids, only lysine residues of the internal proteins were seen to be in close contact with the DNA phosphates, arguing for specific internal protein DNA structures. Electrostatic contributions from internal proteins and interactions of polyamines with DNA entering the prohead to the packaging motor were proposed to account for the higher packaging rates achieved by the phage T4 packaging machine when compared to that of Bacillus subtiis phi29 and E. coli λ phages.
IV. Display on Capsid Using Hoc and Soc Proteins
In addition to the essential capsid proteins, gp23, gp24, and gp20, the T4 capsid is decorated with two nonessential outer capsid proteins: Hoc (highly antigenic outer capsid protein), a dumbbell-shaped monomer at the center of each gp23 hexon, up to 155 copies per capsid (39 kDa; red subunits); and Soc (small outer capsid protein), a rod-shaped molecule that binds between gp23 hexons, up to 870 copies per capsid (10 kDa; white subunits)(Fig. 1). Both Hoc and Soc are dispensable and bind to the capsid after the completion of capsid assembly (Ishii and Yanagida, 1977; Ishii et al., 1978). Null (amber or deletion) mutations in either or both the genes do not affect phage production, viability, or infectivity.
The structure of Soc has been determined (Qin et al., 2009). The 77 amino acid RB69 Soc is a tadpole-shaped molecule with two binding sites for gp23*. Interaction of Soc to the two gp23 molecules glues adjacent hexons. Trimerization of bound Soc molecules results in clamping of three hexons, and 270 such clamps form a cage reinforcing the capsid structure (Fig. 1). Soc assembly thus provides great stability to phage T4 to survive under hostile environments such as extreme pH (pH 11), high temperature (60°C), and a host of denaturing agents. Soc-minus phage lose viability at pH 10.6, and the addition of Soc enhances its survival by ∼104-fold.
Hoc does not provide significant additional stability. A monomer of Hoc is present at the center of each hexameric gp23 capsomer. Hoc consists of a string of four domains, three immunoglobulin (Ig)-like domains and one non-Ig domain at the C terminus. The capsid-binding site is localized to the C-terminal 25 amino acids, which are well conserved in T4-related bacteriophages (Sathaliyawala et al., 2010). The crystal structure of the first three Ig domains of RB49 Hoc shows a linear arrangement of the Ig domains (Fokine et al., 2011). The Ig-like fold of each domain resembles ones found frequently in cell attachment molecules of higher organisms. Decorating the virus with long fibers of Hoc molecules may provide survival advantages to the virus. Hoc might be able to attach the phage capsids loosely to bacterial surface molecules, allowing the virus to stay attached to the cell while its tail fibers find their receptors.
The aforementioned properties of Hoc and Soc are uniquely suited to engineer the T4 capsid surface by arraying pathogen antigens. Recombinant vectors have been developed that allow fusion of pathogen antigens to the N or C termini of Hoc and Soc (Jiang et al., 1997; Ren and Black, 1998; Ren et al., 1996, 1997). The fusion proteins were expressed in E. coli and, upon infection with hoc-soc- phage, the fusion proteins assembled on the capsid. Phages purified from the infected extracts are decorated with the pathogen antigens. Alternatively, the fused gene can be transferred into the T4 genome by recombinational marker rescue, and infection with the recombinant phage expresses and assembles the fusion protein on the capsid as part of the infection process. Short peptides or protein domains from a variety of pathogens—Neisseria meningitides (Jiang et al., 1997), polio virus (Ren et al., 1996), human immunodeficiency virus (HIV) (Sathaliyawala et al., 2006), swine fever virus (Wu et al., 2007), and foot and mouth disease virus (Ren et al., 2008)—have been displayed on the T4 capsid using this approach. The T4 system can be adapted to prepare bipartite libraries of randomized short peptides displayed on T4 capsid Hoc and Soc and use these libraries to “fish out” peptides that interact with the protein of interest (Malys et al., 2002). Biopanning of libraries by the T4 large packaging protein gp17 selected peptides that match with the sequences of proteins thought to interact with gp17. Of particular interest was the selection of a peptide that matched with the T4 late sigma factor, gp55. The gp55-deficient extracts packaged concatemeric DNA about 100-fold less efficiently, suggesting that the gp17 interaction with gp55 helps in loading the packaging terminase onto the viral genome (Black and Peng, 2006; Malys et al., 2002).
An in vitro display system has been developed that takes advantage of the high-affinity interactions between Hoc or Soc and the capsid (Fig. 5) (Li et al., 2007; Shivachandra et al., 2006). In this system, the pathogen antigen fused to Hoc or Soc with a hexa-histidine tag was overexpressed in E. coli and purified. The purified protein was assembled on the hoc-soc-phage by simply mixing the purified components. This system has certain advantages over the in vivo display: (i) a functionally well-characterized and conformationally homogeneous antigen is displayed on the capsid; (ii) the copy number of the displayed antigen can be controlled by altering the ratio of antigen to capsid binding sites; and (iii) multiple antigens can be displayed on the same capsid. This system was used to display full-length antigens from HIV (Sathaliyawala et al., 2006) and anthrax (Li et al., 2007; Shivachandra et al., 2006) that are as large as 90 kDa. All 155 Hoc-binding sites can be filled with anthrax toxin antigens, protective antigen (PA, 83 kDa), lethal factor (LF, 89 kDa), or edema factor (EF, 90 kDa; Shivachandra et al., 2007). Fusion to the N terminus of Hoc did not affect the apparent binding constant (Kd) or the copy number per capsid (Bmax), but fusion to the C terminus reduced the Kd by 500-fold (Jiang et al., 1997; Shivachandra et al., 2007). All 870 copies of Soc-binding sites can be filled with Soc-fused antigens but the size of the fused antigen must be ∼30 kDa or less; otherwise, the copy number is reduced significantly (Li et al., 2007). For example, the 20-kDa PA domain-4 and the 30-kDa LFn domain fused to Soc can be displayed to full capacity. An insoluble Soc-HIV gp120 V3 loop domain fusion protein with a 43 amino acid C-terminal addition could be refolded and bound with ∼100% occupancy to mature phage head type polyheads (Ren et al., 1996). Large 90-kDa anthrax toxins can also be displayed but the Bmax is reduced to about 300 presumably due to steric constraints. Antigens can be fused to the N terminus, the C terminus, or both termini of Soc simultaneously, without significantly affecting the Kd or Bmax. Thus, as many as 1895 antigen molecules or domains can be attached to each capsid using both Hoc and Soc (Li et al., 2007).
Figure 5.
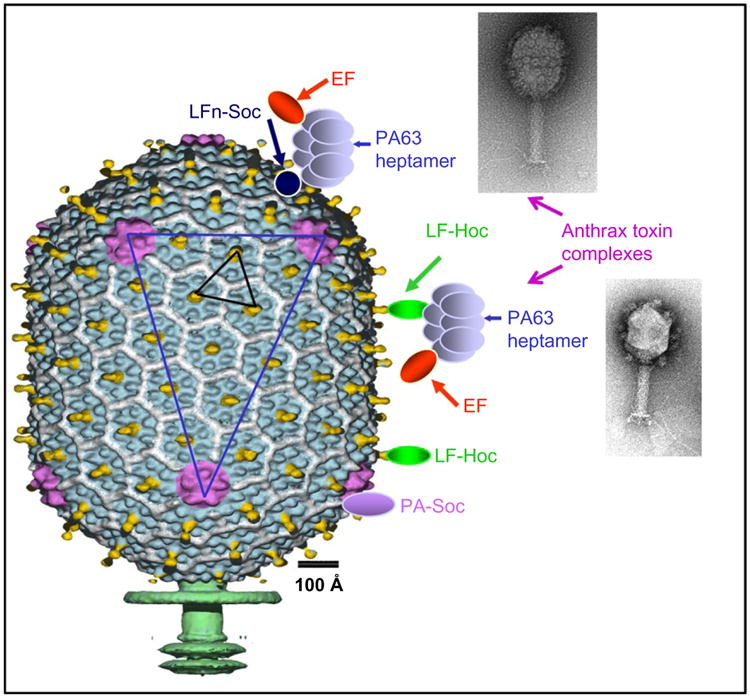
In vitro display of antigens on bacteriophage T4 capsid. Schematic representation of the T4 capsid decorated with large antigens, PA (83 kDa) and LF (89 kDa), or hetero-oligomeric anthrax toxin complexes through either Hoc or Soc binding (Li et al., 2006, 2007). See text for details. (Insets) Electron micrographs of T4 phage with anthrax toxin complexes displayed through Soc (top) or Hoc (bottom). Note that the copy number of the complexes is lower with the Hoc display than with the Soc display.
The in vitro system offers novel avenues to display macromolecular complexes through specific interactions with the already attached antigens (Li et al., 2006). Sequential assembly was performed by first attaching LF-Hoc and/or LFn-Soc to hoc–soc–phage and exposing the N-domain of LF on the surface. Heptamers of PA were then assembled through interactions between the LFn domain and the N-domain of cleaved PA (domain 1′ of PA63). EF was then attached to the PA63 heptamers, completing assembly of the ∼700-kDa anthrax toxin complex on phage T4 capsid (Fig. 5). CryoEM reconstruction shows that native PA63(7)–LFn(3) complexes are assembled in which three adjacent capsid-bound LFn “legs” support the PA63 heptamers (Fokine et al., 2007). Additional layers of proteins can be built on the capsid through interactions with the respective partners.
One of the main applications of T4 antigen particles is their potential use in vaccine delivery. A number of independent studies showed that the T4-displayed particulate antigens without any added adjuvant elicit strong antibody responses and, to a lesser extent, cellular responses. The 43 amino acid residue V3 loop of HIV gp120 fused to Soc displayed on the T4 phage was highly immunogenic in mice and induced anti-gp120 antibodies (Ren et al.,1996); Soc-displayed IgG anti-EWL was active (Ren and Black, 1998). The Hoc fused 183 amino acid N-terminal portion of the HIV CD4 receptor protein is displayed in active form. Strong anthrax lethal-toxin neutralization titers were elicited upon immunization of mice and rabbits with phage T4-displayed PA either through Hoc or through Soc (Rao, unpublished data; Shivachandra et al., 2006, 2007). When multiple anthrax antigens were displayed, immune responses against all the displayed antigens were elicited (Shivachandra et al., 2007). T4 particles displaying PA and LF or those displaying the major antigenic determinant cluster mE2 (123 amino acids) and the primary antigen E2 (371 amino acids) of the classical swine fever virus elicited strong antibody titers (Wu et al., 2007). Furthermore, rhesus macaques immunized with T4-displayed PA were completely protected against aerosol challenge with lethal anthrax Ames spores (Rao et al., 2011). Mice immunized with the Soc displayed foot and mouth disease virus (FMDV) capsid precursor polyprotein (P1, 755 amino acids) and proteinase 3C (213 amino acids) were completely protected upon challenge with a lethal dose of FMDV (Ren et al., 2008; Wu et al., 2007). Pigs immunized with a mixture of T4-P1 and T4-3C particles were also protected when these animals were cohoused with FMDV-infected pigs. In another type of application, a T4-displayed mouse Flt4 tumor antigen elicited anti-Flt4 antibodies and broke immune tolerance to self-antigens. These antibodies provided antitumor and antimetastasis immunity in mice (Ren et al., 2009).
Other applications have shown that heavy single chain shark antibodies fused to Hoc and Soc bind tightly to phage T4 heads. These tightly arrayed antibodies display high antigen affinity and are attractive as biosensors and as antitoxins to ricin and other lethal agents (Archer and Liu, 2009; Robertson et al., 2011). Moreover, dye labeled DNA containing heads are taken up into cells efficiently through clathrin-mediated endocytosis and show little cytotoxicity (J. L. Liu, personal communication).
The aforementioned studies provide abundant evidence that the phage T4 nanoparticle platform has the potential to engineer human as well as veterinary vaccines, as well as to act in transfer and as a biosensor platform.
V. Packaging Proteins
Two nonstructural terminase proteins, gp16 (18 kDa) and gp17 (70 kDa), link head assembly and genome processing (Black, 1989; Rao and Black, 2005; Rao and Feiss, 2008). These proteins are thought to form a hetero-oligomeric complex, which recognizes the concatemeric DNA and makes an endonucleolytic cut (hence the name “terminase”). The terminase– DNA complex docks on the prohead through gp17 interactions with the special portal vertex formed by the dodecameric gp20, thus assembling a DNA-packaging machine. The ATP-fueled machine translocates DNA into the capsid until the head is full, equivalent to about 1.02 times the genome length (171 kb). The terminase dissociates from the packaged head, makes a second cut to terminate DNA packaging, and attaches the concatemeric DNA to another empty head to continue translocation in a processive fashion. Structural and functional analyses of key parts of the machine—gp16, gp17, and gp20—as described here, led to models for the packaging mechanism.
A. Small terminase gp16
gp16, the 18-kDa small terminase subunit, is dispensable for packaging linear DNA in vitro but is essential in vivo; amber mutations in gene 16 accumulate mostly empty proheads and some partially filled heads (Black et al., 1994; Kondabagil and Rao, 2006).
Mutational and biochemical analyses suggest that gp16 is involved in the recognition of viral DNA (Lin et al., 1997; Lin and Black, 1998) and regulation of gp17 functions (Al-Zahrani et al., 2009). gp16 is predicted to contain three domains: a central domain important for oligomerization and N- and C-terminal domains important for DNA binding, ATP binding, and/or gp17-ATPase stimulation (Al-Zahrani et al., 2009; Mitchell et al., 2002). At least one of the major specificity determinants for gp17 interaction resides in the C-terminal domain. Swapping of this region between T4 and RB49 small terminases leads to switching the ATPase stimulation specificity from T4 gp17 to RB49 gp17 and vice versa (Gao and Rao, 2011; Fig. 6).
Figure 6.
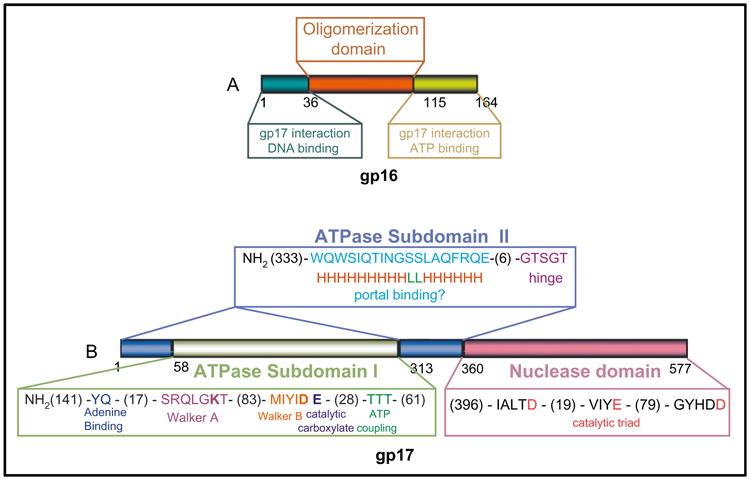
Domains and motifs in phage T4 terminase proteins. Schematic representation of domains and motifs in small terminase protein gp16 (A) and large terminase protein gp17 (B). Functionally critical amino acids are shown in bold. Numbers represent the number of amino acids in the respective coding sequence. For further detailed explanations of the functional motifs, refer to Rao and Feiss (2008) and Al-Zahrani et al. (2009).
Gp16 forms oligomeric single and side-by-side double rings, with each ring having a diameter of ∼8 nm with a ∼2-nm central channel (Lin et al., 1997). Mass spectrometry determination shows that the single and double rings are 11- and 22-mers, respectively (Duijn, 2010). A number of pac site phages produce comparable small terminase subunit multimeric ring structures. Sequence analyses predict two to three coiled coil motifs in gp16 (Kondabagil and Rao, 2006). All T4 family gp16s, as well as other phage small terminases, consist of one or more coiled-coil motifs, consistent with their propensity to form stable oligomers. Oligomerization presumably occurs through parallel coiled-coil interactions between neighboring subunits. Mutations in the long central α-helix of T4 gp16 that perturb coiled-coil interactions lose the ability to oligomerize (Kondabagil and Rao, 2006). Indeed, the X-ray structure shows 11- and 12-mers formed through extensive hydrophobic and electrostatic interactions between the two predicted long helices of the central domain (Sun et al., 2012).
Gp16 oligomerizes, forming a platform for the assembly of the large terminase gp17. This could presumably occur following interaction with the viral DNA concatemer. A predicted helix-turn-helix in the N-terminal domain is thought to be involved in DNA binding (Lin et al., 1997; Mitchell et al., 2002). The corresponding motif in the phage λ small terminase protein, gpNu1, has been well characterized and demonstrated to bind DNA. In vivo genetic studies and in vitro DNA-binding studies show that a 200-bp 3′ end sequence of gene 16 is a preferred “pac” site for gp16 interaction (Lin et al., 1997; Lin and Black, 1998). About 200 bp pieces of DNA are tightly associated with T4 gp16 purified from overexpressed E. coli culture (Kondabagil and Rao, unpublished observations). It was proposed that the stable gp16 double rings were two-turn lock washers that constituted the structural basis for synapsis of two pac site DNAs. This could promote the gp16-dependent gene amplifications observed around the pac site that can be selected in alt- mutants that package more DNA; such synapsis could function as a gauge of DNA concatemer maturation (Black, 1995; Wu and Black, 1995; Wu et al., 1995).
gp16 stimulates the gp17-ATPase activity by >50-fold (Leffers and Rao, 2000), but a high ATPase gp17 multimer does not require continued gp16 association (Baumann and Black, 2003). gp16 also stimulates in vitro DNA packaging activity in the crude system where phage-infected extracts containing all DNA replication/transcription/recombination proteins are present (Leffers and Rao, 2000; Rao and Black, 1988), but inhibits the packaging of linear DNA in the defined system where only two purified components, proheads and gp17, are present (Black and Peng, 2006; Kondabagil et al., 2006). It stimulates in vivo gp17-nuclease activity when T4 transcription factors are also present. It also stimulates gp17-nuclease in vitro in the presence of ATP but inhibits the nuclease in the absence of ATP (Al-Zahrani et al., 2009). gp16 also inhibits the binding of gp17 to DNA (Alam and Rao, 2008). Both N- and C-domains are required for ATPase stimulation or nuclease inhibition (Al-Zahrani et al., 2009). Maximum effects were observed at a ratio of approximately one gp16 oligomer to one gp17 (Kanamaru et al., 2004).
gp16 contains an ATP-binding site with broad nucleotide specificity (Lin et al., 1997; Al-Zahrani et al., 2009); however, it lacks the canonical nucleotide-binding signatures such as Walker A and Walker B (Mitchell et al., 2002). No correlation was evident between nucleotide binding and gp17-ATPase stimulation or gp17-nuclease inhibition. Thus it is unclear what role ATP binding plays in gp16 function. Evidence thus far suggests that gp16 is a regulator of the DNA packaging machine, modulating the nuclease, ATPase, and translocase activities of gp17 for efficient packaging initiation. In one model, a packaging initiation complex consisting of gp16, gp17, and DNA forms at a putative pac site, makes a cut, and delivers the end to the portal. gp16 then stimulates gp17 ATPase and DNA translocation activities to initiate DNA packaging.
B. Large terminase gp17
gp17 is the 70-kDa large subunit of the terminase holoenzyme and the motor protein of the DNA packaging machine. gp17 consists of two functional domains (Fig. 6): an N-terminal ATPase domain having classic ATPase signatures, such as Walker A, Walker B, and catalytic carboxylate, and a C-terminal nuclease domain having a catalytic metal cluster with conserved aspartic and glutamic acid residues coordinating with Mg2+ (Kanamaru et al., 2004).
1. ATPase
gp17 alone is sufficient to package linear DNA in vitro. gp17 exhibits a weak ATPase activity (Kcat = ∼1–2 ATPs hydrolyzed per gp17 molecule/min), which is stimulated by >50-fold by the small terminase protein gp16 (Baumann and Black, 2003; Leffers and Rao, 2000). Any mutation in the predicted catalytic residues of the N-terminal ATPase center results in a loss of stimulated ATPase and DNA packaging activities (Rao and Mitchell, 2001). Even subtle conservative substitutions such as aspartic acid to glutamic acid and vice versa in the Walker B motif resulted in complete loss of DNA packaging, suggesting that this ATPase provides energy for DNA translocation (Goetzinger and Rao, 2003; Mitchell and Rao, 2006).
The ATPase domain also exhibits DNA-binding activity, which may be involved in the DNA cutting and translocation functions of the packaging motor. Genetic evidence shows that gp17 may interact with gp32 (Franklin et al., 1998; Mosig, 1998), but, without other phage components, highly purified preparations of gp17 do not show appreciable affinity for single-stranded DNA or dsDNA. In fact, nontag and full-length purified gp17 has little or no nuclease activity, although is able to cut and package circular plasmid DNAs together with other phage proteins (Baumann and Black, 2003; Black and Peng, 2006). Thus there seem to be complex interactions among the terminase proteins, the concatemeric DNA, and the DNA replication/recombination/repair and transcription proteins that transit DNA metabolism into the packaging phase (Black and Peng, 2006).
One of the ATPase mutants, the DE-ED mutant in which the sequence of Walker B and catalytic carboxylate was reversed, showed tighter binding to ATP than wild-type gp17 but failed to hydrolyze ATP (Mitchell and Rao, 2006). Unlike wild-type gp17 or the ATPase domain that failed to crystallize, the ATPase domain with the ED mutation crystallized readily, probably because it trapped the ATPase in an ATP-bound conformation. The X-ray structure of the ATPase domain was determined up to 1.8 Å resolution in different bound states: apo, ATP bound, and ADP bound (Sun et al., 2007). It is a flat structure consisting of two subdomains: a large subdomain I (NsubI) and a smaller subdomain II (NsubII) forming a cleft in which ATP binds (Fig. 7A). The NsubI consists of the classic nucleotide-binding fold (Rossmann fold), a parallel β sheet of six β strands interspersed with helices. The structure showed that the predicted catalytic residues are oriented into the ATP pocket, forming a network of interactions with bound ATP. These also include an arginine finger proposed to trigger βγ-phosphoanhydride bond cleavage. In addition, the structure showed movement of a loop near the adenine-binding motif in response to ATP hydrolysis, which may be important for the transduction of ATP energy into mechanical motion.
Figure 7.
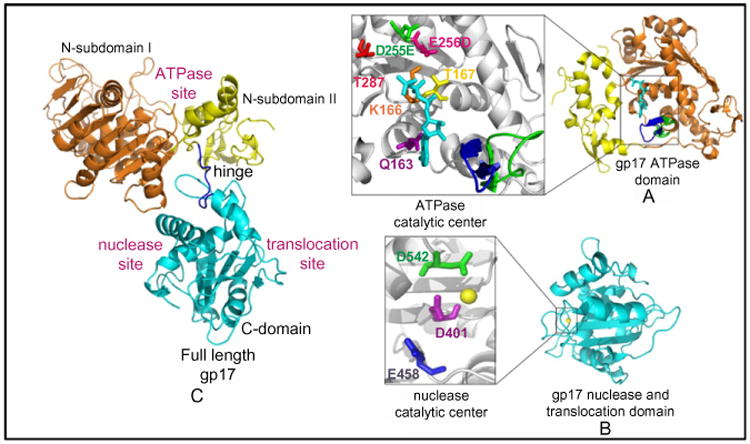
Structures of the T4 packaging motor protein gp17. Structures of the ATPase domain (A), nuclease/translocation domain (B), and full-length gp17 (C). Various functional sites and critical catalytic residues are labeled. For further details, see Sun et al. (2007, 2008).
2. Nuclease
gp17 exhibits a sequence-nonspecific endonuclease activity in vitro, and in vivo upon overexpression in E. coli, apparently producing blunt ends (Bhattacharyya and Rao, 1993, 1994). Biochemical and structural studies suggest that this activity makes packaging initiation and headful termination cuts. In the infected cell, interaction of gp17 with gp16 and other phage components likely controls the frequency and specificity of this nuclease (see later; Alam et al., 2008; Ghosh-Kumar et al., 2011). In the T4-like phage IME08, sequence analysis of the mature DNA ends indicates that its terminase produces ends with a two base overhang at a preferred consensus sequence, TTGGAG (Jiang et al., 2011).
Random mutagenesis of gene 17 and selection of mutants that lost nuclease activity identified a histidine-rich site in the C-terminal domain critical for DNA cleavage (Kuebler and Rao, 1998). Extensive site-directed mutagenesis of this region, combined with sequence alignments, identified a cluster of conserved aspartic acid and glutamic acid residues essential for DNA cleavage (Rentas and Rao, 2003). Unlike ATPase mutants, these mutants retained gp16-stimulated ATPase activity as well as DNA packaging activity as long as the substrate is a linear molecule. However, these mutants fail to package circular DNA as they are defective in cutting DNA that is required for packaging initiation.
The structure of the C-terminal nuclease domain from a T4 family phage, RB49, which has 72% sequence identity to the T4 C-domain, was determined to 1.16 Å resolution (Sun et al., 2008)(Fig. 7B). It has a globular structure consisting mostly of antiparallel β strands forming an RNase H fold that is found in resolvases, RNase Hs, and integrases. As redicted from mutagenesis studies, structures showed that residues D401, E458, and D542 form a catalytic triad coordinating with the Mg2+ ion. In addition, the structure showed the presence of a DNA-binding groove lined with a number of basic residues. The acidic catalytic metal center is buried at one end of this groove. Together, these form the nuclease cleavage site of gp17.
The crystal structure of the full-length T4 gp17 (ED mutant) was determined to 2.8 Å resolution (Fig. 6C; Sun et al., 2008). The N- and C-domain structures of the full-length gp17 superimpose with those solved using individually crystallized domains with only minor deviations. The full-length structure, however, has additional features relevant to the mechanism. A flexible “hinge” or “linker” connects the ATPase and nuclease domains. Previous biochemical studies showed that splitting gp17 into two domains at the linker retained the respective ATPase and nuclease functions but DNA translocation activity was completely lost (Kanamaru et al., 2004). Second, the N- and C-domains have a >1000 square Å complementary surface area consisting of an array of five charged pairs and hydrophobic patches (Sun et al., 2008). Third, the gp17 has a bound phosphate ion in the crystal structure. Docking of B-form DNA guided by shape and charge complementarity with one of the DNA phosphates superimposed on the bound phosphate aligns a number of basic residues, lining what appears to be a shallow translocation groove. Thus the C-domain appears to have two DNA grooves on different faces of the structure—one that aligns with the nuclease catalytic site and another that aligns with the translocating DNA (Fig. 7). Mutation of one of the groove residues (R406) showed a novel phenotype; loss of DNA translocation activity occurs but ATPase and nuclease activities are retained.
It is crucial that the nuclease activity of gp17 be controlled in vivo such that it is active at the initiation and termination steps of DNA packaging but inactive during translocation. Although it is clear that this must involve interactions with gp16, gp20 (portal), and other components, a basic mechanism by which the catalytic activity of the gp17's nuclease center can be controlled was hypothesized from structural and biochemical studies. Comparison of the X-ray structures of gp17 and cryoEM reconstruction of prohead-docked gp17 suggested that gp17 exists in two conformational states: tensed and relaxed (see later). Analysis of these states showed that the residues that line the nuclease groove come closer in the relaxed state, possibly “compressing” the DNA groove by ∼4 Å. One of the mechanisms by which the headful nuclease is regulated might be by relaying conformational signals between the ATPase center to the nuclease center through a “communication track” consisting of residues from subdomain II, hinge, and (β-hairpin (Ghosh-Kumar et al., 2011). During active translocation, subunits would be in the nuclease-inactive relaxed state and unable to form the antiparallel dimer that is essential to make cuts in both strands. However, during the initiation (and termination) step, the free (or freed) gp17 subunits may form a holo-terminase complex with gp16 and other phage proteins to make a cut in the concatemeric viral genome. It is likely that communication with the portal is also essential for making the headful termination.
VI. Packaging Motor
A. Structure
A functional DNA packaging machine could be assembled by mixing proheads and purified gp17. The latter assembles into a packaging motor through specific interactions with the portal vertex (Lin et al., 1999); such complexes, in a bulk in vitro assay, can package the 171-kb phage T4 DNA or any linear DNA (Black and Peng, 2006; Kondabagil et al., 2006). If less than headful length DNA molecules are added as the DNA substrate, the motor shows discontinuous packaging, packaging multiple molecules one molecule after another (Leffers and Rao, 1996). This can lead to head filling when large plasmid DNA molecules are used (∼30 kb)(Leffers and Rao, 1996) but with shorter DNAs, mostly partially filled heads with about six packaged DNA molecules are produced (Sabanayagam et al., 2007; Zhang et al., 2011). Although the unexpanded prohead is likely the true precursor for DNA packaging in vivo, in the in vitro assay, the expanded prohead, the partially full head, or even the full head can assemble the gp17 motor and drive efficient DNA translocation. In fact, packaged DNA of the virion can be emptied and refilled with DNA again (Zhang et al., 2011). Packaging can also be studied in real time either by fluorescence correlation spectroscopy (Sabanayagam et al., 2007) or at the single molecule level by optical tweezers (Fuller et al., 2007). The translocation kinetics of rhodamine (R6G-labeled, 100-bp DNA) was measured by determining the decrease in the diffusion coefficient as the DNA gets confined inside the capsid. Fluorescence resonance energy transfer between green fluorescent protein-labeled proteins within the prohead interior and the translocated rhodamine-labeled DNA confirmed the ATP-powered movement of DNA into the capsid and the packaging of multiple segments per procapsid (Sabanayagam et al., 2007). Analysis of Förster resonance energy transfer (FRET) dye pair end-labeled DNA substrates showed that upon packaging, the two ends of the packaged DNA were held 8–9 nm apart in the procapsid, likely fixed in the portal channel and crown, suggesting that a loop rather than an end of DNA is translocated following initiation at an end (Ray et al., 2010a).
In the optical tweezers system, prohead–gp17 complexes are tethered to a microsphere coated with capsid protein antibody, and the biotinylated DNA is tethered to another microsphere coated with streptavidine. Microspheres are brought together into near contact, allowing the motor to capture the DNA. Single packaging events are monitored, and the dynamics of the T4 packaging process are quantified (Fuller et al., 2007). The T4 motor, like the phi29 DNA packaging motor, generates forces as high as ∼60 pN, which is ∼20–25 times that of myosin ATPase, and a rate as high as ∼2000 bp/sec, among the highest recorded to date. Slips and pauses occur but these are relatively short and rare and the motor recovers and recaptures DNA, continuing translocation. The high rate of translocation is in keeping with the need to package the 171-kb size T4 genome in about 5 minutes. The T4 motor generates enormous power; when an external load of 40 pN is applied, the T4 motor translocates at a speed of ∼380 bp/sec. When scaled up to a macromotor, the T4 motor is approximately twice as powerful as a typical automobile engine.
CryoEM reconstruction of the packaging machine showed two rings of density at the portal vertex (Sun et al., 2008) (Fig. 8). The upper ring is flat, resembling the ATPase domain structure, and the lower ring is spherical, resembling the C-domain structure. This was confirmed by docking of the X-ray structures of the domains into the cryoEM density. The motor has pentamer stoichiometry, with the ATP-binding surface facing the portal and interacting with it. It has an open central channel that is in line with the portal channel, and the translocation groove of the C-domain faces the channel. Minimal contacts exist between the adjacent subunits, suggesting that ATPases may fire relatively independently during translocation.
Figure 8.
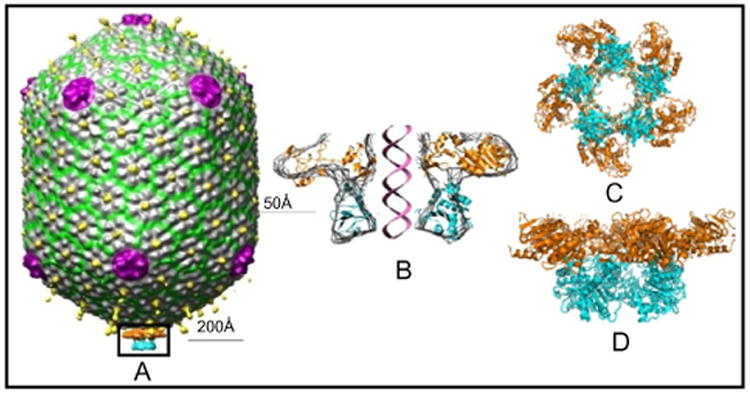
Structure of the T4 DNA packaging machine. (A) CryoEM reconstruction of the phage T4 DNA packaging machine showing the pentameric motor assembled at the special portal vertex. (B–D) Cross section, top, and side views of the pentameric motor, respectively, by fitting X-ray structures of the gp17 ATPase and nuclease/translocation domains into cryoEM density.
Unlike the cryoEM structure where the two lobes (domains) of the motor are separated (“relaxed” state), domains in the full-length gp17 are in close contact (“tensed” state) (Sun et al., 2008). In the tensed state, the subdomain II of ATPase is rotated by 6° and the C-domain is pulled upward by 7 Å, equivalent to 2 bp. The “arginine finger” located between subI and NsubII is positioned toward the βγ-phosphates of ATP and the ion pairs are aligned.
B. Mechanism and dynamics
Of the many models proposed to explain the mechanism of viral DNA translocation, the portal rotation model (Hendrix, 1978) attracted the most attention and was often cited in other contexts in confirmation of the functional biological significance of symmetry mismatch. According to the original rotation model, the portal and DNA are locked like a nut and bolt. The symmetry mismatch between the 5-fold capsid and the 12-fold portal means that there are no reiterated specific portal-capsid subunit interactions, thereby enabling free rotation of the two multimeric interfaces; as proposed, this would allow the portal, or nut, to rotate, powered by ATP hydrolysis, causing the DNA, or bolt, to be translocated into the capsid. X-ray structures of conserved dodecameric Phi29 and SPP1 portals could be interpreted as consistent with the original portal rotation or newer, rotation-incorporating models such as the rotation–compression-relaxation (Simpson et al., 2000), electrostatic gripping (Guasch et al., 2002), and molecular lever (Lebedev et al., 2007) models.
Protein fusions to either the N- or the C-terminal end of the T4 portal protein could be incorporated into up to approximately one-half of the dodecamer positions without loss of prohead function. As compared to the wild type, portals containing C-terminal GFP fusions but not N-terminal GFP fusions (Dixit et al., 2011) lock the proheads into the unexpanded conformation unless terminase packages DNA, suggesting that the portal plays a central role in controlling prohead expansion. Expansion is required to protect the packaged DNA from nuclease but not for packaging itself as measured by fluorescence correlation spectroscopy (Ray et al., 2009). Retention of the DNA packaging function of such portals is inconsistent with the portal rotation model, as rotation would require that bulky C-terminal GFP fusion proteins within the capsid rotate through the densely packaged DNA.
A more direct test tethered the portal to the capsid through Hoc interactions (Baumann et al., 2006). Hoc-binding sites are not present in unexpanded proheads but are exposed following capsid expansion. Unexpanded proheads were first prepared with some of the 12 portal subunits replaced by the N-terminal Hoc–portal fusion proteins. The pro-heads were then expanded in vitro to expose Hoc-binding sites. The Hoc portion of the portal fusion would bind to the center of the nearest hexon, tethering one to five portal subunits to the capsid, thereby protecting Hoc from proteolysis. By this test and also by incorporation of Hoc–gp20 into active phage particles the Hoc was tethered but did not affect DNA packaging. Thus both N- and the C-terminal portal fusion protein portal results strongly suggested that portal rotation does not occur (Baumann et al., 2006). This was supported more recently by single molecule fluorescence spectroscopy of actively packaging phi29 packaging complexes that apparently (to 99% certainty) failed to show rotation (Hugel et al., 2007).
In a second class of models, the terminase not only provides the energy but also translocates DNA actively (Draper and Rao, 2007). Conformational changes in terminase domains cause changes in DNA-binding affinity, resulting in binding and releasing DNA, reminiscent of the inchworm-type translocation by helicases. gp17 and numerous large terminases possess an ATPase coupling motif that is commonly present in helicases and translocases (Draper and Rao, 2007). Mutations in the coupling motif present at the junction of NSubI and NSubII result in a loss of ATPase and DNA packaging activities.
The cryoEM and X-ray structures (Fig. 8), combined with the mutational analyses described earlier, led to the postulation of a terminase-driven packaging mechanism (Sun et al., 2008). The pentameric T4 packaging motor can be considered to be analogous to a five-cylinder engine. It consists of an ATPase center in NsubI, which is the engine that provides energy. The C-domain has a translocation groove, which is the wheel that moves DNA. The smaller NsubII is the transmission domain, coupling the engine to the wheel via a flexible hinge. The arginine finger is a spark plug that fires ATPase when the motor is locked in the firing mode. Charged pairs generate electrostatic force by alternating between relaxed and tensed states (Fig. 9). The nuclease groove faces away from translocating DNA and is activated when packaging is completed.
Figure 9.
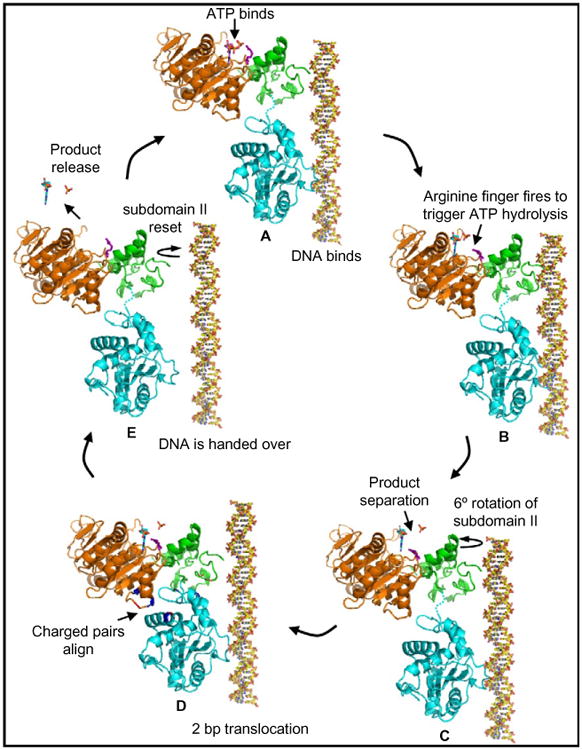
Model for the electrostatic force-driven DNA packaging mechanism. Schematic representation showing the sequence of events that occur in a single gp17 molecule to translocate 2 bp of DNA [for details, see text and Sun et al. (2008)].
In the relaxed conformational state (cryoEM structure), the hinge is extended (Fig. 9). Binding of DNA to the translocation groove and of ATP to NsubI locks the motor in translocation mode (Fig. 9A) and brings the arginine finger into position, firing ATP hydrolysis (Fig. 9B). Repulsion between the negatively charged ADP(3-) and Pi(3-) drives them apart, causing NsubII to rotate by 6° (Fig. 9C), aligning the charge pairs between the N- and C-domains. This generates electrostatic force, attracting the C-domain–DNA complex and causing 7 Å upward movement, the tensed conformational state (X-ray structure) (Fig. 9D). Thus 2 bp of DNA is translocated into the capsid in one cycle. Product release and the loss of six negative charges causes NsubII to rotate back to the original position, misaligning the ion pairs and returning the C-domain to the relaxed state (Fig. 9E).
Translocation of 2 bp would bring the translocation groove of the adjacent subunit into alignment with the backbone phosphates. DNA is then handed over to the next subunit by the matching motor and DNA symmetries. Thus, ATPase catalysis causes conformational changes that generate electrostatic force, which is then converted to mechanical force. The pentameric motor translocates 10 bp (one turn of the helix) when all five gp17 subunits fire in succession, bringing the first gp17 subunit once again in alignment with the DNA phosphates. Synchronized orchestration movements of the motor translocate DNA up to ∼2000 bp/sec.
DNA may not be translocated by a simple linear motion. In vitro packaging experiments with modified DNA substrates support a DNA torsional compression linear translocation mechanism in which the portal grips the DNA while a power stroke is applied by gp17 conformational changes (Oram et al., 2008). This would transiently compress the DNA present in the translocation channel between the portal and the ATPase motor. Short (<200 bp) DNA substrate translocation by gp17 is blocked by nicks or other departures from the B form, suggesting that energy stored as DNA compression may be dissipated by a nick (Figs. 10A–10C). However, longer DNAs containing nicks or other abnormal structures are translocated apparently normally by both T4 and phi29 motors (Aathavan et al., 2009; Oram et al., 2008), presumably by multiple motor cycles. Use of a DNA leader of several kilobases joined to a 90-bp Y-DNA structure showed packaging of the leader segment while the dye containing Y-DNA was nuclease accessible; the Y-junction was arrested in proximity to a prohead portal containing GFP fusions, as evident by FRET transfer between the dye labeled Y-junction and the GFP-labeled portal (Ray et al., 2010b)(Fig. 10D). Comparable stalled Y-DNA substrates containing FRET pair dyes in the Y-stem also suggested that the motor compresses or “crunches” the stem DNA held in the portal channel, as dyes separated by 10 or 14 bp undergo a comparable distance decrease measured by FRET at 22 and 24%, whether the dyes are on the same side or opposite side of the helix, supporting DNA compression rather than bending (Fig. 10E). Transient compression of 10–14 bp DNA in the portal translocation channel is thus proposed to be followed by release by the portal of B form DNA in 10 bp or longer steps. Other evidence in phages T4 and SPP1 also supports an active role for the portal as well as terminase in DNA translocation (Cuervo et al., 2007; Lin et al., 1999; Dixit, Ray, and Black, in preparation).
Figure 10.
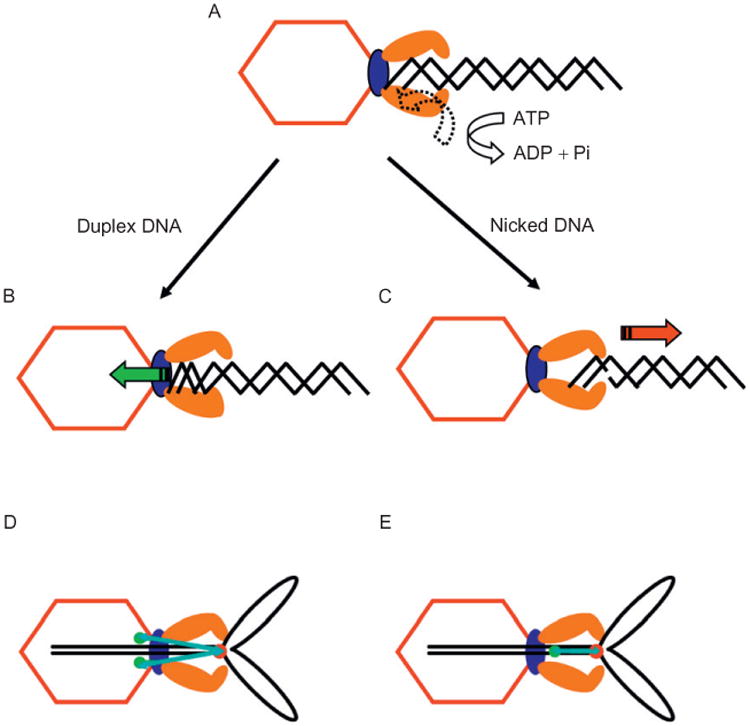
Model for the torsional compression portal-DNA-grip-and-release packaging mechanism (Oram et al., 2008). An ATP-driven conformational change of the terminase to portal interaction is proposed to drive DNA into the prohead by a DNA compression motor stroke. (A–C) Short linear DNAs are packaged by the motor, whereas nicked or other abnormal structures containing DNA substrates are released; (D) kb DNA leader containing Y-DNA substrates is retained by the motor and the Y-junction (red dot) is anchored in the procapsid in proximity to portal GFP fusions; and (E) DNA compression of the Y-stem B two FRET pair dye-containing segment in the stalled complex is observed; fluorophore distances measured by FRET are given by narrow arrows (Ray et al., 2010b).
A portal-bound T4 gp49 Holliday junction resolvase releases packaging-arrested Y- or X-branched structure-containing concatemers, its essential in vivo packaging function (Golz and Kemper, 1999). Consistent with this function and the torsional model is the release of DNA compression in arrested Y-DNA substrates by the addition of purified gp49 enzyme in vitro (Fig. 11A) (Dixit et al., 2011). Active terminases labeled at the N- and C-terminal ends with a single dye molecule showed that the FRET distance between the N-terminal GFP-labeled portal protein and gp17 is 6.9 nm for the N terminus and 5.7 nm for the C terminus (Figs. 11B and 11C). Packaging with a C-terminal fluorescent terminase on a GFP-portal prohead shows a reduction in distance of 0.6 nm in the arrested Y-DNA as compared to linear DNA, and again the reduction is reversed by resolvase treatment, which is consistent with the tensed and relaxed terminase conformational changes (Fig. 9).
Figure 11.
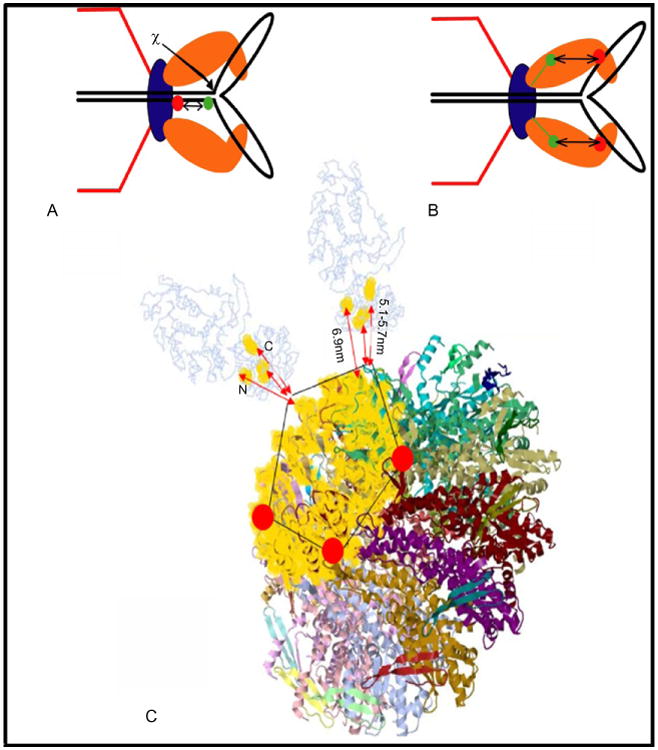
Both motor proteins and DNA undergo major conformational changes during DNA packaging. Schematic of FRET between different dyes within the DNA and site of action of T4 gp49 (χ). Portal-bound Holliday junction resolvase (gp49) action releases compression from the Y-DNA substrates (A). Resolvase release of trapped Y-DNAs is also correlated with increased distance (∼0.6 nm, or 5.1 to 5.7 nm) between the terminase and the portal during packaging (B). Distances are measured from N and C termini of gp17 to the GFP–gp20 portal by FRET (C)(Dixit et al., 2011).
VII. Conclusions and Prospects
Although the aforementioned studies strongly suggest that phage T4 employs a linear motor mechanism to package DNA, some aspects of the mechanism are still speculative. More extensive mutagenesis and single molecule studies need to be performed to determine DNA step size, motor coordination, and specific roles of the structural parts of the terminase and portal proteins in translocation and DNA cutting. Other mechanistic questions that require deeper investigation include whether (i) the DNA structure is altered during normal linear translocation; (ii) to what extent and how the portal contributes to translocation; (iii) whether the DNA rotates during translocation to compensate for winding inside the capsid; and (iv) to what extent does a single conserved mechanism apply among tailed dsDNA phages and comparable portal-containing eukaryotic viruses; for example, herpes viruses.
Despite these remaining questions, it is clear from the discussion given previously that major advances have been made in understanding the phage T4 capsid structure and mechanism of DNA packaging. These advances, by combining genetics and biochemistry with structure and biophysics, set the stage to probe the packaging mechanism with even greater depth and precision. It is reasonable to hope that this would lead to the elucidation of catalytic cycle, mechanistic details, and motor dynamics to near atomic resolution. The accumulated and emerging basic knowledge should also lead to medical applications such as the development of vaccines and phage therapy.
Acknowledgments
The authors thank Aparna Dixit, Bonnie Draper, Vishal Kottadiel, and Alice Kuaban for preparing the figures, references, proofreading, and helpful comments. Research in the authors' laboratories has been funded by the National Science Foundation (VBR: MCB-0923873) and National Institutes of Health (VBR: NIAID-AI081726; LWB: NIAID-AI011676). Special thanks to our present and former lab members for their contributions over the years.
List of Abbreviations
- aa
amino acids
- gp23*
mature gp21 protease processed form of gp23 etc
- EF
edema factor
- EM
electron microscopy
- FCS
fluorescence correlation spectroscopy
- FMDV
foot and mouth disease virus
- FRET
Förster resonance energy transfer
- gp
gene product
- HIV
human immunodeficiency virus
- Hoc
highly antigenic outer capsid protein
- IP
internal protein
- LF
lethal factor
- PA
protective antigen
- Soc
small outer capsid protein
- kb
kilobase
- bp
base pair
References
- Aathavan K, Politzer AT, Kaplan A, Moffitt JR, Chemla YR, Grimes S, Jardine PJ, Anderson DL, Bustamante C. Substrate interactions and promiscuity in a viral DNA packaging motor. Nature. 2009;461:669–673. doi: 10.1038/nature08443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Zahrani AS, Kondabagil K, Gao S, Kelly N, Ghosh-Kumar M, Rao VB. The small terminase, gp16, of bacteriophage T4 is a regulator of the DNA packaging motor. J Biol Chem. 2009;284:24490–24500. doi: 10.1074/jbc.M109.025007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam TI, Draper B, Kondabagil K, Rentas FJ, Ghosh-Kumar M, Sun S, Rossmann MG, Rao VB. The headful packaging nuclease of bacteriophage T4. Mol Microbiol. 2008;69:1180–1190. doi: 10.1111/j.1365-2958.2008.06344.x. [DOI] [PubMed] [Google Scholar]
- Alam TI, Rao VB. The ATPase domain of the large terminase protein, gp17, from bacteriophage T4 binds DNA: Implications to the DNA packaging mechanism. J Mol Biol. 2008;376:1272–1281. doi: 10.1016/j.jmb.2007.12.041. [DOI] [PubMed] [Google Scholar]
- Andreadis JD, Black LW. Substrate mutations that bypass a specific Cpn10 chaperonin requirement for protein folding. J Biol Chem. 1998;273:34075–34086. doi: 10.1074/jbc.273.51.34075. [DOI] [PubMed] [Google Scholar]
- Archer MJ, Liu JL. Bacteriophage T4 nanoparticles as materials in sensor applications: Variables that influence their organization and assembly on surfaces. Sensors. 2009;9:6298–6311. doi: 10.3390/s90806298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bair CL, Black LW. A type IV modification dependent restriction nuclease that targets glucosylated hydroxymethyl cytosine modified DNAs. J Mol Biol. 2007;366:768–778. doi: 10.1016/j.jmb.2006.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bair CL, Rifat D, Black LW. Exclusion of glucosyl-hydroxymethylcytosine DNA containing bacteriophages is overcome by the injected protein inhibitor IPI*. J Mol Biol. 2007;366:779–789. doi: 10.1016/j.jmb.2006.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkes PJ, Faber BW, van Heerikhuizen H, van der Vies SM. The T4-encoded cochaperonin, gp31, has unique properties that explain its requirement for the folding of the T4 major capsid protein. Proc Natl Acad Sci USA. 2005;102:8144–8149. doi: 10.1073/pnas.0500048102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann RG, Black LW. Isolation and characterization of T4 bacteriophage gp17 terminase, a large subunit multimer with enhanced ATPase activity. J Biol Chem. 2003;278:4618–4627. doi: 10.1074/jbc.M208574200. [DOI] [PubMed] [Google Scholar]
- Baumann RG, Mullaney J, Black LW. Portal fusion protein constraints on function in DNA packaging of bacteriophage T4. Mol Microbiol. 2006;61:16–32. doi: 10.1111/j.1365-2958.2006.05203.x. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya SP, Rao VB. A novel terminase activity associated with the DNA packaging protein gp17 of bacteriophage T4. Virology. 1993;196:34–44. doi: 10.1006/viro.1993.1452. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya SP, Rao VB. Structural analysis of DNA cleaved in vivo by bacteriophage T4 terminase. Gene. 1994;146:67–72. doi: 10.1016/0378-1119(94)90834-6. [DOI] [PubMed] [Google Scholar]
- Black L, Thomas JA. In: Condensed Genome Structure in Viral Molecular Machines. Rossmann, Rao, editors. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black LW. DNA packaging in dsDNA bacteriophages. Annu Rev Microbiol. 1989;43:267–292. doi: 10.1146/annurev.mi.43.100189.001411. [DOI] [PubMed] [Google Scholar]
- Black LW. DNA packaging and cutting by phage terminases: Control inphageT4 by a synaptic mechanism. Bioessays. 1995;17:1025–1030. doi: 10.1002/bies.950171206. [DOI] [PubMed] [Google Scholar]
- Black LW, Peng G. Mechanistic coupling of bacteriophage T4 DNA packaging to components of the replication-dependent late transcription machinery. J Biol Chem. 2006;281:25635–25643. doi: 10.1074/jbc.M602093200. [DOI] [PubMed] [Google Scholar]
- Black LW, Showe MK, Steven AC. Morphogenesis of the T4 head. In: Karam JM, editor. Molecular Biology of Bacteriophage T4. ASM Press; Washington, DC: 1994. pp. 218–258. [Google Scholar]
- Calmat S, Hendriks J, van Heerikhuizen H, Schmidt CF, van der Vies SM, Peterman EJ. Dissociation kinetics of the GroEL-gp31 chaperonin complex studied with Forster resonance energy transfer. Biochemistry. 2009;48:11692–11698. doi: 10.1021/bi9013962. [DOI] [PubMed] [Google Scholar]
- Cardarelli L, Lam R, Tuite A, Baker LA, Sadowski PD, Radford DR, Rubinstein JL, Battaile KP, Chirgadze N, Maxwell KL, Davidson AR. The crystal structure of bacteriophage HK97 gp6: Defining a large family of head-tail connector proteins. J Mol Biol. 2010;395:754–768. doi: 10.1016/j.jmb.2009.10.067. [DOI] [PubMed] [Google Scholar]
- Cerritelli ME, Cheng N, Rosenberg AH, McPherson CE, Booy FP, Steven AC. Encapsidated conformation of bacteriophage T7 DNA. Cell. 1997;91:271–280. doi: 10.1016/s0092-8674(00)80409-2. [DOI] [PubMed] [Google Scholar]
- Cheng H, Shen N, Pei J, Grishin NV. Double-stranded DNA bacteriophage prohead protease is homologous to herpesvirus protease. Protein Sci. 2004;13:2260–2269. doi: 10.1110/ps.04726004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clare DK, Bakkes PJ, van Heerikhuizen H, van der Vies SM, Saibil HR. An expanded protein folding cage in the GroEL-gp31 complex. J Mol Biol. 2006;358:905–911. doi: 10.1016/j.jmb.2006.02.033. [DOI] [PubMed] [Google Scholar]
- Clare DK, Bakkes PJ, van Heerikhuizen H, van der Vies SM, Saibil HR. Chaperonin complex with a newly folded protein encapsulated in the folding chamber. Nature. 2009;457:107–110. doi: 10.1038/nature07479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo A, Vaney MC, Antson AA, Tavares P, Oliveira L. Structural rearrangements between portal protein subunits are essential for viral DNA translocation. J Biol Chem. 2007;282:18907–18913. doi: 10.1074/jbc.M701808200. [DOI] [PubMed] [Google Scholar]
- Desplats C, Krisch HM. The diversity and evolution of the T4-type bacteriophages. Res Microbiol. 2003;154:259–267. doi: 10.1016/S0923-2508(03)00069-X. [DOI] [PubMed] [Google Scholar]
- Dixit A, Ray K, Lakowicz JR, Black LW. Dynamics of the T4 bacteriophage DNA packasome motor: Endo VII resolvase release of arrested Y-DNA substrates. J Biol Chem. 2011;286:18878–18889. doi: 10.1074/jbc.M111.222828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper B, Rao VB. An ATP hydrolysis sensor in the DNA packaging motor from bacteriophage T4 suggests an inchworm-type translocation mechanism. J Mol Biol. 2007;369:79–94. doi: 10.1016/j.jmb.2007.03.019. [DOI] [PubMed] [Google Scholar]
- Duijn E. Current limitations in native mass spectrometry based structural biology. J Am Soc Mass Spect. 2010;21:971–978. doi: 10.1016/j.jasms.2009.12.010. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, King J, Harrison SC, Eiserling FA. The structural organization of DNA packaged within the heads of T4 wild-type, isometric and giant bacteriophages. Cell. 1978;14:559–568. doi: 10.1016/0092-8674(78)90242-8. [DOI] [PubMed] [Google Scholar]
- Fokine A, Battisti AJ, Kostyuchenko VA, Black LW, Rossmann MG. Cryo-EM structure of a bacteriophage T4 gp24 bypass mutant: The evolution of pentameric vertex proteins in icosahedral viruses. J Struct Biol. 2006;154:255–259. doi: 10.1016/j.jsb.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Fokine A, Bowman VD, Battisti AJ, Li Q, Chipman PR, Rao VB, Rossmann MG. Cryo-electron microscopy study of bacteriophage T4 displaying anthrax toxin proteins. Virology. 2007;367:422–427. doi: 10.1016/j.virol.2007.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokine A, Chipman PR, Leiman PG, Mesyanzhinov VV, Rao VB, Rossmann MG. Molecular architecture of the prolate head of bacteriophage T4. Proc Natl Acad Sci USA. 2004;101:6003–6008. doi: 10.1073/pnas.0400444101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokine A, Islam MZ, Zhang Z, Bowman VD, Rao VB, Rossmann MG. Structure of the three N-terminal immunoglobulin domains of the highly immunogenic outer capsid protein from a T4-like bacteriophage. J Virol. 2011;85:8141–8148. doi: 10.1128/JVI.00847-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokine A, Leiman PG, Shneider MM, Ahvazi B, Boeshans KM, Steven AC, Black LW, Mesyanzhinov VV, Rossmann MG. Structural and functional similarities between the capsid proteins of bacteriophages T4 and HK97 point to a common ancestry. Proc Natl Acad Sci USA. 2005;102:7163–7168. doi: 10.1073/pnas.0502164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin JL, Haseltine D, Davenport L, Mosig G. The largest (70 kDa) product of the bacteriophage T4 DNA terminase gene 17 binds to single-stranded DNA segments and digests them towards junctions with double-stranded DNA. J Mol Biol. 1998;277:541–557. doi: 10.1006/jmbi.1998.1619. [DOI] [PubMed] [Google Scholar]
- Fuller DN, Raymer DM, Kottadiel VI, Rao VB, Smith DE. Single phage T4 DNA packaging motors exhibit large force generation, high velocity, and dynamic variability. Proc Natl Acad Sci USA. 2007;104:16868–16873. doi: 10.1073/pnas.0704008104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh-Kumar M, Alam TI, Draper B, Stack JD, Rao VB. Regulation by interdomain communication of a headful packaging nuclease from bacteriophage T4. Nucleic Acids Res. 2011;39:2742–2755. doi: 10.1093/nar/gkq1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, Rao VB. Specificity of interactions among the DNA packaging machine components of T4 related bacteriophages. J Biol Chem. 2011;286:3944–3956. doi: 10.1074/jbc.M110.196907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetzinger KR, Rao VB. Defining the ATPase center of bacteriophage T4 DNA packaging machine: Requirement for a catalytic glutamate residue in the large terminase protein gp17. J Mol Biol. 2003;331:139–154. doi: 10.1016/s0022-2836(03)00636-3. [DOI] [PubMed] [Google Scholar]
- Golz S, Kemper B. Association of holliday-structure resolving endonuclease VII with gp20 from the packaging machine of phage T4. J Mol Biol. 1999;285:1131–1144. doi: 10.1006/jmbi.1998.2399. [DOI] [PubMed] [Google Scholar]
- Guasch A, Pous J, Ibarra B, Gomis-Ruth FX, Valpuesta JM, Sousa N, Carrascosa JL, Coll M. Detailed architecture of a DNA translocating machine: The high-resolution structure of the bacteriophage phi29 connector particle. J Mol Biol. 2002;315:663–676. doi: 10.1006/jmbi.2001.5278. [DOI] [PubMed] [Google Scholar]
- Hendrix RW. Symmetry mismatch and DNA packaging in large bacteriophages. Proc Natl Acad Sci USA. 1978;75:4779–4783. doi: 10.1073/pnas.75.10.4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugel T, Michaelis J, Hetherington CL, Jardine PJ, Grimes S, Walter JM, Falk W, Anderson DL, Bustamante C. Experimental test of connector rotation during DNA packaging into bacteriophage phi29 capsids. PLoS Biol. 2007;5:e59. doi: 10.1371/journal.pbio.0050059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii T, Yamaguchi Y, Yanagida M. Binding of the structural protein soc to the head shell of bacteriophage T4. J Mol Biol. 1978;120:533–544. doi: 10.1016/0022-2836(78)90352-2. [DOI] [PubMed] [Google Scholar]
- Ishii T, Yanagida M. The two dispensable structural proteins (soc and hoc) of the T4 phage capsid; their purification and properties, isolation and characterization of the defective mutants, and their binding with the defective heads in vitro. J Mol Biol. 1977;109:487–514. doi: 10.1016/s0022-2836(77)80088-0. [DOI] [PubMed] [Google Scholar]
- Jiang J, Abu-Shilbayeh L, Rao VB. Display of a PorA peptide from Neisseria meningitidis on the bacteriophage T4 capsid surface. Infect Immun. 1997;65:4770–4777. doi: 10.1128/iai.65.11.4770-4777.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Jiang H, Li C, Wang S, Mi Z, An X, Chen J, Tong Y. Sequence characteristics of T4-like bacteriophage IME08 genome termini revealed by high throughput sequencing. Virol J. 2011;8:194. doi: 10.1186/1743-422X-8-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamaru S, Kondabagil K, Rossmann MG, Rao VB. The functional domains of bacteriophage t4 terminase. J Biol Chem. 2004;279:40795–40801. doi: 10.1074/jbc.M403647200. [DOI] [PubMed] [Google Scholar]
- Keppel F, Rychner M, Georgopoulos C. Bacteriophage-encoded cochaperonins can substitute for Escherichia coli's essential GroES protein. EMBO J. 2002;3:893–898. doi: 10.1093/embo-reports/kvf176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondabagil KR, Rao VB. A critical coiled coil motif in the small terminase, gp16, from bacteriophage T4: Insights into DNA packaging initiation and assembly of packaging motor. J Mol Biol. 2006;358:67–82. doi: 10.1016/j.jmb.2006.01.078. [DOI] [PubMed] [Google Scholar]
- Kondabagil KR, Zhang Z, Rao VB. The DNA translocating ATPase of bacteriophage T4 packaging motor. J Mol Biol. 2006;363:786–799. doi: 10.1016/j.jmb.2006.08.054. [DOI] [PubMed] [Google Scholar]
- Krisch HM, Comeau AM. The immense journey of bacteriophage T4: From d'Herelle to Delbruck and then to Darwin and beyond. Res Microbiol. 2008;159:314–324. doi: 10.1016/j.resmic.2008.04.014. [DOI] [PubMed] [Google Scholar]
- Kuebler D, Rao VB. Functional analysis of the DNA-packaging/terminase protein gp17 from bacteriophage T4. J Mol Biol. 1998;281:803–814. doi: 10.1006/jmbi.1998.1952. [DOI] [PubMed] [Google Scholar]
- Lebedev AA, Krause MH, Isidro AL, Vagin AA, Orlova EV, Turner J, Dodson EJ, Tavares P, Antson AA. Structural framework for DNA translocation via the viral portal protein. EMBO J. 2007;26:1984–1994. doi: 10.1038/sj.emboj.7601643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leffers G, Rao VB. A discontinuous headful packaging model for packaging less than headful length DNA molecules by bacteriophage T4. J Mol Biol. 1996;258:839–850. doi: 10.1006/jmbi.1996.0291. [DOI] [PubMed] [Google Scholar]
- Leffers G, Rao VB. Biochemical characterization of an ATPase activity associated with the large packaging subunit gp17 from bacteriophage T4. J Biol Chem. 2000;275:37127–37136. doi: 10.1074/jbc.M003357200. [DOI] [PubMed] [Google Scholar]
- Li Q, Shivachandra SB, Leppla SH, Rao VB. Bacteriophage T4 capsid: a unique platform for efficient surface assembly of macromolecular complexes. J Mol Biol. 2006;363:577–588. doi: 10.1016/j.jmb.2006.08.049. [DOI] [PubMed] [Google Scholar]
- Li Q, Shivachandra SB, Zhang Z, Rao VB. Assembly of the small outer capsid protein, Soc, on bacteriophage T4: A novel system for high density display of multiple large anthrax toxins and foreign proteins on phage capsid. J Mol Biol. 2007;370:1006–1019. doi: 10.1016/j.jmb.2007.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Black LW. DNA requirements in vivo for phage T4 packaging. Virology. 1998;242:118–127. doi: 10.1006/viro.1997.9019. [DOI] [PubMed] [Google Scholar]
- Lin H, Rao VB, Black LW. Analysis of capsid portal protein and terminase functional domains: Interaction sites required for DNA packaging in bacteriophage T4. J Mol Biol. 1999;289:249–260. doi: 10.1006/jmbi.1999.2781. [DOI] [PubMed] [Google Scholar]
- Lin H, Simon MN, Black LW. Purification and characterization of the small subunit of phage T4 terminase, gp16, required for DNA packaging. J Biol Chem. 1997;272:3495–3501. doi: 10.1074/jbc.272.6.3495. [DOI] [PubMed] [Google Scholar]
- Liu J, Mushegian A. Displacements of prohead protease genes in the late operons of double-stranded-DNA bacteriophages. J Bacteriol. 2004;186:4369–4375. doi: 10.1128/JB.186.13.4369-4375.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipińska B, Rao AS, Bolten BM, Balakrishnan R, Goldberg EB. Cloning and identification of bacteriophage T4 gene 2 product gp2 and action of gp2 on infecting DNA in vivo. J Bacteriol. 1989;171:488–497. doi: 10.1128/jb.171.1.488-497.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malys N, Chang DY, Baumann RG, Xie D, Black LW. A bipartite bacteriophage T4 SOC and HOC randomized peptide display library: Detection and analysis of phage T4 terminase (gp17) and late sigma factor (gp55) interaction. J Mol Biol. 2002;319:289–304. doi: 10.1016/S0022-2836(02)00298-X. [DOI] [PubMed] [Google Scholar]
- Miller ES, Kutter E, Mosig G, Arisaka F, Kunisawa T, Ruger W. Bacteriophage T4 genome. Microbiol Mol Biol Rev. 2003;67:86–156. doi: 10.1128/MMBR.67.1.86-156.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell MS, Matsuzaki S, Imai S, Rao VB. Sequence analysis of bacteriophage T4 DNA packaging/terminase genes 16 and 17 reveals a common ATPase center in the large subunit of viral terminases. Nucleic Acids Res. 2002;30:4009–4021. doi: 10.1093/nar/gkf524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell MS, Rao VB. Functional analysis of the bacteriophage T4 DNA-packaging ATPase motor. J Biol Chem. 2006;281:518–527. doi: 10.1074/jbc.M507719200. [DOI] [PubMed] [Google Scholar]
- Mosig G. Recombination and recombination-dependent DNA replication in bacteriophage T4. Annu Rev Genet. 1998;32:379–413. doi: 10.1146/annurev.genet.32.1.379. [DOI] [PubMed] [Google Scholar]
- Mullaney JM, Black LW. Activity of foreign proteins targeted within the bacteriophage T4 head and prohead: Implications for packaged DNA structure. J Mol Biol. 1998;283:913–929. doi: 10.1006/jmbi.1998.2126. [DOI] [PubMed] [Google Scholar]
- Mullaney JM, Thompson RB, Gryczynski Z, Black LW. Green fluorescent protein as a probe of rotational mobility within bacteriophage T4. J Virol Methods. 2000;88:35–40. doi: 10.1016/s0166-0934(00)00166-x. [DOI] [PubMed] [Google Scholar]
- Oram M, Sabanayagam C, Black LW. Modulation of the packaging reaction of bacteriophage T4 terminase by DNA structure. J Mol Biol. 2008;381:61–72. doi: 10.1016/j.jmb.2008.05.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin L, Fokine A, O'Donnell E, Rao VB, Rossmann MG. Structure of the small outer capsid protein, Soc: A clamp for stabilizing capsids of T4-like phages. J Mol Biol. 2009;395:728–741. doi: 10.1016/j.jmb.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao VB, Black LW. Cloning, overexpression and purification of the terminase proteins gp16 and gp17 of bacteriophage T4: Construction of a defined in-vitro DNA packaging system using purified terminase proteins. J Mol Biol. 1988;200:475–488. doi: 10.1016/0022-2836(88)90537-2. [DOI] [PubMed] [Google Scholar]
- Rao VB, Black LW. In: DNA Packaging in Bacteriophage T4 in Viral Genome Packaging Machines, Genetics, Structure, and Mechanism. Catalano CE, editor. Plenum Press; 2005. pp. 40–58. [Google Scholar]
- Rao VB, Feiss M. The bacteriophage DNA packaging motor. Annu Rev Genet. 2008;42:647–681. doi: 10.1146/annurev.genet.42.110807.091545. [DOI] [PubMed] [Google Scholar]
- Rao VB, Mitchell MS. The N-terminal ATPase site in the large terminase protein gp17 is critically required for DNA packaging in bacteriophage T4. J Mol Biol. 2001;314:401–411. doi: 10.1006/jmbi.2001.5169. [DOI] [PubMed] [Google Scholar]
- Rao M, Peachman K, Li Q, Matyas G, Shivachandra S, Borschel, Morthole VI, Fernandez-Prada CR, Alving C, Rao VB. Highly effective generic adjuvant systems for orphan or poverty-related vaccines. Vaccine. 2011;29:873–877. doi: 10.1016/j.vaccine.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray K, Ma J, Oram M, Lakowicz JR, Black LW. Single molecule-and fluorescence correlation spectroscopy-FRET analysis of phage DNA packaging: Co-localization of the packaged phage T4 DNA ends within the capsid. J Mol Biol. 2010a;396:1102–1113. doi: 10.1016/j.jmb.2009.11.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray K, Oram M, Ma J, Black LW. Portal control of viral prohead expansion and DNA packaging. Virology. 2009;391:44–50. doi: 10.1016/j.virol.2009.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray K, Sabanayagam CR, Lakowicz JR, Black LW. DNA crunching by a viral packaging motor: Compression of a procapsid-portal stalled Y-DNA substrate. Virology. 2010b;398:224–232. doi: 10.1016/j.virol.2009.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren SX, Ren ZJ, Zhao MY, Wang XB, Zuo SG, Yu F. Antitumor activity of endogenous mFlt4 displayed on a T4 phage nanoparticle surface. Acta Pharmacol Sin. 2009;30:637–645. doi: 10.1038/aps.2009.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Z, Black LW. Phage T4 SOC and HOC display of biologically active, full-length proteins on the viral capsid. Gene. 1998;215:439–444. doi: 10.1016/s0378-1119(98)00298-4. [DOI] [PubMed] [Google Scholar]
- Ren ZJ, Baumann RG, Black LW. Cloning of linear DNAs in vivo by overexpressed T4 DNA ligase: Construction of a T4 phage hoc gene display vector. Gene. 1997;195:303–311. doi: 10.1016/s0378-1119(97)00186-8. [DOI] [PubMed] [Google Scholar]
- Ren ZJ, Lewis GK, Wingfield PT, Locke EG, Steven AC, Black LW. Phage display of intact domains at high copy number: A system based on SOC, the small outer capsid protein of bacteriophage T4. Protein Sci. 1996;5:1833–1843. doi: 10.1002/pro.5560050909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren ZJ, Tian CJ, Zhu QS, Zhao MY, Xin AG, Nie WX, Ling SR, Zhu MW, Wu JY, Lan HY, Cao YC, Bi YZ. Orally delivered foot-and-mouth disease virus capsid protomer vaccine displayed on T4 bacteriophage surface: 100% protection from potency challenge in mice. Vaccine. 2008;26:1471–1481. doi: 10.1016/j.vaccine.2007.12.053. [DOI] [PubMed] [Google Scholar]
- Rentas FJ, Rao VB. Defining the bacteriophage T4 DNA packaging machine: evidence for a C-terminal DNA cleavage domain in the large terminase/packaging protein gp17. J Mol Biol. 2003;334:37–52. doi: 10.1016/j.jmb.2003.09.028. [DOI] [PubMed] [Google Scholar]
- Repoila F, Tetart F, Bouet JY, Krisch HM. Genomic polymorphism in the T-even bacteriophages. EMBO J. 1994;13:4181–4192. doi: 10.1002/j.1460-2075.1994.tb06736.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rifat D, Wright NT, Varney KM, Weber DJ, Black LW. Restriction endonuclease inhibitor IPI* of bacteriophage T4: A novel structure for a dedicated target. J Mol Biol. 2008;375:720–734. doi: 10.1016/j.jmb.2007.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson KL, Soto CM, Archer MJ, Odoemene O, Liu JL. Engineered T4 viral nanoparticles for cellular imaging and flow cytometry. Bioconjug Chem. 2011;22:595–604. doi: 10.1021/bc100365j. [DOI] [PubMed] [Google Scholar]
- Sabanayagam CR, Oram M, Lakowicz JR, Black LW. Viral DNA packaging studied by fluorescence correlation spectroscopy. Biophys J. 2007;93:L17–L19. doi: 10.1529/biophysj.107.111526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathaliyawala T, Islam MZ, Li Q, Fokine A, Rossmann MG, Rao VB. Functional analysis of the highly antigenic outer capsid protein, Hoc, a virus decoration protein from T4-like bacteriophages. Mol Microbiol. 2010;77:444–455. doi: 10.1111/j.1365-2958.2010.07219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathaliyawala T, Rao M, Maclean DM, Birx DL, Alving CR, Rao VB. Assembly of human immunodeficiency virus (HIV) antigens on bacteriophage T4: A novel in vitro approach to construct multicomponent HIV vaccines. J Virol. 2006;80:7688–7698. doi: 10.1128/JVI.00235-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivachandra SB, Li Q, Peachman KK, Matyas GR, Leppla SH, Alving CR, Rao M, Rao VB. Multicomponent anthrax toxin display and delivery using bacteriophage T4. Vaccine. 2007;25:1225–1235. doi: 10.1016/j.vaccine.2006.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivachandra SB, Rao M, Janosi L, Sathaliyawala T, Matyas GR, Alving CR, Leppla SH, Rao VB. In vitro binding of anthrax protective antigen on bacteriophage T4 capsid surface through Hoc-capsid interactions: A strategy for efficient display of large full-length proteins. Virology. 2006;345:190–198. doi: 10.1016/j.virol.2005.10.037. [DOI] [PubMed] [Google Scholar]
- Simpson AA, Tao Y, Leiman PG, Badasso MO, He Y, Jardine PJ, Olson NH, Morais MC, Grimes S, Anderson DL, Baker TS, Rossmann MG. Structure of the bacteriophage phi29 DNA packaging motor. Nature. 2000;408:745–750. doi: 10.1038/35047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder L, Tarkowski HJ. The N terminus of the head protein of T4 bacterio-phage directs proteins to the GroEL chaperonin. J Mol Biol. 2005;345:375–386. doi: 10.1016/j.jmb.2004.10.052. [DOI] [PubMed] [Google Scholar]
- Stortelder A, Hendriks J, Buijs JB, Bulthuis J, Gooijer C, van der Vies SM, van der Zwan G. Hexamerization of the bacteriophage T4 capsid protein gp23 and its W13V mutant studied by time-resolved tryptophan fluorescence. J Phys Chem B. 2006;110:25050–25058. doi: 10.1021/jp064881t. [DOI] [PubMed] [Google Scholar]
- Sun S, Gao S, Kondabagil K, Xiang Y, Rossmann MG, Rao VB. Structure and function of the small terminase component of the DNA packaging machine from T4 like bacteriophages. Proc Natl Acad Sci USA. 2012;109(3):817–822. doi: 10.1073/pnas.1110224109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Kondabagil K, Draper B, Alam TI, Bowman VD, Zhang Z, Hegde S, Fokine A, Rossmann MG, Rao VB. The structure of the phage T4 DNA packaging motor suggests a mechanism dependent on electrostatic forces. Cell. 2008;135:1251–1262. doi: 10.1016/j.cell.2008.11.015. [DOI] [PubMed] [Google Scholar]
- Sun S, Kondabagil K, Gentz PM, Rossmann MG, Rao VB. The structure of the ATPase that powers DNA packaging into bacteriophage T4 procapsids. Mol Cell. 2007;25:943–949. doi: 10.1016/j.molcel.2007.02.013. [DOI] [PubMed] [Google Scholar]
- Tetart F, Desplats C, Krisch HM. Genome plasticity in the distal tail fiber locus of the T-even bacteriophage: Recombination between conserved motifs swaps adhesin specificity. J Mol Biol. 1998;282:543–556. doi: 10.1006/jmbi.1998.2047. [DOI] [PubMed] [Google Scholar]
- Wang GR, Vianelli A, Goldberg EB. Bacteriophage T4 self-assembly: In vitro reconstitution of recombinant gp2 into infectious phage. J Bacteriol. 2000;182:672–679. doi: 10.1128/jb.182.3.672-679.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikoff WR, Liljas L, Duda RL, Tsuruta H, Hendrix RW, Johnson JE. Topologically linked protein rings in the bacteriophage HK97 capsid. Science. 2000;289:2129–2133. doi: 10.1126/science.289.5487.2129. [DOI] [PubMed] [Google Scholar]
- Wu CH, Black LW. Mutational analysis of the sequence-specific recombination box for amplification of gene 17 of bacteriophage T4. J Mol Biol. 1995;247:604–617. [PubMed] [Google Scholar]
- Wu CH, Lin H, Black LW. Bacteriophage T4 gene 17 amplification mutants: Evidence for initiation by the T4 terminase subunit gp16. J Mol Biol. 1995;247:523–528. [PubMed] [Google Scholar]
- Wu J, Tu C, Yu X, Zhang M, Zhang N, Zhao M, Nie W, Ren Z. Bacteriophage T4 nanoparticle capsid surface SOC and HOC bipartite display with enhanced classical swine fever virus immunogenicity: A powerful immunological approach. J Virol Methods. 2007;139:50–60. doi: 10.1016/j.jviromet.2006.09.017. [DOI] [PubMed] [Google Scholar]
- Yu TY, Schaefer J. REDOR NMR characterization of DNA packaging in bacteriophage T4. J Mol Biol. 2008;382:1031–1042. doi: 10.1016/j.jmb.2008.07.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Kottadiel VI, Vafabakhsh R, Dai L, Chemla YR, Ha T, Rao VB. A promiscuous DNA packaging machine from bacteriophage T4. PLoS Biol. 2011;9:e1000592. doi: 10.1371/journal.pbio.1000592. [DOI] [PMC free article] [PubMed] [Google Scholar]


