Abstract
In the biosynthetic pathway of the spinosyn insecticides, the tailoring enzyme SpnF performs a [4+2]-cycloaddition on a 22-membered macrolactone to forge an embedded cyclohexene ring. To learn more about this reaction, which could potentially proceed through a Diels-Alder mechanism, the 1.50 Å-resolution crystal structure of SpnF bound to S-adenosylhomocysteine (SAH) was determined. This sets the stage for advanced experimental and computational studies to determine the precise mechanism of SpnF-mediated cyclization.
Polyketides generated by iterative polyketide synthases (PKSs) commonly contain carbocycles, whereas those generated by processive PKSs typically do not1,2. The intramolecular carbon-carbon bond-forming reactions that lead to carbocyclic products are often facilitated by the reactive nature of poly β-keto intermediates generated by iterative Type I, Type II, and Type III PKSs (Supplementary Results, Supplementary Figure 1). These cyclizations are catalyzed by cyclases incorporated within the multidomain synthases of Type I pathways and by standalone cyclases in Type II pathways3,4. The active sites of these cyclases position the corresponding substrates to undergo cyclization through regiospecific carbon-carbon bond-forming condensations. Iterative Type I PKSs and their associated tailoring enzymes are also known to generate and cyclize highly unsaturated polyene intermediates into complex carbon skeletons, such as those of calicheamicin and ikarugamycin5,6.
The spinosyn biosynthetic pathway is unusual in that it utilizes a processive Type I PKS yet generates a carbocyclic product7,8. The key reactions in the construction of the tetracyclic spinosyn core are the carbocyclization reactions catalyzed by SpnF and SpnL (Figure 1 and Supplementary Figure 2)9. Catalysis by SpnL likely follows a Rauhut-Currier-type mechanism, while the [4+2]-cyclization catalyzed by SpnF has been hypothesized to proceed through a Diels-Alder-like mechanism10. Both SpnF and SpnL resemble S-adenosylmethionine (SAM)-dependent methyltransferases (MTs), yet whether SAM participates in these cyclization reactions is unclear. Although other biosynthetic pathways (e.g., those of lovastatin, equisetin, solanapyrones, spirotetronates, riboflavin, and thiocillin I) may also involve [4+2]-cyclases11, SpnF is the first enzyme to be verified whose sole function is to catalyze such a [4+2]-cycloaddition reaction9. Macrophomate synthase had been proposed to catalyze [4+2]-cycloaddition through a route that includes a Diels-Alder reaction; however, subsequent studies show that this enzyme most likely catalyzes a two-step Michael-aldol sequence11–14. If the cyclization catalyzed by SpnF indeed proceeds through a Diels-Alder mechanism, the features it employs will be of fundamental interest to synthetic chemists and enzymologists alike15.
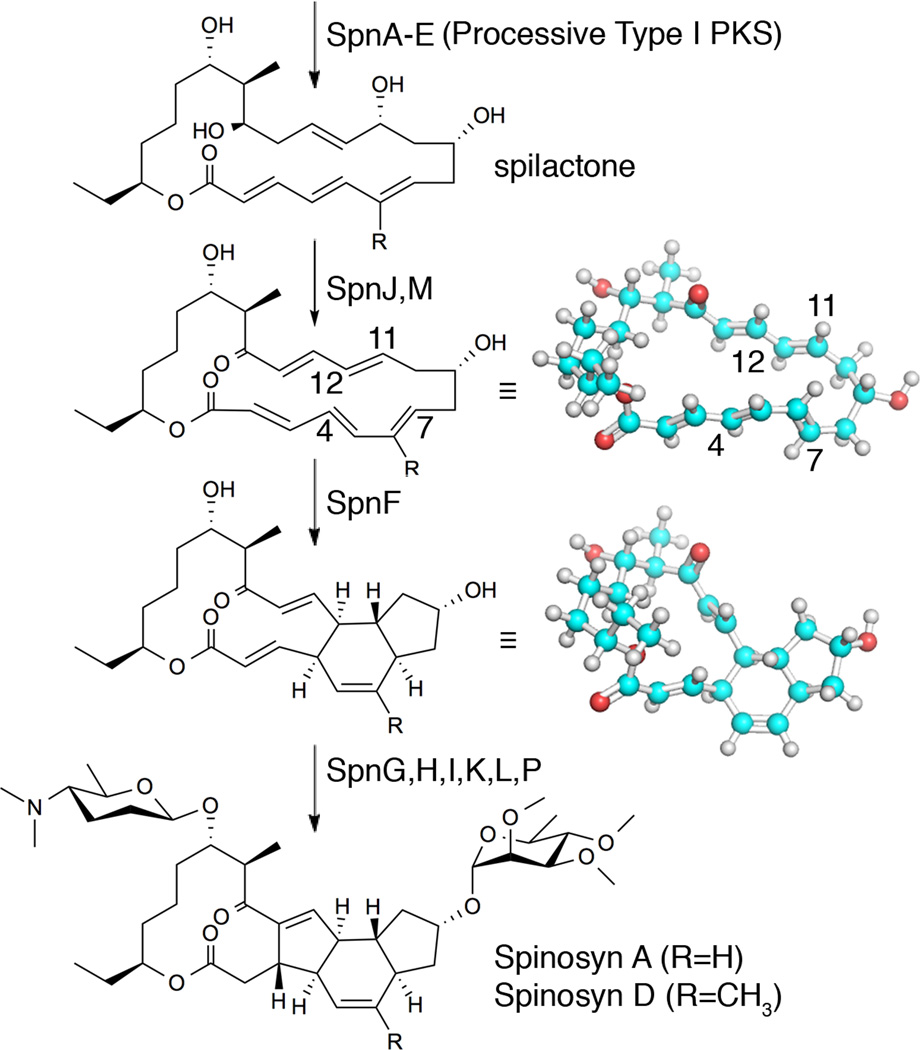
Figure 1.
Reactions mediated by SpnF and accompanying tailoring enzymes. The processive spinosyn Type I PKS generates spilactone, which is processed by several tailoring enzymes into spinosad (Spinosyns A and D). SpnF performs a [4+2]-cyclization, possibly through a Diels-Alder mechanism, between the C4–C7 diene (C5–C6 bond in s-cis geometry) and the C11–C12 dienophile, embedding a cyclohexene ring in the product (conformations minimized with the MMFF94s force field).
The spilactone ring produced by the spinosyn Type I PKS is relatively unreactive, with a conjugated polyene on one side of the macrolactone, similar to the structure of pimaricin. The oxidation and 1,4-dehydration catalyzed by SpnJ and SpnM, respectively, introduce a conjugated π-system on the opposite side of the macrolactone, create ring strain, and enable a [4+2]-cycloaddition that can be accelerated 500-fold by SpnF (kcat vs. knon)9. All the inter-olefinic bonds of the SpnM product (the SpnF substrate), including C5–C6 of the reactive diene, have been determined by 1H-NMR to be in s-trans conformations16. The low-energy conformations of the SpnM product can thus be represented by a macrocycle that resembles an elongated triangle in which the two longest sides are constituted by stretches of approximately coplanar atoms (7- and 10-atoms long). While the SpnM product with its C5–C6 in the s-trans conformation is lower in energy than the s-cis conformation, conversion to the s-cis conformer is a necessary step for the [4+2]-cycloaddition. An equilibrium between these conformers can be readily established in solution since cyclization can occur without enzyme to yield the same product generated by SpnF9. However, within the SpnF active site it is unknown what fraction of substrate molecules is in the active C5–C6 s-cis conformation and whether the interconversion of s-trans and s-cis conformations of C5–C6 occurs while the substrate is bound.
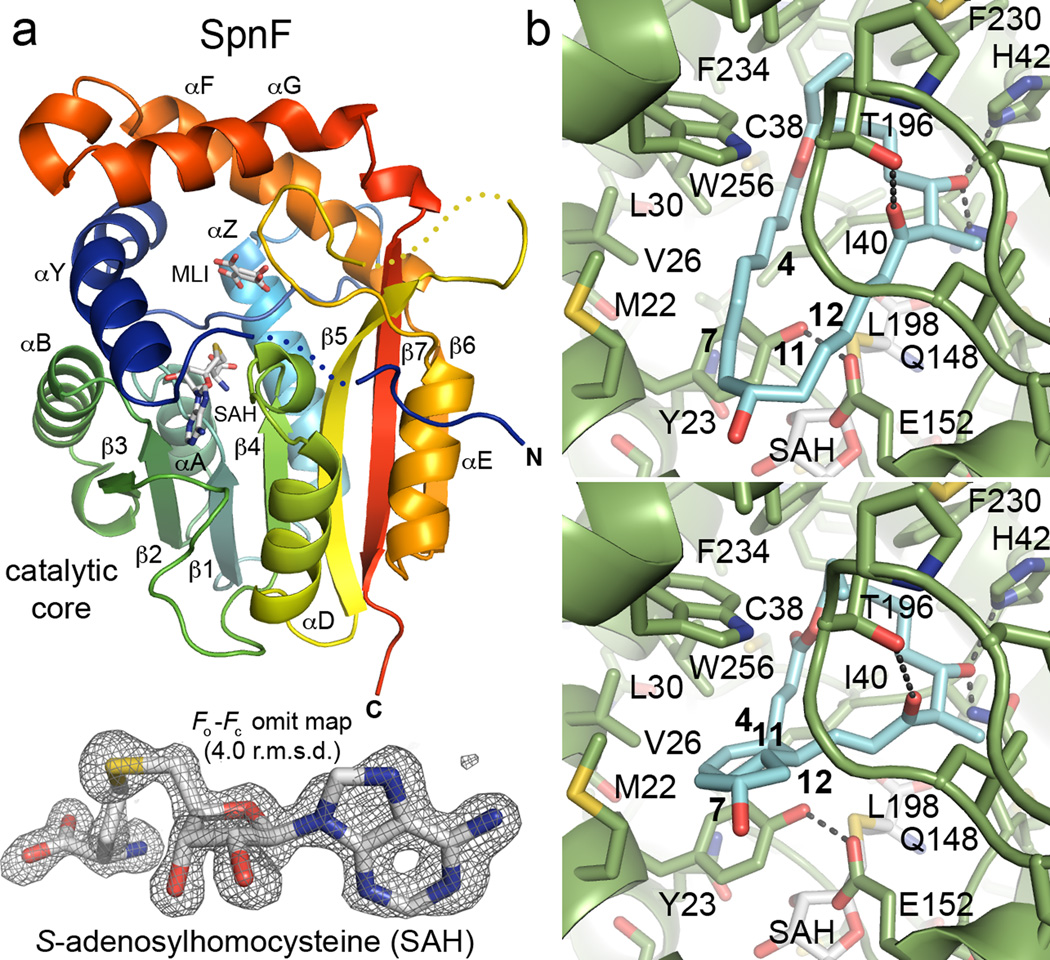
To learn more about how cyclization is catalyzed, the crystal structure of SpnF was determined (Figure 2a and Supplementary Table 1). The catalytic core of RebM (the rebeccamycin MT, PDB code: 3BUS) was used as a molecular replacement search model. Two monomers of SpnF were identified in the asymmetric unit within space group P21. These monomers align well (0.13 Å r.m.s.d. over 206 Cα atoms), with the pitch of helix αG being the only significant difference in conformation (Supplementary Figure 3). A few portions of the polypeptide chains are not well-resolved: the histidine tag, the N-terminus (residues 1–2 and 1–3 in monomers A and B, respectively), and two loop regions (residues 9–12, 186–191 in monomer A; 8–12, 184–185 in monomer B) are invisible, while only weak density is present for the C-terminal glycine (residue 282).
Figure 2.
SpnF structure and consensus docking of its substrate and product. a) SpnF adopts a SAM-dependent methyltransferase (MT) fold, with a catalytic core and a cover like most natural product MTs. The electron density maps reveal the presence of two small molecules bound to each SpnF monomer – SAH and what appears to be malonate (MLI) in two conformations. The Fo−Fc omit map, contoured at 4.0 r.m.s.d., shows SAH from one of the two SpnF monomers in the asymmetric unit. b) The programs AutoDock Vina and DOCK6 were used to dock SpnF substrate and product into the substrate cavity (structures minimized with the MMFF94s force field are shown here; see Supplementary Table 2 for more detail). A top orientation for both molecules in both programs is shown. The majority of contacts are hydrophobic although the C17-OH forms hydrogen bonds with both H42 and Q148. The C11–C12 π-bond may be polarized by T196 through its hydrogen bond to the C15 keto group to react with the C4–C7 diene. SAH does not make contact with either the substrate or product.
Based on the Fo−Fc omit maps, SAH, not SAM, is predominantly bound to each monomer in a 490-Å3 pocket, resembling those of class I SAM-dependent MTs17. SpnF makes numerous contacts with SAH (likely co-purified with the enzyme; Supplementary Figure 4). The H41 side chain and the I40 backbone NH form charged hydrogen bonds with the SAH carboxylate, while the backbone carbonyls of G80 and M147 form charged hydrogen bonds with the homocysteine amino group of SAH. The T102 and Q107 side chains, as well as the G82 NH, form hydrogen bonds with the SAH ribose hydroxyl groups. The D130 side chain and the A131 NH form hydrogen bonds with the SAH adenine. Hydrogen-bonded to one another, the Y23 and E152 side chains form a lid for the SAH/SAM binding pocket that essentially separates it from the substrate cavity. The H42 and Q148 side chains bind a molecule of malonate (present in the crystallization buffer) in each monomer, apparently in two alternate conformations (Supplementary Figure 5). As malonate was found to significantly enhance crystal quality and bind in the same cavity in which the SpnF substrate is anticipated to bind, it may stabilize the closed conformation observed for SpnF.
Similar to many natural product MTs17, the catalytic core (a 7-stranded β-sheet (β1–β7) surrounded by 5α-helices (αA, αB, αD, αE, αZ)) of SpnF is embellished with an N-terminal extension (αY) and a “cover” formed by a loop (residues 181–202, inserted between β5 and αE) and 2α-helices (αF and αG, inserted between β6 and β7). The closest structural homologues identified by the DALI server18 are a sarcosine dimethylglycine MT, RebM, a geranyl diphosphate MT, and a mycolic acid cyclopropane synthase (PDB codes 2O57, 3BUS, 4F86, and 1KPG with Z-scores 28.9, 28.5, 28.4, and 28.0, respectively). The structures of geranyl diphosphate MT and cyclopropane synthase provide examples of closed, ternary complexes in which both SAM and the substrate are isolated from solvent (PDB codes 4F86 and 1KPG). In contrast, the structure of RebM provides an example of an MT in an open state (PDB code 3BUS).
The 860-Å3 volume of the substrate cavity located between the SpnF catalytic core, αY, and the cover is large enough to completely enclose the SpnF substrate. BusF is the SpnF homologue in the biosynthetic pathway of butenyl-spinosyns, which contain a butenyl substituent at C21 in place of the ethyl substituent of the spinosyns19. While 23 out of 275 residues differ between the two enzymes, 22 are surface residues. As the one buried substitution (M147) is distant from the active site, the BusF and SpnF substrate cavities are formed by an identical set of residues.
Since natural product MTs are known to close around their substrates in complementary fashion and SpnF was captured in a closed conformation, we attempted to dock the SpnF substrate and product into the active site. To obtain representative structures of these molecules, their conformations were investigated using two force fields (UFF and MMFF94s)20,21. The restraints imposed by the two extended conjugated systems of the substrate greatly reduce the available conformational space compared to macrocycles of similar size. With the C5–C6 bond fixed in the s-cis state, the conformations of other rotatable bonds (e.g., s-cis or s-trans at C1–C2, C3–C4, C12–13, and C14–C15) were investigated. The conformer in which the carbonyls point in the same directions as those of the spinosyns (i.e., with the enone C14–C15 also in the s-cis orientation as displayed in Figure 1), was predicted by both force fields to be lowest in energy. To obtain structures for the product, the bonds of the substrate were adjusted to those of the cyclized product and the resulting molecule was energy-minimized. The UFF and MMFF94s force fields yielded similar structures for both the SpnF substrate and product.
Consensus docking of the SpnF substrate and product was performed using the programs AutoDock Vina22 and DOCK6.623 (Supplementary Table 2). The top solutions represent poses that have the best shape complementarity to the active site cavity. An orientation was observed for the substrate structures and the product structures that not only fits well in the substrate cavity but also forms favorable hydrophobic and hydrophilic contacts (while docking simulations are routinely performed and the consensus solutions reported here seem probable, the presented substrate and product orientations should be interpreted with caution; Figure 2b and Supplementary Figure 6). Hydrophobic contact is made with V19, M22, Y23, V26, L30, C38, I40, W53, E152, V178, T196, G197, L198, F230, M231, F234, W256, V260, and M270. Hydrogen bonds are formed between the C17-OH and the side chains of H42 and Q148 where a malonyl carboxylate binds in the crystal structure. The substrate and product appear to rest on a platform created on the catalytic core by Y23, I40, H42, Q148, and E152 and to be blanketed by the cover residues T196, G197, L198, F230, F234, and W256. Neither substrate nor product makes contact with SAH.
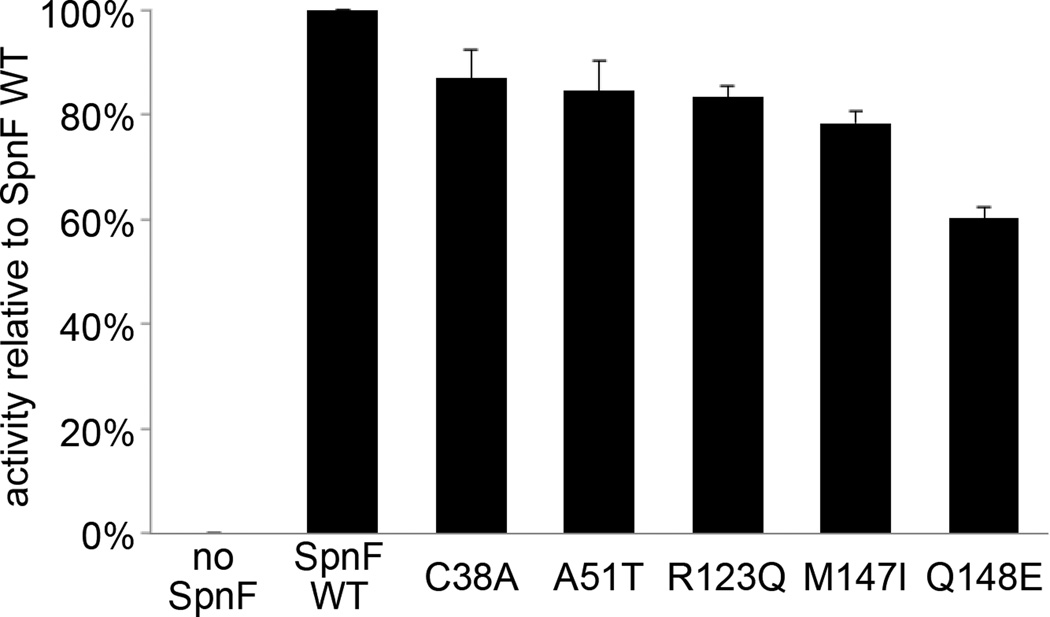
To investigate which residues contribute to SpnF activity, expression plasmids encoding mutations of residues lining the substrate cavity (Y23A, Y23F, C38A, H42A, H42Q, Q148A, Q148E, Q148L, E152A, E152L, E152Q, T196S, and T196A) were generated. Size-exclusion chromatography indicated that only the C38A and Q148E substrate cavity-altered mutants were well-folded, while all three mutants designed with benign substitutions (A51T, R123Q, and M147I, based on a sequence alignment of SpnF and BusF) were well-folded (Supplementary Figure 7). The activity of each mutant was compared to that of wild-type SpnF through a cyclization assay that measures tricyclic product formation after an incubation of the SpnM substrate with SpnM and the SpnF mutant (Figure 3)9. Each of the mutants showed only a small loss in activity compared to wild-type SpnF.
Figure 3.
Cyclization assays of SpnF mutants. Point mutants were generated and compared to wild-type SpnF through a cyclization assay with triplicate measurements (data represent mean values ± s.d.). That Q148E showed the greatest loss in activity may be attributed to the Q148 side chain amino group making contact with the SpnF substrate during catalysis.
While computational studies support a concerted, highly-asynchronous Diels-Alder mechanism for SpnF24, non-Diels-Alder routes (e.g., dipolar and biradical) are not easily disproven. SpnF may facilitate cycloaddition through a combination of 1) removing water molecules surrounding the substrate, 2) stabilizing the reactive geometry, perhaps by lowering the energy of the C5–C6 s-cis conformation relative to those of other conformations, and 3) enhancing the reactivity of the dienophile — T196 is in position to form a hydrogen bond with the C15-carbonyl and could facilitate withdrawing of electron density through the C11–C15 π-system. A comparison of SpnF with MTs solved in the absence of SAM/SAH (e.g., PDB code 2O57) suggests that SAM, the coenzyme most likely bound in vivo, primarily plays a structural role.
Carbocycles within polyketides are generally derived from intramolecular cyclization of reactive, π-enriched intermediates. Different from iterative Type I, Type II, and Type III PKS pathways, the spinosyn pathway introduces reactive π-bonds after the core polyketide backbone has been assembled by the PKS machinery. This may avoid byproducts generated by side reactions of the conjugated π-systems of intermediates bound to the slower, processive Type I PKS enzymes (kcat ≈ 1 min−1 for the erythromycin PKS)25. This unique strategy relies on a previously uncharacterized type of [4+2]-cyclase, SpnF, whose structure is now resolved.
ONLINE METHODS
Materials
Isopropyl β-d-1-thiogalactopyranoside (IPTG) was purchased from Carbosynth Limited, SAM was bought from Ark Parm, Inc., and all other chemicals were acquired from Sigma-Aldrich or Fisher Scientific. All chemicals were used without further purification. Ni-NTA resin was a product from Expedeon. Amicon centrifugal units were obtained from Millipore. PCR primers were ordered from Sigma-Aldrich.
Cloning
Previously, expression and purification of SpnF led to insoluble protein, and the chaperone proteins GroEL/ES were required to obtain soluble protein9. Reexamining the spnF sequence led to the observation of a new start codon upstream of the spnF open reading frame annotated by NCBI. The longer spnF gene construct was amplified from the genomic DNA of Saccharopolyspora spinosa strain NRLL18537 using the primers 5′-CCGGAATTCCATATGCAGAGATCAGGCATACCGGTG-3′ and 5′-CCAAGCTTTTAGCCGACCGGCTTCCGCGCCGTC-3′. The amplicon was then inserted into a pET28b(+) vector (Novagen) between the NdeI and HindIII sites. Escherichia coli TOP10 cells (Invitrogen) were transformed with the ligated plasmid and plated on Luria Broth (LB)-agar supplemented with kanamycin (50 µg/mL), and a desired expression plasmid was obtained from a single colony.
Protein expression and purification
The expression plasmid was transformed into the E. coli BL21(DE3) expression host. The transformed host was grown in overnight cultures (4 mL of LB with 50 µg/mL kanamycin) that were used to inoculate 6 × 1 L LB (50 mg/L kanamycin). The cultures were incubated at 37 °C with shaking until OD600 of 0.6, at which point the temperature was lowered to 15 °C. When cultures reached 15 °C, IPTG was added to a final concentration of 0.5 mM. The cultures were grown for an additional 16 h. Cells were harvested via centrifugation (5000 ×g for 10 min) and resuspended in lysis buffer (20% v/v glycerol, 500 mM NaCl, 30 mM HEPES, pH 7.5). Cells were lysed with a sonicator and centrifuged (30,000 ×g for 30 min). The lysate was loaded onto a Ni-NTA column equilibrated with lysis buffer. The column was washed with 20 column volumes of 20 mM imidazole in lysis buffer, and protein was eluted with 2 column volumes of 200 mM imidazole in lysis buffer. SpnF was further purified on a gel filtration column (Superdex 200, GE Healthcare Life Sciences) equilibrated with 5% v/v glycerol, 150 mM NaCl, and 10 mM HEPES, pH 7.5. A centrifugal concentrator was employed to yield 23 mg/mL SpnF. Aliquots were flash-frozen in liquid nitrogen and stored at −80°C until further use.
Crystallization, data processing, and structure determination
Crystals were grown over 6 months at 25 °C with the sitting-drop vapor-diffusion method. Drops consisted of 2:1 ratio of SpnF (23 mg/mL, 2 mM SAM, 5% v/v glycerol, 150 mM NaCl, 10 mM HEPES, pH 7.5) and crystallization buffer (1.8 M ammonium sulfate, 2% v/v PEG 400, 100 mM HEPES, pH 7.5, 100 mM sodium malonate, pH 7.0). Crystals were briefly soaked in 35% v/v PEG 400, 50 mM HEPES, pH 7.5 before they were flash-frozen in liquid nitrogen. Data for SpnF were collected at 100 K and 12.000 keV at Advanced Photon Source Beamline 23-ID-D and processed and scaled in XDS (Supplementary Table 1)26. Data truncation was guided by I/σ(I) and CC1/2 values27. Free-R flags were added to 5% of reflections in Uniqueify of the CCP4 software suite28. The phases were solved with molecular replacement using the catalytic core of the rebeccamycin MT, RebM (PDB code: 3BUS), as the search model in the program Phaser29 of CCP4. The solution was submitted to the ARP/wARP server for automated refinement30 before several rounds of refinement were carried out in Coot31, Refmac5 of CCP432, and phenix.refine of the PHENIX software package33. Default NCS restraints were enabled in Refmac5 and phenix.refine. No Ramachandran outliers are observed, and 97.6% of residues fall within the favored region. Slightly elevated Rwork and Rfree values may be attributed to the presence of multiple lattices within the SpnF crystals, readily observable in the diffraction images. All protein images were generated in PyMOL (Schrödinger). Cavity volumes were calculated with 3V34.
SpnF activity assay
Assay mixtures of wild-type or mutant SpnF (15 µL) contained 2 mM SpnM substrate and 1.25 µL SpnM in a total volume of 50 µL Tris HCl buffer (50 mM, pH 8)9. The reaction was incubated for 15 min at 30 °C before quenching with 80 µL of cold ethanol. The sample was centrifuged and the supernatant was analyzed by HPLC using a Varian Microsorb-MV 100-5 C18 250 × 4.6 mm column. The following method was used: linear gradient of 31–36% B over 60 min, 36–70% B over 1 min, 70% B over 13 min, 70–31% B over 1 min, and 31% B for 10 min (solvent A, water; solvent B, acetonitrile; flow-rate, 1 mL/min; detection at 254 nm). The assays were performed in triplicate.
Construction and purification of SpnF mutants
SpnF mutants (Y23A, Y23F, C38A, H42A, H42Q, A51T, R123Q, M147I, Q148A, Q148E, E152A, E152L, E152Q, T196A, T196S) were engineered by site-directed mutagenesis (QuikChange, Stratagene) using primers 5′-CAGGTTGGGCAGAT-GGCTGACCTGGTCACGCC-3′ and 5′-GGCGTGACCAGGTCAGCCATCTGCCCAACCTG-3′ for Y23A, 5′-GCAGCAGGTTGGGCAGATGTTTGACCTGGTCA-3′ and 5′-TGACCAGGTCAAACATCTGCCCAACCTGCTGC-3′ for Y23F, 5′-CGGGCGGCCCCGCCGCCATCCACC-3′ and 5′-GGTGGATGGCGGCGGG-GCCGCCCG-3′ for C38A, 5′-CTGCGCCATCCACGCCGGCTACTGGGAG-3′ and 5′-CTCCCAGTAGCCGGCGTGGATGGCGCAG-3′ for H42A, 5′-CTGCGCCATCCACCAAG-GCTACTGGGA-3′ and 5′-TCCCAGTAGCCTTGGTGGATGGCGCAG-3′ for H42Q, 5′-GAGAACGACGGGCGGACTTCCTGGCAGCAGG-3′ and 5′-CCTGCTGCCAGGAAGTC-CGCCCGTCGTTCTC-3′ for A51T, 5′-GCGGACTAAGCCACCAGGTGGACTTCTCGTG-3′ and 5′-CACGAGAAGTCCACCTGGTGGCTTAGTCCGC-3′ for R123Q, 5′-CGACGCCG-CCTGGGCCATCCAGTCGCTGTTGGAGATG-3′ and 5′-CATCTCCAACAGCGACTGGA-TGGCCCAGGCGGCGTCG-3′ for M147I, 5′-GACGCCGCCTGGGCCATGGCGTCGCTGT-TGGAGATGTC-3′ and 5′-GACATCTCCAACAGCGACGCCATGGCCCAGGCGGCGTC-3′ for Q148A, 5′-GCCTGGGCCATGGAGTCGCTGTTGG-3′ and 5′-CCAACAGCGACTCC-ATGGCCCAGGC-3′ for Q148E, 5′-GCAGTCGCTGTTGGCGATGTCCGAACCGG-3′ and 5′-CCGGTTCGGACATCGCCAACAGCGACTGC-3′ for E152A, 5′-CATGCAGTCGCTGTTG-CTGATGTCCGAACCGGAC-3′ and 5′-GTCCGGTTCGGACATCAGCAACAGCGACTGCA-TG-3′ for E152L, and 5′-TGCAGTCGCTGTTGCAGATGTCCGAACCG-3′ and 5′-CGGTTCGGACATCTGCAACAGCGACTGCA-3′ for E152Q, 5′-GGGGACAGGTGGCC-GTCCGGCCTTCGGATCT-3′ and 5′-AGATCCGAAGGCCGGACGGCCACCTGTCCCC-3′ for T196A, 5′-GGGGACAGGTGGCCGGCCGGCCTTCGGATCT-3′ and 5′-AGATCCG-AAGGCCGGCCGGCCACCTGTCCCC-3′ for T196S. Expression and purification of these mutant proteins followed the protocol described for wild-type SpnF.
Energy minimization of the SpnF substrate and product
Representative substrate and product structures were obtained using Avogadro 1.1.035. Energies were minimized using the steepest descent algorithm with the UFF and MMFF94s force fields.
Consensus docking
All heteroatoms and solvent atoms were removed from the SpnF coordinates. Docking calculations were performed with AutoDock Vina22 and DOCK 6.623. For AutoDock Vina, nonpolar H atoms were merged and a cubic grid (15 Å per side) centered on coordinates 6.98, 1.77, 15.23 was drawn around the manually docked SpnF product with AutoDockTools 1.5.6. For DOCK6, molecules were processed with Dock Prep, charges were adjusted with Add Charge, and after deleting hydrogen atoms the surface of SpnF was calculated with Write DMS (all features of UCSF Chimera 1.8.1). Spheres were generated in sphgen of DOCK6 and those within 6 Å of every atom of the manually docked SpnF product were accepted (any spheres outside of the substrate cavity were rejected). A scoring grid with 0.2 Å spacing was generated around the spheres with an additional 1.0 Å on each side, resulting in a box of 18.94 Å× 18.32 Å× 15.45 Å centered on coordinates 8.065, 0.447, 13.745. In each program, docking was performed with the optimized defaults. Top docking modes (highlighted) are chemically reasonable, fully contained within the binding pocket, and in agreement between AutoDock Vina and DOCK6 (Supplementary Table 2).
Supplementary Material
ACKNOWLEDGMENTS
We thank Mark Ruszczycky for helpful discussions. We thank Kay Diederichs for assistance in processing diffraction data. We thank the National Institutes of Health (GM106112, A.T.K. and GM035906, H.-w.L.) and the Welch Foundation (F-1712, A.T.K. and F-1511, H.-w.L.) for financial support. Instrumentation and technical assistance for this work were provided by the Macromolecular Crystallography Facility, with financial support from the College of Natural Sciences, the Office of the Executive Vice President and Provost, and the Institute for Cellular and Molecular Biology at the University of Texas at Austin. Use of the Advanced Photon Source, an Office of Science User Facility operated for the U.S. Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the U.S. DOE under Contract No. DE-AC02-06CH11357.
Footnotes
AUTHOR CONTRIBUTIONS
C.D.F. and E.A.I. performed the crystallography and generated mutants for activity assays. C.D.F. managed the consensus docking studies. D.T.W. assessed the stability of SpnF mutants. Y.L. conducted the cyclization assays. All authors analyzed and discussed the results. C.D.F., E.A.I., H.-w.L., and A.T.K. prepared the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Accession code
Atomic coordinates for the SpnF/SAH complex have deposited in the Protein Data Bank under the code 4PNE.
REFERENCES
- 1.Keatinge-Clay AT. The structures of Type I polyketide synthases. Nat. Prod. Rep. 2012;29:1050–1073. doi: 10.1039/c2np20019h. [DOI] [PubMed] [Google Scholar]
- 2.Hertweck C, Luzhetskyy A, Rebets Y, Bechthold A. Type II polyketide synthases: gaining a deeper insight into enzymatic teamwork. Nat. Prod. Rep. 2007;24:162–190. doi: 10.1039/b507395m. [DOI] [PubMed] [Google Scholar]
- 3.Crawford JM, Korman TP, Labonte JW, Vagstad AL, Hill EA, Kamari-Bidkorpeh O, Tsai SC, Townsend CA. Structural basis for biosynthetic programming of fungal aromatic polyketide cyclization. Nature. 2009;461:1139–1143. doi: 10.1038/nature08475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ames BD, Korman TP, Zhang W, Smith P, Vu T, Tang Y, Tsai SC. Crystal structure and functional analysis of tetracenomycin ARO/CYC: implications for cyclization specificity of aromatic polyketides. Proc. Natl. Acad. Sci. U.S.A. 2008;105:5349–5354. doi: 10.1073/pnas.0709223105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahlert J, Shepard E, Lomovskaya N, Zazopoulos E, Staffa A, Bachmann BO, Huang K, Fonstein L, Czisny A, Whitwam RE, Farnet CM, Thorson JS. The calicheamicin gene cluster and its iterative type I enediyne PKS. Science. 2002;297:1173–1176. doi: 10.1126/science.1072105. [DOI] [PubMed] [Google Scholar]
- 6.Zhang G, Zhang W, Zhang Q, Shi T, Ma L, Zhu Y, Li S, Zhang H, Zhao YL, Shi R, Zhang C. Mechanistic insights into polycycle formation by reductive cyclization in ikarugamycin biosynthesis. Angew. Chem. Int. Ed. Engl. 2014;53:4840–4844. doi: 10.1002/anie.201402078. [DOI] [PubMed] [Google Scholar]
- 7.Waldron C, Madduri K, Crawford K, Merlo DJ, Treadway P, Broughton MC, Baltz RH. A cluster of genes for the biosynthesis of spinosyns, novel macrolide insect control agents produced by Saccharopolyspora spinosa. Antonie Van Leeuwenhoek. 2000;78:385–390. doi: 10.1023/a:1010289901631. [DOI] [PubMed] [Google Scholar]
- 8.Kirst H. The spinosyn family of insecticides: realizing the potential of natural products research. J. Antibiot. 2010;62:101–111. doi: 10.1038/ja.2010.5. [DOI] [PubMed] [Google Scholar]
- 9.Kim HJ, Ruszczycky MW, Choi S-h, Liu Y-n, Liu H-w. Enzyme-catalysed [4+2] cycloaddition is a key step in the biosynthesis of spinosyn A. Nature. 2011;473:109–112. doi: 10.1038/nature09981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diels O, Alder K. Synthesis in the hydroaromatic series, IV. Announcement: the rearrangement of malein acid anhydride on arylated diene, triene, and fulvene. Chemische Berichte. 1929;62:2081–2087. [Google Scholar]
- 11.Kim HJ, Ruszczycky MW, Liu H-w. Current developments and challenges in the search of a naturally selected Diels-Alderase. Curr. Opin. Chem. Biol. 2012;16:124–131. doi: 10.1016/j.cbpa.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ose T, Watanabe K, Mie T, Honma M, Watanabe H, Yao M, Oikawa H, Tanaka I. Insight into a natural Diels-Alder reaction from the structure of macrophomate synthase. Nature. 2003;422:185–189. doi: 10.1038/nature01454. [DOI] [PubMed] [Google Scholar]
- 13.Guimarães CR, Udier-Blagović M, Jorgensen WLJ. Macrophomate synthase: QM/MM simulations address the Diels-Alder versus Michael-Aldol reaction mechanism. J. Am. Chem. Soc. 2005;127:3577–3588. doi: 10.1021/ja043905b. [DOI] [PubMed] [Google Scholar]
- 14.Serafimov JM, Gillingham D, Kuster S, Hilvert D. The putative Diels-Alderase macrophomate synthase is an efficient aldolase. J. Am. Chem. Soc. 2008;130:7798–7799. doi: 10.1021/ja8017994. [DOI] [PubMed] [Google Scholar]
- 15.Preiswerk N, Beck T, Schulz JD, Milovník P, Mayer C, Siegel JB, Baker D, Hilvert D. Impact of scaffold rigidity on the design and evolution of an artificial Diels-Alderase. Proc. Natl. Acad. Sci. U.S.A. 2014;111:8013–8018. doi: 10.1073/pnas.1401073111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim HJ, Choi S-h, Jeon B-s, Kim N, Pongdee R, Wu Q, Liu H-w. Chemoenzymatic synthesis of spinosyn A. Angew. Chem. Int. Ed. 2014;53:13553–13557. doi: 10.1002/anie.201407806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liscombe DK, Louie GV, Noel JP. Architectures, mechanisms and molecular evolution of natural product methyltransferases. Nat. Prod. Rep. 2012;29:1238–1250. doi: 10.1039/c2np20029e. [DOI] [PubMed] [Google Scholar]
- 18.Holm L, Rosenström P. Dali server: conservation mapping in 3D. Nucl. Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hahn DR, Gustafson G, Waldron C, Bullard B, Jackson JD, Mitchell J. Butenyl-spinosyns, a natural example of genetic engineering of antibiotic biosynthetic genes. J. Ind. Microbiol. Biotechnol. 2006;33:94–104. doi: 10.1007/s10295-005-0016-9. [DOI] [PubMed] [Google Scholar]
- 20.Rappe AK, Casewit CJ, Colwell KS, Goddard WA, Skiff WM. UFF, a full periodic table force field for molecular mechanics and molecular dynamics simulations. J. Am. Chem. Soc. 1992;114:10024–10035. [Google Scholar]
- 21.Halgren T. MMFF VI. MMFF94s option for energy minimization studies. J. Comput. Chem. 1999;20:720–729. doi: 10.1002/(SICI)1096-987X(199905)20:7<720::AID-JCC7>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 22.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brozell SR, Mukherjee S, Balius TE, Roe DR, Case DA, Rizzo RC. Evaluation of DOCK 6 as a pose generation and database enrichment tool. J. Comp-Aided Mol. Des. 2012;26:749–773. doi: 10.1007/s10822-012-9565-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hess BA, Jr, Smentek L. Concerted, highly asynchronous, enzyme-catalyzed [4 + 2] cycloaddition in the biosynthesis of spinosyn A; computational evidence. Org. Biomol. Chem. 2012;10:7503–7509. doi: 10.1039/c2ob25827g. [DOI] [PubMed] [Google Scholar]
- 25.Khosla C, Tang Y, Chen AY, Schnarr NA, Cane DE. Structure and mechanism of the 6-deoxyerythronolide B synthase. Annu. Rev. Biochem. 2007;76:195–221. doi: 10.1146/annurev.biochem.76.053105.093515. [DOI] [PubMed] [Google Scholar]
- 26.Kabsch W. XDS. Acta Crystallogr. D. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Diederichs K, Karplus PA. Better models by discarding data? Acta Crystallogr. D. 2013;69:1215–1222. doi: 10.1107/S0907444913001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Crystallogr. D. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated molecular building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. A. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 32.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 33.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Voss NR, Gerstein M. 3V: cavity, channel and cleft volume calculator and extractor. Nucleic Acids Res. 2010;38:W555–W562. doi: 10.1093/nar/gkq395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hanwell MD, Curtis DE, Lonie DC, Vandermeersch T, Zurek E, Hutchison GR. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012;4:17. doi: 10.1186/1758-2946-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.