Summary
The human neocortex differs from that of other great apes in several notable regards including altered cell cycle, prolonged corticogenesis, and increased size [1–5]. While these evolutionary changes likely contributed to the origin of distinctively human cognitive faculties, their genetic basis remains almost entirely unknown. Highly conserved non-coding regions showing rapid sequence changes along the human lineage are candidate loci for the development and evolution of uniquely human traits. Several studies have identified human-accelerated enhancers [6–14], but none have linked an expression difference to a specific organismal trait. Here we report the discovery of a human-accelerated regulatory enhancer (HARE5) of FZD8, a receptor of the Wnt pathway implicated in brain development and size [15, 16]. Using transgenic mice, we demonstrate dramatic differences in human and chimpanzee HARE5 activity, with human HARE5 driving early and robust expression at the onset of corticogenesis. Similar to HARE5 activity, FZD8 is expressed in neural progenitors of the developing neocortex [17–19]. Chromosome conformation capture assays reveal HARE5 physically and specifically contacts the core Fzd8 promoter in the mouse embryonic neocortex. To assess the phenotypic consequences of HARE5 activity, we generated transgenic mice in which Fzd8 expression is under control of orthologous enhancers (Pt-HARE5::Fzd8 and Hs-HARE5::Fzd8). In comparison to Pt-HARE5::Fzd8, Hs-HARE5::Fzd8 mice showed marked acceleration of neural progenitor cell cycle and increased brain size. Changes in HARE5 function unique to humans thus alter cell cycle dynamics of a critical population of stem cells during corticogenesis, and may underlie some distinctive anatomical features of the human brain.
Results
Identification of human-accelerated enhancer loci in the developing neocortex
The dramatic expansion of the neocortex during hominoid evolution is proposed to underlie the emergence of our uniquely human cognitive abilities [20–22], although strong genetic correlations between these traits have remained elusive [23]. The evolution of human cortical features, such as enlarged brain size, has been attributed to cellular changes including neuron number and neural progenitor cell cycle [1–5, 15]. However the genetic basis for these traits, which so markedly distinguish humans from other primates, remains poorly understood. Mutations within regulatory elements have been proposed to play a significant role in the evolution of human-specific traits [24, 25]. Recent genomic studies support this notion, and have collectively identified highly conserved non-coding regions that are rapidly evolving along the human lineage [6–10]. Of note, these human-accelerated noncoding loci are frequently located nearby genes implicated in brain development and function [11, 26, 27]. Together, these studies suggest the evolution of human neocortical traits may have occurred through modification of cis-regulatory enhancers involved in brain development. Yet to date just a handful of human-accelerated regions have been shown to function as forebrain enhancers [11–13] and none have been shown to impact neocortical expansion. Here we sought to discover human-accelerated regulatory loci important for corticogenesis, in order to gain insights into the genetic basis for the evolution of uniquely human brain features.
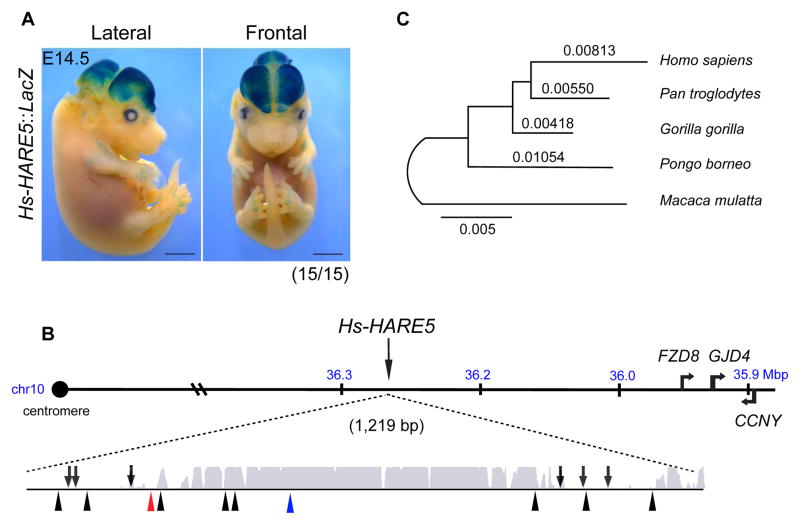
We identified HARE5 from an in silico screen for rapidly evolving human noncoding regions predicted to function as developmental enhancers in the mammalian neocortex (Figure S1A, Table S1, Supplemental Experimental Procedures)[6–8, 28, 29]. Using a standard mouse transient transgenic assay [11, 14], HARE5 reporter activity was robust in the lateral neocortex and dorso-lateral midbrain (15/15 embryos) (Figures 1A, S1C). HARE5 was prioritized due to this enhancer activity and its chromosomal location adjacent to FRIZZLED8 (FZD8), a receptor for the Wnt signaling pathway implicated in neocortical development (Figure 1B)[15–18, 30, 31]. The Homo sapiens (Hs) HARE5 orthologue contains 16 changes compared to Pan troglodytes (Pt). Based on outgroup comparison, 10 mutations were fixed on the human branch and 6 on the chimpanzee branch since the latest common ancestor (Figure 1B). A phylogenetic analysis of the 1.2 Kb HARE5 locus across several great ape species revealed a longer branch for the Hs orthologue compared to that of Pt (Figure 1C). This is consistent with the original signature of positive selection detected in the human relative to chimpanzee lineage [7]. Analysis of predicted transcription factor binding sites across the HARE5 locus revealed differences, particularly at human-derived mutations, for key transcription factors relevant to corticogenesis (see Table S2) [32]. Together these results support the prediction that Hs-HARE5 acquired unique enhancer activity since diverging from the common chimpanzee lineage.
Figure 1. Identification of Hs-HARE5 as a human-accelerated neocortical enhancer.
(A) Representative E14.5 Hs-HARE5::LacZ embryo stained for β-galactosidase (LacZ) activity. (B) Schematic of Hs-HARE5 locus on human chromosome 10 (hg19). The 1,219 bp long HARE5 genomic locus with enhancer activity includes the original 619 bp human-accelerated sequence and flanking 5′ and 3′ sequences. Represented below is a PhastCons conservation track for the HARE5 locus, shown with the region of high conservation (grey). Also shown are lineage-specific mutations for chimpanzee (6, arrows, above line), and human (10, arrowheads, bottom), including 1 Denisovan (red) and 1 currently identified human polymorphism (blue). (C) Maximum likelihood phylogenetic tree for the HARE5 orthologous locus from five anthropoid primates. Scale bars, 2 mm (A). See also Figure S1 and Table S1.
Distinct enhancer activity of human and chimpanzee HARE5 in the developing neocortex
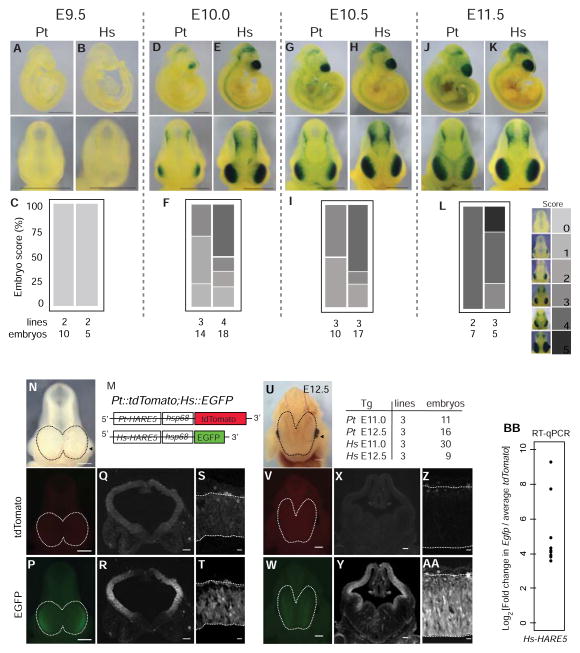
We postulated that human and chimpanzee HARE5 might differentially regulate gene expression during corticogenesis. To test this we generated independent stable mouse transgenic lines (Pt-HARE5::LacZ and Hs-HARE5::LacZ). Corticogenesis initiates at embryonic day (E) 9.5 and continues to E18.5 [2]. At E9.5, both Pt-HARE5 and Hs-HARE5 enhancer activity were undetectable (Figures 2A–C). However within a half day of development at E10.0, Hs-HARE5 activity was rapidly and robustly upregulated in the lateral telencephalon (Figures 2E,F). In contrast, Pt-HARE5 activity in the E10.0 telencephalon was markedly weaker and limited to more lateral regions (Figures 2D,F). This spatial difference in enhancer activity was sustained at E10.5, as evidenced by both whole mount embryos and coronal brain sections (Figures 2G–I, S2A–D). By E11.5, species-specific differences in HARE5-driven LacZ activity were still evident, although far less dramatic (Figures 2J–L). These results indicate that HARE5 orthologues drive expression in the developing lateral telencephalon. However, relative to chimpanzee, the human enhancer has considerably earlier and robust activity during corticogenesis.
Figure 2. Hs-HARE5 activity drives robust, early enhancer activity relative to Pt-HARE5 during corticogenesis.
(A–L) Developmental time-series of Pt-HARE5::LacZ (A,D,G,J) and Hs-HARE5::LacZ (B,E,H,K) reporter activity from stable transgenic lines. Representative images of LacZ stained embryos from lateral (top) and anterior (bottom) views. (C,F,I,L) Enhancer activity was qualitatively scored in the telencephalon, using the indicated scoring schema shown on the right, on a scale from no reporter activity (score 0) to full telencephalic activity (score 5). Number of embryos and independent transgenic lines analyzed for each stage are listed below. Embryos were scored blindly and independently by at least three individuals. (M) Schematic of destabilized reporter constructs drawn to scale. (N-AA) Representative embryos from dual reporter transgenic Pt-HARE5::tdTomato; Hs-HARE5::EGFP E11.0 (N–T) and E12.5 (U-AA) embryos detected by brightfield (N,U), and endogenous fluorescence for tdTomato (O,Q,S,V,X,Z) and EGFP (P,R,T,W,Y,AA) channels. Dotted lines demarcate dorsal neocortices of whole mount embryos (N–P, U–W). (Q,R,X,Y) Coronal sections from mid-cortex (plane indicated by arrowhead in N,U) in tdTomato (Q,X) and EGFP (R,Y) channels. (S,T,Z,AA) High-magnification images of the lateral telencephalon for tdTomato (S,Z) and EGFP (T,AA). The number of embryos and lines for each analysis is listed beside U. Endogenous fluorescence images were captured using identical exposure conditions. (BB) Graph depicting log fold changes for RT-qPCR from E12.5 neocortices. Each data point is the average fold change for an individual Hs-HARE5::EGFP embryo relative to the aggregated average for all Pt-HARE5::tdTomato embryos. mRNA input levels were normalized to Gapdh. n=4 technical replicates per embryo; n=9 embryos from 3 transgenic lines from each genotype. Scale bars, 1 mm (A–K), 500 μm (N–P, U–W); 150 μm (Q,R,X,Y); 25 μm (S,T,Z,AA). See also Figure S2 and Table S2.
Having established spatial and temporal differences in chimpanzee and human HARE5 enhancer activity, we next sought a more sensitive and dynamic readout of HARE5 transcriptional activity. The LacZ protein is stable for at least 48 hours whereas destabilized fluorescent proteins with PEST motifs are only stable for 2 hours post-translation [33]. We generated new stable transgenic mouse lines, Pt-HARE5::tdTomatoPEST and Hs-HARE5::EGFP-PEST, and compared native fluorescence in embryos co-expressing the reporters (Figures 2M,N). Both orthologues drove enhancer activity in the E11.0 neocortex, however Hs-HARE::EGFP was considerably brighter than Pt-HARE5::tdTomato, despite tdTomato having intrinsically brighter fluorescent emission than EGFP (Figures 2N–T) [33]. This reporter difference was sustained at E12.5, though the chimpanzee enhancer remained active (Figures 2U-AA, S2E–H). We quantified enhancer activity by RT-qPCR measurement of reporter transcript levels in E12.5 neocortices. Hs-HARE5::EGFP embryos showed 10–30 fold higher transcript levels than Pt-HARE5::tdTomato (Figure 2BB). Hence multiple independent reporter lines (LacZ and fluorescent) demonstrate that compared to chimpanzee HARE5, human HARE5 drives dramatically higher enhancer activity in the telencephalon.
In the E10.5 telencephalon, the predominant neural progenitor populations are neuroepithelial cells, and by E12.5 these are replaced by radial glia (termed neural stem cells) [2]. At E10.5, both enhancers were active in the majority (about 75%) of Pax6-positive neuroepithelial cells and in some TuJ1-positive neurons (Figures S3I–U). At E12.5, reporter activity was highest in the ventricular zone (VZ) (Figure 2SE–H), where radial glial cells reside. Thus both human and chimpanzee HARE5 enhancers are active in neural progenitors of the developing neocortex.
Chromosome conformation capture (3C) detects HARE5-Fzd8 interactions
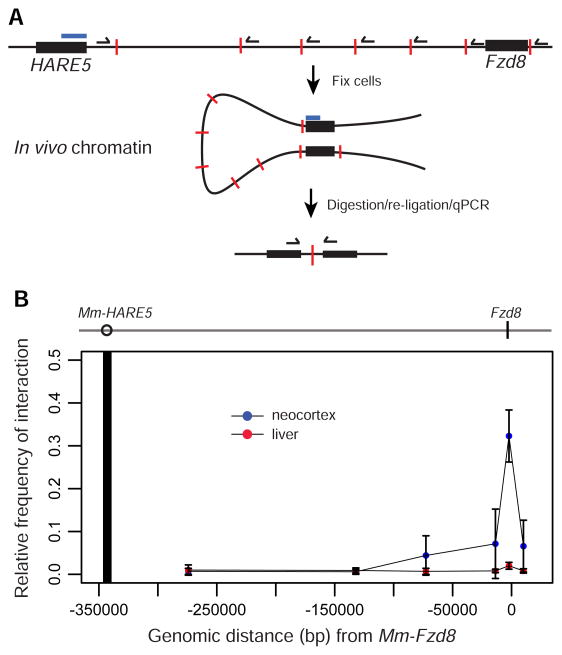
Having established HARE5 activity within the lateral telencephalon, we next sought to identify the likely target gene. The most proximal gene, Hs-FZD8, is located 307,758 bps downstream from HARE5 and was an obvious candidate due to its expression in the developing human and mouse neocortex [17–19, 30, 31]. LacZ reporter activity and Fzd8 in situ hybridization showed similar expression patterns in E10.5 and E11.5 whole mount embryos and neocortical sections (Figure S3)(developingmouse.brain-map.org and www.emouseatlas.org) [31]. We used chromosome conformation capture (3C) assays [34] to test for physical association between endogenous mouse (Mm) HARE5 and the core Fzd8 promoter within E12.5 mouse neocortices (Figure 3A). In neocortices we observed a strong peak of interaction between Mm-HARE5 and the proximal Fzd8 promoter, compared to flanking loci (Figure 3B). In contrast, no interactions were evident between Mm-HARE5 and Fzd8 in age-matched liver, which lacks detectable HARE5 activity and Fzd8 expression. These data indicate HARE5 physically and specifically associates with the core Fzd8 promoter in the developing mouse neocortex. Given the cis-regulatory activity of HARE5 orthologues, we propose HARE5 functions as a distal-acting enhancer of FZD8 during early human neocortical development.
Figure 3. 3C analysis showing HARE5 physically contacts the Fzd8 promoter.
(A) Schematic of 3C protocol showing HARE5 and Fzd8 loci (black bars), with indicated TaqMan probe (blue bar), test primers (black half arrows), and HindIII restriction sites (red lines). (B) 3C assay of E12.5 mouse neocortices (blue dots) and liver control tissue (red dots). Dark vertical line indicates location of TaqMan probe and constant primer anchored within the Mm-HARE5 locus. The 0 position indicates ATG of Fzd8 coding sequence. The graph depicts the relative frequency of interactions between Mm-HARE5 and 6 genomic locations. See also Figure S3.
Human HARE5 accelerates neural progenitor cell cycle and impacts neocortical size
We next assessed the functional consequences of chimpanzee and human HARE5 activities during corticogenesis. We generated new independent transgenic mouse lines in which Hs-HARE5 or Pt-HARE5 drove expression of a MYC-tagged mouse Fzd8 coding sequence (Pt-HARE5::Fzd8 and Hs-HARE5::Fzd8) (Figure 4A). Expression of MYC in embryonic neocortices was confirmed by western blot analysis (Figure S4A). We postulated that Fzd8 expression driven by the HARE5 enhancer would impact the cell cycle state of neural progenitors based upon the following rationale. First, both Hs-HARE5 and Pt-HARE5 drive expression in neural progenitors. Second, modulating Fzd8 levels impacts neural progenitor cell cycle in the retina [18]. Third, overexpression of stabilized β-catenin, a Wnt signaling component downstream of Frizzled, induces an expanded and gyrencephalic brain and slows cell cycle exit of neural stem cells in mice [15]. Fourth, cell cycle length is critical for corticogenesis and is postulated as a likely mechanism for the evolutionary expansion of the primate neocortex [35, 36].
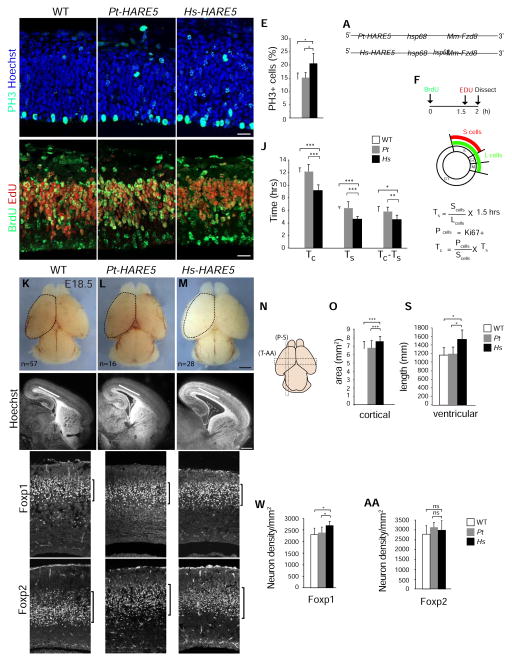
Figure 4. Hs-HARE5 driven expression of Fzd8 accelerates cell cycle of neural progenitors, and increases neuron number and neocortical size.
(A) Schematic of Pt-HARE5::Fzd8 and Hs-HARE5::Fzd8 constructs. (B–I) Images of coronal sections from E12.5 WT littermate (B,G), Pt-HARE::Fzd8 (C,H), and Hs-HARE5::Fzd8 (D,I) transgenic cortices. Sections stained for (B–D) PH3 (green) and Hoechst (blue), (G–I) BrdU (green) and EdU (red). (E) Graph of WT (white), Pt-HARE::Fzd8 (grey), and Hs-HARE5::Fzd8 (black) depicting percentage of all cells that are PH3-positive. (F) Paradigm for analysis of cell cycle length using double pulse of BrdU and EdU. Nucleotide analogs were injected at indicated time-points and overall cell cycle length (Tc) and S phase length (Ts) were calculated as shown. (J) Graph of WT (white), Pt-HARE::Fzd8 (grey), and Hs-HARE5::Fzd8 (black) cell cycle lengths of cycling progenitors. (K–M) Whole mount E18.5 brains from indicated genotypes with n=number of brains examined. A dotted line was drawn on WT cortex in K to indicate dorsal cortical area, and then superimposed on transgenic cortices in L and M. (N) Schematic cartoon representation of E18.5 brain with indicated regions of analyses for sagittal sections (P–S) and coronal sections (T-AA). (O) Graph of WT (white), Pt-HARE::Fzd8 (grey), and Hs-HARE5::Fzd8 (black) dorsal cortical area measurements. Note a 12% increase was seen in Hs-HARE5::Fzd8 cortical area. (P–R) Sagittal E18.5 sections from brains of indicated genotypes. A line drawn on WT cortex in P indicates ventricular length, and was superimposed on transgenic cortices in Q and R. Note no evidence of cortical gyrification was seen. (S) Graph depicting ventricular length for indicated genotypes. (T–V, X–Z) Coronal E18.5 sections from neocortices of indicated genotypes and stained for Foxp1 (T–V) and Foxp2 (X–Z). Note no significant apoptosis was observed. (W, AA) Graphs depicting densities of Foxp1 (W) and Foxp2 (AA) neurons in radial columns of neocortical sections. The following were analyzed for each genotype: for B–E, n=5 embryos each from 3 transgenic lines; for F–J, 5–7 embryos each from 2–3 transgenic lines; K–O, 16–57 embryos each from 2–3 transgenic lines; P–S, 4–5 embryos each (2–5 sections per embryo) from 2–3 transgenic lines; T-AA, 5–6 embryos each (2–4 sections per embryo) from 2–3 transgenic lines. All analyses were done blind to genotype. Error bars, s.d., *, P < 0.05, **, P<0.01, ***, P<0.001. Scale bars, 25 μm (B–I), 1 mm (K–M), 500 μm (P–R), and 100 μm (T–Z). See also Figure S4 and Table S3.
We measured the cell cycle state of progenitors at E12.5, predicting that species-specific differences in HARE5 activity would be evident within two days of onset of enhancer activity. At this stage, radial glial progenitors primarily undergo symmetric divisions to expand laterally, but a subset divides asymmetrically to produce excitatory neurons [2]. Quantification of G2/M phases using phospho-histoneH3 (PH3) staining revealed a significant 1.3-fold increase in the proportion of total PH3+ cells in Hs-HARE5::Fzd8 brains relative to both Pt-HARE5::Fzd8 and non-transgenic wild-type (WT) littermates (Figures 4B–E). We also observed a trend towards more Pax6-positive radial glia in Hs-HARE5::Fzd8 brains, with a significant increase relative to WT (Figure S4B). These snapshot measurements indicate that at E12.5 Hs-HARE5 driven expression of Fzd8 alters the proliferating population. More G2/M positive progenitors may indicate a faster overall cell cycle with similar G2/M phases, or alternatively an identical cell cycle with longer G2/M.
To help discriminate between these possibilities, we quantified cell cycle duration at E12.5. We used a paradigm of 2 hour BrdU exposure and 30 minute EdU exposure coupled with Ki67 staining, as previously described [37] (Figures 4F). Both WT and Pt-HARE5::Fzd8 progenitors cycled for about 12 hours, as previously reported for this age [37, 38]. In contrast Hs-HARE5 driven Fzd8 expression significantly accelerated both the total cell cycle (to approximately 9.2 hours) and S phase, by 25% (Figures 4G–J, Table S3). These cell cycle differences correspond to a 23% shorter G1/G2/M duration (Tc-Ts) of Hs-HARE5::Fzd8 progenitors compared to Pt-HARE5::Fzd8 (P=0.003). Taken together, this functional analysis reveals that relative to both WT and Pt-HARE5::Fzd8, human HARE5 directed expression of Fzd8 accelerates neural progenitor cell cycle.
Increased proliferation of neural progenitors is frequently associated with changes in brain size. Therefore, we measured the cortical dimensions of transgenic E18.5 brains. Compared to Pt-HARE5::Fzd8 and WT, the dorsal area of Hs-HARE5::Fzd8 cortices was significantly larger by 12% (Figures 4K–O). Across 5 additional measurements, Hs-HARE5::Fzd8 cortices were consistently larger than both Pt-HARE5::Fzd8 and WT (Figures S4F–H). As larger cortical area could be due to increased cortical thickness or tangential length, we quantified these dimensions in sagittal and coronal sections (Figures 4P–S). Hs-HARE5::Fzd8 brains were thinner than Pt-HARE5::Fzd8 and WT, although differences were only significant in comparison to WT (Figure S4I). In contrast, compared to both Pt-HARE5::Fzd8 and WT, Hs-HARE5::Fzd8 brains showed significantly longer tangential distance along the cortical VZ (Figure 4S). As seen in other mutants with longer tangential growth, Hs-HARE5::Fzd8 brains also showed enlarged ventricles. The increased tangential length phenotype is often associated with greater progenitor proliferation and larger cortical size, as evidenced in mouse embryonic brains mis-expressing β-catenin or FGF2 [15, 39]. These data indicate that tangential expansion is a likely contributing factor for the increased cortical area.
We predicted faster progenitor proliferation would ultimately be associated with more neurons. To test this, we quantified the densities of FoxP1-positive neurons (mid-layers III–V), born between E13.5–E16.5, and FoxP2 neurons (deep-layer VI), born around E12.5 (Figures 4T-AA) within radial columns of E18.5 brains [40, 41]. Compared to chimpanzee, Hs-HARE5::Fzd8 brains showed a significant 14% increase in the density of FoxP1 neurons, but no difference in FoxP2 neurons, nor any notable apoptosis. Thus, Hs-HARE5::Fzd8 brains contain a higher density of neurons that are produced beginning around E13.5. Together these data indicate that, compared to Pt-HARE5, Hs-HARE5 promotes faster progenitor cell cycle, which is ultimately associated with increased Foxp1 excitatory neuron density, and overall larger cortical size.
Discussion
The neocortex expanded spectacularly during human evolution, giving rise to distinctively human anatomical and cognitive capabilities [1, 2, 20–22]. Yet to date, just a handful of genetic loci have been associated with human-specific brain traits [3, 5, 25], and none have been shown to functionally impact corticogenesis in an evolutionarily divergent fashion. In this study we report the discovery of the first human-accelerated enhancer that functions in brain development. We demonstrate dramatic temporal and spatial differences in activity of human and chimpanzee enhancers of FZD8 during early corticogenesis, and show these differences impact neural progenitor cell cycle and brain size. Our study suggests the intriguing hypothesis that evolutionary changes in HARE5 sequence and activity contributed to the origin of unique features of the human brain.
The evolutionarily divergent activities of HARE5 support a model proposed 16 years ago by Pasko Rakic, that species differences in progenitor proliferation may contribute to distinctions in brain size between humans and non-human primates. The proposed radial unit hypothesis predicts that the number and proliferative capacity of progenitor cells drives the evolution of brain cytoarchitecture and explains species differences in neocortical size and structure [36]. Indeed both empirical and predicted measurements of neural progenitor cell cycle reveal stark differences between humans, non-human primates, and mice [1, 36, 42]. In non-human primates, distinct G1 phase durations are associated with unique brain cytoarchitecture [35]. Moreover genetic evidence strongly supports a causal link between neural stem cell proliferation and human brain size [43].
How might a faster cell cycle impact human brain size? We speculate that in the context of extended human corticogenesis and gestation, HARE5 increases progenitor proliferation, which expands the progenitor pool during early corticogenesis. Increased progenitor expansion would ultimately produce more neurons and a larger neocortex. This could involve either altering progenitor cell cycle exit and/or the division state of progenitors from neurogenic to proliferative. In E14.5 mice, proliferating and neurogenic neural progenitors have distinct S phase durations [44]. Experimentally shortening G1 phase in mice promotes proliferative divisions in lieu of neurogenic divisions, impacting neuron production [45, 46]. Our study implicates shorter G1 as a potential mechanism, as the Tc-Ts fraction was shorter in human transgenic brains. Follow-up studies of the Hs-HARE5::Fzd8 mouse will clarify the detailed relationship between altered cell cycle and brain size, and elucidate if modifications in structural and behavioral traits exist.
We have shown that a key target gene of HARE5 activity in the neocortex is FZD8, which encodes a Wnt receptor. Given the neurogenesis roles of β-catenin and Lef/Tcf, it is likely that FZD8 acts via canonical Wnt signaling [16]. FZD8 expression in the neonatal human brain is highest in cortical areas at 9 pcw (post conception weeks) (brainspan.org)[19] when neural stem cells are rapidly expanding during early corticogenesis [2], but markedly lower in non-cortical areas. The FZD8 expression pattern correlates strongly with neural stem cell markers SOX2 and PAX6 (r > 0.90)[19, 47]. Hence the pattern of HARE5 activity and FZD8 expression is consistent with a functional relationship in neural stem cell regulation in humans. Although chimpanzee expression data are not available, developing rhesus macaque (Macaca mulatta) neocortical data are (www.blueprintnhpatlas.org). Relative to 10 common transcripts of human and macaque developing neocortices, FZD8 was more abundant in humans. As RNA expression data becomes available [48], it may become possible to more directly compare FZD8 levels in human and non-human primates.
In addition to its requirement for early mouse corticogenesis, Wnt signaling is implicated in human brain traits. In 2002 Chenn et al. showed that expression of stabilized β-catenin induced a larger, gyrencephalic phenotype reminiscent of the human brain [15]. However, evidence for the involvement of this pathway in human brain evolution has remained elusive until now. Our identification of HARE5 highlights the transcriptional regulation of Wnt signaling components as a new avenue to explore for understanding the evolutionary origin of human specific anatomical and cognitive traits. With the ability to identify regulatory elements active during development [49], we are now poised for the discovery of additional loci and pathways whose modification provided the underpinnings for the evolution of the human brain.
Supplementary Material
Highlights.
Discovery of a human-accelerated enhancer functioning in the developing neocortex
Compared to chimpanzee, human HARE5 drives earlier and more robust brain expression
The HARE5 locus physically contacts the core promoter of the WNT receptor, Fzd8
HARE5::Fzd8 mice have accelerated neural progenitor cell cycle and enlarged brains
Acknowledgments
We thank the following: Dr. Len Pennacchio, Dr. Jérôme Collignon, Dr. Jeremy Nathans and Dr. Yanshu Wang for sharing reagents; Meilang Flowers and Cheryl Bock (Duke Transgenic Mouse Facility) for generating mouse transgenics; Autumn Rorrer for assistance with mouse husbandry; Dr. Hiro Matsunami for reading the manuscript; members of the DLS and GAW labs for helpful discussions; Han-Yu Shih for advice on 3C analysis; EM for assistance with western blotting; DLS lab members for assistance in blind scoring of phenotypes. Funding from a Research Incubator grant from the Duke Institute for Brain Sciences (to DLS and GAW), by R01NS083897 (to DLS), and by NSF HOMIND BCS-08-27552 (to GAW).
Footnotes
Author Contributions
JLB, GAW, and DLS conceived the study and wrote the paper. JLB, SLS, JPR, LJP, TB, RG, and DLS performed and analyzed experiments.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Geschwind DH, Rakic P. Perspective. Neuron. 2013;80:633–647. doi: 10.1016/j.neuron.2013.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lui JH, Hansen DV, Kriegstein AR. Development and evolution of the human neocortex. Cell. 2011;146:18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Enard W, Gehre S, Hammerschmidt K, HOlter SM, Blass T, Somel M, BrUckner MK, Schreiweis C, Winter C, Sohr R, et al. A Humanized Version of Foxp2 Affects Cortico-Basal Ganglia Circuits in Mice. Cell. 2009;137:961–971. doi: 10.1016/j.cell.2009.03.041. [DOI] [PubMed] [Google Scholar]
- 4.Herculano-Houzel S. The remarkable, yet not extraordinary, human brain as a scaled-up primate brain and its associated cost. Proceedings of the National Academy of Sciences. 2012;109(Suppl 1):10661–10668. doi: 10.1073/pnas.1201895109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dennis MY, Nuttle X, sudmant PH, antonacci F, Graves TA, Nefedov M, Rosenfeld JA, Sajjadian S, malig M, Kotkiewicz H, et al. Evolution of Human-Specific Neural SRGAP2 Genes by Incomplete Segmental Duplication. Cell. 2012:1–11. doi: 10.1016/j.cell.2012.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prabhakar S, Noonan JP, Pääbo S, Rubin EM. Accelerated evolution of conserved noncoding sequences in humans. Science. 2006;314:786. doi: 10.1126/science.1130738. [DOI] [PubMed] [Google Scholar]
- 7.Bird CP, Stranger BE, Liu M, Thomas DJ, Ingle CE, Beazley C, Miller W, Hurles ME, Dermitzakis ET. Fast-evolving noncoding sequences in the human genome. Genome Biol. 2007;8:R118. doi: 10.1186/gb-2007-8-6-r118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lindblad-Toh K, Garber M, Zuk O, Lin MF, Parker BJ, Washietl S, Kheradpour P, Ernst J, Jordan G, Mauceli E, et al. A high-resolution map of human evolutionary constraint using 29 mammals. Nature. 2011;478:476–482. doi: 10.1038/nature10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bush EC, Lahn BT. A genome-wide screen for noncoding elements important in primate evolution. BMC Evol Biol. 2008;8:17. doi: 10.1186/1471-2148-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pollard KS, Salama SR, King B, Kern AD, Dreszer T, Katzman S, Siepel A, Pedersen JS, Bejerano G, Baertsch R, et al. Forces shaping the fastest evolving regions in the human genome. PLoS Genet. 2006;2:e168. doi: 10.1371/journal.pgen.0020168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Capra JA, Erwin GD, McKinsey G, Rubenstein JLR, Pollard KS. Many human accelerated regions are developmental enhancers. Philosophical Transactions of the Royal Society B: Biological Sciences. 2013;368:20130025. doi: 10.1098/rstb.2013.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamm GB, López-Leal R, Lorenzo JR, Franchini LF. A fast-evolving human NPAS3 enhancer gained reporter expression in the developing forebrain of transgenic mice. Philosophical Transactions of the Royal Society B: Biological Sciences. 2013;368:20130019. doi: 10.1098/rstb.2013.0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oksenberg N, Stevison L, Wall JD, Ahituv N. Function and regulation of AUTS2, a gene implicated in autism and human evolution. PLoS Genet. 2013;9:e1003221. doi: 10.1371/journal.pgen.1003221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prabhakar S, Visel A, Akiyama JA, Shoukry M, Lewis KD, Holt A, Plajzer-Frick I, Morrison H, FitzPatrick DR, Afzal V, et al. Human-Specific Gain of Function in a Developmental Enhancer. Science. 2008;321:1346–1350. doi: 10.1126/science.1159974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chenn A, Walsh CA. Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science. 2002;297:365–369. doi: 10.1126/science.1074192. [DOI] [PubMed] [Google Scholar]
- 16.Freese JL, Pino D, Pleasure SJ. Wnt signaling in development and disease. Neurobiology of Disease. 2010;38:148–153. doi: 10.1016/j.nbd.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischer T, Guimera J, Wurst W, Prakash N. Distinct but redundant expression of the Frizzled Wnt receptor genes at signaling centers of the developing mouse brain. Neuroscience. 2007;147:693–711. doi: 10.1016/j.neuroscience.2007.04.060. [DOI] [PubMed] [Google Scholar]
- 18.Liu C, Bakeri H, Li T, Swaroop A. Regulation of retinal progenitor expansion by Frizzled receptors: implications for microphthalmia and retinal coloboma. Hum Mol Genet. 2012;21:1848–1860. doi: 10.1093/hmg/ddr616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller JA, Ding S-L, Sunkin SM, Smith KA, Ng L, Szafer A, Ebbert A, Riley ZL, Royall JJ, Aiona K, et al. Transcriptional landscape of the prenatal human brain. Nature. 2014:1–19. doi: 10.1038/nature13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berwick RC, Friederici AD, Chomsky N, Bolhuis JJ. Evolution, brain, and the nature of language. Trends in Cognitive Sciences. 2013;17:91–100. doi: 10.1016/j.tics.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 21.Whiten A. The scope of culture in chimpanzees, humans and ancestral apes. Philosophical Transactions of the Royal Society B: Biological Sciences. 2011;366:997–1007. doi: 10.1098/rstb.2010.0334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shettleworth SJ. Modularity, comparative cognition and human uniqueness. Philosophical Transactions of the Royal Society B: Biological Sciences. 2012;367:2794–2802. doi: 10.1098/rstb.2012.0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schoenemann PT. Evolution of the Size and Functional Areas of the Human Brain. Annu Rev Anthropol. 2006;35:379–406. [Google Scholar]
- 24.King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 25.Bae BI, Tietjen I, Atabay KD, Evrony GD, Johnson MB, Asare E, Wang PP, Murayama AY, Im K, Lisgo SN, et al. Evolutionarily dynamic alternative splicing of GPR56 regulates regional cerebral cortical patterning. Science. 2014;343:764–768. doi: 10.1126/science.1244392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haygood R, Babbitt CC, Fedrigo O, Wray GA. Contrasts between adaptive coding and noncoding changes during human evolution. Proceedings of the National Academy of Sciences. 2010;107:7853–7857. doi: 10.1073/pnas.0911249107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson MB, Kawasawa YI, Mason CE, Krsnik Z, Coppola G, BogdanoviC D, Geschwind DH, Mane SM, State MW, Sestan N. Functional and Evolutionary Insights into Human Brain Development through Global Transcriptome Analysis. Neuron. 2009;62:494–509. doi: 10.1016/j.neuron.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proceedings of the National Academy of Sciences. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim AS, Lowenstein DH, Pleasure SJ. Wnt receptors and Wnt inhibitors are expressed in gradients in the developing telencephalon. Mech Dev. 2001;103:167–172. doi: 10.1016/s0925-4773(01)00342-2. [DOI] [PubMed] [Google Scholar]
- 31.Summerhurst K, Stark M, Sharpe J, Davidson D, Murphy P. 3D representation of Wnt and Frizzled gene expression patterns in the mouse embryo at embryonic day 11.5 (Ts19) Gene Expression Patterns. 2008;8:331–348. doi: 10.1016/j.gep.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berger MF, Philippakis AA, Qureshi AM, He FS, Estep PW, Bulyk ML. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat Biotechnol. 2006;24:1429–1435. doi: 10.1038/nbt1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 34.Hagège H, Klous P, Braem C, Splinter E, Dekker J, Cathala G, de Laat W, Forné T. Quantitative analysis of chromosome conformation capture assays (3C-qPCR) Nat Protoc. 2007;2:1722–1733. doi: 10.1038/nprot.2007.243. [DOI] [PubMed] [Google Scholar]
- 35.Lukaszewicz A, Savatier P, Cortay V, Giroud P, Huissoud C, Berland M, Kennedy H, Dehay C. G1 phase regulation, area-specific cell cycle control, and cytoarchitectonics in the primate cortex. Neuron. 2005;47:353–364. doi: 10.1016/j.neuron.2005.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rakic P. Specification of cerebral cortical areas. Science. 1988;241:170–176. doi: 10.1126/science.3291116. [DOI] [PubMed] [Google Scholar]
- 37.Martynoga BS, Morrison H, Price DJ, Mason JO. Foxg1 is required for specification of ventral telencephalon and region-specific regulation of dorsal telencephalic precursor proliferation and apoptosis. Dev Biol. 2005;283:113–127. doi: 10.1016/j.ydbio.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 38.Takahashi T, Nowakowski RS, Caviness VS. The cell cycle of the pseudostratified ventricular epithelium of the embryonic murine cerebral wall. J Neurosci. 1995;15:6046–6057. doi: 10.1523/JNEUROSCI.15-09-06046.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rash BG, Tomasi S, Lim HD, Suh CY, Vaccarino FM. Cortical Gyrification Induced by Fibroblast Growth Factor 2 in the Mouse Brain. Journal of Neuroscience. 2013;33:10802–10814. doi: 10.1523/JNEUROSCI.3621-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ferland RJ, Cherry TJ, Preware PO, Morrisey EE, Walsh CA. Characterization of Foxp2 and Foxp1 mRNA and protein in the developing and mature brain. J Comp Neurol. 2003;460:266–279. doi: 10.1002/cne.10654. [DOI] [PubMed] [Google Scholar]
- 41.Greig LC, Woodworth MB, Galazo MJ, Padmanabhan H, Macklis JD. Molecular logic of neocortical projection neuron specification, development and diversity. Nature Publishing Group. 2013;14:755–769. doi: 10.1038/nrn3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kornack DR, Rakic P. Changes in cell-cycle kinetics during the development and evolution of primate neocortex. Proc Natl Acad Sci USA. 1998;95:1242–1246. doi: 10.1073/pnas.95.3.1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mirzaa GM, Parry DA, Fry AE, Giamanco KA, Schwartzentruber J, Vanstone M, Logan CV, Roberts N, Johnson CA, Singh S, et al. De novo CCND2 mutations leading to stabilization of cyclin D2 cause megalencephaly-polymicrogyria-polydactyly-hydrocephalus syndrome. Nat Genet. 2014;46:510–515. doi: 10.1038/ng.2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arai Y, Pulvers JN, Haffner C, Schilling B, Nüsslein I, Calegari F, Huttner WB. Neural stem and progenitor cells shorten S-phase on commitment to neuron production. Nature Communications. 2011;2:154. doi: 10.1038/ncomms1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pilaz LJ, Patti D, Marcy G, Ollier E, Pfister S, Douglas RJ, Betizeau M, Gautier E, Cortay V, Doerflinger N, et al. Forced G1-phase reduction alters mode of division, neuron number, and laminar phenotype in the cerebral cortex. Proceedings of the National Academy of Sciences. 2009;106:21924–21929. doi: 10.1073/pnas.0909894106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lange C, Huttner WB, Calegari F. Cdk4/cyclinD1 overexpression in neural stem cells shortens G1, delays neurogenesis, and promotes the generation and expansion of basal progenitors. Cell stem cell. 2009;5:320–331. doi: 10.1016/j.stem.2009.05.026. [DOI] [PubMed] [Google Scholar]
- 47.Lui JH, Nowakowski TJ, Pollen AA, Javaherian A, Kriegstein AR, Oldham MC. Radial glia require PDGFD–PDGFRβ signalling in human but not mouse neocortex. Nature. 2014;515:264–268. doi: 10.1038/nature13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pollen AA, Nowakowski TJ, Shuga J, Wang X, Leyrat AA, Lui JH, Li N, Szpankowski L, Fowler B, Chen P, et al. Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nat Biotechnol. 2014;32:1053–1058. doi: 10.1038/nbt.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Visel A, Taher L, Girgis H, May D, Golonzhka O, Hoch RV, McKinsey GL, Pattabiraman K, Silberberg SN, Blow MJ, et al. A High-Resolution Enhancer Atlas of the Developing Telencephalon. Cell. 2013;152:895–908. doi: 10.1016/j.cell.2012.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.