Summary
The cytoplasmic functions of Wiskott-Aldrich Syndrome family (WAS) proteins are well established and include roles in cytoskeleton reorganization and membrane-cytoskeletal interactions important for membrane/vesicle trafficking, morphogenesis, immune response and signal transduction. Mis-regulation of these proteins is associated with immune deficiency and metastasis [1-4]. Cytoplasmic WAS proteins act as effectors of Rho family GTPases and polymerize branched actin through the Arp2/3 complex [1, 5]. Previously, we identified Drosophila washout (wash) as a new member of the WAS family with essential cytoplasmic roles in early development [6, 7]. Studies in mammalian cells and Dictyostelium suggest that WASH functions primarily in a multiprotein complex that regulates endosome shape and trafficking in an Arp2/3-dependent manner [8-11]. However, roles for classically cytoplasmic proteins in the nucleus are beginning to emerge, in particular as participants in the regulation of gene expression [12, 13]. Here we show that Drosophila Wash is present in the nucleus where it plays a key role in global nuclear organization. wash mutant and knockdown nuclei disrupt sub-nuclear structures/organelles and exhibit the abnormal wrinkled morphology reminiscent of those observed in diverse laminopathies [14-16]. We find that nuclear Wash interacts with B-type Lamin (Lamin Dm0), and like Lamin, Wash associates with constitutive heterochromatin. Wash knockdown increases chromatin accessibility of repressive compartments and results in a global redistribution of repressive histone modifications. Thus, our results reveal a novel role for Wash in modulating nucleus morphology and in the organization of both chromatin and non-chromatin nuclear sub-structures.
Keywords: Wash, Wiskott Aldrich Syndrome, nuclear organization, Lamin, genome organization, chromatin accessibility, Drosophila
Results and Discussion
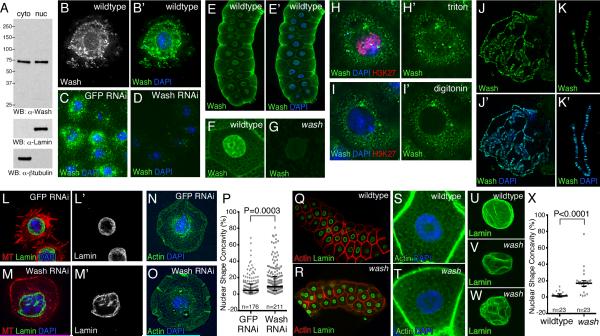
While examining cytoplasmic functions of Wash, we found that Wash is present in fly cell nuclear extracts (Figures 1A). In addition to localizing in the cytoplasm, Wash is present in the nucleus in Drosophila S2R+ cells and many embryo/larval tissues (e.g., salivary glands) (Figures 1B-1H’; data not shown) [7, 17]. Nuclear Wash is distributed in a punctuate pattern (Figures 1B-1C, 1F). This nuclear Wash staining is not observed in Wash knockdowns cells, wash mutant tissues, or when digitonin is used to permeabilize only the plasma membrane (Figures 1D, 1G, 1H-1I’, and S1A-S1A’). Although Wash lacks a DNA binding domain, polytene chromosome staining shows that Wash associates (directly or indirectly) with ~500 chromatin regions (Figures 1J-1K’).
Figure 1.
Wash is in the nucleus and disrupts nuclear morphology. (A) Wash is expressed in both the nucleus and cytoplasm as shown by western blot analysis of nuclear and cytoplasmic Drosophila Kc167 cell extracts. Extract specificity shown by western blot analysis with Lamin (nuclear) and β-tubulin (cytoplasmic). (B-B’) Micrographs of immunostained S2R+ cells (single focal plane) showing Wash is both cytoplasmic and nuclear. (C-D) Wash staining in S2R+ cells treated with dsRNA to GFP (control; C) or Wash (D) showing specificity of the Wash antibody. (E-E’) Confocal micrograph of larval salivary glands (projection) showing Wash is present in both the cytoplasm and the nucleus. (F-G) Nuclear and cytoplasmic Wash staining in wildtype salivary gland cells (F) and its absence in washΔ185 mutants (G). (H-I’) S2R+ cells treated with 5mg/ml digitonin (to permeabilize only the plasma membrane; I-I’) or 0.2% triton X-100 (to permeabilize both the plasma and nuclear membranes; H-H’), then stained for Wash and H3K27me3. Nuclear Wash and H3K27me3 staining are not detected when the nuclear membrane is not permeabilized (I-I’). (J-K’) Wash associates with specific regions on third-instar larval polytene chromosomes. (L-O’) wash knockdown mutants exhibit morphological alterations in nuclear shape. Confocal projections of S2R+ cells treated with dsRNA for GFP (L-L’, N-N’) or Wash (M-M’, O-O’) then stained for Lamin (L-M’), microtubules (MT) (L-M), actin (N-O) and DNA (DAPI; L-M, N-O) showing that Wash knockdown disrupts nuclear morphology in addition to cytoplasmic architecture. (P) Quantification of nuclear shape concavity in S2R+ cells treated with dsRNA to GFP (9.8±1.2%, n=176) and Wash (17.0±1.6%, n=211) (P=0.0003). Scatterplot with median ± IQR shown. (Q-V) Confocal projections of wildtype versus washΔ185 mutant salivary glands stained for actin and Lamin show that Wash affects nuclear morphology without gross cytoplasmic defects. Whole salivary glands (Q-R), salivary gland cells (S-T), and salivary gland nuclei (U-V). (X) Quantification of nuclear shape concavity in wildtype (2.7±0.68%, n=23) and washΔ185 (18.9±3.2%, n=23) salivary gland nuclei (P<0.0001). Scatterplot with median ± IQR shown. See also Figure S1.
To determine if nuclear Wash is biologically significant, we examined nuclear morphology in Wash RNAi-treated S2R+ cells. The effectiveness of Wash knockdown was established by western blot (Figure S1B) and confirmed by the disorganization of the cytoplasm of knockdown cells (Figures 1L-1O’ and S1C-S1D’). Importantly, Wash depletion caused irregularly shaped nuclei in S2R+ cells without affecting expression of the nuclear lamina protein Lamin (nuclear shape concavity: 9.8±1.2% GFP RNAi versus 17.0±1.6% Wash RNAi, P=0.0003; Figures 1L-1P and S1C-S1E). This altered nuclear morphology phenotype is not due to cell culture manipulation or an indirect effect of cytoplasmic Wash knockdown, as salivary gland nuclei from washΔ185 mutants stained for Lamin also show dramatic morphological alterations in nuclear shape while retaining overall cell and gland shape (nuclear shape concavity: 2.7±0.68% wildtype versus 18.9±3.2% wash, P<0.0001; Figures 1Q-1X). Thus, Wash plays a key role in modulating nuclear shape in Drosophila.
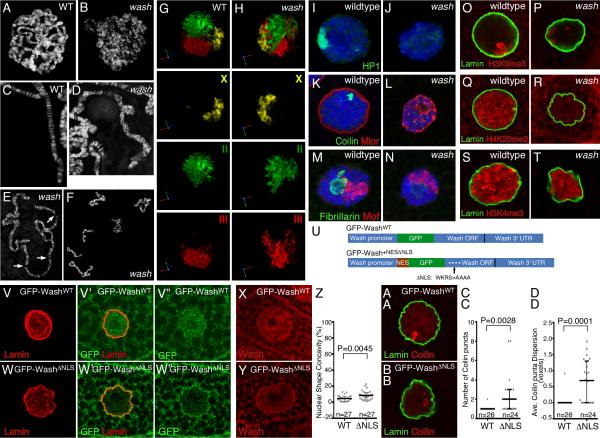
We next explored if lack of Wash affects nuclear organization. wash mutants exhibit reduced nuclear volume (8243±473 μm3 wildtype, 2797±146 μm3 wash, P<0.0001) and lower DNA content (Figures S1F-S1H). DAPI staining of wildtype and wash polytene chromosome spreads showed that chromosomes in wash mutants are less organized (Figures 2A-2B), appear more twisted (Figures 2C-2D), and tend to easily break and/or fragment upon physical stress (Figures 2E-2F). We performed FISH on salivary glands using chromosome paints for the three larger chromosomes. While wildtype chromosomes occupy well-defined spaces (chromosome territories), chromosomes of wash mutants are positioned closer to the nuclear periphery, with chromosome 3 in particular tending to be more dispersed (Figures 2G-2H and S1I-S1K; Movie S1). We also examined the organization of diverse non-chromatin nuclear sub-structures, as well as different chromatin modifications. Strikingly, HP1, which accumulates at and marks the chromocenter, is present in the nucleus but accumulates only weakly at the chromocenter in wash mutants (Figures 2I-2J and S1L-S1M). Similarly, Coilin and Fibrillarin (markers of cajal bodies and the nucleolus, respectively) and the nuclear envelope component Mtor exhibit non-specific nuclear localization in the absence of Wash (Figures 2K-2N and S1L-S1M). Importantly, these mis-localizations are specific, as lack of Wash does not affect localization of the dosage compensation complex (Mof) (Figures 2M-2N). The alterations of HP1, Coilin, Fibrillarin, and Mtor are likely due to their nuclear disorganization rather than protein loss, as protein levels remain roughly the same in wash mutants or knockdown cells (with the exception of Coilin; Figure S1L-S1M). We also examined histone modifications associated with repressive or active chromatin. Consistent with our previous results, H3K9me3, a histone modification recognized by HP1, is reduced in wash mutants (Figures 2O-2P and S1N-O). The repressive mark H4K20me2 and the active mark H3K4me3 are also lower in wash mutant nuclei (Figures 2Q-2T and S1P-S1S).
Figure 2.
Wash disrupts nuclear and genome organization. (A-F) Confocal projections of wildtype and washΔ185 mutant salivary gland polytene chromosomes; washΔ185 chromosomes show aberrant alignment and banding (D) and are extremely fragile (E-F). (G-H) Fluorescent in situ hybridization of chromosome-specific BAC pools hybridized to the X (yellow), second (green) and third (red) chromosome in wildtype and washΔ185 mutant salivary gland nuclei shows less compact chromosome territories in wash mutants. (I-N) Confocal micrographs of wildtype and washΔ185 mutant salivary gland nuclei stained for DNA and nuclear markers. HP1 (green) chromocenter localization is lost in washΔ185 nuclei (I-J). Coilin (green) cajal body localization is disrupted in washΔ185 nuclei while Mtor (red) remains localized to the periphery of salivary gland nuclei (K-L). Fibrillarin (green) localization at the nucleolus is disrupted in washΔ185 nuclei (M-N). While MOF localizes properly to the X chromosome in both wildtype and washΔ185 nuclei, it highlights the disruption to chromosome territory compaction observed in wash mutants (M-N). (O-T) Confocal projections of histone modifications in wildtype and washΔ185 salivary gland nuclei. Both repressive histone marks (H3K9me3 (O-P) and H4K20me2 (Q-R)) and the active histone mark H3K4me3 (S-T) are reduced in wash nuclei. (U) Schematic diagram of the Wash constructs used to generate the transgenic lines indicating the position of the added NES, GFP fusion, and the substitution mutations (WKRS>AAAA) in the Wash NLS (not drawn to scale). (V-DD) Specific reduction of Wash in the nucleus results in disrupted nuclear shape and sub-nuclear structure/organelle organization. Confocal projections of salivary gland nuclei from washΔ185 P{GFP-WashWT} (V-V’”, X, AA) and washΔ185 P{GFP-Wash+NESΔNLS} (W-W’”, Y, BB) stained for Lamin (V-V’, W-W’, AA-BB), GFP (V’-V”, W’-W”), Wash (X-Y), and Coilin (AA-BB). Quantification of nuclear shape concavity in washΔ185 P{GFP-WashWT} (WT: 4.74±0.67%; n=27) or washΔ185 P{GFP-Wash+NESΔNLS} (ΔNLS: 8.58±1.09%; n=27) transgenic salivary gland nuclei (P=0.0045) (Z). Scatterplot with median ± IQR shown. Quantification of the number of Coilin puncta in washΔ185 P{GFP-WashWT} (WT: 1.04±0.04; n=26) or washΔ185 P{GFP-Wash+NESΔNLS} (ΔNLS: 2.38±0.40; n=24) transgenic salivary gland nuclei (P=0.0028) (CC). Quantification of average dispersion of Coilin puncta in washΔ185 P{GFP-WashWT} (WT: 0.04±0.04; n=26) or washΔ185 P{GFP-Wash+NESΔNLS} (ΔNLS: 0.69±0.14; n=24) transgenic salivary gland nuclei (P=0.0001) (DD). Boxplot graphs show the median and 25% and 75% percentile measures. The whiskers indicate variability outside the upper and lower quartiles. See also Figure S1 and Movie S1.
To further examine the requirement for Wash in the nucleus, we expressed a GFP-Wash fusion protein with and without its conserved nuclear localization sequence mutated under the control of its own endogenous Wash promoter in a washΔ185 mutant background (Figure 2U; see Supplemental Experimental Procedures). The wildtype version of these transgenics (washΔ185 P{GFP-WashWT}) is expressed in both the nucleus and cytoplasm, rescues the washΔ185 mutant to viability, and does not exhibit nuclear phenotypes (Figure 2V-V”, 2X, 2Z, 2AA, 2CC-DD and S1T). In transgenics with the version lacking the nuclear localization signal (washΔ185 P{GFP-Wash+NESΔNLS}), the GFP-Wash+NESΔNLS fusion protein is expressed in the cytoplasm while largely excluded from the nucleus (Figure 2W’-W”, 2Y, 2Z, 2BB, 2CC-DD and S1T). These transgenics exhibit similar nuclear phenotypes as washΔ185 mutants, including disruption of nuclear morphology (Figure 2V-2Z and S1T) and disorganization of sub-nuclear structures/organelles (Figure 2AA-DD). Thus, Wash’s nuclear activity is required during development, where it plays a key role in global nuclear organization.
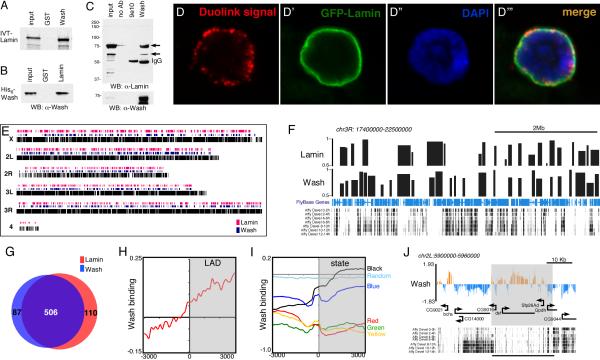
We identified a strong interaction between Wash and the Drosophila B-type lamin, Lamin Dm0, in a yeast two-hybrid screen (data not shown). Lamins are intermediate filament proteins that form a mesh lining the inner nuclear membrane and function in diverse roles including nuclear shape, aging, genome organization, and gene expression [18, 19]. Lamin Dm0 (hereafter referred to as Lamin) is present throughout development in all cells (except mature sperm). Using GST pulldowns with in vitro translated and/or bacterially purified proteins, we show that Wash interacts directly with Lamin (Figures 3A-3B). Co-immunoprecipitation experiments using nuclear enriched extracts of 0-12h Drosophila embryos show that this nuclear Wash-Lamin interaction occurs in vivo (Figure 3C). To confirm this interaction in vivo, we performed a Duolink in situ Proximity Ligation Assay (PLA) for Wash and GFP in salivary glands expressing a GFP-Lamin fusion protein. Fluorescent signal was observed and localized exclusively around the DNA confirming the interaction between Wash and GFP-Lamin (Figure 3D-3D’”). Thus, nuclear Wash interacts physically with Lamin.
Figure 3.
Wash interacts with lamin and associates with LADs. (A-B) Wash interacts with Lamin directly by GST-pulldown assays using IVT (A) or bacterially expressed proteins (B). (C) Wash and Lamin interact in vivo. Western blot of immunoprecipitations from embryo nuclear extracts with Wash, no primary antibody, or with an unrelated antibody (9e10). (D) Duolink Proximity Ligation Assay performed with antibodies recognizing Wash and GFP in GFP-Lamin expressing salivary glands. Duolink signal is only observed if the two antibodies are within 30 nm. (E) Genome-wide chromatin profile of Lamin (red) and Wash (blue) bound regions. (F) Chromosome-3R region aligned to developmental expression showing a significant overlap of LADs with Wash-associated regions at transcriptionally silent regions. Track with squished view of genes is shown in blue. (G) Venn-diagram comparing LADs to Wash associated chromatin regions (P<1×10−6). (H) LADs were aligned at their ends (+/− 3 Kb) and the normalized DamID probe signals were averaged in 150-bp bins for Wash occupancy. (I) DamID-based 5 color chromatin states and random sequences were aligned at their ends and the normalized DamID probe signals were averaged in 150-bp bins for Wash occupancy. YELLOW and RED chromatin contain proteins and histone modifications characteristic of active chromatin. BLUE chromatin contains H3K27me3 and PcG proteins, GREEN chromatin contains HP1 and Su(var)3-9, and BLACK chromatin contains Lamin and histone H1. (J) Wash chromatin profile on a 60Kb region of chromosome 2L showing an inverse correlation with developmental gene expression (similar results were obtained with the RNAseq data for Kc167 cells). See also Figure S2.
While Lamin does not exhibit a banding pattern on polytene chromosomes (Figures S2A-S2A”), genome-wide studies have shown that Lamin associates with specific genomic regions termed Lamin-associated domains (LADs) [20, 21]. These regions exhibit low gene density and are usually associated with repressive chromatin. As Wash associates with discrete regions on polytene chromosomes, we asked whether these regions correspond to LADs. Using DamID chromatin profiling in Kc cells, we identified 593 large continuous Wash-associated domains spread along the Drosophila genome with an average size of 40 Kb (Figures 3E-3F). Wash domains mainly localize with non-transcribed regions during early embryogenesis (Figure 3F). We performed similar DamID profiling for Lamin and identified 616 LADs (Figures 3E-3G). These domains show ~93% overlap with LADs previously described in Kc167 cells using a different microarray platform (385 of 412 LADs identified by [21]) (Figure S2B). Importantly, Wash-associated regions indeed coincide with LADs (Pearson correlation=0.72). Meta-analysis of the Wash DamID signal to LADs boundaries confirms the genomic co-localization between Wash and Lamin (Figures 3H and S2C).
To establish whether Wash domains correspond to functional domains defined by factor binding and epigenetic marks, we performed a meta-analysis of Wash binding to the boundaries of the 5-chromatin types classification (Yellow, Red, Blue, Green & Black) established previously in Kc cells [22]. Wash association with chromatin is mainly enriched in Black chromatin, which is the predominant type of repressive chromatin containing silent or low-expressing genes and co-localizing with Lamin-enriched nuclear envelope. Wash DamID signal is depleted from other chromatin types and coding regions (except Blue (Polycomb chromatin)) (Figure 3I-3J), suggesting that Wash targets transcriptionally silent chromatin regions in a similar fashion to Lamin [21, 22]. Interestingly, Wash also targets expressed developmental-regulated domains, suggesting that Wash binding does not necessarily repress transcription (Figure S2D-S2F).
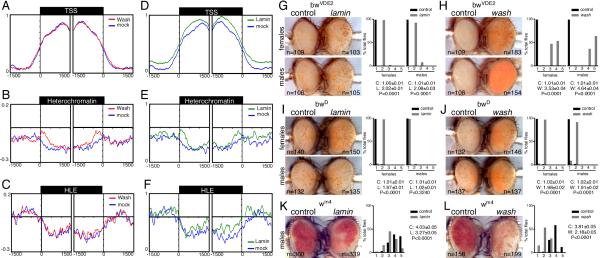
The alteration of histone modifications and sub-nuclear structures in wash mutants strongly suggests that Wash directly impacts chromatin organization. As chromatin accessibility is the main organizing feature that explains chromatin organization [23], we evaluated the effect of Wash knockdown in S2R+ cells on chromatin accessibility using an M.SssI-based approach [24]. Meta-analysis of M.SssI-methylated accessible chromatin of the 9 modENCODE consortium-defined chromatin states revealed higher accessibility in promoters and enhancers than in heterochromatic regions (Figures 4A-4C and S4A-S4H) [25]. Nevertheless, and consistent with its association to BLACK chromatin, Wash knockdown increased chromatin accessibility at the borders of constitutive heterochromatin, but not promoter or heterochromatin-like euchromatin (Figures 4A-4C). MeDIP-qPCR experiments confirmed these structural alterations on constitutive heterochromatin upon Wash knockdown, but not in promoter regions (Figure S3I). Notably, although no effect was observed on promoter, open chromatin, male X-linked genes, or heterochromatin-like euchromatic regions upon Wash knockdown, we observed increased accessibility on enhancers and reduced accessibility on Polycomb, elongating, enhancer, and basal interchromatin regions (Figures S3B-S3H), suggesting that absence of Wash indirectly impacts particular functional domains of the genome.
Figure 4.
Wash and Lamin affect chromatin accessibility at heterochromatic regions and Position Effect Variegation. (A-F) Distribution of M.SssI-based chromatin accessibility around active promoter, constitutive heterochromatin and heterochromatin-like euchromatin (HLE) regions. modENCODE Consortium generated chromatin states were aligned at their 5’ and 3’ (+/− 1.5 Kb) and the normalized probe signals were averaged in 50-bp bins. The y-axis in each plot represents the relative enrichment of M.SssI-methylated DNA for mock and Wash knockdown (A-C) and for mock and Lamin knockdown (D-F) in S2R+ cells. Statistical significance (determined using the two-sample KS test): TSS: Wash, P=0.823, Lamin P<4×10−3; Heterochromatin: Wash, P<2×10−3, Lamin P<4×10−3; HLE: Wash, P= 0.256, Lamin P<2×10−3. (G-J) wash and lamin suppress brown-mediated PEV. Wash and Lamin mediate suppression of the classical PEV allele bwVDE2, a chromosomal rearrangement that juxtaposes the bw gene near 2R heterochromatin (G-H). wash (male & female) and lamin (female only) mediate suppression of the non-classical PEV allele bwD, which inserts heterochromatin into the bw gene and silences the homolog (I-J). (K-L) wash and lamin enhance white mediated centromeric PEV (w+ gene inserted in proximal X chromosome heterochromatin). Bar plots showing percentage of flies falling into each expression quintile (flies sorted into one of five bins based on the percentage of ommatidia expressing the w+ marker or bw gene (bin 1 = 0-20% to bin 5 = 80-100%). The median±SEM and P-values are given in each panel. Eyes shown are representative of the average phenotype for each genotype and each pair is an age-matched, sibling pair (G-L). The number of eyes scored (N) is given beside each eye. See also Figures S3 and S4.
Similar to Wash, knockdown of Lamin alters nuclear architecture and correlates with alterations on heterochromatin organization of repressive histone modifications (e.g. H3K9m3) [26, 27]. Despite exhibiting a higher global background (Figure S3B), adjusted chromatin accessibility reveals that, similar to Wash, Lamin depletion causes an increase in accessibility of heterochromatic regions (Figures 4D-4F). Nevertheless, unlike Wash, Lamin knockdown also increased the accessibility of promoter regions (Figure 4D), suggesting that Lamin has more widespread roles in chromatin organization whereas Wash is particularly relevant for heterochromatin architecture, which is consistent with recent reports suggesting that loss of LMNB1 results in decondensation of chromosome territories [28].
Since Wash modifies genome organization by impairing chromatin accessibility of silent regions, we hypothesized that the wash mutant should behave as a suppressor of position effect variegation (PEV). Several Drosophila PEV models have been used as in vivo reporters of a particular protein’s ability to modulate chromatin in different contexts [29]. Using the brown-based PEV model [30], we found that lack of wash suppresses both bwVDE2 (classical) and bwD (non-classical) variegation (less heterochromatic repression; Figures 4H and 4J), which is consistent with our genomic data. Importantly, similar results were obtained using the laminsz18 mutant, however, this lamin-dependent effect is sex-specific in the case of bwD (Figures 4G and 4I). Strikingly, when the washΔ185 and laminsz18 alleles were tested in the presence of white-based centromeric, heterochromatic, and telomeric PEV [31, 32], we found that both mutants behave as enhancers of centromeric and heterochromatic PEV (Figure 4K-4L and S4A-S4D), while showing no affect on telomeric PEV (Figures S4E-S4H). This is consistent with a previous report for lamin effects on centromeric PEV [33]. It is worth noting that both brown and white are found in LADs, but while brown is located in a gene-rich region flanked by LADs, the white gene is embedded in a gene poor region that is part of a LAD (Figures S4I-S4K). To our knowledge this is the first time that the two PEV models have yielded different results when assaying a given mutation, suggesting that these PEV models differentially affect heterochromatic domains.
Classically cytoplasmic proteins are emerging as significant players in the nucleus. Two WAS family members, WASp and WAVE1, have been identified as positive regulators of gene expression [12, 13]. Indeed, before Wash was recognized as a cytoplasmic cytoskeletal factor, it was identified as part of the nuclear TRF2 complex [34] and, more recently, a required regulator of the NURF complex on the c-Myc locus in hematopoesis [35]. Despite this potential role in regulating gene expression, our findings link the nuclear presence of Wash with global nuclear organization, suggesting that Wash is required for the formation, maintenance, and/or movements of specific nuclear domains, organelles, and/or machineries. We also identify Wash as a Lamin-interacting protein and a key component of “BLACK” chromatin. Lamins and lamina-interacting proteins mainly compose the nuclear envelope, which plays key roles in nuclear structure, chromosome organization, DNA repair, transcriptional control, and like its plasma membrane counterpart, is the site for signal sensing, molecule trafficking, and inter/intra-nuclear attachments. Consistent with this, mutations in Lamins and lamina-interacting proteins such as emerin lead to phenotypically diverse diseases including muscular dystrophies, cardiomyopathies, and premature aging syndromes (collectively called laminopathies) [26]. Interestingly, the abnormal wrinkled morphology in both wash mutant and knockdown nuclei resembles that observed in diverse laminopathies [14-16], including the Hutchinson-Gilford progeria syndrome. It is possible that Wash dynamically modifies specific compartments of the nuclear envelope to adapt it to different types of stimuli. It is known that tissue-specific differential expression of lamin isoforms reinforce the nucleus against physical stress by stabilizing the nuclear envelope and chromatin [36, 37]. We have previously shown that, like other WAS family members, Wash requires Arp2/3 for actin nucleation [6, 7]. Recently, both actin and the WAS family-interacting protein Arp2/3 were implicated in nuclear envelope fragmentation in starfish oocytes [38]. Although it is not yet clear whether the Wash-Lamin functional link requires Arp2/3, Wash could potentially be the connection point for coordinating the remodeling of the Lamin-based meshwork along with the cytoplasmic cytoskeleton in response to physical stress. Alternatively, in vitro data suggest that Lamin monomers tend to self-assemble into filamentous fibers under particular salt conditions [39, 40], Wash could modulate Lamin self-assembly in vivo based on its molecular functions with other structural proteins. Additionally, Wash is involved in endocytosis by modulating vesicle formation and trafficking (cf. [4, 41]), these regulatory activities could be useful at the nuclear envelope for the dynamic remodeling of the lamin meshwork in response to nuclear shape changes required for cells to squeeze through small spaces or when undergoing metastasis. An alternate possibility relies on the fact that actin and actin-binding proteins have been found in several chromatin remodeling factors (cf. [42]): Wash could modulate actin functions and/or its mobility during its association with these chromatin remodelers to maintain a dynamic chromatin structure. Our data highlights Wash as a key regulator of cytoplasmic and nuclear processes by maintaining microfilament (actin), intermediate filament, and microtubule homeostasis, and whose mutations lead to alterations in both cellular compartments. In the future, it will be exciting to further explore Wash’s nuclear roles, and to determine if and how closely these roles parallel those in the cytoplasm.
Experimental Procedures
Please refer to the Supplemental Information for detailed experimental procedures.
The raw and normalized datasets and DamID domains can be found in the following GEO dataset: GSE59854.
Supplementary Material
Acknowledgements
We thank Steve Henikoff, Amir Orian, and members of the Parkhurst and Groudine labs for their interest and comments on the manuscript. We thank K. Barry, E. Rodriguez-Mesa, and T. Ragoczy for help at various stages of the project and J. Henikoff, R. Basom and J. Young for bioinformatics help. We also thank A. Akhtar, J. Gall, S. Henikoff, G. Karpen, A. Minoda, L. Pagie, P. Talbert, B. van Steensel, T. Tsukiyama, L. Wallrath, L. Wayner, the M.J. Murdoch Charitable Trust, the Bloomington Stock Center, and the Developmental Studies Hybridoma Bank for antibodies, flies, software, microscope, and other reagents used in this study. This work was supported by NIH grants T32CA009657 (to HR-A), DK44746 and HL65440 (to MG), and GM097083 (to SMP).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Takenawa T, Suetsugu S. The WASP-WAVE protein network: connecting the membrane to the cytoskeleton. Nature reviews. Molecular cell biology. 2007;8:37–48. doi: 10.1038/nrm2069. [DOI] [PubMed] [Google Scholar]
- 2.Rottner K, Hanisch J, Campellone KG. WASH, WHAMM and JMY: regulation of Arp2/3 complex and beyond. Trends in cell biology. 2010;20:650–661. doi: 10.1016/j.tcb.2010.08.014. [DOI] [PubMed] [Google Scholar]
- 3.Massaad MJ, Ramesh N, Geha RS. Wiskott-Aldrich syndrome: a comprehensive review. Annals of the New York Academy of Sciences. 2013;1285:26–43. doi: 10.1111/nyas.12049. [DOI] [PubMed] [Google Scholar]
- 4.Burianek LE, Soderling SH. Under lock and key: spatiotemporal regulation of WASP family proteins coordinates separate dynamic cellular processes. Seminars in cell & developmental biology. 2013;24:258–266. doi: 10.1016/j.semcdb.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campellone KG, Welch MD. A nucleator arms race: cellular control of actin assembly. Nature reviews. Molecular cell biology. 2010;11:237–251. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Linardopoulou EV, Parghi SS, Friedman C, Osborn GE, Parkhurst SM, Trask BJ. Human subtelomeric WASH genes encode a new subclass of the WASP family. PLoS genetics. 2007;3:e237. doi: 10.1371/journal.pgen.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu R, Abreu-Blanco MT, Barry KC, Linardopoulou EV, Osborn GE, Parkhurst SM. Wash functions downstream of Rho and links linear and branched actin nucleation factors. Development. 2009;136:2849–2860. doi: 10.1242/dev.035246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Derivery E, Sousa C, Gautier JJ, Lombard B, Loew D, Gautreau A. The Arp2/3 activator WASH controls the fission of endosomes through a large multiprotein complex. Developmental cell. 2009;17:712–723. doi: 10.1016/j.devcel.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 9.Gomez TS, Billadeau DD. A FAM21-containing WASH complex regulates retromer-dependent sorting. Developmental cell. 2009;17:699–711. doi: 10.1016/j.devcel.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duleh SN, Welch MD. WASH and the Arp2/3 complex regulate endosome shape and trafficking. Cytoskeleton (Hoboken) 2010;67:193–206. doi: 10.1002/cm.20437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park L, Thomason PA, Zech T, King JS, Veltman DM, Carnell M, Ura S, Machesky LM, Insall RH. Cyclical action of the WASH complex: FAM21 and capping protein drive WASH recycling, not initial recruitment. Developmental cell. 2013;24:169–181. doi: 10.1016/j.devcel.2012.12.014. [DOI] [PubMed] [Google Scholar]
- 12.Miyamoto K, Teperek M, Yusa K, Allen GE, Bradshaw CR, Gurdon JB. Nuclear Wave1 is required for reprogramming transcription in oocytes and for normal development. Science. 2013;341:1002–1005. doi: 10.1126/science.1240376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sadhukhan S, Sarkar K, Taylor M, Candotti F, Vyas YM. Nuclear role of WASp in gene transcription is uncoupled from its ARP2/3-dependent cytoplasmic role in actin polymerization. J Immunol. 2014;193:150–160. doi: 10.4049/jimmunol.1302923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hutchison CJ. B-type lamins in health and disease. Seminars in cell & developmental biology. 2014;29:158–163. doi: 10.1016/j.semcdb.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dechat T, Shimi T, Adam SA, Rusinol AE, Andres DA, Spielmann HP, Sinensky MS, Goldman RD. Alterations in mitosis and cell cycle progression caused by a mutant lamin A known to accelerate human aging. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:4955–4960. doi: 10.1073/pnas.0700854104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haithcock E, Dayani Y, Neufeld E, Zahand AJ, Feinstein N, Mattout A, Gruenbaum Y, Liu J. Age-related changes of nuclear architecture in Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:16690–16695. doi: 10.1073/pnas.0506955102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodriguez-Mesa E, Abreu-Blanco MT, Rosales-Nieves AE, Parkhurst SM. Developmental expression of Drosophila Wiskott-Aldrich Syndrome family proteins. Dev Dyn. 2012;241:608–626. doi: 10.1002/dvdy.23742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spann TP, Goldman AE, Wang C, Huang S, Goldman RD. Alteration of nuclear lamin organization inhibits RNA polymerase II-dependent transcription. The Journal of cell biology. 2002;156:603–608. doi: 10.1083/jcb.200112047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilson KL, Zastrow MS, Lee KK. Lamins and disease: insights into nuclear infrastructure. Cell. 2001;104:647–650. [PubMed] [Google Scholar]
- 20.Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 21.Pickersgill H, Kalverda B, de Wit E, Talhout W, Fornerod M, van Steensel B. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nature genetics. 2006;38:1005–1014. doi: 10.1038/ng1852. [DOI] [PubMed] [Google Scholar]
- 22.Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, Brugman W, de Castro IJ, Kerkhoven RM, Bussemaker HJ, et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143:212–224. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bell O, Schwaiger M, Oakeley EJ, Lienert F, Beisel C, Stadler MB, Schubeler D. Accessibility of the Drosophila genome discriminates PcG repression, H4K16 acetylation and replication timing. Nature structural & molecular biology. 2010;17:894–900. doi: 10.1038/nsmb.1825. [DOI] [PubMed] [Google Scholar]
- 25.Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, Liu T, Yip KY, Robilotto R, Rechtsteiner A, Ikegami K, et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science. 2010;330:1775–1787. doi: 10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dittmer TA, Misteli T. The lamin protein family. Genome biology. 2011;12:222. doi: 10.1186/gb-2011-12-5-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sadaie M, Salama R, Carroll T, Tomimatsu K, Chandra T, Young AR, Narita M, Perez-Mancera PA, Bennett DC, Chong H, et al. Redistribution of the Lamin B1 genomic binding profile affects rearrangement of heterochromatic domains and SAHF formation during senescence. Genes & development. 2013;27:1800–1808. doi: 10.1101/gad.217281.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Camps J, Wangsa D, Falke M, Brown M, Case CM, Erdos MR, Ried T. Loss of lamin B1 results in prolongation of S phase and decondensation of chromosome territories. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2014;28:3423–3434. doi: 10.1096/fj.14-250456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elgin SC, Reuter G. Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila. Cold Spring Harbor perspectives in biology. 2013;5:a017780. doi: 10.1101/cshperspect.a017780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sass GL, Henikoff S. Comparative analysis of position-effect variegation mutations in Drosophila melanogaster delineates the targets of modifiers. Genetics. 1998;148:733–741. doi: 10.1093/genetics/148.2.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dorer DR, Henikoff S. Expansions of transgene repeats cause heterochromatin formation and gene silencing in Drosophila. Cell. 1994;77:993–1002. doi: 10.1016/0092-8674(94)90439-1. [DOI] [PubMed] [Google Scholar]
- 32.Cryderman DE, Cuaycong MH, Elgin SC, Wallrath LL. Characterization of sequences associated with position-effect variegation at pericentric sites in Drosophila heterochromatin. Chromosoma. 1998;107:277–285. doi: 10.1007/s004120050309. [DOI] [PubMed] [Google Scholar]
- 33.Bao X, Girton J, Johansen J, Johansen KM. The lamin Dm0 allele Ari3 acts as an enhancer of position effect variegation of the wm4 allele in Drosophila. Genetica. 2007;129:339–342. doi: 10.1007/s10709-006-0012-7. [DOI] [PubMed] [Google Scholar]
- 34.Hochheimer A, Zhou S, Zheng S, Holmes MC, Tjian R. TRF2 associates with DREF and directs promoter-selective gene expression in Drosophila. Nature. 2002;420:439–445. doi: 10.1038/nature01167. [DOI] [PubMed] [Google Scholar]
- 35.Xia P, Wang S, Huang G, Zhu P, Li M, Ye B, Du Y, Fan Z. WASH is required for the differentiation commitment of hematopoietic stem cells in a c-Myc-dependent manner. The Journal of experimental medicine. 2014 doi: 10.1084/jem.20140169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swift J, Ivanovska IL, Buxboim A, Harada T, Dingal PC, Pinter J, Pajerowski JD, Spinler KR, Shin JW, Tewari M, et al. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science. 2013;341:1240104. doi: 10.1126/science.1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buxboim A, Swift J, Irianto J, Spinler KR, Dingal PC, Athirasala A, Kao YR, Cho S, Harada T, Shin JW, et al. Matrix elasticity regulates lamin-A,C phosphorylation and turnover with feedback to actomyosin. Current biology : CB. 2014;24:1909–1917. doi: 10.1016/j.cub.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mori M, Somogyi K, Kondo H, Monnier N, Falk HJ, Machado P, Bathe M, Nedelec F, Lenart P. An Arp2/3 nucleated F-actin shell fragments nuclear membranes at nuclear envelope breakdown in starfish oocytes. Current biology : CB. 2014;24:1421–1428. doi: 10.1016/j.cub.2014.05.019. [DOI] [PubMed] [Google Scholar]
- 39.Sasse B, Lustig A, Aebi U, Stuurman N. In vitro assembly of Drosophila lamin Dm0--lamin polymerization properties are conserved. European journal of biochemistry / FEBS. 1997;250:30–38. doi: 10.1111/j.1432-1033.1997.t01-1-00030.x. [DOI] [PubMed] [Google Scholar]
- 40.Herrmann H, Foisner R. Intermediate filaments: novel assembly models and exciting new functions for nuclear lamins. Cellular and molecular life sciences : CMLS. 2003;60:1607–1612. doi: 10.1007/s00018-003-3004-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rotty JD, Wu C, Bear JE. New insights into the regulation and cellular functions of the ARP2/3 complex. Nature reviews. Molecular cell biology. 2013;14:7–12. doi: 10.1038/nrm3492. [DOI] [PubMed] [Google Scholar]
- 42.Dion V, Shimada K, Gasser SM. Actin-related proteins in the nucleus: life beyond chromatin remodelers. Current opinion in cell biology. 2010;22:383–391. doi: 10.1016/j.ceb.2010.02.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.