Abstract
When using cell lines to study cancer, phenotypic similarity to the original tumor is paramount. Yet, little has been done to characterize how closely Merkel cell carcinoma (MCC) cell lines model native tumors. To determine their similarity to MCC tumor samples, we characterized MCC cell lines via gene expression microarrays. Using whole transcriptome gene expression signatures and a computational bioinformatic approach, we identified significant differences between variant cell lines (UISO, MCC13, and MCC26) and fresh frozen MCC tumors. Conversely, the classic WaGa and Mkl-1 cell lines more closely represented the global transcriptome of MCC tumors. When compared to publicly available cancer lines, WaGa and Mkl-1 cells were similar to other neuroendocrine tumors, but the variant cell lines were not. WaGa and Mkl-1 cells grown as xenografts in mice had histological and immunophenotypical features consistent with MCC, while UISO xenograft tumors were atypical for MCC. Spectral karyotyping and short tandem repeat analysis of the UISO cells matched the original cell line’s description, ruling out contamination. Our results validate the use of transcriptome analysis to assess the cancer cell line representativeness and indicate that UISO, MCC13, and MCC26 cell lines are not representative of MCC tumors, whereas WaGa and Mkl-1 more closely model MCC.
Introduction
Cancer cell lines are essential tools for modeling human malignancy. However, many factors can alter the representativeness of a cultured cell line. Some differences to the native tumor are expected with cells grown in culture due to the absence of vascular stroma and tumor architecture. Additional discrepancies can arise due to the evolution of atypical subclones that possess in vitro growth advantages, genomic instability associated with repeated passaging, alterations secondary to microbial infections in culture, and contamination from other cell lines (Barallon et al., 2010; Capes-Davis et al., 2010). Phenotypic dissimilarity to the original tumor impacts the relevance of a cell line as an experimental model. Thus, systematic comparison of cell lines to the cancers they are modeling is critical.
Microarray expression profiles have been used to compare cancer cell lines to native tumors for a number of cancer types (Barretina et al., 2012; Carlson et al., 2007; Wang et al., 2006; Gillet et al., 2011; Khan et al., 2001; Li et al., 2008). In some cases, extensive transcriptomic differences were found between cell lines and primary tumors (Li et al., 2008; Wang et al., 2006). These studies illustrate the utility of microarray expression profiling in characterizing cancer cell lines, however the implications of expression profile differences on the biological representativeness of cells lines were not assessed.
Merkel cell carcinoma (MCC) is an aggressive neuroendocrine skin cancer (Toker, 1972; Maricich et al., 2009, 2012). The risk of developing MCC is associated with ultraviolet light exposure, advanced age, and immunosuppression (Hodgson, 2005; Becker, 2010). Approximately 80% of MCC tumors have DNA of the Merkel cell polyomavirus (MCV) clonally integrated into their genome (Feng et al., 2008). The MCV T antigen is thought to be important in MCC carcinogenesis (Houben et al., 2010), however, it is unclear if the presence or absence of integrated MCV alters the outcome or course of MCC.
Like most solid malignancies, diagnosis of MCC relies on histopathological examination of tumor tissue. MCC shares a number of histopathological characteristics with other tumors of neuroendocrine origin. For example, both MCC and small cell lung cancer (SCLC) are frequently composed of morphologically similar, small, round cells that frequently express chromogranin (CHGA), synaptophysin (SYP), and neuron-specific enolase (ENO2). In contrast, immunostaining for cytokeratin 20 (KRT20) is more specific for MCC, whereas thyroid transcription factor 1 (TTF1) is a marker of SCLC (Leech et al., 2001; Pulitzer et al., 2009).
A growing number of MCC tumor cell lines are available (Krasagakis et al., 2001; Leonard et al., 1993; Rosen et al., 1987; Leonard et al., 1995; Moll et al., 1994; Ronan et al., 1993). Most MCC cell lines grow in suspension, express MCC immunomarkers, and demonstrate dense core granules on ultrastructural analysis. MCC cell lines with these typical features, such as WaGa and Mkl-1, are termed “classic” (Ronan et al., 1993; Leonard et al., 1993; Van Gele et al., 2004). In contrast, “variant” MCC cell lines like UISO-MCC-1 (henceforth referred to as UISO) (Ronan et al., 1993), MCC13, and MCC26 (Leonard et al., 1995) grow adherently in culture, lack typical markers on immunostaining, and have reduced numbers of neurosecretory granules. The majority of classic MCC cell lines test positive for MCV, whereas most variant cell lines are MCV negative. Little has been done to assess the representativness of either classic or variant MCC cell lines.
Here we apply computational and experimental methods to compare MCC cell line transcriptomes to each other, to fresh tumor tissue, and to other cancer cells in order to examine their representativeness. Using these approaches, we find that three variant cell lines are not characteristic of MCC whereas three classic MCC cell lines more closely represent native MCC tumors. We validate the non-representative biology of UISO cells by demonstrating atypical in vivo growth in a xenograft tumor model.
Results
Variant cell lines clusters distinctly from MCC tumors and classic MCC cell lines
In order to test how well they modeled MCC, we analyzed global RNA expression in six MCC lines: the variant lines UISO, MCC13, and MCC26, and the classic lines WaGa (Houben et al., 2010), Mkl-1 (Rosen et al., 1987), and a newly established MCC cell line SK-MC01 (hereafter MC01). We also analyzed global RNA expression from 23 MCC and 9 SCLC fresh frozen tumor samples.
We performed hierarchical clustering to identify groups of samples with similar global gene expression profiles (Figure 1A). The variant cell line samples formed a group divergent from the MCC tumors, SCLC tumors, and the classic WaGa, Mkl-1, and MC01 samples, indicating that these cell lines are distinct from both native tumors and the more typical MCC cells. Furthermore, the UISO samples clustered separately from the other variant cell lines, MCC13 and MCC26. In contrast to the variant lines, WaGa, Mkl-1, and MC01 cells clustered with the MCC tumor samples. These differences in gene expression can also be visualized in a principal components analysis (PCA) projection (Figure 1B, Figure S1). In the PCA, expected differences due to in vitro growth appear to contribute to the separation between cultured cells and fresh frozen tumor samples along the second principal component.
Figure 1. Variant cell lines cluster separately from MCC tumors and classic MCC cell lines.
A. Hierarchical clustering of microarray expression data from MCC cell lines and MCC and SCLC frozen tumor samples. Average linkage was applied for merging clusters to the variance-filtered probe set expression values. One minus the Spearman correlation was used as a dissimilarity metric. Legend: MCV positive, red; MCV negative, blue; not tested, black. B. Principal components analysis of microarray expression data from MCC cell lines and MCC and SCLC tumor samples was performed with variance-filtered probe set expression values for each sample. The variance in the expression data accounted for by the first three principal components are 26%, 22%, and 7%.
Global gene expression differences between the MCC cell lines and tumor samples
To identify RNA expression differences, we performed differential expression analysis between the MCC tumor samples and each group of cell lines: classic (WaGa, Mkl-1, and MC01), variant (MCC13 and MCC16), and UISO. Figure 2 depicts a Venn diagram of the differentially expressed probe sets in the comparison of each group to the tumor samples. In total, 1023 probe sets showed common differential expression between the tumor samples and all three groups. In line with our prior results, many more probe sets were uniquely differentially expressed between the MCC tumor samples and UISO cells (4223) or the other variant lines (4103) than between the MCC tumors and the classic cell lines (938). To quantify the overall similarity of cell line expression profiles to MCC tumor samples, Spearman’s rank correlation coefficients between individual expression profiles of MCC cell lines and tumor samples were computed (Figure S2). The classic samples were similar to tumor samples (median correlation = 0.83). UISO cells were less similar (median correlation = 0.66), as were the other variant lines (median correlation = 0.68).
Figure 2. Compared to MCC tumors, variant MCC cell lines have more differentially expressed genes than classic MCC cell lines.
A Venn diagram showing the number of probe sets commonly differentially expressed when comparing the MCC tumor samples to UISO, other variant (MCC13 and MCC26), and classic (WaGa and Mkl-1) cell lines. Only probe sets with an absolute fold change greater than 2 and a q-value less than 0.05 are counted (total: 13,329 probe sets).
Virus status of MCC tumors does not influence gene expression comparisons
Despite having varied clinical features, the MCC tumor samples analyzed in this study were remarkably homogeneous at the RNA expression level. We observed no significantly differentially expressed probe sets when comparing samples based on clinically relevant phenotypes such as tumor site (primary skin vs. metastasis), if the patient ultimately had a recurrence, or tumor stage (stage 1–2 vs. Stage 3, coded as early vs. late; Table S1). We found relatively few (214) significantly differentially expressed probe sets between MCV positive and negative tumors (Table S2), including previously described differences in TP53 and RB1 expression (Harms et al., 2013; Sihto et al., 2011; Bhatia et al., 2010; Waltari et al., 2011). Consistent with this high degree of similarity, there was little difference in the correlation values for each cell line when compared independently to MCV positive or MCV negative tumors (Table S3). These observations suggest that clinical differences and MCV status are not driving the divergent expression profile observed in the MCV-negative (Guastafierro et al., 2013) variant cell lines.
Predicted functional differences between MCC cell lines and tumor samples
Having identified significant differences between the variant MCC cell lines and the tumor samples at the single gene level, we sought to identify functional differences at the pathway level. To identify pathways that are significantly differentially expressed between MCC cell lines and tumor samples, we applied Gene Set Enrichment Analysis (GSEA) (Subramanian et al., 2005) using the differential expression data and pathway gene sets taken from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Kanehisa and Goto, 2000). We assessed pathway enrichment in the differentially expressed genes comparing MCC tumors to UISO cells, variant cell lines, and classic cell lines (Table S4).
Although enrichment of any given pathway requires experimental validation, GSEA analysis often accurately reveals biological schemes. It is therefore noteworthy that the gene expression differences between UISO cells and MCC tumors show concordant changes in processes central to oncogenesis such as DNA replication and RNA processing (spliceosome). Despite having a similarly large number of differentially expressed probe sets as UISO, the other variant cell lines had very few differentially expressed pathways, with only the proteasome pathway showing high enrichment. On the other hand, highly differentially expressed pathways between MCC tumors and the classic cell lines could be attributed to cultured cells lacking both stromal interactions (focal adhesion and ECM interactions) and an immune infiltrate (multiple autoimmune, inflammatory, and infectious pathways). Taken together, the GSEA results further suggest that UISO cells are less appropriate to use as a model for MCC, but give little insight into the large differences between MCC tumors and the other variant cell lines.
Classification of MCC cell lines using tumor samples
Since our analyses suggested that variant cell lines were similarly distinct from both skin and lung neuroendocrine tumors, we sought to test if the variant lines would be classified as MCC or SCLC using a random forest machine learning classifier trained on expression signatures of the fresh frozen tumor samples (see Supplementary Methods). This classifier was applied to the expression signatures of the MCC cell lines. The trained model strongly classified all classic cell lines as MCC (over 80% probability, Table 1). None of the variant cell lines were strongly classified as MCC or SCLC, indicating they resemble SCLC as much as they resemble MCC.
Table 1. Random forest classification of MCC cell lines.
A random forest classifier was trained using the microarray probe set expression data for the 23 MCC and 9 SCLC tumor samples. The classifier was then applied to the MCC cell lines, and the average class prediction was determined for each of the MCC cell lines (over sample replicates). The class assignment (MCC and SCLC) probability is given as the fraction of trees in the random forest voting for each class. The values shown are the average of six replicates for Mkl-1 WaGa, and UISO, 3 replicates for MCC13 and MCC16, and 1 replicate of MC01.
| MCC | SCLC | |
|---|---|---|
| WaGa | 0.89 | 0.11 |
| Mkl-1 | 0.85 | 0.15 |
| MC01 | 0.83 | 0.17 |
| MCC13 | 0.43 | 0.57 |
| MCC26 | 0.40 | 0.60 |
| UISO | 0.43 | 0.57 |
Classification of MCC cell lines using the Cancer Cell Line Encyclopedia
Since the gene expression of variant cell lines did not resemble MCC tumors or more typical classic MCC cell lines, we assessed their distinctness using data from the Cancer Cell Line Encyclopedia (CCLE) (Barretina et al., 2012) to compare MCC cell line expression data to a broad set of cancer cell lines. We selected a set of eight cancer types from the CCLE that were common malignancies or shared some feature with MCC (from the skin, neuroendocrine, or neuroectodermal) to analyze with the MCC cell lines.
A random forest classifier was constructed using the selected CCLE lines and the Mkl-1 and WaGa cell lines (grouped as an MCC class) to classify the variant cell lines (see Supplementary Methods). The resulting random forest did not classify the UISO samples as MCC (1% probability). They were classified with nearly equal probability for a number of tumor types, including melanoma, hepatocellular, breast, and lung carcinomas (Table S5). The variant samples MCC13 and MCC26 were classified more strongly as MCC (up to 18% probability), but were similarly classified to other cancer types as UISO. Held out WaGa or Mkl-1 samples were strongly classified as MCC (70% probability, Table S6). Using PCA visualization, we observed that WaGa and Mkl-1 samples clustered near neuroendocrine (SCLC) and neuroectodermal (Ewings sarcoma) samples. As expected, UISO samples clustered away from the neuroendocrine tumors, while the MCC13 and MCC26 samples cluster in between the UISO and classic samples (Figure 3). These findings reinforce that UISO, MCC13, and MCC26 do not resemble typical MCC cell lines.
Figure 3. Among multiple cancer cell lines, variant cell lines are distinct from classic MCC cell lines.
as well as other neuroendocrine lines. Principal components analysis of microarray expression data from MCC cell lines and cell lines from the Cancer Cell Line Encyclopedia was computed from variance-filtered probe set expression values for each sample. The variance in the expression data accounted for by components one and two are 8% and 5%, respectively.
Immunohistochemical analysis of MCC xenograft tumors
To confirm that the distinct expression profile observed in cultured UISO cells correlates with an atypical tumor phenotype in vivo, we grew xenograft tumors with UISO, WaGa, and Mkl-1 cells in immunocompromised mice. Histologically, WaGa tumors showed features consistent with MCC: sheets of tumor cells with scant cytoplasm, rounded nuclei, inconspicuous nucleoli, stippled chromatin patterning, and a brisk mitotic rate (Figure 4). WaGa xenograft tissue also resembled the immunophenotype of typical MCC tumors with positive immunostainng for the neuroendocrine markers chromogranin, NCAM1 (CD56), and ENO2. The MCC marker KRT20 and pan-cytokeratin (AE1/AE3) both stained with a characteristic paranuclear dot pattern in WaGa tumors, and the SCLC marker TTF1 was negative. As a MCV positive cell line (Houben et al., 2010), WaGa xenografts stained with CM2B4, an anti-MCV T antigen antibody. Mkl-1 tumors showed immunohistochemical features similar to WaGa tumors, consistent with typical MCC tumors (data not shown).
Figure 4. UISO xenograft tumors are histologically atypical for MCC.
Representative images of hematoxylin and eosin staining (H&E) and immunohistochemical staining of WaGa and UISO xenograft tumors in NOD/Scid mice. Scale bar = 100μm.
In contrast, UISO xenografts did not resemble typical MCC tumors. The UISO tumors had abundant fibrovascular stroma separating islands of tumor cells with large, irregular, hyperchromatic nuclei, irregular dendritic processes, and perinuclear clearing. UISO tumors lacked staining for MCC markers such as chromogranin, KRT20, and pan-cytokeratin. However, there was focal staining for NCAM1 and ENO2. Staining for TTF1 and CM2B4 were negative, as expected. Staining was also negative for S100, CD34, and KRT7 (data not shown). Overall, UISO xenograft tumors were not diagnostic for MCC and were most consistent with a poorly-differentiated neuroendocrine tumor possessing sarcomatoid features.
The UISO cell line is not contaminated
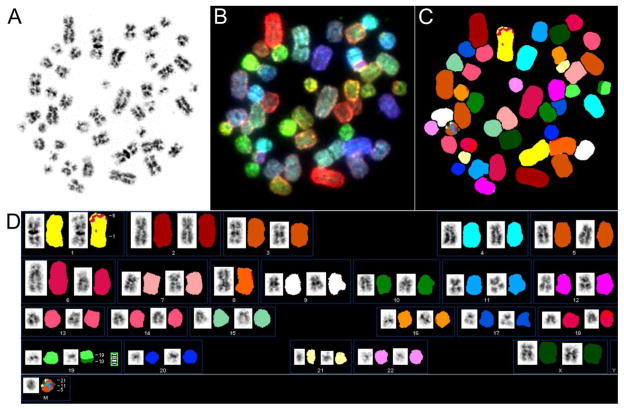
Cross contamination with another cancer cell line could explain why UISO cells are not representative of MCC. Fortunately, early cytogenetic characterizations of UISO cells and the availability of low-passage cells allowed us to confirm the provenance of the contemporary cell line. We performed spectral karyotyping (SKY) on UISO cells to compare with previously reported cytogenetic changes (Van Gele et al., 2002, 1998). On metaphase spreads, several recurrent chromosomal changes were identified consistent with prior studies, including a small insertion at 1p36.2, a cryptic insertion or duplication on 6q, a dicentric chromosome 8, and a small heterogeneous ring chromosome (Figure 5). A new t(10;19) translocation was also observed. Conservation of these features strongly supports the present day cell line being of the same origin as the UISO cells used in earlier publications.
Figure 5. Spectral karyotyping (SKY) of metaphase chromosomes confirms the identity of the UISO cell line.
A. Chromosomes stained with DAPI and converted into G-banding-like appearance. B. Chromosomes in display colors. C. Chromosomes in classification colors. D. UISO spectral karyotype (with DAPI and classification images of each chromosome side by side). This cell has 46 chromosomes and contains previously described rearrangements, including an insertion at 1p36.2, a cryptic insertion or duplication on 6q, a dicentric chromosome 8, and a small heterogeneous ring chromosome.
To confirm this, we obtained archived early passage cells directly from the laboratory that established the UISO cell line (Ronan et al., 1993). Short tandem repeat (STR) profiling (Masters et al., 2001) was performed using 16 markers (see Supplementary Methods) on all of the MCC cell lines used here. The STR fingerprint of the UISO cell line was identical to the early passage UISO cells (Table S7). Moreover, none of the cell lines were similar to any cell line available in the major cell line databases (ATCC, DSMZ, JCRB), indicating no cross contamination with an STR characterized cell line occurred.
Discussion
Characterization of tumor cell lines is important in developing appropriate and representative in vitro models for the study of cancer. Here we found that three variant MCC cell lines were not representative of MCC tumors based on multiple comparative analyses of global gene expression and predicted gene set function. Furthermore, machine learning approaches failed to classify UISO cells as MCC, whereas other variant cell lines were more similar to classic lines, but were not strongly classified as MCC. UISO also had an atypical histopathologic phenotype when grown as xenograft tumors in mice. At the same time, we found that the WaGa and Mkl-1 cell lines more closely resembled MCC tumors in gene expression and in vivo growth.
In light of these findings, we speculate that an atypical MCC tumor gave rise to the UISO cell line. UISO cells were derived from a tumor arising on the thigh of a patient much younger than the mean age for MCC presentation (46 versus 72 years) (Ronan et al., 1993). The primary tumor uncharacteristically failed to show chromogranin and MAK-6 pan-cytokeratin staining, but did have ENO2 and focal CAM5.2 staining. KRT20 staining was not assessed, as the antibody was not in widespread use at the time. This immunophenotype in a skin tumor is consistent with MCC, but would not be considered diagnostic by today’s standards. We found UISO xenografts to be chromogranin negative, cytokeratin negative by AE1/AE3 staining, negative for KRT20 staining, and possessing sarcomatoid histologic features. The focal NCAM1 and ENO2 staining in UISO xenografts were consistent with a neuroendocrine tumor, possibly a poorly differentiated MCC. UISO xenograft tumors did express CD99 (data not shown), which is seen in peripheral primitive neuroectodermal tumors. However, UISO cells did not have the canonical t(11;22)(q24;q12) translocation associated with primitive neuroectodermal tumors (Figure 5 and data not shown)(Turc-Carel et al., 1988). Given the atypical nature of UISOs tumor of origin, it is not surprising that the cell line is atypical and possesses a transcriptome and in vivo biology that is non-representative of MCC tumors in general.
An atypical tumor of origin may not account for all the uncharacteristic features of UISO or other variant MCC cell lines. Even in typical MCC tumors, a minority sub-population of aberrant cells could exhibit a selective growth advantage in culture. A cell line derived from such cells would not reflect the average gene expression or biology of MCC tumors. In such a scenario, the sub-population cells could still be biologically important to tumor behavior, and having a cell culture model of the atypical cells could potentially be instructive. Therefore, the UISO cell line and other non-representative cell lines may still prove useful in the study of cancer biology.
In contrast to the variant lines, WaGa and Mkl-1 cells were more representative of MCC tumors. Expression differences relative to tumor tissue did exist with WaGa and Mkl-1 cells, which are expected due to the lack of stroma, vasculature, and hematopoietic cells in cultured cancer cells. As a model system to study the biology of MCC and predict the behavior of native tumors, it appears that WaGa and Mkl-1 are preferable to UISO or other variant lines.
A previous study comparing MCC cell lines using expression microarrays with only 1083 genes found that UISO cells clustered with classic MCC lines, such as Mkl-1 (Van Gele et al., 2004). The limited scope of the genes analyzed and the lack of MCC tumor samples likely contributed to this finding that conflicts with our results. In contrast, using a comprehensive set of more than 40,000 human transcripts, we have demonstrated through multiple analyses that UISO samples cluster separately from both classic cell lines as well as MCC tumor samples.
It is interesting to note that on a transcriptome level, the set of MCC tumor samples analyzed in this study are quite homogeneous. Clustering of the expression data failed to identify subclasses of MCC with strongly distinct gene expression signatures. We identified few differences in gene expression between MCV positive and negative tumor samples. This contrasts with a recent publication comparing virus-positive and virus-negative MCC tumors in which many more significant differences were found (Harms et al., 2013). The reason for these differing results is unknown. However, in their study, only 46% of the tumors analyzed were found to be virus positive, whereas most reports indicate 70–80% of MCC tumors are virus positive (Pulitzer et al., 2009; Feng et al., 2008). This is more consistent with the 87% virus positive tumors we observed here. It is not clear if this reflects a difference in virus detection methods, or a true difference in the patient populations. Consistent with most prior studies, our cohort had only a small number of virus negative tumors. Although we employed a robust technique for differential gene expression detection that works well with small sample sizes (Smyth, 2004, 2005), we cannot entirely rule out effects due to sample size and class imbalance in our analysis. Regardless, there was no concordance between the virus status of the tumors and their dissimilarity to the variant cell lines or their similarity to WaGa and Mkl-1, indicating that the differences observed in the variant cell lines are not driven by viral status.
The independence of tumor viral status on cell line similarity is an important observation, as variant MCC cell lines are predominantly MCV-negative, and classic lines tend to be MCV-positive. All three variant cell lines used in this study are MCV-negative, whereas the classic lines are MCV-positive. By definition, variant lines lack expression of certain differentiation markers, and our data suggest that variant cell lines may be divergent from MCC tumors on a more general level. However, the fact that MCV-negative native tumors are not similarly distinct from MCV-positive tumors and classic MCC cell lines suggests that MCV status alone is not the driver of atypical gene expression in the variant cell lines. We have demonstrated that the question of cell line representativeness can be answered (in part) by the use of gene expression microarray data. Straightforward computational methods and publicly available data can be applied to compare cell lines with native tumor tissue and other cell lines to assess cancer type concordance. These types of techniques can, and should, be applied when using existing lines or developing a new one. With respect to MCC, our analysis suggests that representative cell lines such as WaGa or Mkl-1 should be favored over UISO, MCC13, or MCC16 as an in vitro experimental model.
Materials and Methods
Ethics statement
All human research was conducted in accordance with approved IRB protocols. Patient specimens were collected at MSKCC under IRB protocol #00-144 A. All patients provided written informed consent for use of their sample for general research purposes, and did not specify limitations that restricted use of their samples for this study. Analysis of samples was conducted under NCI Protocol #13CN024 without obtaining further consent, as patients had provided prior consent and the samples were analyzed anonymously. This study was performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the National Cancer Institute’s Animal Care and Use Committee (Protocol DB-091).
MCV detection by real time quantitative PCR
Primers were designed to amplify a region of the MCV T antigen (coordinates 444–579 in the “MCV350” sequence GenBank Accession EU375803) to detect MCV DNA in MCC tumor samples by qPCR. Primers for β-actin (ACTB), a gene on chromosome 7, were used as a reference on all MCC tumor samples.
Cell lines
The MCC cell lines Mkl-1 (Rosen et al., 1987), WaGa (Houben et al., 2010), and UISO (Ronan et al., 1993), and MCC13 and MCC26 (Leonard et al., 1995) have been described previously. SK-MC01 cells were generated at the Memorial Sloan Kettering Cancer Center (MSKCC).
cDNA array hybridization of tumor samples and cell lines
Fresh frozen tumor specimens were supplied by the MSKCC tumor bank. Total RNA was extracted from tumor specimens and hybridized to Human Genome U133A 2.0 Array (Affymetrix) GeneChips.
Total RNA was extracted from three to six replicates of each cell line (single replicate of MC01) and hybridized to the Human Genome U133A 2.0 Array (Affymetrix). Three replicates of all cell lines (excluding MC01) were also hybridized to the GeneChip Human Genome U133 Plus 2.0 Array (Affymetrix).
Microarray analysis
For MCC tumors, SCLC tumors, and MCC cell lines on the Affymetrix U133A 2.0 platform, normalization, filtering, and analysis of microarray expression data were performed using R (details in Supplemental Methods). Raw CEL files for CCLE data were downloaded directly from GEO (accession GSE36133) and were normalized as above with MCC cell line Affymetrix U133 Plus 2.0 array data. We call probe sets to be differentially expressed at a fold change >= 2 and q <= 0.05.
Classification of MCC cell lines with MCC and SCLC tumors
A random forest classifier was trained using the microarray expression data for the 23 MCC and 9 SCLC tumor samples. The classifier was then applied to the MCC cell lines, and the average class prediction was determined for each of the MCC cell lines across sample replicates.
Classification of MCC cell lines with CCLE cell lines
A random forest classifier was trained using the microarray expression data for MCC cell lines (except for UISO) along with samples from the CCLE dataset. The classifier was then applied to the variant (UISO, MCC13, and MCC26) samples.
STR Profiling
Genomic DNA was isolated from all cell lines and sent to the Johns Hopkins Fragment Analysis Facility for STR profiling on all cell lines except MCC13 and MCC26 using the AmpFt’STR Identifiler PCR Amplification Kit (Applied Biosystems), which profiles 16 STR markers (Table S7). MCC13 and MCC26 were profiled using the Promega PowerPlex 18D Kit, which profiles the same 16 markers plus two others.
Mouse xenografts
Five to eight-week-old female NOD/scid mice were obtained from the NCI Animal Production Program and housed under specific pathogen-free conditions. Tumors were induced by subcutaneous injection of 107 WaGa cells, 2×107 Mkl-1 cells, or 106 UISO cells in 200μL sterile saline into the posterior lateral flank of the mice. Tumor tissue was collected and fixed in neutral buffered formalin after tumors were clinically detectable.
Immunohistochemistry of mouse xenograft tumors
Paraffin-embedded tissue sections (5μm) of fixed xenograft tumors on glass slides were stained on an automated immunostainer (Ventana Medical System), according to the manufacturer’s instructions. The primary antibodies and antigen retrieval methods used are indicated in Table S8.
Spectral karyotyping of the UISO cell line
UISO cells were harvested after Colcemid (Life Technologies) treatment (0.025μg/mL: 12 hours) and processed by standard cytogenetic methods. Breakpoints on the SKY-painted chromosomes were determined by comparison with corresponding inverted DAPI banding. We report SKY results by using the International System for Human Cytogenetic Nomenclature (ISCN, 2013).
Data access
All tumor sample and cell line microarray data have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 2002) and is accessible through GEO Series accession number GSE50451. All analysis code, including generation of all figures, tables, and supplementary tables and figures, are available in reference Daily and Brownell (2014) (DOI 10.6084/m9.figshare.1030618). See the Supplementary Methods for details.
Supplementary Material
Acknowledgments
This research was supported by the NIH Intramural Research Program, Center of Cancer Research, National Cancer Institute. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. We thank the NCI CCR Informatics Core for assistance. We thank Dr. Tapas K. Das Gupta for providing early passage UISO cells. We thank Dr. Patrick Moore and Dr. Jürgen Becker for providing WaGa, Mkl-1, and late-passage UISO cells. We thank Dr. Patrick Moore for providing MCC13 and MCC26 cells. We thank Dr. Jedd Wolchock for assistance in generating SK-MC01 cells. We thank Dr. Mark Raffeld and Cindy Harris for immunohistochemical assays. We thank Dr. Anna Roschke and Kenneth Nakahara for the SKY analysis. We thank Dr. Sean Davis for critical reading of the manuscript and methodological insights.
This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the NIH (http://biowulf.nih.gov).
Abbreviations
- MCC
Merkel cell carcinoma
- MCV
Merkel cell polyomavirus
- SCLC
small cell lung cancer
- CCLE
Cancer Cell Line Encyclopedia
Footnotes
Conflicts of Interest
The authors state no conflict of interest.
References
- Barallon R, Bauer SR, Butler J, et al. Recommendation of short tandem repeat profiling for authenticating human cell lines, stem cells, and tissues. In vitro cellular & Developmental Biology – Animal. 2010;46:727–732. doi: 10.1007/s11626-010-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretina J, Caponigro G, Stransky N, et al. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker JC. Merkel cell carcinoma. Annals of Oncology. 2010;21:vii81–vii85. doi: 10.1093/annonc/mdq366. [DOI] [PubMed] [Google Scholar]
- Bhatia K, Goedert JJ, Modali R, et al. Merkel cell carcinoma subgroups by Merkel cell polyomavirus DNA relative abundance and oncogene expression. Int J Cancer. 2010;126:2240–6. doi: 10.1002/ijc.24676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capes-Davis A, Theodosopoulos G, Atkin I, et al. Check your cultures! a list of cross-contaminated or misidentified cell lines. Int J Cancer. 2010;127:1–8. doi: 10.1002/ijc.25242. [DOI] [PubMed] [Google Scholar]
- Carlson M, Iyer V, Marcotte E. Quantitative gene expression assessment identifies appropriate cell line models for individual cervical cancer pathways. BMC Genomics. 2007;8:117+. doi: 10.1186/1471-2164-8-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daily K, Brownell I. Reproducible Analysis for Assessment of Cancer Cell Line Representativeness using Microarrays for Merkel Cell Carcinoma. figshare. 2014 doi: 10.1038/jid.2014.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Research. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H, Shuda M, Chang Y, et al. Clonal integration of a polyomavirus in human merkel cell carcinoma. Science. 2008;319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillet JP, Calcagno AM, Varma S, et al. Redefining the relevance of established cancer cell lines to the study of mechanisms of clinical anti-cancer drug resistance. Proceedings of the National Academy of Sciences. 2011;108:18708–18713. doi: 10.1073/pnas.1111840108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guastafierro A, Feng H, Thant M, et al. Characterization of an early passage merkel cell polyomavirus-positive merkel cell carcinoma cell line, MS-1, and its growth in NOD scid gamma mice. Journal of Virological Methods. 2013;187:6–14. doi: 10.1016/j.jviromet.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms PW, Patel RM, Verhaegen ME, et al. Distinct gene expression profiles of viral- and Nonviral-Associated merkel cell carcinoma revealed by transcriptome analysis. Journal of Investigative Dermatology. 2013;133:936–945. doi: 10.1038/jid.2012.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson NC. Merkel cell carcinoma: changing incidence trends. Journal of Surgical Oncology. 2005;89:1–4. doi: 10.1002/jso.20167. [DOI] [PubMed] [Google Scholar]
- Houben R, Shuda M, Weinkam R, et al. Merkel cell Polyomavirus-Infected merkel cell carcinoma cells require expression of viral t antigens. Journal of Virology. 2010;84:7064–7072. doi: 10.1128/JVI.02400-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Research. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan J, Wei JS, Ringnér M, et al. Classification and diagnostic prediction of cancers using gene expression profiling and artificial neural networks. Nature Medicine. 2001;7:673–679. doi: 10.1038/89044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasagakis K, Almond-Roesler B, Geilen C, et al. Growth and characterization of a cell line from a human primary neuroendocrine carcinoma of the skin (merkel cell carcinoma) in culture and as xenograft. Journal of Cellular Physiology. 2001;187:386–391. doi: 10.1002/jcp.1086. [DOI] [PubMed] [Google Scholar]
- Leech SN, Kolar AJ, Barrett PD, et al. Merkel cell carcinoma can be distinguished from metastatic small cell carcinoma using antibodies to cytokeratin 20 and thyroid transcription factor 1. Journal of Clinical Pathology. 2001;54:727–729. doi: 10.1136/jcp.54.9.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard JH, Bell JR, Kearsley JH. Characterization of cell lines established from merkel-cell (‘small-cell’) carcinoma of the skin. Int J Cancer. 1993;55:803–810. doi: 10.1002/ijc.2910550519. [DOI] [PubMed] [Google Scholar]
- Leonard JH, Dash P, Holland P, et al. Characterisation of four merkel cell carcinoma adherent cell lines. Int J Cancer. 1995;60:100–107. doi: 10.1002/ijc.2910600115. [DOI] [PubMed] [Google Scholar]
- Li A, Walling J, Kotliarov Y, et al. Genomic changes and gene expression profiles reveal that established glioma cell lines are poorly representative of primary human gliomas. Molecular Cancer Research. 2008;6:21–30. doi: 10.1158/1541-7786.MCR-07-0280. [DOI] [PubMed] [Google Scholar]
- Maricich SM, Morrison KM, Mathes EL, et al. Rodents rely on merkel cells for texture discrimination tasks. The Journal of Neuroscience. 2012;32:3296–3300. doi: 10.1523/JNEUROSCI.5307-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maricich SM, Wellnitz SA, Nelson AM, et al. Merkel cells are essential for Light-Touch responses. Science. 2009;324:1580–1582. doi: 10.1126/science.1172890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters JR, Thomson JA, Daly-Burns B, et al. Short tandem repeat profiling provides an international reference standard for human cell lines. Proceedings of the National Academy of Sciences. 2001;98:8012–8017. doi: 10.1073/pnas.121616198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll I, Bohnert E, Herbst C, et al. Establishment and characterization of two merkel cell tumor cultures. Journal of Investigative Dermatology. 1994;102:346–353. doi: 10.1111/1523-1747.ep12371794. [DOI] [PubMed] [Google Scholar]
- Pulitzer MP, Amin BD, Busam KJ. Merkel cell carcinoma: review. Advances in Anatomic Pathology. 2009;16:135–144. doi: 10.1097/PAP.0b013e3181a12f5a. [DOI] [PubMed] [Google Scholar]
- Ronan S, Green A, Shilkaitis A, et al. Merkel cell carcinoma: In vitro and in vivo characteristics of a new cell line. Journal of the American Academy of Dermatology. 1993;29:715–722. doi: 10.1016/0190-9622(93)70236-m. [DOI] [PubMed] [Google Scholar]
- Rosen ST, Gould VE, Salwen HR, et al. Establishment and characterization of a neuroendocrine skin carcinoma cell line. Laboratory Investigation; A Journal of Technical Methods and Pathology. 1987;56:302– 312. [PubMed] [Google Scholar]
- Sihto H, Kukko H, Koljonen V, et al. Merkel cell polyomavirus infection, large T antigen, retinoblastoma protein and outcome in Merkel cell carcinoma. Clin Cancer Res. 2011;17:4806–13. doi: 10.1158/1078-0432.CCR-10-3363. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Statistical Applications in Genetics and Molecular Biology. 2004;3 doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Smyth GK. limma: Linear models for microarray data. In: Gentleman R, Carey V, Huber W, Irizarry R, Dudoit S, editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor, Statistics for Biology and Health. Vol. 23. Springer New York; New York: 2005. pp. 397–420. [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toker C. Trabecular carcinoma of the skin. Archives of Dermatology. 1972;105:107–110. [PubMed] [Google Scholar]
- Turc-Carel C, Aurias A, Mugneret F, et al. Chromosomes in ewing’s sarcoma. i. an evaluation of 85 cases of remarkable consistency of t(11;22)(q24;q12) Cancer Genetics and Cytogenetics. 1988;32:229–238. doi: 10.1016/0165-4608(88)90285-3. [DOI] [PubMed] [Google Scholar]
- Van Gele M, Boyle GM, Cook AL, et al. Gene-expression profiling reveals distinct expression patterns for classic versus variant merkel cell phenotypes and new classifier genes to distinguish merkel cell from small-cell lung carcinoma. Oncogene. 2004;23:2732–2742. doi: 10.1038/sj.onc.1207421. [DOI] [PubMed] [Google Scholar]
- Van Gele M, Leonard JH, Van Roy N, et al. Combined karyotyping, CGH and M-FISH analysis allows detailed characterization of unidentified chromosomal rearrangements in merkel cell carcinoma. Int J Cancer. 2002;101:137–145. doi: 10.1002/ijc.10591. [DOI] [PubMed] [Google Scholar]
- Van Gele M, Speleman F, Vandesompele J, et al. Characteristic pattern of chromosomal gains and losses in merkel cell carcinoma detected by comparative genomic hybridization. Cancer Research. 1998;58:1503– 1508. [PubMed] [Google Scholar]
- Waltari M, Sihto H, Kukko H, et al. Association of Merkel cell polyomavirus infection with tumor p53, KIT, stem cell factor, PDGFR-alpha and survival in Merkel cell carcinoma. Int J Cancer. 2011;129:619–28. doi: 10.1002/ijc.25720. [DOI] [PubMed] [Google Scholar]
- Wang H, Huang S, Shou J, et al. Comparative analysis and integrative classification of NCI60 cell lines and primary tumors using gene expression profiling data. BMC Genomics. 2006;7:166+. doi: 10.1186/1471-2164-7-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.