Summary
Transcription factors related to the insect sex determination gene Doublesex (DMRT proteins) control sex determination and/or sexual differentiation in diverse metazoans, and are implicated in transitions between sex-determining mechanisms during vertebrate evolution [1]. In mice Dmrt1 is required for male gonadal differentiation in somatic cells and germ cells [2-4]. DMRT1 also maintains male gonadal sex: its loss, even in adults, can trigger sexual fate reprogramming in which male Sertoli cells transdifferentiate into their female equivalents – granulosa cells – and testicular tissue reorganizes to a more ovarian morphology [5]. Here we use a conditional Dmrt1 transgene to show that Dmrt1 is not only necessary but also sufficient to specify male cell identity in the mouse gonad. DMRT1 expression in the ovary silenced the female sex-maintenance gene Foxl2 and reprogrammed juvenile and adult granulosa cells into Sertoli-like cells, triggering formation of structures resembling male seminiferous tubules. DMRT1 can silence Foxl2 even in the absence of the testis-determining genes Sox8 and Sox9. mRNA profiling found that DMRT1 activates many testicular genes and downregulates ovarian genes and single cell RNA-seq in transdifferentiating cells identified dynamically expressed candidate mediators of this process. Strongly upregulated genes were highly enriched on chromosome X, consistent with sexually antagonistic functions. This study provides an in vivo example of single gene reprogramming of cell sexual identity. Our findings suggest a reconsideration of mechanisms involved in human disorders of sexual development (DSD) and empirically support evolutionary models where loss or gain of Dmrt1 function promotes establishment of new vertebrate sex determination systems.
Keywords: cell fate reprogramming, transdifferentiation, sex determination, DMRT1, testis, ovary, FOXL2
Results
Ectopic DMRT1 induces formation of Sertoli-like cells in the ovary
Mammalian sex is determined in the fetal gonad by presence or absence Y-linked Sry gene [6]. In genetic males, bipotential precursors become Sertoli cells while in females the same cells become granulosa cells. These pivotal gonadal cells trigger a cascade of events leading to body-wide sexual differentiation and later provide essential support for developing germ cells. The Sertoli vs. granulosa cell fate decision is not necessarily permanent: loss of a single transcription factor (Dmrt1 in males or Foxl2 in females) can trigger direct transdifferentiation between the two cell types, even in adults [5, 7]. Dmrt1 and Foxl2 therefore are essential components of antagonistic regulatory networks actively maintaining sex in differentiated cells retaining latent plasticity [8].
While neither Dmrt1 nor Foxl2 is required for fetal sex determination in mammals, Dmrt1 orthologs determine sex in other vertebrates [9-12]. Thus Dmrt1 can play an instructive role in determining sexual cell fates. Moreover, Dmrt1 orthologs in such species appear to have undergone mutational events causing either loss or gain of function, suggesting that altered Dmrt1 activity helped drive evolutionary transitions leading to distinct genetic sex determination systems [1]. To help evaluate this possibility we asked whether gain-of-function in Dmrt1 can determine male fate in the mouse ovary.
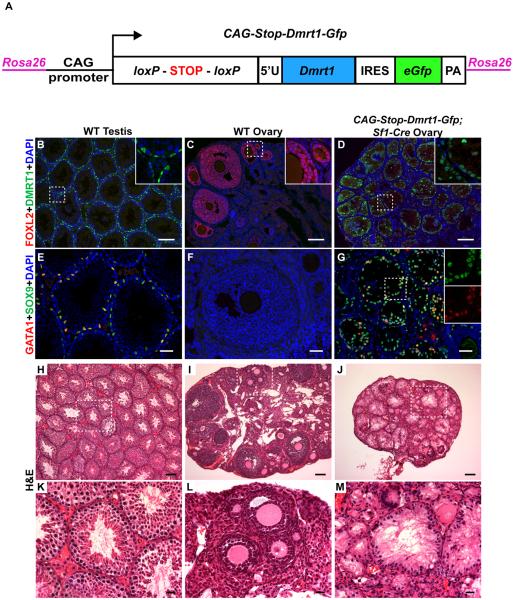
To conditionally express DMRT1 we generated mice with the construct CAG-Stop-Dmrt1-Gfp integrated into the Rosa26 locus (Figure 1A, Figure S1A-B). Cre-mediated removal of a transcriptional stop cassette generates CAG-Dmrt1-Gfp, which expresses a bicistronic mRNA encoding DMRT1 and eGFP. We tested whether CAG-Dmrt1-Gfp can functionally replace the Dmrt1 gene by activating CAG-Stop-Dmrt1-Gfp while deleting Dmrt1 with Sf1-Cre, which is expressed in somatic gonadal cells from embryonic day 10.5 (E10.5) [13]. CAG- DMRT1 expression from Dmrt1-Gfp was comparable to wild type and rescued Sertoli differentiation sufficiently to support complete male spermatogenesis (Figure S1C-K).
Figure 1. Ectopic DMRT1 in the ovary causes granulosa cell to Sertoli-like cell differentiation.
(A) Schematic diagram of conditional DMRT1 expression transgene CAG-Stop-Dmrt1-Gfp, which is transcribed to express DMRT1 and GFP upon Cre-mediated deletion of a floxed “STOP” cassette (for additional detail see Figure S1A). (B-D) Immunofluorescence (IF) of gonads from 8-10 week old mice showing that activation of CAG-Dmrt1-Gfp in somatic cells of the fetal ovary by Sf1-Cre activates DMRT1, silencing the ovarian granulosa cell transcription factor FOXL2. Dashed boxes indicate areas shown in higher magnification insets. (E-G) IF of adult gonads showing that activation of CAG-Dmrt1-Gfp also activates the Sertoli cell determinant SOX9 and the Sertoli cell differentiation factor GATA1. Dashed boxes indicate areas shown in higher magnification insets. (H-M) Hematoxylin and Eosin (H&E) stained sections of adult testes, ovaries, and CAG-Dmrt1-Gfp expressing ovaries, at low and high magnification (dashed boxes indicate magnified areas shown in K-M). Ovaries expressing DMRT1 show tubule-like morphology typical of testes, with polarized Sertoli-like cells lining the periphery and extending cytoplasmic veils into a central lumen. Scale bars: 100 μm (B-D, H-J); 40 μm (E-G); 20 μm (K-M). See also Figure S1.
DMRT1 is expressed in both sexes until about embryonic day 13.5 (E13.5) and then becomes testis-specific [14-17]. To determine the effect of ectopic DMRT1 in the ovary we examined adult mice with CAG-Stop-Dmrt1-Gfp activated by Sf1-Cre. Doubly transgenic CAG-Stop-Dmrt1-Gfp;Sf1-Cre ovaries had widespread DMRT1 and few FOXL2-positive granulosa cells (Figure 1B-D). DMRT1+ cells often were at the periphery of follicle remnants (Figure 1D), similar to DMRT1-positive Sertoli cells in wild-type testis tubules (Figure 1B), and most expressed the Sertoli cell markers SOX9 and GATA1 (Figure 1E-G). The switch from FOXL2+ to SOX9+/GATA1+ suggested granulosa cells were re-specified as Sertoli-like cells. Hematoxylin and eosin (H&E) staining (Figure 1H-M) confirmed that transformed cells had typical Sertoli morphology, including cell polarization with cytoplasmic veils (Figure 1M) and often organized in a seminiferous tubule-like arrangement surrounding a central lumen (Figure 1J).
DMRT1 induces postnatal sexual transdifferention
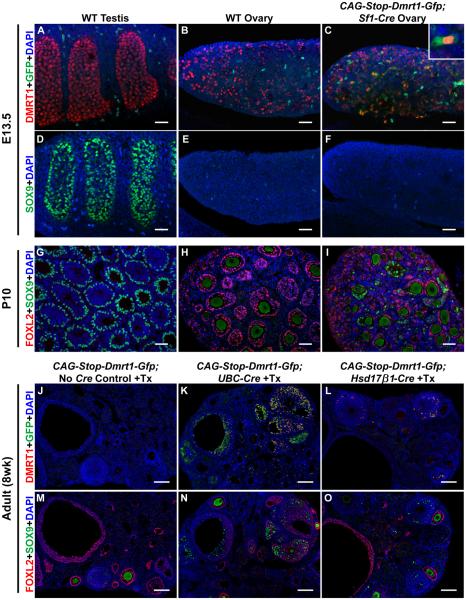
We next asked whether DMRT1 expression induces male sex determination or sexual transdifferentiation. Although Sf1-Cre is active by the time of sex determination (about E11-E12) [13], transgenic XX animals were born female, with ovaries containing oocytes in diplotene arrest (Figure S2A-D), suggesting fetal gonads were functionally female. By E13.5 CAG-Stop-Dmrt1-Gfp;Sf1-Cre ovaries expressed ectopic DMRT1 (Figure 2A-C), but SOX9 was not detectable until about postnatal day 10 (P10) (Figure 2D-I). We conclude that fetal DMRT1 expression causes transdifferentiation rather than primary sex reversal and fetal granulosa cells are refractory to SOX9 activation by DMRT1.
Figure 2. DMRT1 expression triggers postnatal granulosa cell transdifferentiation.
(A-C) Activation of CAG-Dmrt1-Gfp in the fetal gonad. Confocal images of whole mount IF on E13.5 gonads showing normal expression of DMRT1 in testis (A) and ovarian germ cells (B) and activation of CAG-Dmrt1-Gfp in ovarian somatic cells (C) as indicated by cytoplasmic GFP (example is shown in higher magnification inset). Dispersed green cells lacking DMRT1 in wild type gonads are autofluorescent cells of unknown type. (D-F) SOX9 expression in the fetal gonad. IF showing that SOX9 is strongly expressed in pre-Sertoli cells of wild type testes at E13.5 (D) but is not detected in wild type fetal ovaries (E) or CAG-Stop-Dmrt1-Gfp;Sf1-Cre transgenic ovaries (F). (G-I) Postnatal expression of SOX9 and FOXL2. IF showing that wild type testes at P10 express SOX9 and not FOXL2 (G), wild type ovaries express FOXL2 and not SOX9 (H), and CAG-Stop-Dmrt1-Gfp;Sf1-Cre transgenic ovaries have cells expressing each protein (I), indicating the onset of transdifferentiation. (J-O) Transdifferentiation in the adult ovary. (J-L) Control tamoxifen-injected ovaries from adults carrying CAG-Stop-Dmrt1-Gfp but lacking a Cre transgene do not express DMRT1 or GFP (J), but ovaries from animals also containing UBC-CreERT2 or Hsd17β1-Cre have cells expressing both proteins (K,L). (M-O) Somatic cells from control adult ovaries express FOXL2 but not SOX9 (L), whereas animals with UBC-CreERT2 or Hsd17β1-Cre have cells expressing each protein. (SOX9 IF in adult oocytes is thought to be a non-specific antibody artifact). Scale bars: 40 μm (A-I); 100 μm (J-O). See also figure S2.
In case ectopic DMRT1 was not present early enough to induce SOX9, we expressed DMRT1 before sex determination using Wt1-CreERT2, which is expressed by E9.5 [18]. Earlier activation did not masculinize the fetal gonad (Figure S2E-H) suggesting DMRT1 cannot cause primary sex reversal in the fetal ovary.
DMRT1 expression reprograms adult granulosa cells into Sertoli-like cells
Adult granulosa cells can transdifferentiate into Sertoli-like cells when Foxl2 is deleted, and one of the earliest changes is activation of DMRT1 [7]. We therefore asked whether activating DMRT1 alone is sufficient to induce transdifferentiation in the adult ovary. Using ubiquitously expressed UBC-CreERT2 [19] to activate DMRT1 silenced FOXL2 and resulted in SOX9-positive Sertoli-like cells (Figure 2J,K,M,N). Expression of DMRT1 mainly in granulosa cells of growing adult follicles using Hsd17b1-CreERT2 [20], also caused transdifferentiation, though less efficiently (Figure 2L,O)). We conclude that DMRT1 can trigger either juvenile or adult granulosa cells to transdifferentiate.
DMRT1 expression masculinizes the ovarian transcriptome
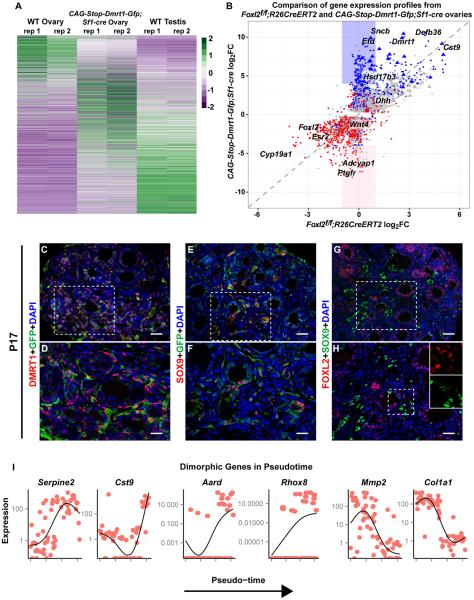
We used RNA-seq to compare transcriptomes of wild type adult testes and ovaries to DMRT1-expressing ovaries (CAG-Stop-Dmrt1-Gfp;Sf1-Cre). DMRT1-expressing ovaries had many differences in mRNA abundance relative to wild type ovaries (Figure 3A), with 2430 mRNAs overexpressed and 2078 underexpressed by two-fold or more (P < 0.05; DESeq2 Wald test; Table S1A). DMRT1-expressing ovaries had highly elevated expression of the testicular Leydig cell marker Insl3 (>16-fold) suggesting Leydig-like cells also may have been induced [21].
Figure 3. DMRT1 masculinizes the ovarian transcriptome.
(A) Heat map comparing mRNA expression in adult wild type testis and ovary with CAG--Dmrt1-Gfp;Sf1-Cre ovaries. Columns are from RNA-seq of two gonads (rep1, rep 2) of each genotype. Genes differentially expressed in wild type ovary and DMRT1-expressing ovary (>4-fold ; p<0.05, Table S1A) are shown in rows that are sorted based on high expression in the wild type ovary (top) to high expression in the testis (bottom). Each gene was normalized to a range of −2 (violet) to +2 (green). (B) Scatter plot comparing gene expression in adult Foxl2 conditionally mutant ovaries (data from [7]) and CAG-Stop-Dmrt1-Gfp;Sf1-Cre ovaries. Blue indicates mRNAs with 4-fold or greater expression in wild-type testis vs wild-type ovary and red indicates those with 4-fold or greater expression in wild-type ovary vs wild-type testis. Grey indicates mRNAs not differing significantly between testis and ovary. Identities of some of the most strongly affected mRNAs are indicated. Triangles denote X-linked genes and blue and pink boxes highlight mRNAs strongly up or down-regulated, respectively, in CAG-Dmrt1-Gfp expressing ovaries but not in Foxl2 mutant ovaries. (C-H) IF showing that P17 CAG-Stop-Dmrt1-Gfp;Sf1-Cre transgenic ovaries have a mix of GFP+ cells expressing DMRT1, SOX9, or FOXL2. Scale bars: 40 μm (C,E,G); 20 μm (D,F,H). (I) Expression levels (FPKM) of select mRNAs in single cells from P17 CAG-Stop-Dmrt1-Gfp;Sf1-Cre transgenic ovaries, ordered by pseudotime along the x-axis. See also Figure S3 and Table S1.
Foxl2 deletion and DMRT1 expression cause similar remodeling of the ovarian transcriptome
Foxl2 deletion or Dmrt1 activation triggers transdifferentiation in the ovary, suggesting that repressing DMRT1 is a primary means by which FOXL2 maintains ovarian cell fates. To examine this possibility we compared mRNA profiles of Foxl2 mutant versus DMRT1-expressing ovaries. We compared microarray data from Foxl2 conditional mutants [7] with RNA-seq data from CAG-Stop-Dmrt1-Gfp;Sf1-Cre ovaries, plotting mRNAs represented in both datasets (Figure 3B). Ovary- and testis-enriched genes are highlighted in red and blue respectively and constituted the majority of strongly-affected genes. Nearly all genes strongly affected by loss of Foxl2 were affected by gain of DMRT1 (Figure 3B, diagonal line), suggesting that ectopic DMRT1 can account for most of the Foxl2 mutant phenotype. Some mRNAs were significantly affected in CAG-Stop-Dmrt1-Gfp;Sf1-Cre ovaries but not in Foxl2 mutants (Figure 3B, blue and pink boxes). This difference may reflect the higher expression of DMRT1 in transgenic ovaries relative to Foxl2 mutant ovaries (Figure 3B).
One-fifth of highly differentially expressed genes (>32-fold) in DMRT1 expressing ovaries are X-linked (58/288, triangles in Figure 3B; also Figure S3A-D, Table S1B). As discussed below, this X-linkage is consistent with sexual antagonism.
In spermatogonia DMRT1 inhibits meiosis by suppressing retinoic acid (RA) signaling [22]. We found significant misexpression of RA-regulated genes [23] in transgenic ovaries (Table S1B), suggesting DMRT1 also may regulate RA-dependent gene expression in somatic transdifferentiation.
Single cell transcriptome profiling identifies candidate mediators of transdifferentiation
Analysis of intact gonads reveals the extent of transdifferentiation. However transdifferentiation initiates asynchronously, so whole gonad transcriptomes provide limited insights into the dynamics of the process. We therefore turned to single-cell transcriptomes. We used ovaries from CAG-Stop-Dmrt1-Gfp;Sf1-Cre females at 17 days, when GFP and DMRT1 are widely expressed (Figure 3C,D) and many cells express FOXL2, SOX9, or more rarely both (Figure 3E-H). We isolated live GFP-positive cells by fluorescence-activated cell sorting (FACS), and used the Fluidigm C1 system for capture and cDNA synthesis. We sequenced cDNA libraries from 68 live cells and used the Monocle dimension reduction algorithm [24] to place them into a “pseudo-temporal” transdifferentiation pathway. This method infers a sequence of gene expression accounting for the heterogeneity observed between cells. Ordering was based on expression of 32 abundant mRNAs selected because of differential expression in the adult testis vs. ovary RNA-seq and presence in at least half of profiled cells (SI). These mRNAs included Serpine2 and Cst9 (Figure 3I, left). We used a Bayesian probabilistic model (SCDE [25]) to identify genes differentially expressed between cells in the first third and last third of pseudotime. Many mRNAs not used for ordering showed significant variation from ovary-like to testis-like levels in single cells over pseudotime (e.g., Aard, corrected Z-score=1.22; Rhox8, corrected Z-score = 0.9, Figure 3I, middle), suggesting that ordering at least partially reflects actual temporal change. Single cell analysis also detected mRNAs varying in pseudotime but not detected by analysis of intact DMRT1-expressing ovaries (Mmp2 corrected Z-score = −1.0 and Col1a1 corrected Z-score=-2.4, Figure 3I, right). The ordering of cells in pseudotime may indicate similarity between transdifferentiation and fetal gonad development, as 56 of the 777 genes differentially expressed between the first and last third of pseudotime also exhibit sexually dimorphic expression in fetal gonads [26] (Table S1C). The mRNAs that differed over transdifferentiation pseudotime will be important subjects for future investigation.
DMRT1 can silence Foxl2 without SOX8 and SOX9
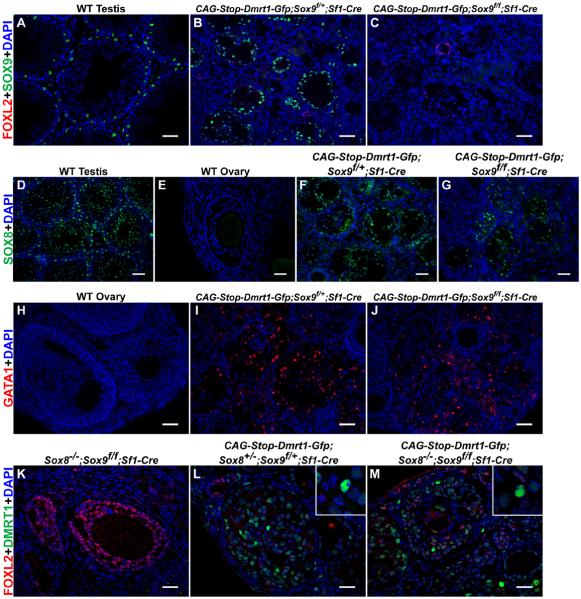
Cell fate maintenance involves antagonism between DMRT1 and FOXL2. DMRT1 can activate or repress transcription [27] and ChIP experiments suggest it directly represses Foxl2 [5]. DMRT1 also activates the male-determining gene Sox9, which helps maintain male fates [5, 28]. An important question is whether DMRT1 and FOXL2 antagonize each other directly or by competing to regulate SOX9. We therefore asked whether DMRT1 can silence Foxl2 in the absence of Sox9.
Adult ovaries with DMRT1 activated and Sox9 deleted using Sf1-Cre (Figure 4A-C) had many cells lacking FOXL2 and expressing Sertoli markers SOX8 (Figure 4D-G) and GATA1 (Figure 4H-J), suggesting DMRT1 can cause transdifferentiation without Sox9. However, Sox9 is partially redundant with Sox8 [29, 30], which was elevated in DMRT1-expressing ovaries (not shown). We therefore repeated the experiment in ovaries lacking Sox8 and Sox9. Again most DMRT1-positive cells had reduced or no FOXL2 (Figure 4K-M), indicating that DMRT1 does not require SOX9 or SOX8 to silence FOXL2.
Figure 4. DMRT1 can silence FOXL2 without SOX8 and SOX9.
(A-C) CAG-Stop-Dmrt1-Gfp can silence FOXL2 in Sox9 conditional mutant ovaries. IF showing that SOX9 is expressed in Sertoli cells of wild-type testes and in Sertoli-like cells of control conditional Sox9/+ DMRT1-expressing ovaries. FOXL2 is almost completely silenced in DMRT1-expressing ovaries conditionally deleted for one (B) or both (C) copies of Sox9 with Sf1-Cre. (D-G) CAG-Dmrt1-Gfp can activate SOX8 in SOX9 mutant granulosa cells. IF showing that SOX8 is expressed in Sertoli cells in wild-type adult testes (D) and is not detectable in wild type adult ovaries (E). CAG-Dmrt1-Gfp can activate SOX8 in ovaries conditionally deleted for one copy (F) or both copies (G) of Sox9 in somatic cells using Sf1-Cre. (H-J) CAG-Dmrt1-Gfp can activate the mature Sertoli cell marker GATA1 in Sox9 mutant granulosa cells. IF showing that GATA1 is not expressed in wild type adult ovaries (H) but is expressed in ovaries conditionally deleted for one (I) or two (J) copies of Sox9 in somatic cells using Sf1-Cre. (K-M) DMRT1 silences FOXL2 in granulosa cells lacking both Sox8 and Sox9. IF showing that Sox8;Sox9 double mutant ovaries have normal FOXL2 expression, normal morphology and lack DMRT1 (K). Activation of CAG-Dmrt1-Gfp in ovaries heterozygous for Sox8 and Sox9 (L) or homozygous mutant for both genes in somatic cells (M) can induce DMRT1 expression and silence FOXL2. Scale bars: 40 μm (A-D,F-J); 20 μm (E,K-M)
Discussion
We found that ovarian expression of DMRT1 alone can silence FOXL2 and trigger sexual transdifferentiation. Thus DMRT1 controls a regulatory network that can sexually reprogram differentiated cells in vivo. Ectopic DMRT1 activity can account for virtually all transcriptome changes resulting from deletion of Foxl2, suggesting that silencing Dmrt1, directly or indirectly, is the primary means by which FOXL2 blocks sexual transdifferentiation.
Specification of Sertoli or granulosa cells is the crucial first event in mammalian sex determination and involves activating the male-promoting gene Sox9 or, alternatively, a female-promoting Wnt/b-catenin pathway involving Wnt4 and Rspo1 [31]. DMRT1 is dispensable for sex determination in mice [15]. However XY humans hemizygous for DMRT1 can be born phenotypically female [32-34], so DMRT1 may play a role in human gonadal sex determination. We found that even in mice the presence or absence of DMRT1 can toggle a switch between the Sertoli and granulosa cell fates. While this switching occurs postnatally it nevertheless reveals that DMRT1 can control the choice between the two cell types that underlie mammalian sex. Human disorders of sex differentiation (DSD) have been presumed to result from incomplete gonadal differentiation but our findings suggest that some cases of DSD could involve transdifferentiation resulting from mutations causing later gain or loss of DMRT1.
The ability of ectopic DMRT1 expression to specify Sertoli cell fates also has potential significance for understanding evolution of genetic sex-determining mechanisms. In therian mammals, Y-linked Sry serves to activate Sox9 in the male gonad and Sox9 expression can determine sex even without Sry [30, 35-38]. Other vertebrates lack Sry and control gonadal sex by different means. DMRT1 regulates sex determination in an expanding group of non-mammalian vertebrates. In some species a DMRT1 homolog acts as a dominant Y-linked masculinizing gene (fish) [9], a dose-dependent Z-linked masculinizing gene (birds) [11, 12], or a dominant W-linked feminizing gene (amphibians) [10] (reviewed in [1, 39]). Molecular and genetic analysis suggests acquisition of these genetic sex determination mechanisms was tied to dominant gain-of-function, recessive loss-of-function, or dominant-negative Dmrt1 alleles, respectively [10-12, 40-42]. These new DMRT1 alleles may have triggered formation of new sex determination mechanisms or merely arisen coincident with them. Regardless, the ability of DMRT1 to toggle Sertoli/granulosa cell fate empirically supports models in which of loss- or gain-of-function mutations in Dmrt1 can elevate it into a sex-determining role [1, 39]. Such mutations would help promote the major changes between genetic sex determination mechanisms that are commonly observed among vertebrates [43, 44].
DMRT1, with other testicular transcription factors (SOX9, GATA4, NR5A1/SF1, WT1), can reprogram cultured iPS cells into Sertoli-like cells in vitro, but cannot reprogram them on its own [45]. In vivo, by contrast, DMRT1 can reprogram granulosa cells into Sertoli-like cells. This difference presumably reflects the greater similarity of granulosa cells to Sertoli cells. Indeed, although very different morphologically, both express NR5A1/SF1, GATA4, and WT1. Also, they arise from a common genital ridge precursor cell and thus may be separated by relatively small epigenetic barriers. Differences between in vivo and in vitro conditions may be important; however, DMRT1 cannot reprogram other cells to Sertoli cells in vivo. For example, pre-granulosa cells and cells of the blastocyst inner cell mass normally express DMRT1 without adopting Sertoli fates ([15] and unpublished). Moreover, broad expression of DMRT1 did not induce SOX9 expression or apparent Sertoli fates in extragonadal tissues (not shown).
X-linked genes were highly over-represented among those strongly activated by DMRT1 (Figures 3B and S2). X chromosomes are predicted to accumulate female-antagonistic regulators of sexual dimorphism - genes benefitting males but compromising females [46]. This is because rare male-advantageous recessive alleles on X get expressed much more frequently in males than females and thus are likely to be fixed even if they have a significant female fitness cost. There is perhaps no more direct sexual antagonism than during gonadal development, and our data suggest the X-linked targets of DMRT1 include genes with significant roles in testicular differentiation.
The ability of DMRT1 to induce cell fate reprogramming provides a window into transdifferentiation mechanisms but puzzles remain, for example why postnatal but not fetal granulosa cells can be reprogrammed. Here mammals differ from birds, where DMRT1 can induce SOX9 and male differentiation at the normal time of sex determination [12]. Resistance to fetal sex reversal in mice is also seen in estrogen receptor and Wnt/β-catenin pathway mutants [47, 48], and it has been suggested that granulosa cells must first undergo a germ cell-dependent maturation step [48]. We suggest that resistance to Sox9 induction in fetal pre-granulosa cells may provide a protective mechanism, as these cells normally express DMRT1 during sex determination: resisting inappropriate Sox9 activation by DMRT1 would ensure the fidelity of sex determination during a critical developmental interval. Such a mechanism also might contribute to the observed evolutionary stability of the mammalian sex determination mechanism.
Experimental Procedures: See Supplementary Information
Supplementary Material
Acknowledgements
We thank M. Wegner for SOX8 antibodies, F. Poulat for SOX9 antibodies, R. Behringer, E. Brown, E. Casanova, M. Lewandoski, K. Parker, G. Scherer, M. Treier and M. Wegner for mice, R. Behringer for advice, C. Corcoran for technical assistance, and M. Murphy for helpful discussions. This work was supported by the NIH (5 R01 GM59152 and 1 F32 GM106484), Minnesota Medical Foundation, University of Minnesota Medical School and College of Biological Sciences, and University of Minnesota Genomics Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Matson CK, Zarkower D. Sex and the singular DM domain: insights into sexual regulation, evolution and plasticity. Nat Rev Genet. 2012;13:163–174. doi: 10.1038/nrg3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raymond CS, Murphy MW, O'Sullivan MG, Bardwell VJ, Zarkower D. Dmrt1, a gene related to worm and fly sexual regulators, is required for mammalian testis differentiation. Genes Dev. 2000;14:2587–2595. doi: 10.1101/gad.834100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim S, Bardwell VJ, Zarkower D. Cell type-autonomous and non-autonomous requirements for Dmrt1 in postnatal testis differentiation. Dev Biol. 2007;307:314–327. doi: 10.1016/j.ydbio.2007.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matson CK, Murphy MW, Griswold MD, Yoshida S, Bardwell VJ, Zarkower D. The mammalian doublesex homolog DMRT1 is a transcriptional gatekeeper that controls the mitosis versus meiosis decision in male germ cells. Dev Cell. 2010;19:612–624. doi: 10.1016/j.devcel.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matson CK, Murphy MW, Sarver AL, Griswold MD, Bardwell VJ, Zarkower D. DMRT1 prevents female reprogramming in the postnatal mammalian testis. Nature. 2011;476:101–104. doi: 10.1038/nature10239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koopman P, Gubbay J, Vivian N, Goodfellow P, Lovell-Badge R. Male development of chromosomally female mice transgenic for Sry. Nature. 1991;351:117–121. doi: 10.1038/351117a0. [DOI] [PubMed] [Google Scholar]
- 7.Uhlenhaut NH, Jakob S, Anlag K, Eisenberger T, Sekido R, Kress J, Treier AC, Klugmann C, Klasen C, Holter NI, et al. Somatic sex reprogramming of adult ovaries to testes by FOXL2 ablation. Cell. 2009;139:1130–1142. doi: 10.1016/j.cell.2009.11.021. [DOI] [PubMed] [Google Scholar]
- 8.Herpin A, Schartl M. Sex determination: switch and suppress. Current biology : CB. 2011;21:R656–659. doi: 10.1016/j.cub.2011.07.026. [DOI] [PubMed] [Google Scholar]
- 9.Matsuda M, Shinomiya A, Kinoshita M, Suzuki A, Kobayashi T, Paul-Prasanth B, Lau EL, Hamaguchi S, Sakaizumi M, Nagahama Y. DMY gene induces male development in genetically female (XX) medaka fish. Proc Natl Acad Sci U S A. 2007;104:3865–3870. doi: 10.1073/pnas.0611707104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoshimoto S, Okada E, Umemoto H, Tamura K, Uno Y, Nishida-Umehara C, Matsuda Y, Takamatsu N, Shiba T, Ito M. A W-linked DM-domain gene, DM-W, participates in primary ovary development in Xenopus laevis. Proc Natl Acad Sci U S A. 2008;105:2469–2474. doi: 10.1073/pnas.0712244105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith CA, Roeszler KN, Ohnesorg T, Cummins DM, Farlie PG, Doran TJ, Sinclair AH. The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature. 2009;461:267–271. doi: 10.1038/nature08298. [DOI] [PubMed] [Google Scholar]
- 12.Lambeth LS, Raymond CS, Roeszler KN, Kuroiwa A, Nakata T, Zarkower D, Smith CA. Over-expression of DMRT1 induces the male pathway in embryonic chicken gonads. Dev Biol. 2014;389:160–172. doi: 10.1016/j.ydbio.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bingham NC, Verma-Kurvari S, Parada LF, Parker KL. Development of a steroidogenic factor 1/Cre transgenic mouse line. Genesis. 2006;44:419–424. doi: 10.1002/dvg.20231. [DOI] [PubMed] [Google Scholar]
- 14.Raymond CS, Kettlewell JR, Hirsch B, Bardwell VJ, Zarkower D. Expression of Dmrt1 in the genital ridge of mouse and chicken embryos suggests a role in vertebrate sexual development. Dev Biol. 1999;215:208–220. doi: 10.1006/dbio.1999.9461. [DOI] [PubMed] [Google Scholar]
- 15.Raymond CS, Murphy MW, O'Sullivan MG, Bardwell VJ, Zarkower D. Dmrt1, a gene related to worm and fly sexual regulators, is required for mammalian testis differentiation. Genes Dev. 2000;14:2587–2595. doi: 10.1101/gad.834100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lei N, Hornbaker KI, Rice DA, Karpova T, Agbor VA, Heckert LL. Sex-specific differences in mouse DMRT1 expression are both cell type- and stage-dependent during gonad development. Biol Reprod. 2007;77:466–475. doi: 10.1095/biolreprod.106.058784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Grandi A, Calvari V, Bertini V, Bulfone A, Peverali G, Camerino G, Borsani G, Guioli S. The expression pattern of a mouse doublesex-related gene is consistent with a role in gonadal differentiation. Mech Dev. 2000;90:323–326. doi: 10.1016/s0925-4773(99)00282-8. [DOI] [PubMed] [Google Scholar]
- 18.Hu YC, Okumura LM, Page DC. Gata4 is required for formation of the genital ridge in mice. PLoS Genet. 2013;9:e1003629. doi: 10.1371/journal.pgen.1003629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruzankina Y, Pinzon-Guzman C, Asare A, Ong T, Pontano L, Cotsarelis G, Zediak VP, Velez M, Bhandoola A, Brown EJ. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell. 2007;1:113–126. doi: 10.1016/j.stem.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grabner B, Blaas L, Musteanu M, Hoffmann T, Birbach A, Eferl R, Casanova E. A mouse tool for conditional mutagenesis in ovarian granulosa cells. Genesis. 2010;48:612–617. doi: 10.1002/dvg.20664. [DOI] [PubMed] [Google Scholar]
- 21.Ivell R, Wade JD, Anand-Ivell R. INSL3 as a biomarker of Leydig cell functionality. Biol Reprod. 2013;88:147. doi: 10.1095/biolreprod.113.108969. [DOI] [PubMed] [Google Scholar]
- 22.Matson CK, Murphy MW, Griswold MD, Yoshida S, Bardwell VJ, Zarkower D. The mammalian doublesex homolog DMRT1 is a transcriptional gatekeeper that controls the mitosis versus meiosis decision in male germ cells. Developmental cell. 2010;19:612–624. doi: 10.1016/j.devcel.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Balmer JE, Blomhoff R. Gene expression regulation by retinoic acid. Journal of lipid research. 2002;43:1773–1808. doi: 10.1194/jlr.r100015-jlr200. [DOI] [PubMed] [Google Scholar]
- 24.Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nature biotechnology. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kharchenko PV, Silberstein L, Scadden DT. Bayesian approach to single-cell differential expression analysis. Nature methods. 2014;11:740–742. doi: 10.1038/nmeth.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Munger SC, Natarajan A, Looger LL, Ohler U, Capel B. Fine time course expression analysis identifies cascades of activation and repression and maps a putative regulator of mammalian sex determination. PLoS Genet. 2013;9:e1003630. doi: 10.1371/journal.pgen.1003630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murphy MW, Sarver AL, Rice D, Hatzi K, Ye K, Melnick A, Heckert LL, Zarkower D, Bardwell VJ. Genome-wide analysis of DNA binding and transcriptional regulation by the mammalian Doublesex homolog DMRT1 in the juvenile testis. Proc Natl Acad Sci U S A. 2010;107:13360–13365. doi: 10.1073/pnas.1006243107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minkina A, Matson CK, Lindeman RE, Ghyselinck NB, Bardwell VJ, Zarkower D. DMRT1 protects male gonadal cells from retinoid-dependent sexual transdifferentiation. Dev Cell. 2014;29:511–520. doi: 10.1016/j.devcel.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barrionuevo F, Georg I, Scherthan H, Lecureuil C, Guillou F, Wegner M, Scherer G. Testis cord differentiation after the sex determination stage is independent of Sox9 but fails in the combined absence of Sox9 and Sox8. Dev Biol. 2009;327:301–312. doi: 10.1016/j.ydbio.2008.12.011. [DOI] [PubMed] [Google Scholar]
- 30.Chaboissier MC, Kobayashi A, Vidal VI, Lutzkendorf S, van de Kant HJ, Wegner M, de Rooij DG, Behringer RR, Schedl A. Functional analysis of Sox8 and Sox9 during sex determination in the mouse. Development. 2004;131:1891–1901. doi: 10.1242/dev.01087. [DOI] [PubMed] [Google Scholar]
- 31.Maatouk DM, DiNapoli L, Alvers A, Parker KL, Taketo MM, Capel B. Stabilization of beta-catenin in XY gonads causes male-to-female sex-reversal. Hum Mol Genet. 2008;17:2949–2955. doi: 10.1093/hmg/ddn193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raymond CS, Shamu CE, Shen MM, Seifert KJ, Hirsch B, Hodgkin J, Zarkower D. Evidence for evolutionary conservation of sex-determining genes. Nature. 1998;391:691–695. doi: 10.1038/35618. [DOI] [PubMed] [Google Scholar]
- 33.Veitia R, Nunes M, Brauner R, Doco-Fenzy M, Joanny-Flinois O, Jaubert F, Lortat-Jacob S, Fellous M, McElreavey K. Deletions of distal 9p associated with 46,XY male to female sex reversal: definition of the breakpoints at 9p23.3-p24.1. Genomics. 1997;41:271–274. doi: 10.1006/geno.1997.4648. [DOI] [PubMed] [Google Scholar]
- 34.Tannour-Louet M, Han S, Corbett ST, Louet JF, Yatsenko S, Meyers L, Shaw CA, Kang SH, Cheung SW, Lamb DJ. Identification of de novo copy number variants associated with human disorders of sexual development. PloS one. 2010;5:e15392. doi: 10.1371/journal.pone.0015392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lavery R, Lardenois A, Ranc-Jianmotamedi F, Pauper E, Gregoire EP, Vigier C, Moreilhon C, Primig M, Chaboissier MC. XY Sox9 embryonic loss-of-function mouse mutants show complete sex reversal and produce partially fertile XY oocytes. Dev Biol. 2011;354:111–122. doi: 10.1016/j.ydbio.2011.03.029. [DOI] [PubMed] [Google Scholar]
- 36.Qin Y, Kong LK, Poirier C, Truong C, Overbeek PA, Bishop CE. Long-range activation of Sox9 in Odd Sex (Ods) mice. Hum Mol Genet. 2004;13:1213–1218. doi: 10.1093/hmg/ddh141. [DOI] [PubMed] [Google Scholar]
- 37.Vidal VP, Chaboissier MC, de Rooij DG, Schedl A. Sox9 induces testis development in XX transgenic mice. Nat Genet. 2001;28:216–217. doi: 10.1038/90046. [DOI] [PubMed] [Google Scholar]
- 38.Jakob S, Lovell-Badge R. Sex determination and the control of Sox9 expression in mammals. FEBS J. 2011;278:1002–1009. doi: 10.1111/j.1742-4658.2011.08029.x. [DOI] [PubMed] [Google Scholar]
- 39.Kopp A. Dmrt genes in the development and evolution of sexual dimorphism. Trends in genetics : TIG. 2012;28:175–184. doi: 10.1016/j.tig.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsuda M, Nagahama Y, Shinomiya A, Sato T, Matsuda C, Kobayashi T, Morrey CE, Shibata N, Asakawa S, Shimizu N, et al. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature. 2002;417:559–563. doi: 10.1038/nature751. [DOI] [PubMed] [Google Scholar]
- 41.Nanda I, Kondo M, Hornung U, Asakawa S, Winkler C, Shimizu A, Shan Z, Haaf T, Shimizu N, Shima A, et al. A duplicated copy of DMRT1 in the sex-determining region of the Y chromosome of the medaka, Oryzias latipes. Proc Natl Acad Sci U S A. 2002;99:11778–11783. doi: 10.1073/pnas.182314699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen S, Zhang G, Shao C, Huang Q, Liu G, Zhang P, Song W, An N, Chalopin D, Volff JN, et al. Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat Genet. 2014;46:253–260. doi: 10.1038/ng.2890. [DOI] [PubMed] [Google Scholar]
- 43.Bull JJ. Evolution of sex determining mechanisms. Benjamin/Cummings Pub. Co.; Menlo Park, Calif.: 1983. Advanced Book Program. [Google Scholar]
- 44.Bachtrog D, Mank JE, Peichel CL, Kirkpatrick M, Otto SP, Ashman TL, Hahn MW, Kitano J, Mayrose I, Ming R, et al. Sex determination: why so many ways of doing it? PLoS Biol. 2014;12:e1001899. doi: 10.1371/journal.pbio.1001899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Buganim Y, Itskovich E, Hu YC, Cheng AW, Ganz K, Sarkar S, Fu D, Welstead GG, Page DC, Jaenisch R. Direct reprogramming of fibroblasts into embryonic Sertoli-like cells by defined factors. Cell Stem Cell. 2012;11:373–386. doi: 10.1016/j.stem.2012.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rice WR. Sex-Chromosomes and the Evolution of Sexual Dimorphism. Evolution. 1984;38:735–742. doi: 10.1111/j.1558-5646.1984.tb00346.x. [DOI] [PubMed] [Google Scholar]
- 47.Couse JF, Hewitt SC, Bunch DO, Sar M, Walker VR, Davis BJ, Korach KS. Postnatal sex reversal of the ovaries in mice lacking estrogen receptors alpha and beta. Science. 1999;286:2328–2331. doi: 10.1126/science.286.5448.2328. [DOI] [PubMed] [Google Scholar]
- 48.Maatouk DM, Mork L, Chassot AA, Chaboissier MC, Capel B. Disruption of mitotic arrest precedes precocious differentiation and transdifferentiation of pregranulosa cells in the perinatal Wnt4 mutant ovary. Dev Biol. 2013;383:295–306. doi: 10.1016/j.ydbio.2013.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.