Abstract
Although the pattern of BAP1 inactivation in ocular melanoma specimens and in the BAP1 cutaneous/ocular melanoma (CM/OM) predisposition syndrome suggests a tumor suppressor function, the specific role of this gene in the pathogenesis of cutaneous melanoma is not fully understood. We thus set out to characterize BAP1 in cutaneous melanoma and discovered an unexpected pro-survival effect of this protein. Tissue and cell lines analysis showed that BAP1 expression was maintained, rather than lost, in primary melanomas compared to nevi and normal skin. Genetic depletion of BAP1 in melanoma cells reduced proliferation and colony forming capability, induced apoptosis and inhibited melanoma tumor growth in vivo. On the molecular level, suppression of BAP1 led to a concomitant drop in the protein levels of survivin a member of anti-apoptotic proteins and a known mediator of melanoma survival. Restoration of survivin in melanoma cells partially rescued the growth-retarding effects of BAP1 loss. In contrast to melanoma cells, stable overexpression of BAP1 into immortalized but non-transformed melanocytes did suppress proliferation and reduce survivin. Taken together, these studies demonstrate that BAP1 may play a growth-sustaining role in melanoma cells, but that its impact on ubiquitination underpins a complex physiology which is context and cell dependent.
Keywords: cutaneous melanoma, BAP1, cancer genetics
Introduction
The BRCA1-associated protein 1 (BAP1) gene is a recent addition to the canon of high-risk melanoma susceptibility genes. Many groups have described germline BAP1 alterations in families predisposed to cutaneous and ocular melanoma among other malignancies (Abdel-Rahman et al., 2011; Harbour et al., 2010; Njauw et al., 2012; Wiesner et al., 2011). To date, both heritable and acquired mutations in BAP1 have been deleterious with loss-of-heterozygosity described in melanoma tumor specimens (Njauw et al., 2012; Wiesner et al., 2011). This genetic pattern suggests a tumor suppressor function for the BAP1 protein. However, unlike ocular melanomas, cutaneous melanomas do not commonly harbor BAP1 mutations outside of the familial context (Harbour et al., 2010; Njauw et al., 2012; Wiesner et al., 2011). More importantly, the role of BAP1 in the pathogenesis of cutaneous melanoma has yet to be fully characterized.
Functional analyses of BAP1 have yielded conflicting results. Early experiments found that BAP1 enhanced BRCA1-mediated inhibition of breast cancer cell growth (Jensen et al., 1998). BAP1 has also been reported to be inactivated in about 15% of renal cell carcinomas (RCC) and has been shown to be growth suppressive in functional assays (Pena-Llopis et al., 2012). With the recognition that BAP1 directly interacts with HCF-1, there were hints that BAP1 could also play a positive proliferative role (Machida et al., 2009). These findings were further substantiated by genome-wide RNA suppression screens which established BAP1 as an essential proliferation and cell cycle gene (Kittler et al., 2007; Schlabach et al., 2008). Even among uveal melanomas, where the prevalence of deleterious mutations remains the highest, recent studies suggest that BAP1 is not functionally suppressive and that the biology of this deubiquitinase is highly complex (Matatall et al., 2013).
We thus set out to better understand the role of BAP1 in cutaneous melanoma- one of the signature cancers in the BAP1 tumor predisposition syndrome. Using a combination of genetic and functional studies, we provide evidence that BAP1 may be an important growth sustaining protein that is linked to the regulation of survivin- a known anti-apoptotic factor in melanoma (McKenzie and Grossman, 2012).
Results
Basal expression level of BAP1 in melanomas
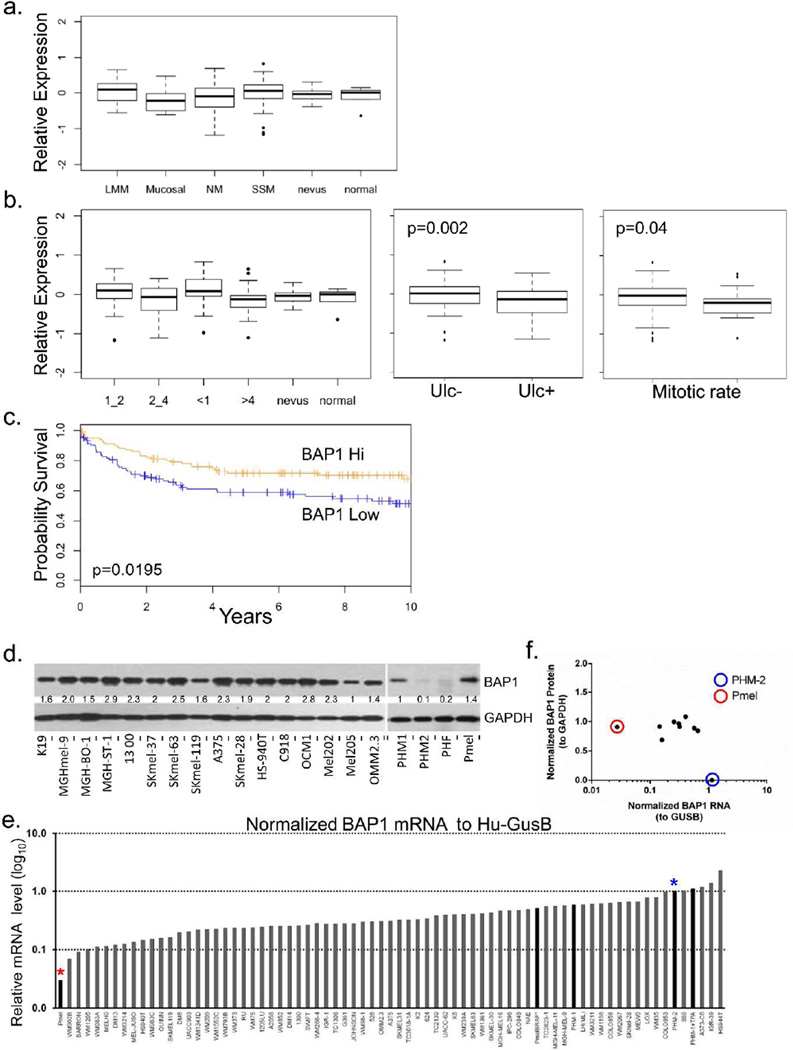
We first examined the mRNA levels of BAP1 in primary melanocytic tumors using a set of 223 melanomas, 11 nevi and 6 samples of normal tissue (Harbst et al., 2012) and found no significant difference in relative BAP1 expression (Fig 1a) though levels may be slightly lower in mucosal melanomas. When stratified by tumor features (Fig 1b), BAP1 expression was increased in thin melanomas (<1mm) and decreased in thick melanomas (>4mm) compared to nevi and normal tissue; however, the difference across all groups was non-significant. Lower BAP1 expression was observed in primary melanomas with ulceration (p=0. 002) and higher mitotic rate (≥6 mitoses vs <6 mitoses; p=0.04). Finally, when supervised by outcome, primary melanomas with low BAP1 expression exhibited a worse prognosis than those with high BAP1 levels (Fig 1c) although these results were likely confounded by its association with ulceration and high mitotic rate. We then examined BAP1 levels in an independent set of metastatic melanomas (Gene Expression Omnibus GDS1375) and identified a significant increase in the levels of BAP1 among metastatic cases compared to normal tissue and nevi (Fig S1a).
Figure 1. BAP1 expression in primary cutaneous melanomas and melanoma lines.
(a) BAP1 expression in histological types of melanoma. (b) BAP1 expression melanomas stratified by Breslow thickness, BAP1 expression in relation to ulceration and mitotic rate. (c) Survival differences between BAP1 high (>mean gene expression across the melanomas) and BAP1 low tumors (≤ mean gene expression across the melanomas). (d) Western blot analysis showing relative protein levels of BAP1 in a collection of 16 melanoma cell lines, primary human melanocytes (PHMs), primary human fibroblasts (PHF), and an immortalized non-transformed human melanocytes (Pmel). (e) Ranked BAP1 RNA levels (normalized to human GUSB) in a panel of melanoma lines and PHM-1, PHM-2 and Pmel. (f) Correlation between normalized BAP1 protein (to GAPDH) and normalized BAP1 RNA (to Hu-GUSB). Pmel (red circle and red asterisk) shows low RNA levels but strong protein expression while PHM-2 (blue circle and blue asterisk) shows high RNA levels with a near absence of detectable protein.
The levels of BAP1 in proliferating melanoma cells were then determined. We first assessed BAP1 protein levels in 16 melanoma lines, 2 independent primary human melanocyte lines (PHM1 and PHM2), a primary human fibroblast (PHF) line and an immortalized, but non-transformed, melanocyte line (Pmel) (Fig 1d). Except for PHM-2 and the PHF, there was a robust protein expression in all samples. RNA expression was then examined in a broader panel of lines by qPCR (Fig 1e) and was found to be well sustained in all the melanoma lines. Interestingly, protein levels were generally constant despite a gradient of RNA expression (Fig 1e). One of the primary melanocyte lines, PHM-2, showed negligible protein expression (Fig 1f) even with relatively high RNA content (blue asterisk, blue circle) while the immortalized melanocyte line, Pmel, demonstrated strong BAP1 protein levels despite low RNA expression (red asterisk, red circle). These findings indicate that melanoma cells, but not necessarily primary cells, preserve the amount of intracellular BAP1. To replicate these findings and to put melanoma in the context of other proliferating cancer cells, we used the Cancer Cell Line Encyclopedia (N=1,036 cancer lines) and found that the median expression of BAP1 in 61 melanoma lines ranked 9th among the 37 cancer cell types (Fig S1b) and was significantly higher compared to all non-melanoma lines (Fig S1b; 7.59 vs. 7.33; p<0.001, Student T test). Thus, BAP1 appears central to the survival of melanoma cells, though its role in primary cells is less clear.
BAP1 depletion abrogates melanoma growth
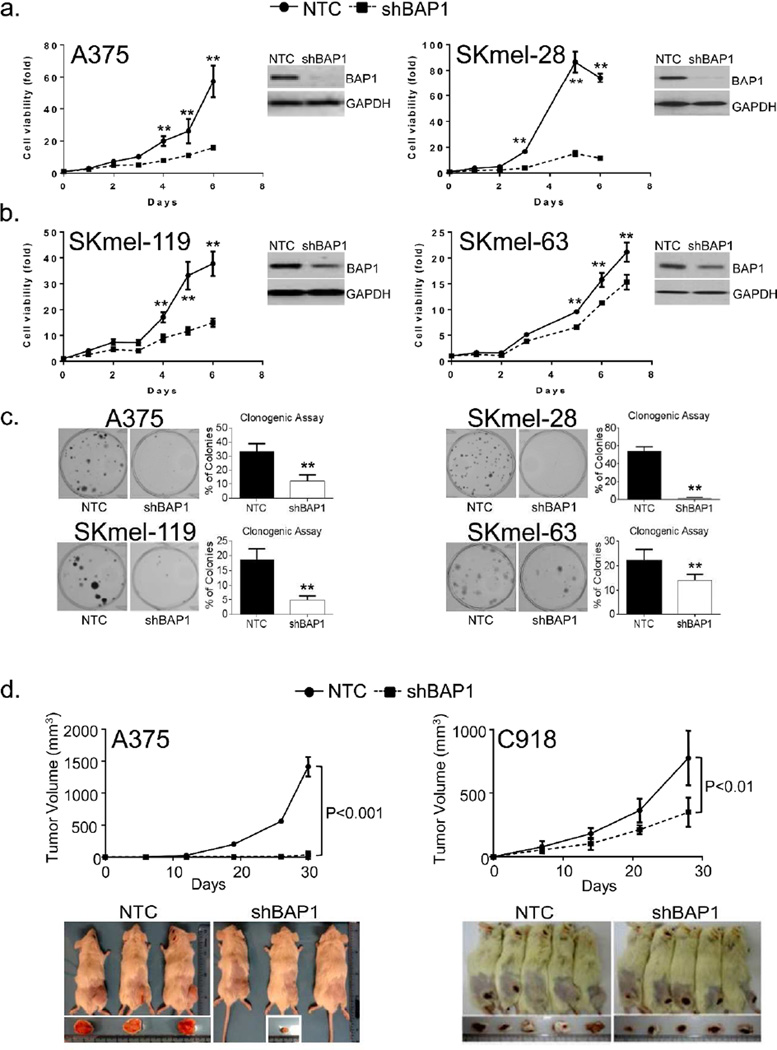
To test the hypothesis that BAP1 contributes to the melanoma cell maintenance, we examined the effects of BAP1 depletion on the growth kinetics of melanoma cells. As shown in Figure 2, depletion of BAP1 in two BRAF(V600E)-mutant lines (A375 and SKmel-28, Fig 2a) and two NRAS(Q61R)-mutant lines (SKmel-119 and SKmel-63, Fig 2b) led to dramatic reductions in melanoma proliferation. These were also accompanied by significant decreases in the colony forming capacity of the BAP1-depleted cells (Fig 2c). Lastly, we investigated the effects of BAP1 loss on tumor growth in vivo using two sh(BAP1)-suppressed lines (A375 and C918). As shown in Figure 2d, BAP1 depletion diminished the tumorigenicity of melanoma xenografts in immunocompromised mice. Examination of the tumor specimens demonstrated less Ki67 and more TUNEL staining in the two sh(BAP1) tumors compared to the control tumors (Fig S2). This suggests that BAP1 loss can produce similar anti-proliferative and pro-apoptotic effects in vivo as found in vitro.
Figure 2. BAP1 depletion leads to melanoma growth suppression.
The shRNA-mediated suppression of BAP1 results in reduced in vitro proliferation in 4 cutaneous melanoma cell lines. (a) A375 and SKmel-28 harbor BRAF (V600E) mutations while, (b) SKmel-119 and SKmel-63 contain NRAS (Q61R) and NRAS (Q61K) mutations, respectively. Error bars represent SEM from triplicate samples. (c) Loss of BAP1 is also associated with diminished colony forming capability. Error bars represent ±S.D., from at least 3 independent experiments. *p<0.05, **p<0.01 by student T test. (d) Effects of BAP1 depletion on tumor growth in a xenograft model. One million A375(NTC), A375(shBAP1), C918(NTC), and C918(shBAP1) cells were implanted subcutaneously in NOD-SCID-IL2G-null mice with matrigel in a 1:1 ratio and observed over the indicated time period. With A375 (cutaneous melanoma), 3 animals were used in each arm while for C918 (uveal melanoma), 5 animals were used in each arm. Error bars represent ± S.D., in tumor volume. *p<0.05, **p<0.01 by student T test.
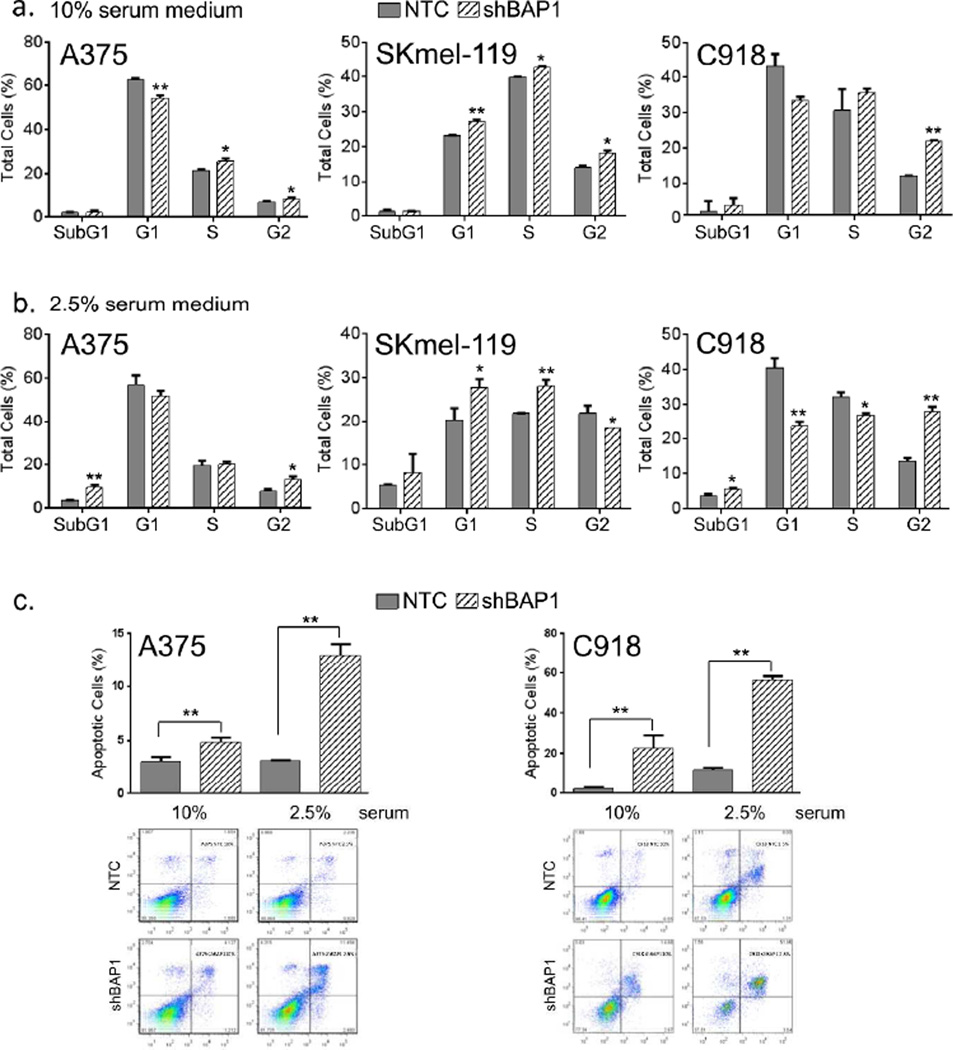
For the cell cycle and apoptosis assays (Fig 3), A375 [BRAF(V600E)], SKmel-119 [NRAS(Q61R)] and C918 (uveal melanoma) cells were used. In 10% serum (Fig 3a), suppression of BAP1 led to G2/M arrest with modest G1/S effects. However, in 2.5% serum (Fig 3b), there were appreciable increases in apoptosis as measured by subG1 fractionation; this apoptotic response was independently confirmed using FITC-Annexin staining (Fig 3c). These results indicate that both cell cycle arrest and apoptosis may contribute, in part, to the proliferative shut down observed with BAP1 suppression in vitro.
Figure 3. BAP1 depletion causes cell cycle arrest and apoptosis.
Analysis of cell cycle progression and apoptosis were performed at 6 or 7th day following shRNA-mediated BAP1 silencing. Cell cycle analysis in (a) 10% and (b) 2.5% serum medium using control cells (gray shading) and shBAP1 knockdown cells (hatched shading). Error bars represent SEM from triplicate samples and 3 experimental replicates are shown. A375 and SKmel-119 are cutaneous melanomas and C918 is an ocular melanoma used for comparison; *p<0.05, **p<0.01 by student T test. (c) FITC-Annexin staining of cultured control and BAP1-depleted cells in both 10% and 2.5% serum. DNA fragmentation may underestimate the level of apoptosis, especially if the total DNA content is elevated from G2/M arrest. Error bars represent SEM from triplicate samples both replicates shown; **p<0.01 by student T test.
BAP1 regulates survivin
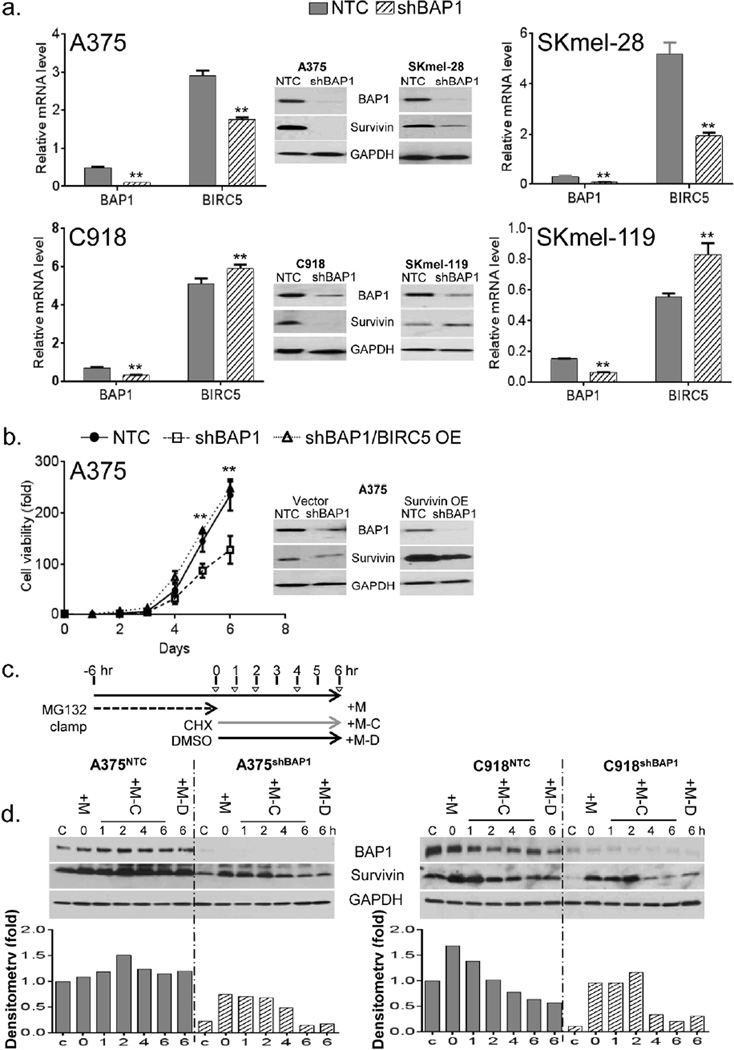
We set out to identify potential mediators of BAP1-dependent survival. Since one of the IAP family members- BIRC5, or survivin- has been implicated both as a viability factor in melanoma and a target of BAP1 regulation in U2OS cells (Yu et al., 2010), we hypothesized that BAP1 depletion may have an effect on survivin levels. As shown in Figure 4a, BAP1 suppression led to a dramatic loss of survivin protein levels in 3 of 4 cell lines examined (A375, SKmel-28 and C918); qPCR confirmed a concomitant loss of BIRC5 mRNA in A375 and SKmel-28, but not in C918, melanoma cells. In one cell line (SKmel-119), there was a rise in BIRC5 mRNA and survivin protein levels. Thus, BAP1 effects are cell context dependent and may exist at both transcriptional and post-transcriptional levels. In order to determine if the loss of survivin was a necessary component of the growth inhibition observed with BAP1 depletion, we overexpressed survivin in the context of BAP1 loss. As shown in Figure 4b, there was a consistent rescue of cell growth in the A375(shBAP1) cells when survivin was overexpressed (Fig 4b; A375 (shBAP1/BIRC5-OE) vs A375 (shBAP1)). In addition, if BIRC5 is under BAP1 regulation, then one might expect a direct relationship between the levels of the two genes. As shown in Figure S3a, there was indeed a significant correlation between BIRC5 and BAP1 mRNA levels in the CCLE melanoma dataset (p=0.0026; N=62). Moreover, BIRC5 loss was similarly associated with a worsened outcome in the 223 primary melanomas (Fig S3b; p=0.0009). Taken together, these findings suggest that BAP1 loss appears to have a negative impact on survival factors and that down regulation of species such as survivin may be one component of the BAP1 effect.
Figure 4. BAP1 suppression is associated with survivin depletion.
(a) RNA and protein levels of BIRC5 (survivin) upon BAP1 suppression in 4 melanoma lines. There is evidence of near total survivin loss at the protein level in A375, SKmel-28 and C918 but not SKmel-119. Error bars represent SEM from triplicate samples; *p<0.05 **p<0.01 by student T test. (b) Survivin over-expression (OE) in A375 cells led to the rescue of the in vitro growth arrest induced by shBAP1. (c) Schematic diagram of experimental design for ubiquitination assay. Indicated cell lines were incubated with MG132 (25 QM) for 6 hours, followed by removal of MG132 and the addition of cycloheximide (CHX, 25 Qg/ml) for the designated time intervals. “C” represents control cells that were exposed to neither MG132 nor CHX. “+M” indicates cells that were exposed to only MG132 and not CHX; for these cells, lysate was collected at time=0. “+M-C” represents cells that were exposed to MG132 and then switched to CHX; lysates were collected at 1, 2, 4 and 6 hrs post-CHX switch. “+M-D” represent cells that were exposed to MG132 and then DMSO control for 6 hours. (d) The effect of BAP1 depletion on survivin and GAPDH protein levels as measured by western blotting. If BAP1 directly deubiquitinated survivin, then survivin decay should be accelerated with BAP1 depletion. Even though the absolute levels of BAP1 appear to be lower in the shBAP1 lines, survivin degradation appears similar. Error bars represent SEM of triplicate samples. The experiments were performed 3 times with similar results; **p<0.01 by student T test.
To examine more specific role of ubiquitination, we performed a proteasome/protein clamp experiment using A375 and C918 cell lines, both of which showed a loss of survivin protein levels with BAP1 depletion and yet contrasting effects on the BIRC5 mRNA level. Briefly, control and BAP1 depleted A375 and C918 cells (i.e. A375 (NTC) /A375 (shBAP1) and C918 (NTC) /C918 (shBAP1)) were subjected to 6-hours pre-treatment with proteasome inhibitor MG132 (25 QM) in order to enrich for ubiquitinated proteins. As shown in Figure S4, total protein ubiquitination was dramatically increased by the MG132 treatment. The cells were then removed from MG132 exposure and treated with the protein synthesis inhibitor, cycloheximide (CHX 25 Qg/ml; transfer time=0). The levels and decay of BAP1 regulated protein(s) were then analyzed at 1, 2, 4 and 6 hours after CHX exposure using western blotting (Fig 4d). In untreated A375 and C918 cells, the loss of BAP1 was associated with a dramatic reduction in survivin levels (Fig 4d, control “C” lanes), consistent with the prior analysis. MG132 treatment alone (“+M” vs “C”) led to increased survivin suggesting that proteasomal degradation modulates survivin levels. Both A375(shBAP1) and C918(shBAP1) lines exhibited appropriate survivin decay upon release of MG132 and inhibition of new protein synthesis by CHX despite having attenuated survivin protein levels at baseline. Since BAP1 depletion led to diminished resting levels of survivin but did not abrogate survivin decay upon MG132/CHX treatment, BAP1 likely participates in the homeostatic maintenance of survivin levels through other mechanisms beyond simple deubiquitination. Furthermore, co-immunoprecipitation experiments were done, but did not demonstrate direct protein-protein interaction between BAP1 and survivin (data not shown).
BAP1 suppresses non-transformed melanocytes
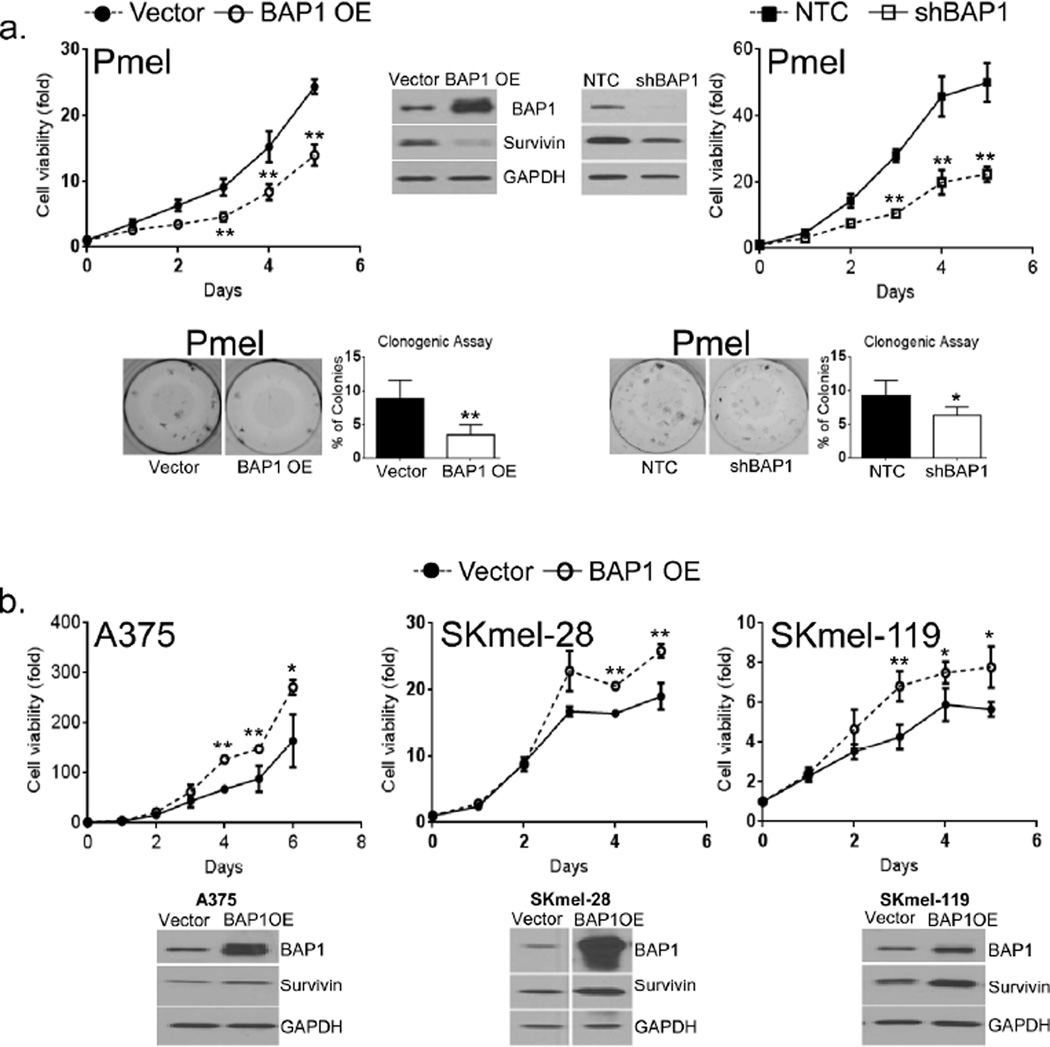
Our findings so far support a role for BAP1 in melanoma cell viability. However, BAP1’s effects may be different in non-transformed cells. We thus chose to restore BAP1 in cells that appear to exist in relative BAP1 deficiency. Studies were first initiated in PHM-2, which lacked BAP1 protein, but neither control vector nor BAP1-expressing cells could be sufficiently recovered for experimentation. We next turned to an immortalized but non-transformed melanocyte line, Pmel, which had lower RNA levels of BAP1 than any of the melanoma cells. As shown in Figure 5a, stable introduction of BAP1 into Pmel cells (i.e. Pmel(BAP1)) reduced proliferation, colony forming capacity and survivin levels. Interestingly, Pmel(shBAP1) cells also exhibited a drop in survivin levels, albeit less dramatic than in Pmel(BAP1), and a similar growth-suppressed phenotype (Fig 5a). On the other hand, ectopic expression of BAP1 in three distinct melanoma cell lines led to a modest increase in survivin and proliferation (Fig 5b).
Figure 5. BAP1 overexpression in Pmel and melanomas.
(a) Effects of BAP1 overexpression (BAP1 OE) and depletion (shBAP1) on Pmel proliferation, survivin levels and colony forming capacity. Both BAP1 elevation and suppression in Pmel cells were associated with a decrease in colony formation, proliferation and levels of survivin. (b) Overexpression of BAP1 in 3 melanoma lines had more stimulatory effects on proliferation and survivin. The experiments were performed 3 times with similar results; *p<0.05, **p<0.01 by student T test.
Discussion
Although inactivating germline mutations of BAP1 have been described in families prone to cutaneous and ocular melanoma (Abdel-Rahman et al., 2011; Njauw et al., 2012; Wiesner et al., 2011), the role of BAP1 in the pathogenesis of cutaneous melanoma outside of the familial context is not fully known. Somatic mutations of BAP1 occur in only 5% of sporadic primary melanomas (Wiesner et al., 2011) suggesting that sustained BAP1 activity may play a critical function in tumor maintenance. In this study, we discovered that melanoma cells appear to defend their BAP1 protein levels despite a wide range of BAP1 RNA content. Furthermore, BAP1 appears to play a role in regulating levels of survival genes, such as BIRC5/survivin, and in maintaining the growth of at least a subset of melanomas. BAP1 mRNA levels do not appear to be significantly diminished in primary melanomas compared to nevi and normal skin, though lower expression levels do correlate with adverse features, such as mitotic rate and ulceration, and a worse outcome. Among metastatic cases, BAP1 mRNA levels appear to be higher than those in found in nevi and normal skin. Using immunohistochemistry, Murali et al. also recently reported that BAP1 loss occurred in only 5% of primary melanomas and was associated with worsened survival (Murali et al., 2013)- a finding that resonates with our results. Interestingly, reduced BAP1 was observed more often in the desmoplastic variant of melanomas where there is greater fibroblastic investment. Though preliminary, primary dermal fibroblasts (PHF) in our survey (Fig 1) expressed no BAP1 and may therefore account for the lower observed staining. Overall, BAP1 is rarely mutated in primary CM and its expression is generally sustained, which is in sharp contrast to primary uveal melanomas where over half of the cases exhibit a total absence of BAP1 (Shah et al., 2013). Thus, the biologic function of BAP1 may be different between the two types of melanoma cells.
A second, somewhat unexpected finding is that BAP1 appears to be a survival factor in melanoma cells. BAP1 depletion diminished proliferation, and enhanced apoptosis, both in vitro and in vivo, and phenotypically inhibited tumor growth in mice xenografts. These functional studies may explain the consistent high level expression in nearly all melanoma lines. Others have also observed growth arrest in non-melanoma cancer cells (Dalinghaus et al., 1991; Kittler et al., 2007; Machida et al., 2009, Testa JR et al., 2011) with loss of BAP1. However, BAP1 is not universally required for cellular viability as some primary human melanocytes and primary human fibroblasts do not appear to express appreciable levels of BAP1 protein. Paradoxically, recent findings have confirmed that BAP1 also promotes growth and differentiation even in uveal melanomas where loss of BAP1 expression is a common event (Matatall et al., 2013). Germline Bap1 deletion in mice is lethal during embryogenesis indicating that fetal growth and development requires this gene (Dey et al., 2012). Thus, the specific phenotype that results from BAP1 disruption may be dictated by the balance of growth-restricting and growth-promoting effects that are regulated by BAP1. This context-specific effect is supported by our early results with the Pmel cells, suggesting that BAP1 may harbor altogether different properties in non-transformed cells.
Our studies reveal that BAP1 also impinges on the intracellular metabolism of survivin though the precise mechanistic details are still under investigation. In a microarray profiling experiment, survivin mRNA was also reported to be significantly decreased in U2OS cells that have been depleted of BAP1 (Yu et al., 2010). Gene expression studies, however, may not fully uncover all BAP1 targets. In our hands, C918 uveal melanoma cells show sustained, if not slightly higher, BIRC5 mRNA levels upon BAP1 suppression despite a near absence of BAP1 protein levels (Fig 4a). Thus, it appears that BAP1 could regulate target proteins at both transcriptional and the post-transcriptional levels (Carbone et al., 2013; Bott M et al; Yu et al., 2010).
There are several limitations to our study. Since BAP1 is differentially expressed in primary melanocytes, the role of BAP1 in non-malignant cells may be functionally distinct from that in their malignant counterparts. This could resolve the apparent paradox between germline predisposition and cellular dependence. Studies are underway to examine the cooperative effects of BAP1 loss, or gain, with other oncogenic alleles in human melanocytes. In addition, the type and position of the reported mutations may bear on the suppressive or oncogenic nature of the altered BAP1 proteins. We are in the process of performing a more refined genotype-phenotype correlation on the cellular level. Lastly, the biology of BAP1 will likely require a full annotation of BAP1 targets, both as direct enzymatic substrates and as an indirect co-regulators of transcription.
In summary, we provide early, but provocative evidence that BAP1 plays an important growth-sustaining role in many cutaneous melanomas and that some of the growth retarding effects of BAP1 loss may be mediated by viability factors such as survivin. These studies also suggest that BAP1 could play a different role in non-malignant cells and highlight the complex nature of BAP1 biology genetic and functional attributes.
Material and Methods
Cellular proliferation, colony formation, cell cycle and apoptosis assays
These assays were performed as previously described by our laboratory (Ji et al., 2013; Ji et al., 2012; Udayakumar et al., 2011). The PrestoBlueR cell viability assay was performed as per manufacturer’s instruction (Life Technologies). After trypsinization and trypan Blue staining (Sigma-Aldrich), viable cells were plated at a density of 103 cells per well in black 96-well plates. Cell proliferation assays were performed at 24-hour intervals for up to 6 days. Briefly, PrestoBlue® dye was added, at 1/10 of the culture medium volume and incubated for 10 mins at 37°C. The reaction was stopped with 15 µl/well of 1× SDS (Life Technologies), and the fluorescence was measured at 540 nm excitation and 590 nm emission using a microplate reader (SpectraMaxplus 1311, Sunnyvale, CA). Raw fluorescence values were subtracted from the background of the No-cell control wells for each experimental well. All experiments were performed at least three times in triplicates under each condition.
For the colony formation assay, shRNA-BAP1 and NTC transduced melanoma cells were plated at 100–200 cells/ml/well into 12-well plates and kept in a humidified CO2 incubator at 37oC for 15–20 days (Ji et al., 2013; Ji et al., 2012; Udayakumar et al., 2011). Cells were then washed with cold 1× PBS and fixed in 100% methanol for 30 min at room temperature and stained with crystal violet 0.5% W/V for 30 min. The stained colonies were counted under the stereo microscope and compared with a non-targeted control. Colony-forming capacity (CFC) or plating efficiency (PE) was expressed as a ratio of the number of colonies consisting of ≥50 cells, to the number of cells seeded.
Cells were processed for cell cycle and apoptosis experiments concurrently. Cell cycle analyses were performed to evaluate the distribution of cells in various cell cycle phases (subG1, G1, S, and G2/M) by measuring the DNA content of nuclei labeled with propidium iodide (Life Technologies). Briefly, BAP1 depleted and control A375, SKmel-119, and C918 viable cells were plated at 0.3×106 cells/well in 6-well tissue culture plates and incubated for 6–7 days at 37°C in 5% CO2. At 6–7 days post lentiviral infection, the cells were trypsinized then fixed with ice-cold ethanol final 70% (v/v) at −20°C overnight. Cells were centrifuged and washed twice with cold 1× PBS and re-suspended in 0.5ml of solution (20 µg/ml of PI and 200 µg/ml of RNase in 0.1%Triton X-100 in phosphate buffered saline (PBS) (RNAse: Becton Dickinson, San Jose, CA, USA) and incubated at room temperature in the dark for 30 min. Samples were then subjected to FACS (BD FACS Calibur flow cytometer, BD Biosciences). Using the FlowJo 7.6.5 software, the Watson model was used to calculate the percentages of cells in various cell cycle phases. All experiments were performed at least three times in triplicate.
For apoptosis, subG1 fractionation of cells with reduced DNA content was determined at the time of cell cycle analysis. In addition, apoptosis of shRNA(BAP1) and NTC-transduced melanoma cells (grown in complete (10%) or reduced (2.5%) serum media) were determined using Alexa Fluor 488 annexin-V conjugate detection kit as per manufacturer’s instructions (Life Technologies). Briefly, melanoma cells (A375 and C918) were plated (0.3 × 106 cells/well/ml) into 6-well plates and incubated in a 5% CO2 incubator at 37° C for 6–7 days after transduction. Cells were then trypsinized, centrifuged at 300g for 5 minutes and stained with Alexa Fluor 488 annexin-V and PI for 15 min at room temperature in the dark. 10,000 events were analyzed for each sample on a flow cytometer FACS Verse (Becton Dickinson), and results were analyzed using FlowJo 7.6.5 software. The cell cycle and apoptosis assays were performed in parallel and in triplicates for each condition.
Xenograft tumor growth assay
Nod-SCID-gamma (NSG) mice (Jackson Laboratory, Bar Harbor, ME) were injected subcutaneously with NTC-transduced or shRNA (BAP1) transduced A375 or C918 cells. The cells were mixed with matrigel in 1:1 ratio (1×106 cells/mice in three to five mice per group). Animal body weights and tumor development were monitored and dimensions were measured by a Mitutoyo caliper (MSC, Melville, NY) once to twice per week. Tumor volume was calculated using mm3= length × width2 × 0.5. Animals were maintained in well-ventilated animal facility and tested in accordance with the MGH Animal Care and Use Committee guidelines. Data were expressed as mean ± SEM. Tumor histology was confirmed through hematoxylin/eosin staining of formalin-fixed tissue.
Animal Material
The mice experiments were performed in accordance with a protocol approved by the Institutional Animal Care and Use Committee (IACUC) of MGH.
Patient Material
The experiments on patient specimens were approved by the local ethics committee of the Lund University, Lund, Sweden (Katja H et al., 2012).
Statistical analysis
Data from different experiments were represented as means ± standard deviation (SD) from at least three independent experiments. To analyze cell viability linear regression analysis was performed using GraphPad Prism 6.0 (GraphPad Software, La Jolla, CA). Significance was established at p<0.05, as customary.
Supplementary Material
Acknowledgements
This work was supported in part by the generous donors of the Massachusetts General Hospital, the American Skin Association (to H.T.) and the Swedish Cancer Society and Swedish Research Council (both to G.J.). Mentorship during the performance of this research was supported by an NIH K24 CA149202 award (to H.T.).
Footnotes
Conflict of Interest
None of the authors have a conflict of interest related to the content of this manuscript.
References
- Abdel-Rahman MH, Pilarski R, Cebulla CM, et al. Germline BAP1 mutation predisposes to uveal melanoma, lung adenocarcinoma, meningioma, and other cancers. J Med Genet. 2011 doi: 10.1136/jmedgenet-2011-100156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bott M, Brevet M, Taylor BS, et al. The nuclear deubiquitinase BAP1 is commonly inactivated by somatic mutations and 3p21.1 losses in malignant pleural mesothelioma. Nat Genet. 2011 Jun 5;43(7):668–672. doi: 10.1038/ng.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone M, Yang H, Pass HI, et al. BAP1 and cancer. Nature reviews Cancer. 2013;13:153–159. doi: 10.1038/nrc3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalinghaus M, Rudolph CD, Rudolph AM. Effects of maternal fasting on hepatic gluconeogenesis and glucose metabolism in fetal lambs. Journal of developmental physiology. 1991;16:267–275. [PubMed] [Google Scholar]
- Dey A, Seshasayee D, Noubade R, et al. Loss of the tumor suppressor BAP1 causes myeloid transformation. Science. 2012;337:1541–1546. doi: 10.1126/science.1221711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garraway LA, Widlund HR, Rubin MA, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- Harbour JW, Onken MD, Roberson ED, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330:1410–1413. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbst K, Staaf J, Lauss M, et al. Molecular profiling reveals low- and high-grade forms of primary melanoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:4026–4036. doi: 10.1158/1078-0432.CCR-12-0343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen DE, Proctor M, Marquis ST, et al. BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene. 1998;16:1097–1112. doi: 10.1038/sj.onc.1201861. [DOI] [PubMed] [Google Scholar]
- Ji Z, Kumar R, Taylor M, et al. Vemurafenib synergizes with nutlin-3 to deplete survivin and suppresses melanoma viability and tumor growth. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19:4383–4391. doi: 10.1158/1078-0432.CCR-13-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Njauw CN, Taylor M, et al. p53 rescue through HDM2 antagonism suppresses melanoma growth and potentiates MEK inhibition. The Journal of investigative dermatology. 2012;132:356–364. doi: 10.1038/jid.2011.313. [DOI] [PubMed] [Google Scholar]
- Katja H, Johan S, Martin L, et al. Molecular Profiling Reveals Low- and High-Grade Forms of Primary Melanoma. Clin Cancer Res. 2012 Aug 1;18(15):4026–4036. doi: 10.1158/1078-0432.CCR-12-0343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittler R, Pelletier L, Heninger AK, et al. Genome-scale RNAi profiling of cell division in human tissue culture cells. Nature cell biology. 2007;9:1401–1412. doi: 10.1038/ncb1659. [DOI] [PubMed] [Google Scholar]
- Machida YJ, Machida Y, Vashisht AA, et al. The deubiquitinating enzyme BAP1 regulates cell growth via interaction with HCF-1. The Journal of biological chemistry. 2009;284:34179–34188. doi: 10.1074/jbc.M109.046755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matatall KA, Agapova OA, Onken MD, et al. BAP1 deficiency causes loss of melanocytic cell identity in uveal melanoma. BMC cancer. 2013;13:371. doi: 10.1186/1471-2407-13-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie JA, Grossman D. Role of the apoptotic and mitotic regulator survivin in melanoma. Anticancer Res. 2012;32:397–404. [PubMed] [Google Scholar]
- Murali R, Wilmott JS, Jakrot V, et al. BAP1 expression in cutaneous melanoma: a pilot study. Pathology. 2013;45:606–609. doi: 10.1097/PAT.0b013e3283653818. [DOI] [PubMed] [Google Scholar]
- Njauw CN, Kim I, Piris A, et al. Germline BAP1 Inactivation Is Preferentially Associated with Metastatic Ocular Melanoma and Cutaneous-Ocular Melanoma Families. PloS one. 2012;7:e35295. doi: 10.1371/journal.pone.0035295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena-Llopis S, Vega-Rubin-de-Celis S, Liao A, et al. BAP1 loss defines a new class of renal cell carcinoma. Nat Genet. 2012;44:751–759. doi: 10.1038/ng.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlabach MR, Luo J, Solimini NL, et al. Cancer proliferation gene discovery through functional genomics. Science. 2008;319:620–624. doi: 10.1126/science.1149200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah AA, Bourne TD, Murali R. BAP1 protein loss by immunohistochemistry: a potentially useful tool for prognostic prediction in patients with uveal melanoma. Pathology. 2013;45:651–656. doi: 10.1097/PAT.0000000000000002. [DOI] [PubMed] [Google Scholar]
- Testa JR, Cheung M, Pei J, et al. Germline BAP1 mutations predispose to malignant mesothelioma. Nat Genet. 2011 Aug 28;43(10):1022–1025. doi: 10.1038/ng.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao H, Zhang X, Fowlkes K, et al. Relative reciprocity of NRAS and PTEN/MMAC1 alterations in cutaneous melanoma cell lines. Cancer research. 2000;60:1800–1804. [PubMed] [Google Scholar]
- Udayakumar D, Zhang G, Ji Z, et al. Epha2 is a critical oncogene in melanoma. Oncogene. 2011;30:4921–4929. doi: 10.1038/onc.2011.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesner T, Obenauf AC, Murali R, et al. Germline mutations in BAP1 predispose to melanocytic tumors. Nat Genet. 2011;43:1018–1021. doi: 10.1038/ng.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Mashtalir N, Daou S, et al. The ubiquitin carboxyl hydrolase BAP1 forms a ternary complex with YY1 and HCF-1 and is a critical regulator of gene expression. Molecular and cellular biology. 2010;30:5071–5085. doi: 10.1128/MCB.00396-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.