Abstract
This study was conducted to investigate the association between ompK36 variants and international high-risk clones in Klebsiella pneumoniae. Fifty-nine sequence types (STs) divided into four ompK36 allele groups (groups A to D) were identified among 185 K. pneumoniae isolates. The major high-risk clones (29 ST11, 13 ST15, 7 ST37 and 1 ST147 isolates) were assigned to group A, while 6 STs (15 ST23, 2 ST65, 3 ST86, 1 ST163, 1 ST373 and 2 ST375 isolates) associated with pyogenic liver abscess were assigned to group C. The genotyping assay developed in this study may be useful for screening of epidemic STs.
Keywords: High-risk clones, Klebsiella pneumoniae, multilocus sequence typing, pyogenic liver abscess, sequence types
Antibiotic-resistant bacterial clones with high interhost transmission and colonization efficiency may play an important role in the dissemination of antimicrobial resistance [1]. Several international high-risk resistant clones of Klebsiella pneumoniae have been identified by multilocus sequence typing (MLST) [1]. The major epidemic clones belong to sequence type (ST) 11 (ST11), ST15, ST147 and ST258 [1–9]. ST11 and ST258 are included in clonal group 258 (CG258), while ST15 and ST147 belong to two different CGs. ST11, ST15 and ST37 are the most common STs among carbapenem-nonsusceptible K. pneumoniae isolates in Taiwan [6].
The OmpK36 porin contributes to carbapenem susceptibilities and virulence in K. pneumoniae[10–12]. The correlation of OmpK36 variants with specific STs has been described [13]. It is therefore likely that sequence heterogeneity of ompK36 is driven by selective pressures created by both host immunity and antimicrobial use. The present study was conducted to investigate the relationship between ompK36 types and high-risk clones in K. pneumoniae in Taiwan.
Firstly, MLST was performed as described by Diancourt et al. [14] using 35 previously characterized K. pneumoniae isolates with decreased carbapenem susceptibilities and intact ompK36 sequences (Table 1) [15–17]. Twelve STs were obtained, of which ST11 was the most common (16 isolates, 45.7%). Nucleotide sequences of ompK36 were compared using the GCG SeqWeb software (University of Wisconsin Biotechnology Center, Madison, WI, USA), and multiple sequence alignments and a phylogenetic tree were generated using the Pileup and Growtree programs, respectively. Thirteen ompK36 alleles were obtained; they were divided into four clusters, designated groups A, B, C and D (Fig. 1a). The major differences between the allele groups were located between nucleotide position 500 and 1000 in the coding region (Online Supplemental Fig. S1). Groups A, B, C and D accounted for 68.6%, 2.9%, 17.1% and 11.4% of the 35 isolates, and included 5, 1, 4 and 2 STs, respectively. All isolates of high-risk clones ST11, ST15 and ST37 belonged to group A. Pulsed-field gel electrophoresis was performed as described [16]. The result was consistent with the MLST analysis and showed genetic unrelatedness among isolates of different CGs with similar ompK36 sequences (Fig. 1b).
Table 1.
Results of ompK36 typing, β-lactamase typing and multilocus sequence typing for 81 Klebsiella pneumoniae isolates collected from the National Cheng Kung University in 2010
| ompK36 group (no. of isolates) | Extended-spectrum β-lactamases or AmpC | ST (no. of isolates) |
|---|---|---|
| A (14) | DHA-1 | ST11 (1) |
| DHA-1 | ST15 (1) | |
| SHV, DHA-1 | ST15 (1) | |
| CTX-M-15 | ST37 (1) | |
| SHV | ST147 (1) | |
| SHV | ST152 (1) | |
| SHV | ST1440 (2) | |
| Negative | ST15 (1), ST661 (1), ST950 (1), ST1536 (1), untypeable (2) | |
| B (9) | CTX-M-15 | ST711 (1) |
| ST361 (1), ST605 (1), ST929 (1), ST1180 (1), untypeable (4) | ||
| C (39) | SHV, DHA-1 | ST1 (1) |
| SHV, DHA-1 | ST39 (1) | |
| SHV | ST39 (1) | |
| SHV | ST48 (1) | |
| CTX-M-3 | Untypeable (1) | |
| Negative | ST23 (13), ST25 (1), ST35 (1), ST39 (1), ST43 (1), ST65 (2), ST86 (3), ST373 (1), ST375 (2), ST660 (1), ST1049 (2), untypeable (6) | |
| D (17) | SHV | ST133 (2) |
| SHV, DHA-1 | ST196 (1) | |
| Negative | ST36 (2), ST107 (1), ST133 (1), ST268 (2), ST298 (1), ST397 (1), ST420 (1), ST528 (1), ST776 (1), ST873 (1), ST1027 (1), ST1590 (1), untypeable (2) | |
| Untypeable (2) | Negative | Untypeable (2) |
ST, sequence type.
Fig. 1.
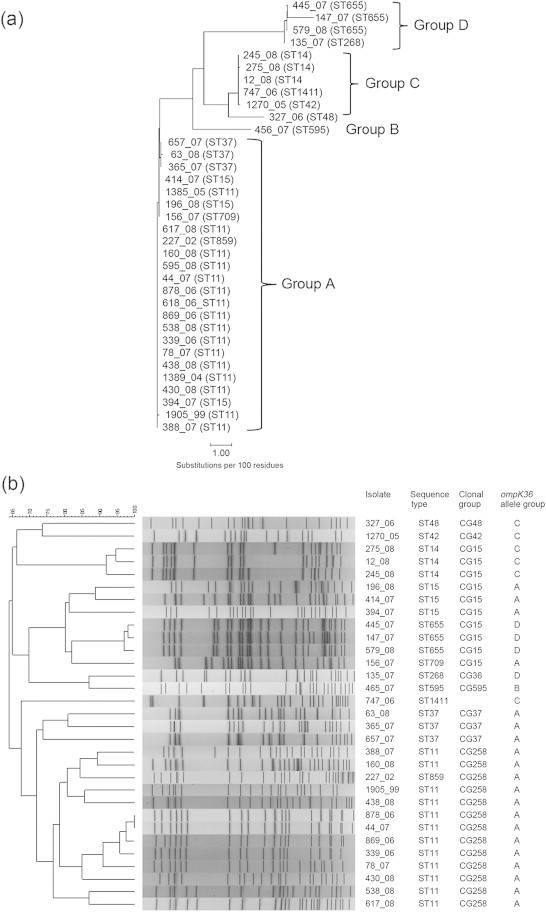
Phylogenetic analysis of 35 previously characterized K. pneumoniae isolates with decreased carbapenem susceptibilities. (a) Phylogenetic tree based on nucleotide sequences of ompK36 genes. The unrooted dendrogram was obtained with Growtree with uncorrected distances and neighbor joining. (b) Pulsed-field gel electrophoresis of XbaI-digested genomic DNA. Four isolates were not typeable probably due to DNA lysis. The profiles were compared using the BioNumerics program (Bio-Rad Laboratories). A dendrogram was generated from the distance matrix by the unweighted pair-group method with arithmetic mean, and genetic relatedness was calculated based on the Dice coefficient.
A PCR-based method using ompK36-targeted group-specific primer pairs was then developed, and 69 previously characterized extended-spectrum β-lactamase (ESBL)-producing and/or AmpC-producing K. pneumoniae isolates from 7 medical centres were examined [18]. The primer sequences and amplicon sizes are as follows: 5′-GAAGGCGCTCTGTCTCCTA-3′ and 5′-TGCCATCATAGATGTCATAGG-3′ for group A, 97 bp; 5′-CGGTCGTGGCGCGCAGAAA-3′ and 5′-GGTTGTTCTGATCGTCGGTA-3′ for group B, 125 bp; 5′-CAACAACGGTCGTGGTTGGA-3′ and 5′-CCCAGTGCCGGAACACTATT-3′ for group C, 144 bp; 5′-GAAGGTACTTCTCCGACCAA-3′ and 5′-AATCAGATTCTCCGTTGCCG-3′ for group D, 283 bp. Simplex PCR experiments were performed under the PCR conditions as follows: 1 cycle of 94°C for 10 min; 35 cycles of 94°C for 30 seconds, 60°C for 30 seconds and 72°C for 60 seconds, with a final extension at 72°C for 10 minutes. PCR conditions were the same for all primer pairs. Of the 69 isolates, 35 (50.7%), 6 (8.7%), 23 (33.3%) and 5 (7.2%) isolates were assigned to groups A, B, C and D, respectively. Twelve ST11, 7 ST15, 3 ST37, 1 ST273, 3 ST395, 2 ST709 and 2 ST833 isolates were included in group A. The ST11, ST15, and ST37 isolates were collected from 5, 4 and 2 hospitals, respectively. Group B comprised 3 ST17, 1 ST414, 1 ST1465 and 1 ST1535 isolates. In group C, 10 STs (ST1, ST23, ST25, ST35, ST39, ST42, ST48, ST101, ST163 and ST416) were identified, of which each included 1 or 2 isolates. Group D comprised 1 ST321, 2 ST378, 1 ST528 and 1 ST655 isolates. Currently available alleles or STs were not obtained for 7 isolates.
The prevalence of each ompK36 group among 81 K. pneumoniae bloodstream isolates collected at the National Cheng Kung University Hospital in 2010 was determined by the PCR-based ompK36 typing tests. Phenotyping and genotyping of β-lactamases were performed as described previously [15–17]. Fourteen (17.3%), 9 (11.1%), 39 (48.1%) and 17 (21.0%) of the 81 isolates belonged to groups A, B, C and D, respectively, and 2 isolates could not be typed. The MLST analysis showed 41 STs and 13 unassignable types. The major international clones (ST11, ST15, ST37 and ST147) were uncommon (6 isolates, 7.4%). The virulent clone ST23 in group C was the most common ST (n = 13). Seventeen (21.0%) of the 81 isolates carried blaESBL, blaDHA-1 (6 isolates) or both (4 isolates), and groups A, B, C and D accounted for 8 (47.1%), 1 (5.9%), 5 (29.4%) and 3 (29.4%) of the 17 isolates, respectively.
In summary, a total of 59 currently available STs were identified, among which the major international high-risk clones (29 ST11, 13 ST15, 7 ST37 and 1 ST147 isolates) were grouped together by ompK36 typing. Six STs reported to be associated with pyogenic liver abscess [19,20] were detected, and of note, all isolates of these STs (15 ST23, 2 ST65, 3 ST86, 1 ST163, 1 ST373 and 2 ST375 isolates) were assigned to the other ompK36 allele group.
In conclusion, this study demonstrated the association of the major high-risk resistant clones and virulent STs with specific ompK36 allele groups. The association might result from the occurrences of convergent evolution driven by the selective pressure created by antimicrobial use and host factors. Active surveillance of the epidemic resistant clones followed by appropriate infection control measures is needed to prevent the spread of antimicrobial resistance, and the ompK36-targeted PCR method developed in this study may be useful for screening of epidemic clones. The major high-risk resistant clones were uncommon. The use of the PCR screening method should therefore be cost-effective despite the lack of specificity, and subsequent sequencing of one or two housekeeping genes for MLST should reduce the number of isolates for complete MLST analysis further. Further large-scale studies are needed to confirm the association identified by ompK36 typing and the usefulness of the typing method.
Conflict of interest
None declared.
Acknowledgements
This work was supported by grants NCKUH-10301001 (National Cheng Kung University Hospital, Taiwan) and NSC 101-2320-B-006-012-MY2 (Ministry of Science and Technology, Taiwan).
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- 1.Woodford N., Turton J.F., Livermore D.M. Multiresistant Gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol Rev. 2011;35:736–755. doi: 10.1111/j.1574-6976.2011.00268.x. [DOI] [PubMed] [Google Scholar]
- 2.Damjanova I., Tóth A., Pászti J. Expansion and countrywide dissemination of ST11, ST15 and ST147 ciprofloxacin-resistant CTX-M-15-type β-lactamase-producing Klebsiella pneumoniae epidemic clones in Hungary in 2005—the new ‘MRSAs’? J Antimicrob Chemother. 2008;62:978–985. doi: 10.1093/jac/dkn287. [DOI] [PubMed] [Google Scholar]
- 3.Samuelsen Ø., Toleman M.A., Hasseltvedt V. Molecular characterization of VIM-producing Klebsiella pneumoniae from Scandinavia reveals genetic relatedness with international clonal complexes encoding transferable multidrug resistance. Clin Microbiol Infect. 2011;17:1811–1816. doi: 10.1111/j.1469-0691.2011.03532.x. [DOI] [PubMed] [Google Scholar]
- 4.Breurec S., Guessennd N., Timinouni M. Klebsiella pneumoniae resistant to third-generation cephalosporins in five African and two Vietnamese major towns: multiclonal population structure with two major international clonal groups, CG15 and CG258. Clin Microbiol Infect. 2013;19:349–355. doi: 10.1111/j.1469-0691.2012.03805.x. [DOI] [PubMed] [Google Scholar]
- 5.Lee M.Y., Ko K.S., Kang C.I., Chung D.R., Peck K.R., Song J.H. High prevalence of CTX-M-15-producing Klebsiella pneumoniae isolates in Asian countries: diverse clones and clonal dissemination. Int J Antimicrob Agents. 2011;38:160–163. doi: 10.1016/j.ijantimicag.2011.03.020. [DOI] [PubMed] [Google Scholar]
- 6.Chiu S.K., Wu T.L., Chuang Y.C. National surveillance study on carbapenem non-susceptible Klebsiella pneumoniae in Taiwan: the emergence and rapid dissemination of KPC-2 carbapenemase. PLOS One. 2013;8:e69428. doi: 10.1371/journal.pone.0069428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qi Y., Wei Z., Ji S., Du X., Shen P., Yu Y. ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J Antimicrob Chemother. 2011;66:307–312. doi: 10.1093/jac/dkq431. [DOI] [PubMed] [Google Scholar]
- 8.Ko K.S., Lee J.Y., Baek J.Y. Predominance of an ST11 extended-spectrum β-lactamase-producing Klebsiella pneumoniae clone causing bacteraemia and urinary tract infections in Korea. J Med Microbiol. 2010;59:822–828. doi: 10.1099/jmm.0.018119-0. [DOI] [PubMed] [Google Scholar]
- 9.Illiaquer M., Caroff N., Bémer P. Occurrence and molecular characterization of Klebsiella pneumoniae ST37 clinical isolates producing plasmid-mediated AmpC recovered over a 3-year period. Diagn Microbiol Infect Dis. 2012;74:95–97. doi: 10.1016/j.diagmicrobio.2012.05.023. [DOI] [PubMed] [Google Scholar]
- 10.Clancy C.J., Hao B., Shields R.K. Doripenem, gentamicin, and colistin, alone and in combinations, against gentamicin-susceptible, KPC-producing Klebsiella pneumoniae strains with various ompK36 genotypes. Antimicrob Agents Chemother. 2014;58:3521–3525. doi: 10.1128/AAC.01949-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsai Y.K., Liou C.H., Fung C.P., Lin J.C., Siu L.K. Single or in combination antimicrobial resistance mechanisms of Klebsiella pneumoniae contribute to varied susceptibility to different carbapenems. PLoS One. 2013;8:e79640. doi: 10.1371/journal.pone.0079640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J.H., Siu L.K., Fung C.P. Contribution of outer membrane protein K36 to antimicrobial resistance and virulence in Klebsiella pneumoniae. J Antimicrob Chemother. 2010;65:986–990. doi: 10.1093/jac/dkq056. [DOI] [PubMed] [Google Scholar]
- 13.Papagiannitsis C.C., Giakkoupi P., Kotsakis S.D. OmpK35 and OmpK36 porin variants associated with specific sequence types of Klebsiella pneumoniae. J Chemother. 2013;25:250–254. doi: 10.1179/1973947813Y.0000000075. [DOI] [PubMed] [Google Scholar]
- 14.Diancourt L., Passet V., Verhoef J., Grimont P.A., Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol. 2005;43:4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee N.Y., Wu J.J., Lin S.H., Ko W.C., Tsai L.H., Yan J.J. Characterization of carbapenem-nonsusceptible Klebsiella pneumoniae bloodstream isolates at a Taiwanese hospital: clinical impacts of lowered breakpoints for carbapenems. Eur J Clin Microbiol Infect Dis. 2012;31:1941–1950. doi: 10.1007/s10096-011-1525-2. [DOI] [PubMed] [Google Scholar]
- 16.Wu J.J., Wang L.R., Liu Y.F., Chen H.M., Yan J.J. Prevalence and characteristics of ertapenem-resistant Klebsiella pneumoniae isolates in a Taiwanese university hospital. Microb Drug Resist. 2011;17:259–266. doi: 10.1089/mdr.2010.0115. [DOI] [PubMed] [Google Scholar]
- 17.Yan J.J., Lee N.Y., Chen H.M. Bloodstream infections caused by IMP-8-producing Enterobacteriaceae isolates: the need for clinical laboratory detection of metallo-β-lactamases? Eur J Clin Microbiol Infect Dis. 2013;32:345–352. doi: 10.1007/s10096-012-1748-x. [DOI] [PubMed] [Google Scholar]
- 18.Yan J.J., Hsueh P.R., Lu J.J. Extended-spectrum β-lactamases and plasmid-mediated AmpC enzymes among clinical isolates of Escherichia coli and Klebsiella pneumoniae from seven medical centers in Taiwan. Antimicrob Agents Chemother. 2006;50:1861–1864. doi: 10.1128/AAC.50.5.1861-1864.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siu L.K., Fung C.P., Chang F.Y. Molecular typing and virulence analysis of serotype K1 Klebsiella pneumoniae strains isolated from liver abscess patients and stool samples from noninfectious subjects in Hong Kong, Singapore, and Taiwan. J Clin Microbiol. 2011;49:3761–3765. doi: 10.1128/JCM.00977-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin J.C., Koh T.H., Lee N. Genotypes and virulence in serotype K2 Klebsiella pneumoniae from liver abscess and non-infectious carriers in Hong Kong, Singapore and Taiwan. Gut Pathog. 2014;6:21. doi: 10.1186/1757-4749-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


