Abstract
Variants associated with blood lipid levels may be population-specific. To identify low-frequency variants associated with this phenotype, population-specific reference panels may be used. Here we impute nine large Dutch biobanks (~35,000 samples) with the population-specific reference panel created by the Genome of the Netherlands Project and perform association testing with blood lipid levels. We report the discovery of five novel associations at four loci (P value <6.61 × 10−4), including a rare missense variant in ABCA6 (rs77542162, p.Cys1359Arg, frequency 0.034), which is predicted to be deleterious. The frequency of this ABCA6 variant is 3.65-fold increased in the Dutch and its effect (βLDL-C=0.135, βTC=0.140) is estimated to be very similar to those observed for single variants in well-known lipid genes, such as LDLR.
 Frequencies of rare variants fluctuate over populations, hampering
gene discovery. Here the authors use a population-specific reference panel, the Genome
of the Netherlands, to discover four novel loci involved in lipid metabolism, including
an exonic variant in ABCA6.
Frequencies of rare variants fluctuate over populations, hampering
gene discovery. Here the authors use a population-specific reference panel, the Genome
of the Netherlands, to discover four novel loci involved in lipid metabolism, including
an exonic variant in ABCA6.
Genome-wide association studies (GWAS) have identified a large number of loci associated with blood lipid levels and analysis suggest there are additional susceptibility loci that have not yet been discovered1,2,3. Despite the fact that rare functional variants are known to play a major role in lipid metabolism1,2,3, there has been limited success in finding such variants in population-based studies using next-generation sequencing. Even if the effect of these variants is expected to be larger than that of common variants, the sample size needed to detect these rare or low-frequency variants increases dramatically with variant rarity. As the frequency of rare variants may increase in certain populations because of drift and founder effects4, the power of searches for rare functional variants may improve by the use of reference sets specific to distinct populations. Such references allow for better quality imputation of rare variants especially those with increased frequency in the population of interest3,5,6. Previous studies have successfully detected rare variants by imputation into larger sets of individuals in isolated populations followed by association testing to detect variants associated with the trait of interest7,8,9.
Here we describe an imputation-based GWAS for circulating lipid levels using a custom-built reference panel for the Dutch population (Genome of the Netherlands, GoNL, http://www.nlgenome.nl/), in which the whole genomes of 250 parent–offspring trios were sequenced at ~13 × coverage5,6. Owing to the trio design, the phasing quality of the reference panel was better than that of the 1000 Genomes (1-kG) Phase 1 panel. In this study we show that using this population-specific reference panel we were able to identify five novel associations at four loci.
Results
Nine large Dutch epidemiological cohorts (comprising 36,000 samples in total) were imputed with the GoNL reference panel (~19.5 million single-nucleotide polymorphisms (SNPs)) on an identical protocol6,10. All cohorts conducted association analysis on the imputed variants assuming an additive genetic effect on high-density lipoprotein cholesterol (HDL-C), low-density lipoprotein cholesterol (LDL-C), total cholesterol (TC) and triglyceride (TG) levels (Methods, Supplementary Methods and Supplementary Table 1), and the results were meta-analysed. We used conditional analysis implemented in GCTA11 to identify variants associated independently with lipid levels.
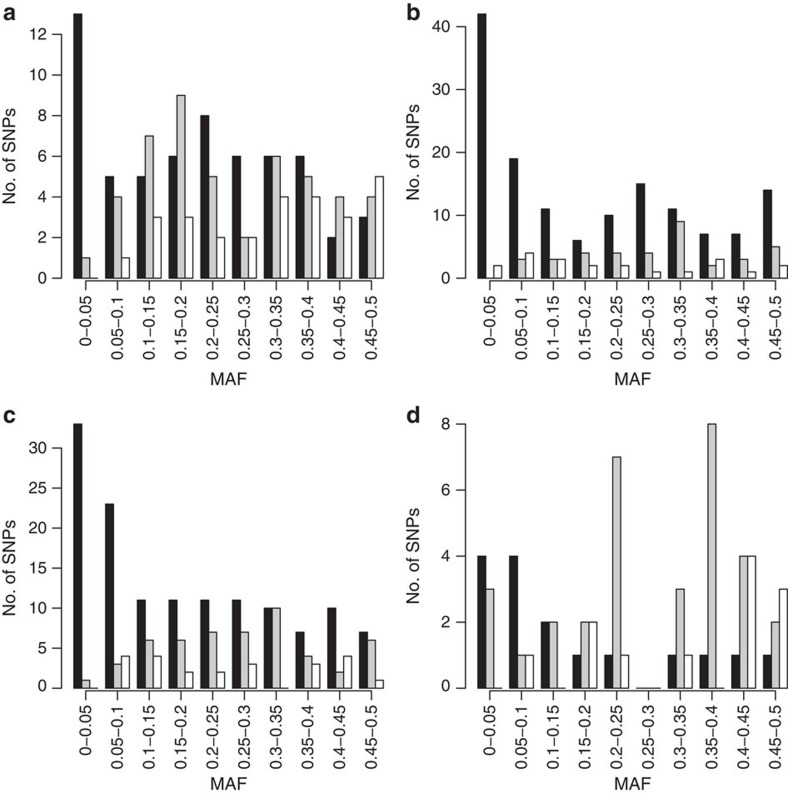
Both rare (minor allele frequency (MAF) <0.01), low (0.01<MAF<0.05) and common variants (MAF>0.05) were associated with HDL-C (N=60 variants), LDL-C (N=142 variants), TC (N=134 variants) and TG (N=16 variants) in both known and novel loci (Methods, Supplementary Tables 2–5 and Supplementary Fig. 1). In Fig. 1 we compare the allele frequencies that reach genome-wide significance in the GCTA analysis (P value <5 × 10−8) to those reported in refs 1, 2 (Fig. 1). The majority of the known HDL-C (31 of 45, 68.9%), LDL-C (24 of 34, 70.6%), TC (33 of 48, 68.6%) and TG (13 of 30, 43.3%) loci described in ref. 1 replicated at a P value <3.18 × 10−4 (Bonferroni correction based on 157 variants; Methods, Supplementary Figs 2 and 3 and Supplementary Tables 6–7). We also confirmed several of the HDL-C (6 of 27, 22.2%), LDL-C (7 of 21, 33.3%), TC (4 of 23, 17.4%) and TG (1 of 12, 8.3%) loci described in ref. 2 at a P value <6.02 × 10−4 (Bonferroni correction based on 83 variants) despite a sample size of ~20% of the other studies.
Figure 1. Identified variants for plasma lipid levels.
Distribution of the variants identified by conditional analysis implemented by GCTA to be independently associated with the lipid traits (a) HDL-C (60 variants), (b) LDL-C (142 variants), (c) TC (134 variants) and (d) TG (16 variants)) over MAF bins after meta-analysis of discovery cohorts (black). The histograms also include loci identified in ref. 1 (grey) and ref. 2 (white).
To identify novel loci associated with blood lipid levels, we selected from the list of variants identified by GCTA, those variants located more than 1 Mb away from previously identified loci. This resulted in six novel associations at five loci (Methods, Tables 1 and 2 and Supplementary Table 8). The five loci are not in linkage disequilibrium (LD) with previously described GWAS loci (Methods and Supplementary Table 9). Conditional analysis in the discovery cohorts showed that these new variants were independent from previously identified loci (Supplementary Table 10 and Supplementary Fig. 4). Of the five loci, three (rs149580368, rs77542162 and rs144984216) have an increased frequency in GoNL compared with 1-kG (Phase 1 integrated release v3, April 2012, all ancestries; Table 1), suggesting that there may have been genetic drift in the Dutch population for these loci4. Yet, as each of these loci has a MAF>0.005, we assumed that these alleles also segregate in other populations of European descent4, such as those of the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium. Therefore, we set out replication in independent samples from the CHARGE cohorts using the 1-kG reference panel (Phase 1 integrated release v3, April 2012, all ancestries). We were able to replicate five out of the six variants using the Bonferroni-corrected P value threshold of 8.33 × 10−3 (Table 2, Methods and Supplementary Table 11).
Table 1. Summary descriptions for the variants associated with HDL-C, LDL-C, TC or TG.
| SNP | Chr | Position | EA | NEA | Gene | MAF GoNL | MAF 1-kG | MAF GoNL /MAF 1-kG (P value for two population proportions) |
|---|---|---|---|---|---|---|---|---|
| rs4752801 | 11 | 47,907,641 | G | A | Close to the NUP160 | 0.347 | 0.338 | 1.027 (0.258) |
| rs149580368 | 17 | 41,874,745 | A | C | Between C17orf105 and MPP3 | 0.029 | 0.015 | 1.923 (<0.0001) |
| rs77542162 | 17 | 67,081,278 | G | A | ABCA6 | 0.030 | 0.008 | 3.647 (<0.0001) |
| rs144984216 | 19 | 20,479,901 | T | C | ZNF826P | 0.028 | 0.011 | 2.555 (<0.0001) |
| rs117162033 | 19 | 8,627,569 | T | C | MYO1F | 0.007 | 0.007 | 0.957 (<0.0001) |
EA, effect allele; GoNL, Genome of the Netherlands; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; MAFGoNL and MAF1 kG, the minor allele frequency of the effect allele in the GoNL reference panel and in the 1-kG reference panel (Phase 1 integrated release v3, April 2012, all ancestries), respectively; NEA, non-effect allele; SNP, single-nucleotide polymorphism; TC, total cholesterol; TG, triglyceride.
Table 2. Results for the variants associated with HDL-C, LDL-C, TC or TG.
Of the replicated variants, rs77542162 is the most interesting variant. This missense variant is associated with both LDL-C and TC (Supplementary Figs 5 and 6) and is located on chromosome 17 within the ABCA6 gene (ATP-binding cassette, subfamily A (ABC1), member 6). The frequency of this variant is 1.31-fold higher in the discovery cohorts than in the replication cohorts and even 3.65-fold higher in the GoNL population than in the 1-kG population. This missense variant changes the amino acid cysteine into arginine at position 1359 (Cys1359Arg) and is predicted to be damaging for the structure and function of the protein by Polyphen2 (ref. 12), MutationTaster13 and LRT14. The effect size of rs77542162 (βLDL-C=0.135 and βTC=0.140) is very similar to those observed for other single variants in well-known lipid genes, such as LDLR and CETP, as reported in ref. 1. The membrane-associated protein encoded by this gene is a member of the superfamily of ATP-binding cassette (ABC) transporters that transport various molecules across extra- and intracellular membranes. This protein is a member of the ABC1 subfamily, which is the only major ABC subfamily found exclusively in multicellular eukaryotes. ABCA6 is clustered with four other ABC1 family members on chromosome 17q24 and appears to play a role in macrophage lipid homeostasis.
One other replicated variant, rs149580368, is also enriched with a 1.92-fold increase in frequency in the Dutch population compared with the 1-kG population. This intergenic variant (Supplementary Fig. 7), without a significant cis-eQTL effect, is located between the protein-coding genes C17orf105 (chromosome 17 open reading frame 105) and MPP3 (membrane protein, palmitoylated 3). Two replicated variants have similar frequencies in the GoNL and 1-kG reference sets: rs4752801 (Supplementary Fig. 8), an new intergenic variant with a high frequency (MAF=0.355) that is located in a region previously identified1, and rs117162033 (Supplementary Fig. 9), an intronic variant in the myosin F (MYO1F)-coding gene. C17orf15, MPP3 and MYO1F have no known impact on lipid levels. As the imputation quality of rs117162033 is lower than the other variants, we validated the imputation of this variant using the same approach as published in ref. 15. We compared in a random sample of 65 participants of the GoNL reference panel their sequence and best-guess GoNL-imputed genotypes and found that the concordance was 100% (all participants were correctly imputed). The association between TG and the intronic variant in the MYO1F gene is remarkable because of the low frequency of the variant. This confirms the conclusions as published before about the GoNL reference panel, that the trio-based phasing contributed significantly to the imputation quality of rare variants5.
In this current study, the GoNL reference panel was used for imputations of the discovery cohorts and the 1-kG reference panel for the imputation of the replication cohorts. Although it would be interesting to impute with a combined reference panel of both the GoNL data, the 1-kG data and other sequence data, this effort is ongoing.
This study shows that the imputation of a population-specific reference panel into large epidemiological cohorts can reveal both low-frequency and rare variants associated with blood lipid levels using classical association testing approaches. The three variants with increased frequency in the Dutch population as compared with the 1-kG population include a rare, predicted to be deleterious missense variant in ABCA6, which has increased frequency 3.65 times larger in the Dutch population. The effect of this variant is comparable to that of variants in the LDLR gene, a gene for which several population-based screening programmes have been initiated. Our findings suggest that next-generation-sequencing effort may yield clinically relevant findings. Our paper further shows that next-generation-sequencing efforts in specific homogeneous populations as the Dutch may yield clinically relevant findings worldwide.
Methods
Study descriptions
The descriptions of the including cohorts can be found in the Supplementary Methods. A written informed consent was obtained from all study participants for all cohorts and local ethical committees at participating institutions approved individual study protocols.
Study samples and phenotypes
A summary of the details of both the discovery and replication cohorts participating in this study can be found in Supplementary Tables 1 and 12.
Only samples of Dutch ancestry were used in the discovery cohorts; the samples in the replication cohorts are from various ancestries (see Supplementary Table 12). In all studies, except MESA Whites, all individuals who used lipid-lowering medication at the time the lipid levels were measured, were excluded. In MESA Whites, the total cholesterol values for individuals on lipid-lowering medication were divided by 0.8. In all studies except for LLS and PREVEND, the subjects were fasting when the lipid levels were measured. In LLS all samples were non-fasted and in PREVEND 2.99% were non-fasted. The LDL-C levels were measured within the ERF, Croatia-Korcula, Croatia-Split, Croatia-Vis, FamHS and Lifelines cohorts, within the other cohorts the Friedewald equation was used to calculate the LDL-C levels16.
The lipid measurements were adjusted for sex, age and age2 in all cohorts. Various methods were used to account for family relationships: in ERF grammar-gamma, GenABEL version 1.7.6 (refs 17, 18) was used; in the Croatia-Korcula, Croatia-Split, Croatia-Vis and Generation Scotland cohorts mmscore (GenABEL)17 was used; and in LLS, qt-assoc was used. In CHS the clinic was used as extra covariate, in Lifelines PC1 and PC2, in FamHS the field centre, the genotyping array (Illumina 550 k, 610 k and 1 M), PC5 only for TC and PC1 only for LDL, in FHS the cohort (offspring and third generation) and PCs, in MESA Whites 2 PCs and study site, in NTR-NESDA PCs and chip effect, in ORCADES the genotyping array and PC1, PC2 and PC3, in PROSPER-Dutch only PC1 and in both PROSPER-Scottish and PROSPER-Irish PC1-PC4.
Genotyping and imputations
Detailed information about genotyping and imputations per cohort can be found in the Supplementary Methods. In summary, all cohorts were genotyped using commercially available Affymetrix or Illumina genotyping arrays, or custom Perlegen arrays. Quality control was performed independently for each study. To facilitate meta-analysis, each replication cohort performed genotype imputation using IMPUTE19 or Minimac20 with reference to the GoNL project data for the discovery cohorts and with reference to the 1-kG project data for the replication cohorts.
GWAS in all discovery cohorts
All nine discovery cohorts ran separate the genome-wide association study for each of the four traits: HDL-C, LDL-C, TC and TG. Supplementary Table 13 shows the genomic control factor λ per trait per cohort and Supplementary Figs 10–13 show the λ per MAF bin per trait per cohort. We therefore used only the SNPs with a R2>0.3, R2<1.1 and expected minor allele count (expMAC=2 × MAF × R2·sample size) >10. Most inflations are observed within the ERF study, especially in the lowest-frequency variants, which is probably caused by the family structure in this cohort.
Meta-analysis of discovery cohorts
The association results of all studies were combined and the s.e.-based weights were calculated using METAL21. This tool also applies genomic control by automatically correcting the test statistics to account for small amounts of population stratification or unaccounted relatedness. METAL also allows for heterogeneity. We used the following filters: 0.3<R2<1.1 and expMAC>10.
After meta-analyses of all available variants, we excluded the variants that are not present in at least six of the nine cohorts. We also excluded all variants that are labelled as being in the inaccessible genome, since the quality of those SNPs cannot be guaranteed22. The remaining variants per trait, see Supplementary Table 14, were used to create Manhattan plots and QQ plots, see Supplementary Figs 14 and 15. The meta-analysis resulted in 1,905 SNPs with a P value less than 5 × 10−8 for HDL-C, 2,626 SNPs for LDL-C, 3,133 SNPs for TC and 1,310 for TG.
Confirmation of known loci
Previously, Teslovich et al1 and Willer et al2 identified 157 loci associated with one of more of the lipids. Teslovich et al1 identified 47, 37, 52 and 32 loci to be associated with HDL-C, LDL-C, TC and TG, respectively. The positions of these loci were reported on human genome build 36; we therefore lifted these positions over to human genome build 37 and checked the association results after the meta-analysis of all discovery cohorts. The effect size of these loci was reported in mg dl−1, whereas in this study we use mmol l−1. We therefore multiplied the effect size for the loci associated with TG with 0.0259 and the other loci with 0.011. Supplementary Fig. 2 and Supplementary Table 6 show the comparison per trait of our meta-analysis of all discovery cohorts with the results of the meta-analysis in ref. 1. We did the same for the loci identified in ref. 2, see Supplementary Fig. 3 and Supplementary Table 7. The effect size of these loci could not be compared with our results, since trait residuals within each study participating in the meta-analysis in ref. 2 were adjusted for sex and age2 and subsequently quantile normalized. Their GWAS was performed with the inverse normal transformed trait values.
Selection of independent variants
In order to select only associated variants that were independent of previous findings, we used the GCTA tool11. This tool performs a stepwise selection procedure to select multiple associated SNPs by a conditional and joint analysis approach using summary-level statistics from a meta-analysis and LD corrections between SNPs estimated from the GoNL reference panel, release 4. This analysis revealed 60 independent variants associated with HDL-C, 142 independent variants associated with LDL-C, 134 independent variants associated with TC and 16 independent variants associated with TG. By using this approach, we were able to identify additional independent variants in known loci. Figure 1 shows that we identified both common and rare variants and more rare variants compared with refs 1, 2. There is an overlap between the genome-wide significant SNPs of the different traits, and also between the independent SNPs of the different traits, as shown in Supplementary Fig. 1.
Identification of potential novel variants
To identify potential novel variants, we first excluded all variants within 1 Mb of a known loci from refs 1, 2. Since the number of loci associated with the four traits differ, we end up with 7,946,245 SNPs for HDL-C, 8,014,693 SNPs for LDL-C, 7,923,530 SNPs for TC and 7,468,790 SNPs for TG. For all traits we do find some genome-wide significant loci, see Supplementary Figs 16 and 17. We used the GCTA tool to select only those variants that are independently associated with the lipid trait. This analysis revealed two novel independent variants associated with HDL-C, one novel independent variant associated with LDL-C, two novel independent variants associated with TC and one novel independent variants associated with TG, see Supplementary Table 8 and Supplementary Fig. 18. We used PLINK to test whether these six variants are in LD with the known loci from refs 1, 2. None of the six variants are in LD with known loci associated with the same trait on the same chromosome (R2<0.14).
Replication of potential novel variants
The six potential novel loci were replicated in 11 cohorts: CHS, Croatia-Korcula, Croatia-Split, Croatia-Vis, FamHS, FHS, Generation Scotland, MESA Whites, ORCADES, PROSPER-Scottish and PROSPER-Irish. The association results of all cohorts were combined and the s.e.-based weights were calculated using METAL21. The Bonferroni correction for multiple testing was 8.33 × 10−3. This resulted in the significant replication of five out of the six variants, see Supplementary Fig. 19 and Supplementary Table 11.
Conditional analysis
Within the discovery cohorts we performed a conditional analysis to see whether the novel variants are independent of the known loci from refs 1, 2. Supplementary Table 10 shows the results within these cohorts with and without adjusting for the known loci for the trait in question, if available in the GoNL reference panel. Since the unadjusted and adjusted results are similar, we conclude that the newly identified variants are independent of the known loci.
Author contributions
E.M.v.L. organized the study and designed the study with substantial input of L.C.K., A.I., P.I.W.d.B. and C.M.v.D. E.M.v.L. drafted the manuscript with substantial input of L.A.C., A.Me, B.M.P., C.W., G.M.P., J.F.W., J.E.H., L.C.F., L.C.K., J.D., P.E.S., D.I.B., J.E.H., H.M., P.M.K., P.I.W.d.B., S.L.P., S.T., C.M.v.D. and G.-J.B.v.O. All authors had the opportunity to comment on the manuscript. Data collection, GWAS and statistical analysis were performed by T.M.B., J.A.B., J.C.B., B.M.P. (CHS); J.E.H., C.H., O.P., V.V., I.R., A.F.W. (CROATIA); E.M.v.L., B.A.O., C.M.v.D. (ERF); C.C.W., L.A.C. (FHS), M.F.F., I.B.B. (FamHS); J.E.H., H.T., L.J.H., D.J.P. (Generation Scotland); G.M.P., Q.D., L.A.L. (JHS); A.Ma., J.I.R., J.C.M., S.S.R. (MESA); A.K., P.D., F.v.D., M.A.S., C.W. (Lifelines); J.D., M.B., A.J.M.C., H.-W.U., P.E.S. (LLS); H.M., G.W., E.J.d.G., Y.M., B.W.J.H.P., J.-J.H., D.I.B. (NTR-NESDA); N.V., I.M.L., P.v.H. (PREVEND); S.T., I.P., N.S., C.J.P., B.M.B., P.M.K., D.J.S., J.W.J. (PROSPER); P.K.J., H.C., J.F.W. (ORCADES); E.M.v.L., C.M.-G., F.R., A.H., O.H.F., E.J.S., A.G.U., C.M.v.D. (Rotterdam Study). D.J.v.E. recruited cohorts. Creation of the GoNL reference panel was carried out by L.C.F., A.Me., S.L.P. and P.D. Design of the GoNL project was made by C.W., M.A.S., C.M.v.D., D.I.B., P.E.S., G.-J.B.O., P.I.W.d.B. E.M.v.L. performed the meta-analysis. Biological association of loci and bioinformatics were carried out by E.M.v.L. and C.M.v.D.
Additional information
How to cite this article: van Leeuwen, E. M. et al Genome of the Netherlands population-specific imputations identify a ABCA6 variant associated with cholesterol levels. Nat. Commun. 6:6065 doi: 10.1038/ncomms7065 (2015).
Supplementary Material
Supplementary Figures 1-19, Supplementary Tables 1-14, Supplementary Note 1, Supplementary Methods and Supplementary References
Acknowledgments
We especially thank all volunteers who participated in our study. This study made use of data generated by the ‘Genome of the Netherlands’ project, which is funded by the Netherlands Organization for Scientific Research (grant no. 184021007). The data were made available as a Rainbow Project of BBMRI-NL. Samples were contributed by LifeLines (http://lifelines.nl/lifelines-research/general), the Leiden Longevity Study (http://www.healthy-ageing.nl; http://www.langleven.net), the Netherlands Twin Registry (NTR: http://www.tweelingenregister.org), the Rotterdam studies (http://www.erasmus-epidemiology.nl/rotterdamstudy) and the Genetic Research in Isolated Populations programme (http://www.epib.nl/research/geneticepi/research.html#gip). The sequencing was carried out in collaboration with the Beijing Institute for Genomics (BGI).
Cardiovascular Health Study: This CHS research was supported by NHLBI contracts HHSN268201200036C, HHSN268200800007C, HHSN268200960009C, N01HC55222, N01HC85079, N01HC85080, N01HC85081, N01HC85082, N01HC85083, N01HC85086; and NHLBI grants HL080295, HL087652, HL105756 and HL103612 with additional contribution from the National Institute of Neurological Disorders and Stroke (NINDS). Additional support was provided through AG023629 from the National Institute on Aging (NIA). A full list of CHS investigators and institutions can be found at http://www.chs-nhlbi.org/pi.htm.
The CROATIA cohorts would like to acknowledge the invaluable contributions of the recruitment teams in Vis, Korcula and Split (including those from the Institute of Anthropological Research in Zagreb and the Croatian Centre for Global Health at the University of Split), the administrative teams in Croatia and Edinburgh and the people of Vis, Korcula and Split. SNP genotyping was performed at the Wellcome Trust Clinical Research Facility in Edinburgh for CROATIA-Vis, by Helmholtz Zentrum München, GmbH, Neuherberg, Germany for CROATIA-Korcula and by AROS Applied Biotechnology, Aarhus, Denmark for CROATIA-Split. They would also like to thank Jared O’Connell for performing the pre-phasing for all cohorts before imputation.
The ERF study as a part of EuroSPAN (European Special Populations Research Network) was supported by European Commission FP-6 STRP grant number 018947 (LSHG-CT-2006-01947) and also received funding from the European Community's Seventh Framework Programme (FP7/2007-2013)/grant agreement HEALTH-F4-2007-201413 by the European Commission under the programme ‘Quality of Life and Management of the Living Resources’ of 5th Framework Programme (no. QLG2-CT-2002-01254). High-throughput analysis of the ERF data was supported by joint grant from the Netherlands Organisation for Scientific Research and the Russian Foundation for Basic Research (NWO-RFBR 047.017.043). This research was financially supported by BBMRI-NL, a Research Infrastructure financed by the Dutch government (NWO 184.021.007). Statistical analyses for the ERF study were carried out on the Genetic Cluster Computer (http://www.geneticcluster.org), which is financially supported by the Netherlands Scientific Organization (NWO 480-05-003 PI: Posthuma) along with a supplement from the Dutch Brain Foundation and the VU University Amsterdam. We are grateful to all study participants and their relatives, general practitioners and neurologists for their contributions and to P. Veraart for her help in genealogy, J. Vergeer for the supervision of the laboratory work and P. Snijders for his help in data collection.
The FamHS is funded by a NHLBI grant 5R01HL08770003, and NIDDK grants 5R01DK06833603 and 5R01DK07568102.
The Framingham Heart Study SHARe Project for GWAS scan was supported by the NHLBI Framingham Heart Study (Contract No. N01-HC-25195) and its contract with Affymetrix Inc for genotyping services (Contract No. N02-HL-6-4278). DNA isolation and biochemistry were partly supported by NHLBI HL-54776. A portion of this research utilized the Linux Cluster for Genetic Analysis (LinGA-II) funded by the Robert Dawson Evans Endowment of the Department of Medicine at the Boston University School of Medicine and Boston Medical Center. We are grateful to Han Chen for conducting the 1000G imputation.
The Family Heart Study was supported by the by grants R01-HL-087700 and R01-HL-088215 from the National Heart, Lung, and Blood Institute (NHLBI).
We would like to acknowledge the invaluable contributions of the families who took part in the Generation Scotland: Scottish Family Health Study, the general practitioners and Scottish School of Primary Care for their help in recruiting them, and the whole Generation Scotland team, which includes academic researchers, IT staff, laboratory technicians, statisticians and research managers. SNP genotyping was performed at the Wellcome Trust Clinical Research Facility in Edinburgh. GS:SFHS is funded by the Scottish Executive Health Department, Chief Scientist Office, grant number CZD/16/6. SNP genotyping was funded by the Medical Research Council, United Kingdom.
We wish to acknowledge the services of the LifeLines Cohort Study, the contributing research centres delivering data to LifeLines and all the study participants.
MESA Whites and the MESA SHARe project are conducted and supported by contracts N01-HC-95159 through N01-HC-95169 and RR-024156 from the NHLBI. Funding for MESA SHARe genotyping was provided by NHLBI Contract N02.HL.6.4278. MESA Family is conducted and supported in collaboration with MESA investigators; support is provided by grants and contracts R01HL071051, R01HL071205, R01HL071250, R01HL071251, R01HL071252, R01HL071258 and R01HL071259. We thank the participants of the MESA study, the Coordinating Center, MESA investigators and study staff for their valuable contributions. A full list of participating MESA investigators and institutions can be found at http://www.mesa-nhlbi.org.
Netherland Twin Register (NTR) and Netherlands Study of Depression and Anxiety (NESDA): Funding was obtained from the Netherlands Organization for Scientific Research (NWO) and MagW/ZonMW grants Middelgroot-911-09-032, Spinozapremie 56-464-14192, Geestkracht programme of the Netherlands Organization for Health Research and Development (Zon-MW, grant number 10-000-1002), Center for Medical Systems Biology (CSMB, NWO Genomics), NBIC/BioAssist/RK(2008.024), Biobanking and Biomolecular Resources Research Infrastructure (BBMRI-NL, 184.021.007), VU University’s Institute for Health and Care Research (EMGO+) and Neuroscience Campus Amsterdam (NCA); the European Science Foundation (ESF, EU/QLRT-2001-01254), the European Community’s Seventh Framework Program (FP7/2007-2013), ENGAGE (HEALTH-F4-2007-201413); the European Science Council (ERC Advanced, 230374); and the European Research Council (ERC-284167). Part of the genotyping and analyses were funded by the Genetic Association Information Network (GAIN) of the Foundation for the National Institutes of Health, Rutgers University Cell and DNA Repository (NIMH U24 MH068457-06), the Avera Institute, Sioux Falls, South Dakota (USA) and the National Institutes of Health (NIH R01 HD042157-01A1, MH081802, Grand Opportunity grants 1RC2 MH089951 and 1RC2 MH089995).
PREVEND genetics is supported by the Dutch Kidney Foundation (Grant E033), the EU project grant GENECURE (FP-6 LSHM CT 2006 037697), the National Institutes of Health (grant 2R01LM010098), The Netherlands Organisation for Health Research and Development (NWO-Groot grant 175.010.2007.006, NWO VENI grant 916.761.70, ZonMw grant 90.700.441) and the Dutch Inter University Cardiology Institute Netherlands (ICIN).
The PROSPER study was supported by an investigator-initiated grant obtained from Bristol-Myers Squibb. J.W.J is an Established Clinical Investigator of the Netherlands Heart Foundation (grant 2001 D 032). Genotyping was supported by the seventh framework programme of the European commission (grant 223004) and by the Netherlands Genomics Initiative (Netherlands Consortium for Healthy Aging grant 050-060-810).
The Rotterdam Study is funded by Erasmus Medical Center and Erasmus University, Rotterdam, Netherlands Organization for the Health Research and Development (ZonMw), the Research Institute for Diseases in the Elderly (RIDE), the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports, the European Commission (DG XII) and the Municipality of Rotterdam. We are grateful to the study participants, the staff from the Rotterdam Study and the participating general practitioners and pharmacists.
The generation and management of GWAS genotype data for the Rotterdam Study is supported by the Netherlands Organisation of Scientific Research NWO Investments (nr. 175.010.2005.011, 911-03-012). This study is funded by the Research Institute for Diseases in the Elderly (014-93-015; RIDE2), the Netherlands Genomics Initiative (NGI)/Netherlands Organisation for Scientific Research (NWO) project no. 050-060-810. We thank Pascal Arp, Mila Jhamai, Marijn Verkerk, Lizbeth Herrera and Marjolein Peters for their help in creating the GWAS database.
Footnotes
B.M.P. served on the DSMB of a clinical trial funded by the manufacturer (Zoll Lifecor) and on the Yale Open Data Access Project funded by Johnson & Johnson. The remaining authors declare no competing financial interests.
Contributor Information
The Genome of the Netherlands Consortium:
Pieter B.T. Neerincx, Clara C. Elbers, Pier Francesco Palamara, Itsik Pe'er, Abdel Abdellaoui, Wigard P. Kloosterman, Mannis van Oven, Martijn Vermaat, Mingkun Li, Jeroen F.J. Laros, Mark Stoneking, Peter de Knijff, Manfred Kayser, Jan H. Veldink, Leonard H. van den Berg, Heorhiy Byelas, Johan T. den Dunnen, Martijn Dijkstra, Najaf Amin, K. Joeri van der Velde, Jessica van Setten, Mathijs Kattenberg, Barbera D.C. van Schaik, Jan Bot, Isaäc J. Nijman, Hailiang Mei, Vyacheslav Koval, Kai Ye, Eric-Wubbo Lameijer, Matthijs H. Moed, Jayne Y. Hehir-Kwa, Robert E. Handsaker, Shamil R. Sunyaev, Mashaal Sohail, Fereydoun Hormozdiari, Tobias Marschall, Alexander Schönhuth, Victor Guryev, H. Eka D. Suchiman, Bruce H. Wolffenbuttel, Mathieu Platteel, Steven J. Pitts, Shobha Potluri, David R. Cox, Qibin Li, Yingrui Li, Yuanping Du, Ruoyan Chen, Hongzhi Cao, Ning Li, Sujie Cao, Jun Wang, and Jasper A. Bovenberg
References
- Teslovich T. M. et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466, 707–713 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Global Lipids Genetics Consortium. et al. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 45, 1274–1283 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer C. J. & Mohlke K. L. Finding genes and variants for lipid levels after genome-wide association analysis. Curr. Opin. Lipidol. 23, 98–103 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo L. M., MacKay I., Oostra B., van Duijn C. M. & Aulchenko Y. S. The effect of genetic drift in a young genetically isolated population. Ann. Hum. Genet. 69, 288–295 (2005). [DOI] [PubMed] [Google Scholar]
- Genome of the Netherlands Consortium. Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat. Genet. 46, 818–825 (2014). [DOI] [PubMed] [Google Scholar]
- Boomsma D. I. et al. The Genome of the Netherlands: design, and project goals. Eur. J. Hum. Genet. 22, 221–227 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson T. et al. A mutation in APP protects against Alzheimer’s disease and age-related cognitive decline. Nature 488, 96–99 (2012). [DOI] [PubMed] [Google Scholar]
- Holm H. et al. A rare variant in MYH6 is associated with high risk of sick sinus syndrome. Nat. Genet. 43, 316–320 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styrkarsdottir U. et al. Nonsense mutation in the LGR4 gene is associated with several human diseases and other traits. Nature 497, 517–520 (2013). [DOI] [PubMed] [Google Scholar]
- Deelen P. et al. Improved imputation quality of low-frequency and rare variants in European samples using the ’Genome of The Netherlands’. Eur. J. Hum. Genet. 22, 1321–1326 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Lee S. H., Goddard M. E. & Visscher P. M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88, 76–82 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei I. A. et al. A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz J. M., Rödelsperger C., Schuelke M. & Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods 7, 575–576 (2010). [DOI] [PubMed] [Google Scholar]
- Chun S. & Fay J. C. Identification of deleterious mutations within three human genomes. Genome Res. 19, 1553–1561 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott L. J. et al. A genome-wide association study of type 2 diabetes in finns detects multiple susceptibility variants. Science 316, 1341–1345 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnick G. R., Knopp R. H., Fitzpatrick V. & Branson L. Estimating low-density lipoprotein cholesterol by the Friedewald equation is adequate for classifying patients on the basis of nationally recommended cutpoints. Clin. Chem. 36, 15–19 (1990). [PubMed] [Google Scholar]
- Aulchenko Y. S., Ripke S., Isaacs A. & van Duijn C. M. GenABEL: an R library for genome-wide association analysis. Bioinformatics 23, 1294–1296 (2007). [DOI] [PubMed] [Google Scholar]
- Svishcheva G. R., Axenovich T. I., Belonogova N. M., van Duijn C. M. & Aulchenko Y. S. Rapid variance components-based method for whole-genome association analysis. Nat. Genet. 44, 1166–1170 (2012). [DOI] [PubMed] [Google Scholar]
- Howie B. N., Donnelly P. & Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 5, e1000529 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B. et al. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet. 44, 955–959 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer C. J., Li Y., Gonçalo R. & Abecasis G. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning S. R. & Browning B. L. Haplotype phasing: existing methods and new developments. Nat. Rev. Genet. 12, 703–714 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1-19, Supplementary Tables 1-14, Supplementary Note 1, Supplementary Methods and Supplementary References