Abstract
L-Threonine aldolases (TAs) represent a family of homologous pyridoxal 5’-phosphate-dependent enzymes found in bacteria and fungi, and catalyse the reversible cleavage of several l-3-hydroxy-α-amino acids. TAs have great biotechnological potential, since they catalyse the formation of carbon-carbon bonds, and therefore may be exploited for bioorganic synthesis of l-3-hydroxyamino acids that are biologically active or constitute building blocks for pharmaceutical molecules. Many TAs, showing different stereospecificity towards the Cβ configuration, have been isolated. Because of their potential to carry out diastereoselective syntheses, TAs have been the subject of structural, functional and mechanistic studies. Nevertheless, their catalytic mechanism and the structural bases of their stereospecificity have not been elucidated.
In this study, we have determined the crystal structure of low-specificity l-threonine aldolase from Escherichia coli at 2.2 Å resolution, in the unliganded form and co-crystallized with l-serine and l-threonine. Furthermore, several active-site mutants have been functionally characterised in order to elucidate the reaction mechanism and the molecular bases of stereospecificity. No active site catalytic residue was revealed, and a structural water molecule was assumed to act as catalytic base in the retro-aldol cleavage reaction.
Interestingly, the very large active site opening of E. coli TA suggests that a much larger molecule than l-threonine isomers may be easily accommodated, and threonine aldolases may actually play diverse physiological functions in different organisms. Substrate recognition and reaction specificity seem to be guided by the overall microenvironment that surrounds the substrate at the enzyme active site, rather than to one ore more specific residues.
Keywords: threonine aldolase structure, catalytic mechanism, substrate preference, catalytic water, protein crystallography
INTRODUCTION
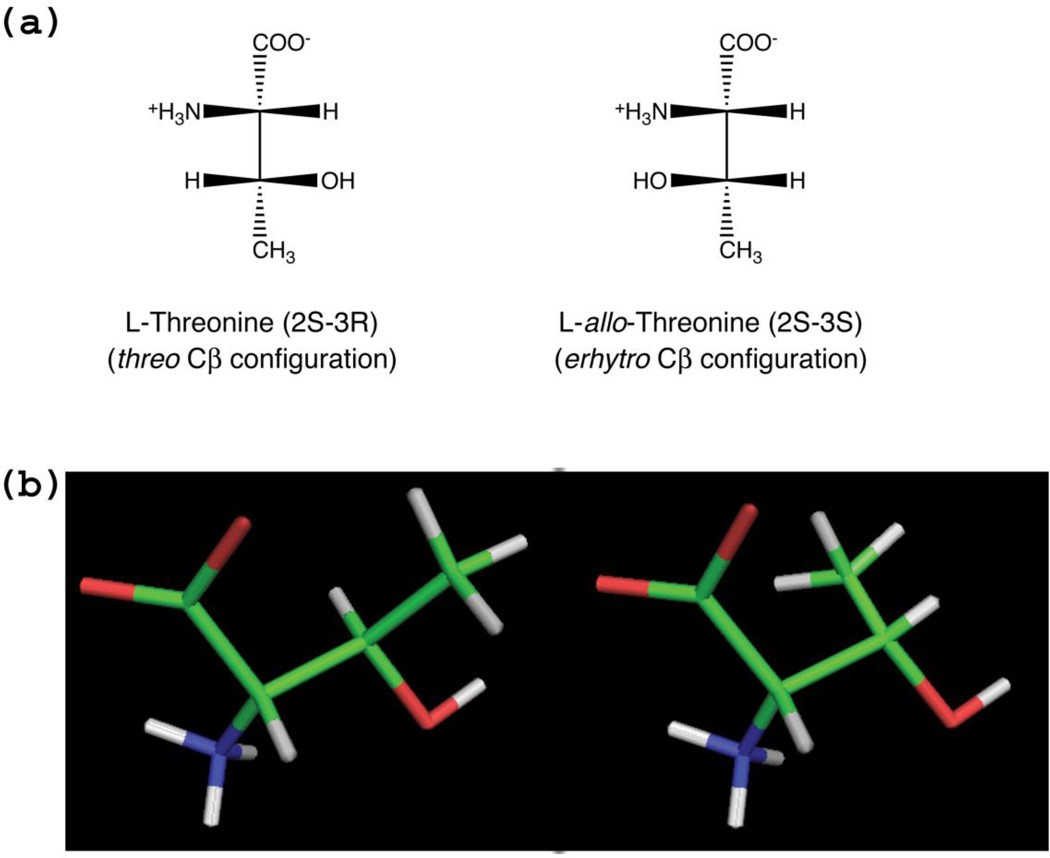
l-Threonine aldolases (l-TAs) make up a family of homologous pyridoxal 5’-phosphate (PLP)-dependent enzymes from a broad number of species of bacteria and fungi that catalyse the reversible cleavage of several different l-3-hydroxy-α-amino acids, comprising l-threonine, l-3-phenylserine and l-3-hydroxytrimethyllysine, to glycine and the corresponding aldehyde (1, 2). These enzymes have a great biotechnological potential, since they can be used to catalyse the formation of carbon-carbon bonds, allowing the synthesis of l-3-hydroxyamino acids that are biologically active or constitute intermediate or building blocks of drugs, such as l-3,4-dihydrohyphenylserine (l-DOPS), 4-hydroxy-l-threonine, l-3-[4-(methylthio)phenylserine] and 3,4,5-trihydroxy-l-aminopentanoic acid (1, 3–5). 3-Hydroxyamino acids contain two chiral centres, one at Cα, which determines the l- or d-configuration, and the second at Cβ, responsible for the erythro or threo configuration (Fig. 1). Many l-TAs with different stereospecificity towards the Cβ configuration have been isolated and characterised (6). Depending on their preference for the erythro or threo configuration, l-TAs are classified into low-specificity l-TAs (EC 4.1.2.48), l-threonine aldolases (EC 4.1.2.5) and l-allo-threonine aldolases (EC 4.1.2.49). Threonine aldolases that are specific for d-3-hydroxyamino acids also exist (7, 8), however, these enzymes are structurally and evolutionary distinct from l-TAs. While l-TAs belong to the aspartate aminotransferase fold-type of PLP-dependent enzymes (fold-type I) (9), d-TAs have a completely different protein fold (fold-type III) (10).
Figure 1.
(a) Fisher projection of l-threonine and l-allo-threonine structures. The nomenclature of threo and erythro configurations is derived from the diastereomeric aldoses threose and erythrose, to which l-threonine and l-allo-threonine can be superimposed, respectively. (b) Three-dimensional PyMol stick representation of L-threonine and L-allo-threonine as they are actually oriented in the eTA-Thr structure (see Fig. 6b), showing that at the enzyme active site the difference between the two diasteroisomers is only found in the position of the methyl group bound to the β-carbon. The figure also clearly shows a synperiplanar conformation of the Oγ-H bond with respect to the Cα-Cβ bond, as present in the enzyme-bound hydroxyamino acids. This periplanar conformation is stereochemically required by the retro-aldol cleavage mechanism (see Scheme 1).
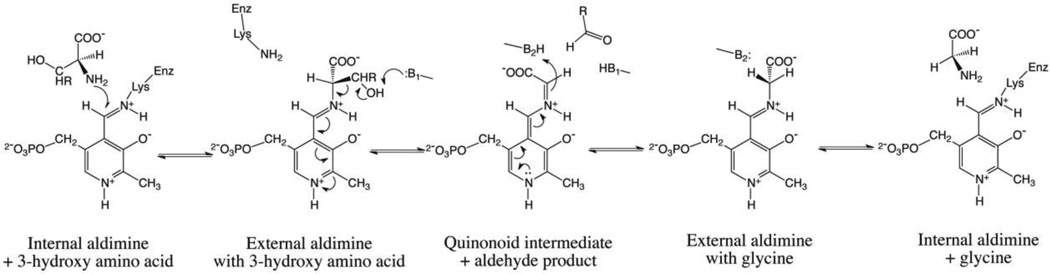
Because of their different stereospecificity and potential to catalyse diastereoselective syntheses, l-TAs have been the subject of structural, functional and mechanistic studies (11–13). Attempts to change or improve stereospecificity of l-TAs have also been carried out (14). Nevertheless, the catalytic mechanism of l-TAs and the structural bases of their stereospecificity have not been elucidated. The aldolase reactions catalysed by l-TAs are likely to proceed through a retro-aldol cleavage mechanism (Scheme 1; (11)). This implies the presence of a catalytic base that abstracts a proton from the hydroxyl group of the l-3-hydroxyamino substrate, which is bound to the enzyme as an external aldimine. Then, Cα has to be protonated before the glycine product can be released. The resolution of Thermotoga maritima l-TA (tTA) crystal structure, in the form of unliganded enzyme and as complex with either l-allo-threonine or glycine, gave interesting clues on the catalytic mechanism of this enzyme, which has a preference for l-allo-threonine over l-threonine, and the structural bases of substrate recognition (12). In tTA, the presence of two histidine residues (His83 and His125) in close proximity to the substrate hydroxyl group led to the hypothesis that two different catalytic bases may be responsible for abstracting the proton from l-allo-threonine and l-threonine, respectively, supporting the retro-aldol cleavage mechanism and providing flexibility in the recognition of Cβ configuration. In particular, in tTA His83 represents a likely candidate for the catalytic base that removes the proton from the l-allo-threonine hydroxyl group. For cleavage of l-threonine, different sterochemical requirements suggest that His125, or a water molecule activated by the negatively charged phosphate PLP group, may function as a catalytic base. Moreover, the analysis of the tTA crystal structure and multiple sequence alignments of l-TAs endowed with different stereospecificity suggested that the side chain of the residue at position 87 (a tyrosine residue in tTA) might determine the degree of stereospecificity of l-TAs for l-allo-threonine versus l-threonine. This is the only variable residue in TAs active site and side chain bulk at this position appears to be correlated with stereospecificity, with larger side chains conferring higher preference for l-allo-threonine (12) (Fig. 2).
Scheme 1.
The hypothesised retro-aldol cleavage mechanism for the reaction catalysed by l-TAs on a generic β-hydroxy-α-amino acid.
Figure 2.
clustalW2 multiple sequence alignment (www.ebi.ac.uk/Tools/msa/clustalw2/) of selected l-threonine aldolases with different specificities: Escherichia coli TA (eTA) and Thermotoga maritima TA (tTA) are low-specificity threonine aldolases with a marked preference for l-allo-threonine; Aeromonas jandaei TA (ajTA) shows an absolute preference for l-allo-threonine; Pseudomonas aeruginosa (paTA) shows no preference for l-threonine isomers. Alignment symbols: “*” conserved residue in all sequences; “:” conserved substitution; “.” semi-conserved substitution. The amino acid residues mutated in the present study are shown in bold red character.
We have already investigated the structural and functional properties of low-specificity Escherichia coli l-TA (eTA) in relation to its cognate enzymes serine hydroxymethyltransferase and fungal alanine racemase (10, 11, 15). E. coli TA, more than tTA, shows a marked preference for l-allo-threonine. Here we present crystallographic and site-directed mutagenesis studies carried out on eTA aimed at determining the identity of the catalytic base (or bases) involved in the retro-aldol cleavage of substrates and the molecular basis of stereospecificity. The following eTA mutant forms were produced and characterized with respect to their catalytic properties: H83N; H83F; H126N; H126F; H83F/H126F double mutant; F87A; F87D; K222A. The mutations concerning the histidine residues were chosen so as to be either conservative (histidine was replaced with asparagine, which could still establish hydrogen bonds) or disruptive from the point of view of the polar interactions the amino acid residue could establish (histidine was replaced with phenylalanine).
RESULTS
Crystallographic studies
Overall structure description of E. coli l-threonine aldolase
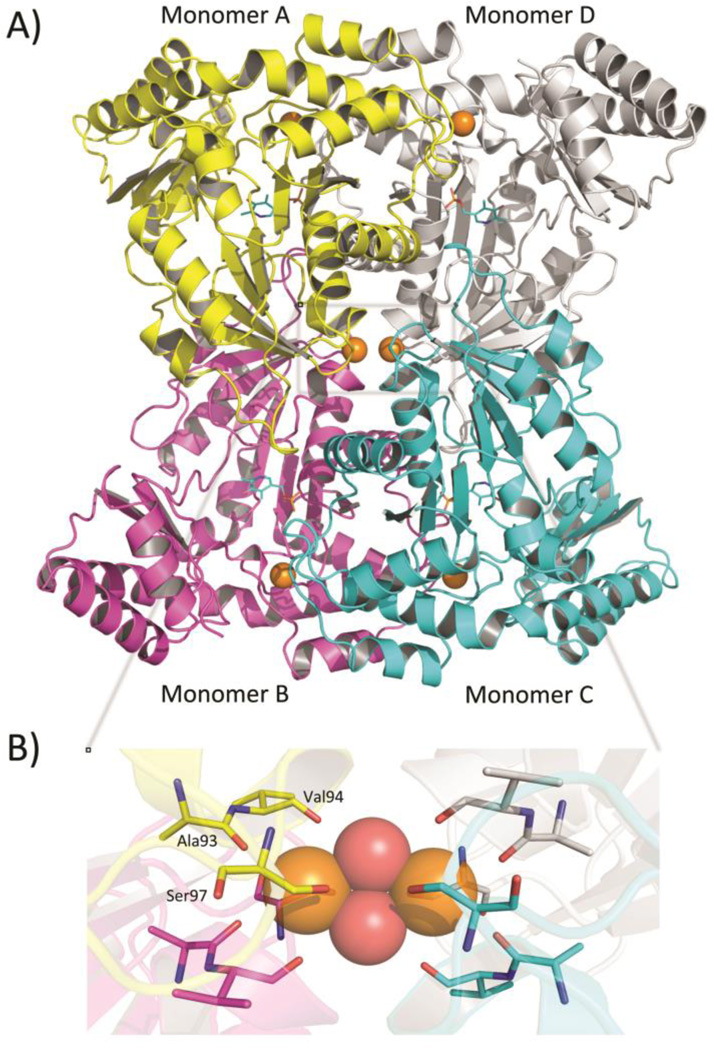
Diffraction data, refinement and structural statistics of the unliganded enzyme (unlig-eTA) and of the binary enzyme forms obtained from co-crystallization with either l-serine (eTA-Ser) or l-threonine (eTA-Thr) are summarized in Table 1. All three crystals belong to space group C2221 and are isomorphous, with typical cell parameters of 77, 101 and 176 Å. The structure of unlig-eTA was first determined using the molecular replacement method, with l-threonine adolase from Thermotoga maritima (tTA; PDB code 1LW5) as search model, and then used to refine the eTA-Ser and eTA-Thr forms. The asymmetric unit contains a homodimer made of monomers A and B. A homotetramer, composed of monomers A, B, C and D (with 222 symmetry), can be generated by application of symmetry element (Fig. 3a). This is consistent with the observation that native eTA, as tTA, is a tetrameric protein (12, 16). However, it should be noted here that the obligate dimer (corresponding to the functional catalytic unit) of fold-type I PLP-dependent enzymes is the AD dimer shown in Fig. 3a. In fold-type I enzymes, the way the obligate dimers are assembled into higher quaternary structure follows different symmetry rules, as seen for example in eukaryotic SHMT (a dimer of dimers; (17, 18)) and prokaryotic glutamate decarboxylase (a trimer of dimers; (19)). In TAs the quaternary structure is different from the latter ones. The overall structure of eTA is very similar to that of tTA (the only other structure available for the TA family (12)), the root-mean-square deviation (rmsd) between the unliganded forms of these enzymes being 1.2 Å and the sequence identity being 48% (12, 16, 20, 21). The dimer buries a total area of 3580 Å2 at the A/D (or B/C) interface, which compares to 3157 Å2 for tTA. The buried areas at the A/B (or C/D) and A/C (or B/D) interfaces are significantly smaller: 1865 Å2 and 1723 Å, respectively. As in all fold-type I PLP-dependent enzymes (9), each monomer of eTA consists of a large domain, with seven β-strand structures flanked by α-helices, and a small domain, composed of three β-strand sheet structures with interlinking α-helices.
Table 1.
Crystallographic data and refinement statistics for unliganded and substrate-bound E. coli threonine aldolase. Values in parentheses refer to the outermost resolution bin.
| eTA-native | eTA-Ser | eTA-Thr | |
|---|---|---|---|
| Data collection statistics | |||
| Space group | C2221 | C2221 | C2221 |
| Cell dimensions (Å) | 76.6, 100.8, 175.7 | 77.2, 101.2, 176.4 | 76.4, 100.9, 176.1 |
| Mols/asymmetric unit | 1 dimer | 1 dimer | 1 dimer |
| Resolution (Å) | 38.2–2.20 (2.28–2.20) | 29.14–2.2 (2.28–2.2) | 29.3–2.1(2.18–2.1) |
| No. of measurements | 365046 (39392) | 99927 (12019) | 168057 (16746) |
| Unique reflections | 32123 (3050) | 30725 (3160) | 37247 (3752) |
| I/sigma I | 15.9 (8.2) | 8.4 (4.3) | 9.8 (4.6) |
| Completeness (%) | 91.8 (88.6) | 86.8 (90.7) | 92.9 (94.9) |
| Rmerge (%)† | 11.1 (31.1) | 10.5 (28.5) | 10.9 (30.5) |
| Structure refinement | |||
| Resolution limit (Å) | 29.3–2.20 (2.28–2.2) | 29.1–2.20 (1.28–2.2) | 29.1–2.10 (2.18–2.1) |
| Sigma cutoff (F) | 0.0 | 0.0 | 0.0 |
| No. of reflections | 30203 (3049) | 30555 (3136) | 37208 (3720) |
| Rfactor (%) | 20.9 (27.0) | 22.6 (29.3) | 18.2 (22.3) |
| Rfree (%)‡ | 27.9 (33.1) | 28.9 (35.6) | 24.4 (27.7) |
| Rmsd standard | |||
| Bond-lengths (Å) | 0.006 | 0.009 | 0.009 |
| Bond- angles (°) | 1.3 | 1.6 | 1.5 |
| Dihedral angles | |||
| Most favored | 89.9 | 88.9 | 91.5 |
| Additional allowed | 10.9 | 10.8 | 8.5 |
| Average B-Factors | |||
| All atoms/ Protein | 27.9/27.5 | 32.4/32.1 | 22.2/21.5 |
| Water/Hepes | 30.2/60.9 | 31.4/72.0 | 27.9/49.5 |
| PLP/Ligand | 26.0 | 39.8/39.9 | 21.6/30.1 |
| Metal | 22.2 | 40.3 | 14.4 |
Rmerge = ∑hkl∑i|Ihkli-〈Ihkli〉|/∑hkl∑i〈Ihkli〉
Rfree calculated with 5% of excluded reflection from the refinement.
Figure 3.
(a) Ribbon representation of the unliganded tetrameric enzyme. One molecule of PLP cofactor (cyan sticks) is bound to each monomer. Six Mg2+ ions (orange spheres) are bound to the tetramer; one is found close to each active site and two are located at the interface of the two dimers. (b) Enlarged view of the Mg2+ binding site located at the two-fold axis of symmetry of the tetramer. Mg2+ ions are shown as orange spheres while water molecules are shown as red spheres. The residues coordinating the Mg2+ ions are shown as sticks. For sake of clarity, only residues contributed from one monomer are labelled.
Structure of the active site in unliganded eTA
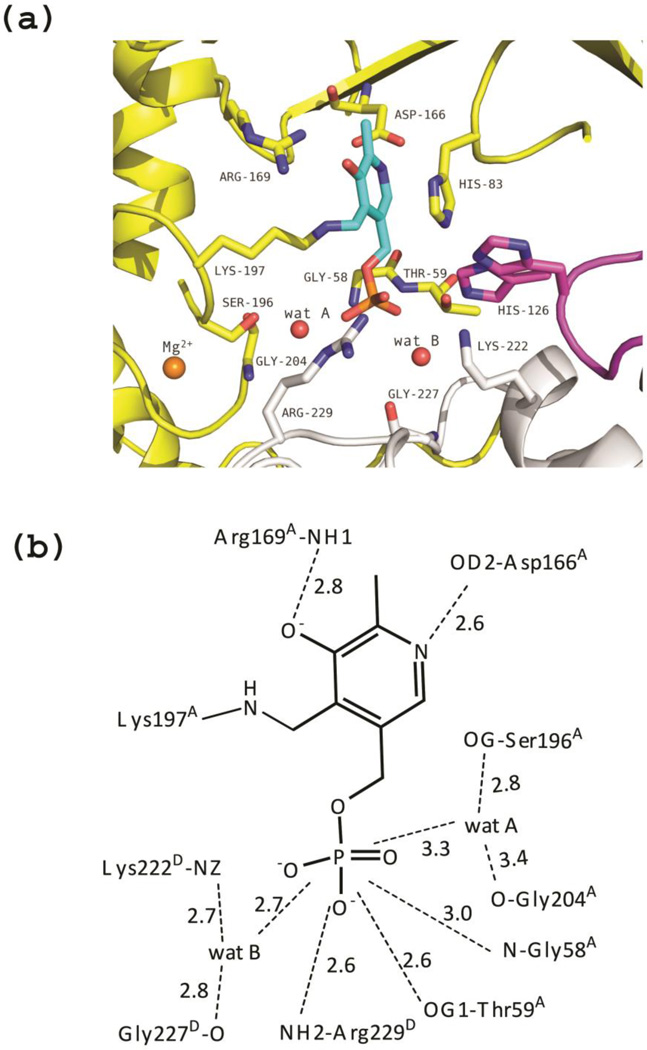
Similarly to tTA, and quite unusually for a PLP-dependent fold-type I enzyme, each of the four active sites in the eTA tetramer is formed from residues belonging to three monomers (Fig 4). This is a peculiar architecture, since the catalytic unit of fold-type I enzymes is typically a dimer, with active sites mainly made by residues of one monomer, interacting with PLP, and by few other residues contributed by the other monomer. The active site in monomer A of the unliganded eTA (unlig-eTA), made by residues from monomers A, B and D, will be used to describe the most relevant interactions between protein and bound PLP. As in all other fold-type I enzymes, each monomer is made by a small and a large domain, with PLP bound at their interface, forming a Schiff base (internal aldimine) with the conserved Lys197A (the A in superscript indicates that this is a residue contributed by monomer A) (Fig. 4a–b). The imidazole of His83A stacks parallel to the re face of PLP ring, while Ala168A makes a hydrophobic contact at the si face. The phenolic oxygen and the pyridine nitrogen of PLP make hydrogen-bond interactions with the guanidinium group of Arg169A and the carboxylate group of Asp166A, respectively. The PLP phosphate group makes hydrogen-bond interactions with the amide nitrogen atoms of Gly58A and Thr59A, and with the side chain of Thr59A. Other contacts from monomer A involve water-mediated interactions between the PLP phosphate group and the hydroxyl groups of Ser196A and Ser205A, and the amide oxygen of Gly204A (wat A in Fig. 4a–b).
Figure 4.
Unliganded eTA. (a) View of the active site of monomer A, showing PLP (in cyan) bound as internal aldimine to Lys197. Three monomers (A, B and D colored as in Fig. 3) contribute to form the active site. Residues of loop 2 from monomer D (222–230) are shown as grey sticks. Loop 3 contributed by monomer B (residues 121–131) is shown in magenta; two different conformations of H126B are shown. (b) Scheme of two-dimensional contacts among PLP, protein residues and the structural water molecules discussed in the text. Dashed lines indicate hydrogen-bond interactions and their length (in Å).
Forming a part of the active site and facing the PLP phosphate group are two loops (loop 1 and 2) from monomer D, made by residues 23–33 and 222–230, respectively (Fig. 4a–b). Loop 1 is located at the active site entrance and projects towards the PLP cofactor, without making any direct interaction with it. From loop 2, only one residue from monomer D (Arg229D) directly interacts with PLP, through its guanidinium group. Other interactions of loop 2 with the PLP phosphate group are mediated by water and are made by the amide oxygen of Gly227D and the amine group of Lys222D (wat B in Fig. 4a–b).
A third loop (loop 3) from monomer B (residues 121–131) also projects into the active site, between monomers A and D, positioning His126B so as to interact with the hydroxyl group of substrates (see next paragraph). Notably, in the structure of unliganded eTA, the His126B imidazole group is mobile and was refined with two alternative conformations (Fig. 4a).
Structure of binary complexes with amino acid ligands
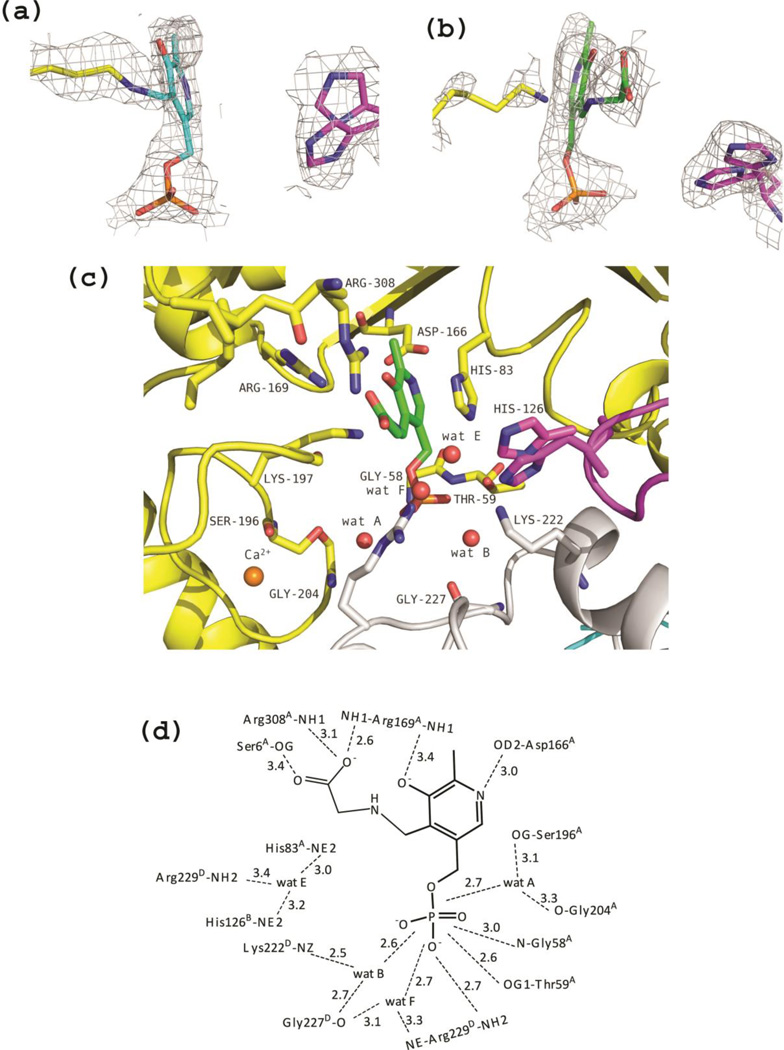
The crystal structures of binary complexes with amino acid ligands were obtained co-crystallizing eTA with either l-serine or l-threonine. In the structure of eTA co-crystallized with l-serine, a mixture of internal aldimine (showing a covalent bond between C4’ of PLP and Lys197A) and external aldimine with glycine (in which the C4’ is bound to the amino group of the ligand), evidently derived from the TA-catalysed cleavage of l-serine into glycine and formaldehyde, is visible in the electron density map at both active sites of the asymmetric unit. It has been shown that eTA slowly catalyses the cleavage of l-serine, although about 60 and 2 × 103 times less efficiently (in terms of kcat/Km ratios) than it cleaves l-threonine and l-allo-threonine, respectively (11). However, the two active sites show different ratios of internal and external aldimines, with monomer A appearing to be predominantly in the internal aldimine form (70% internal aldimine vs. 30% external aldimine), while the opposite is true for monomer B (Fig. 5a–b). As previously reported for tTA (12), formation of external aldimine have little effect on the enzyme overall conformation, the rmsd between monomers of unlig-eTA and eTA-Ser being ~ 0.2 Å. This is equally true for the conformation of the active site, where the side chain of residues in the binary complexes show little or no differences with respect to the unliganded enzyme. In the external aldimine, the amino group of the substrate has displaced the amino group of Lys197A and formed a Schiff base with the C4’ atom of PLP. Breaking of the internal aldimine interaction have resulted in re-orientation of the side chain of Lys197A. Formation of the external aldimine has also led to the characteristic rotation of the PLP ring by 10–30° observed in fold-type I enzymes (22–24). A water molecule (wat E in Fig 5c–d), close to where the hydroxyl group of l-serine ligand may be located, was observed and makes hydrogen-bond interactions with the side-chains of Arg229D, His83A and His126B. Since the density was non-contiguous with the modelled glycine ligand (even at lower contour level), it was refined as water. Also in this structure, the His126B imidazole group is mobile and was refined with two alternative conformations (Fig. 5a–c).
Figure 5.
eTA-Ser complex. (a) View of the electron-density map of monomer A (with coefficients 2Fo-Fc shown at 0.7σ level). In this monomer, PLP is predominantly in the internal aldimine form, as shown by the continuous density between Lys197 and the cofactor. The map is superimposed with the internal aldimine model. (b) View of the electron-density map of monomer B (with coefficients 2Fo-Fc shown at 0.7σ level). Since density is not continuous between PLP and Lys197, the external aldimine form of the cofactor with glycine is predominant in this monomer. The map is superimposed with the model of PLP-glycine external aldimine. (c) Active site of monomer B. Also in this case, two different conformations of H126B are shown. (d) Scheme of two-dimensional contacts among PLP, protein residues and the structural water molecules discussed in the text. Dashed lines indicate hydrogen-bond interactions and their length (in Å).
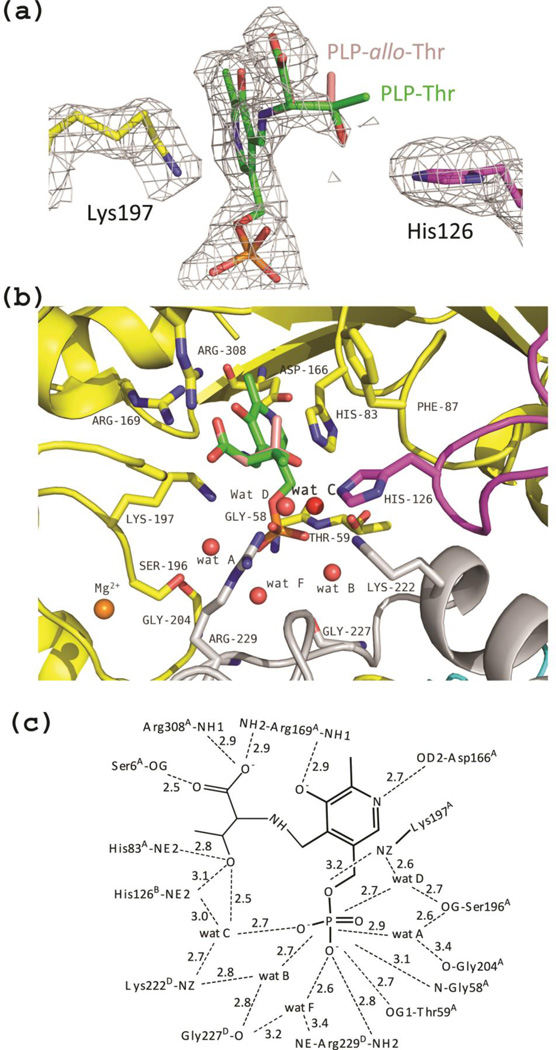
Co-crystallization of eTA with l-threonine resulted in the complete formation of an external aldimine in both active sites of the dimer in the asymmetric unit. However, while monomer B appears to have glycine bound as an external aldimine, in monomer A an additional, relatively well resolved density (30% occupancy) contiguous to Cα of glycine is visible. We interpreted this additional electron density as the Cβ and hydroxyl group of the hydroxyamino acid substrate (Fig. 6a). With l-threonine as substrate, the enzyme catalyses both the forward cleavage reaction and the reverse condensation reaction. Therefore, at equilibrium a mixture of glycine, acetaldehyde and l-threonine/l-allo-threonine will be present (since eTA is a low-specificity TA; (16)). It seems appropriate to notice that also tTA, when co-crystallized with l-threonine, showed the presence of l-allo-threonine bound to the enzyme as an external aldimine (12). As a matter of fact, in the electron density map of the eTA-Thr complex we also observed what appears to be a bound acetaldehyde molecule close to the mouth of the active site. The density was however refined with a network of water molecules (data not shown). According to our interpretation, the hydroxyl group of both l-threonine and l-allo-threonine occupy basically the same position, while the differently oriented methyl groups lack electron density, most likely because of their low occupancy ratio. The known position of the hydroxyl group allowed us to model the methyl groups of both l-allo-threonine and l-threonine (Fig. 6a–b). The modelled l-allo-threonine methyl group is located in a hydrophobic pocket formed by the PLP ring (at about 3.9 Å), His83A (at ~3.3 Å) and Phe87A (Tyr87 in tTA) (at ~4.8 Å). The modelled l-threonine methyl group, which is oriented towards the entrance of the active site, only makes close hydrophobic contacts with His126B (at ~3.4 Å distance), and significantly longer contacts with Tyr30D (at ~5.2 Å distance) and Phe87A (at ~5.5 Å distance). The authors of the tTA structure also reported a hydrophobic contact between the methyl group of l-allo-threonine and Tyr87 at a distance of 3.9 Å, which would lengthen to ~5Å if l-threonine were modelled at the active site, prompting the suggestion that Tyr87 is involved in ensuring preference for l-allo-threonine over l-threonine (12). Even though not mentioned in the tTA paper, the methyl group of l-allo-threonine in the tTA structure is about 3.6 Å from His83, similar to what we observed in the eTA structure.
Figure 6.
eTA-Thr complex. (a) Electron-density map of monomer A (with coefficients Fo-Fc shown at the 2.0σ level) showing that PLP exists as an external aldimine and the presence of the Cβ and the hydroxyl group of the hydroxyamino acid substrates. Eternal aldimines with both l-threonine (green sticks) and l-allo-threonine (salmon sticks) are modeled in the electron-density map. His126B is fixed in a single conformation. (b) View of the active site of monomer A. Both eternal aldimines with l-threonine (green sticks) and l-allo-threonine (salmon sticks) are shown. Wat C is labeled in bold. (c) Scheme of two-dimensional contacts among PLP, protein residues and the structural water molecules discussed in the text. Dashed lines indicate hydrogen-bond interactions and their length (in Å).
An interesting observation concerns the interactions between the substrate and the two active site histidine residues hypothesised in the tTA structure, corresponding to eTA His83A and His126B. In the eTA-Thr structure, we observe direct hydrogen-bond interactions (of ~3 Å length) between the substrate hydroxyl group and the imidazole group of both His83A and His126B (Figure 6b–c). In the active site of monomer B of the eTA-Thr complex, where an external aldimine with glycine is present, a water molecule can be modelled at the hydroxyl position (at lower contour level), making similar hydrogen-bond interactions with the histidine residues. Interestingly, while His126B is well defined in the eTA-Thr complex, making a direct hydrogen-bond to the hydroxyl group of the amino acid ligand and a water-mediated interaction with the PLP phosphate group (wat C in Fig. 6b–c), in both the unliganded and eTA-Ser structures, His126B assumes two different conformations. As noted above, also in the eTA-Ser complex, where an external aldimine with glycine is present, a water molecule (wat E in Fig. 5c–d), which can be modelled so as to occupy the hydroxyl group position of l-serine) makes direct hydrogen-bond contacts with His83A and His126B.
In the eTA-Ser and eTA-Thr forms, the carboxylate group of the substrates bound as external aldimine occupies very similar positions, making hydrogen-bond interactions with Arg169A, Arg308A, and Ser6A side chains (Figures 5c–d and 6b–c). These arginine residues most likely cooperate to neutralise the carboxylate negative charges and help stabilizing the transition state during the enzymatic reaction (25). In the unliganded structure, the positions corresponding to the carboxylate oxygen atoms are occupied by two water molecules, which also make hydrogen-bond interactions with the side chains of Arg308A and Ser6A.
Water Structure at the PLP-binding Site
Several structural water molecules are uniquely found in the eTA structures complexed with substrates. Formation of the external aldimines, even when present in partial occupancy, is accompanied by the presence of a water molecule that mediates interactions between the PLP phosphate and residues Arg229D and Gly227D (wat F in Figures 5c–d, and 6b–c). Another water molecule, which interacts with the PLP phosphate group, the amino group of Lys197A and the hydroxyl group of Ser196A in eTA-Thr complex evidently re-orients the Lys197 side chain when this is eliminated from the internal aldimine as substrates bind (wat D in Fig 6b–c). This water molecule may help explain how the external aldimine conformation of Lys197A is stabilized. In the unliganded structure, where the Lys197A side chain is involved in the internal aldimine with PLP and thus constrained from moving freely, this water molecule is missing.
Importantly, another structural water molecule, which is only present in the eTA-Thr form (wat C in Fig. 6b–c), attracted our attention, since it interacts with the side chains of several crucial residues present at the active site (notably His126B, His83A, and Lys222D; a long hydrogen bond interaction is also established with Arg229D), the substrate hydroxyl group and the PLP phosphate. The authors of the tTA structure also reported similar hydrogen-bond interactions involving the l-allo-threonine hydroxyl group, the His83A side chain, a water molecule and the PLP phosphate group (12). We propose wat C molecule to be important in eTA catalysis, as it will be discussed later.
Binding of divalent ions
Six divalent cations from the respective crystallization buffer are observed in each tetrameric structure (Mg2+ in the unliganded and eTA-Thr structures, and Ca2+ in the eTA-Ser structure. One ion is found close to each active site, about 9 Å from the PLP phosphate, forming a well-ordered octahedral coordination sphere with the amide oxygen atoms of Thr8A, Thr10A, Ser196A and Thr201A, and the side chains of Thr10A and Gln230D (Figs 3, 4a, 5c and 6b). These residues are part or are in close proximity of the active site. In the tTA structure, four calcium ions were also found to bind similarly at the four active sites, with conserved coordinating residues. The two additional metal ions found in the eTA structures are located at the interface of the two dimers, exactly at the two-fold axis of symmetry where all four monomers converge (Fig. 3b). The two ions are separated by ~5 Å and joined together by two water molecules to form a rhombic structure. Each divalent ion is further coordinated by residues from one crystallographic dimer, including the hydroxyl group of Ser97 from monomers A and B, the amide oxygen of Ala93 from monomers A and B, and the amide oxygen of Val94 from monomer A. The other divalent ion makes similar interactions with the dimer composed of monomers C and D. These intricate interactions most likely stabilize the tetrameric structure. Although the tTA structure also contains two additional ions, they are not located at the tetramer interface as described for eTA but are found at the surface of the protein, making interaction with symmetry-related crystal molecules.
Solution studies
Cofactor binding properties of eTA His83 and His126 mutant forms
The absorption spectra of all mutant forms, except those of the His83 mutants, were very similar to that of the wild type enzyme (data not shown; (11)). The presence of an absorption band with a maximum at 418 nm, due to the protonated internal aldimine of PLP, and a 1:1 ratio between cofactor and enzyme subunits demonstrated that these mutations did not affect the cofactor binding properties. This was not the case of the H83N and H83F mutants. Since His83 stacks to the re face of PLP ((12); and present paper), its replacement with either a Phe or an Asn residue was expected to somehow affect the cofactor binding properties. Therefore, the His83 mutants were purified using buffers containing 0.5 mM PLP and 10 mM l-serine, which obscured the absorption spectra of the enzymes. When excess PLP and serine were removed by dialysis, the spectra of the mutants suggested that all of the cofactor had been lost. When in the apoenzyme form, the mutant enzymes precipitated and could not be dissolved again after the addition of PLP. When kept with excess PLP and serine at 4 °C, the purified enzymes where stable for a couple of weeks, then a precipitate became visible and the catalytic activity was progressively lost.
Catalytic properties of eTA His83 and His126 mutant forms
eTA is designated as a low-specificity l-threonine aldolase, since it catalyses the retro-aldol cleavage of both threo and erythro forms of l-threonine, although with a clear preference for the latter isomer (in terms of kcat/Km, which is about 150 fold higher with l-allo-threonine; (11)).
All His83 and His126 mutants showed a measurable aldolase activity with both l-threonine and l-allo-threonine (Table 2), however, the H83N and Hi83F mutants had no activity unless a large excess of PLP over enzyme concentration was included in the assay. With l-allo-threonine, the activity of both His83 mutants showed a hyperbolic dependence on PLP concentration, with apparent dissociation constants of 30 ± 2 µM and of 48 ± 4 µM for H83N and H83F, respectively (data not shown). Therefore, with the His83 mutants, all kinetic measurements were carried out in the presence of 500 µM PLP. Even so, both mutants showed a greatly reduced kcat/Km ratio with respect to wild type eTA, mostly determined by decreased kcat values and increased Km values.
Table 2.
Kinetic parameters of the retro-aldol cleavage reactions catalysed by wild type and mutant eTAs with the threo and erythro isomers of l-threonine.
| Substrate | WT | H126N | H126F | H83N | H83F | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| kcata | Kmb |
|
kcat | Km | kcat | Km | kcat | Km | kcat | Km | |||||||||
| L-allo-Threonine | 213 | 0.24 | 887 | 469 | 0.96 | 488 | 541 | 0.2 | 2705 | 77 | 1.7 | 45 | 3.7 | 7 | 0.53 | ||||
| L-Threonine | 112 | 19.4 | 5.8 | 262 | 61 | 4 | 360 | 1.7 | 212 | 17 | 38 | 0.4 | 1.2 | 7 | 0.17 | ||||
All parameters are the average of three independent determinations, with the range between values being always less than ± 5%.
min−1
mM
Surprisingly, the His126 mutants showed doubled kcat values with respect to wild type eTA, with both erythro and threo substrates. With the H126N mutant, this increase was compensated by the increase of Km for both substrates, so that the specificity constant (kcat/Km) was not much different with respect to the wild type enzyme. However, the H126F mutant catalysed the l-threonine cleavage with a 30-fold higher kcat/Km, since Km for this substrate was also greatly decreased. H126F mutant also exhibited a three-fold increased kcat/Km with l-allo-threonine, mainly accounted by the increase of kcat.
All mutants maintained the capability to catalyse the transamination and racemization of alanine enantiomers. None of the mutations had drastic effects on the rates of these reactions (data not shown).
The kinetic parameters obtained with the mutant enzymes may be better examined if divided by the corresponding parameters obtained with the wild type enzyme, so as to compare relative values (Table 3). It is clear that both His83 mutations had a detrimental effect on the catalytic efficiency of the enzyme, while the H126 mutations had either a neutral (H126N) or a much favourable effect (H126F). What strikes the attention is the outcome of the mutations on stereospecificity. The preference for the erythro over the threo substrate (substrate preference, S.P., in Table 3) may be expressed as the ratio of specificity constants. The substrate preference of wild type enzyme is calculated from Table 2 to be 153 (i.e. 887/5.8). Both the H83N and H126N mutants have very similar substrate preference values (107 and 122, respectively). On the other hand, the H83F and H126F mutants have much lower substrate preference values (12 and 3, respectively), which means that have reduced preference for the erythro substrate. Therefore, with respect to their effect on substrate specificity, mutations may be grouped in two different categories: the His to Asn mutations, that are conservative with respect to substrate preference, and the His to Phe mutations that strongly decrease substrate preference. Interestingly, the H83F and H126F mutants decrease substrate preference following opposite ways. The H126F mutation increases kcat/Km for l-threonine much more than it increases kcat/Km for l-allo-threonine. On the other hand, the H83F mutation decreases kcat/Km for l-allo-threonine much more than it decreases kcat/Km for l-threonine.
Table 3.
Relative kinetic parameters of retro-aldol cleavage reactions catalysed by eTA mutant forms with the threo and erythro isomers of l-threonine.
| Substrate | H126N | H126F | H83N | H83F | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| kcata | Km | S.Pb | kcat | Km | S.P. | kcat | Km | S.P. | kcat | Km | S.P. | ||||||||
| L-allo-Threonine | 2.2 | 4 | 0.5 | 122 | 2.5 | 0.8 | 3 | 12 | 0.4 | 7.1 | 0.05 | 107 | 0.02 | 29.1 | 0.0006 | 3 | |||
| L-Threonine | 2.3 | 3.1 | 0.7 | 3.2 | 0.09 | 36.7 | 0.1 | 1.9 | 0.07 | 0.01 | 0.4 | 0.03 | |||||||
Values in the table are the ratio between the parameters determined with wild type and mutant enzymes, and shown in Table 2 (e.g. kcat mutant enzyme/kcat wild type enzyme). In the table, ratios higher than 2 are in bold, while ratios lower than 0.5 are underlined.
Substrate Preference, expressed as the ratio between the specificity constant (kcat/Km) determined with the erythro substrate and the specificity constant determined with the threo substrate. S.P. of the wild type enzyme is 153.
Catalytic properties of the H83F/H126F double mutant, of F87A, F87D and K222A mutants
Since both H83 and H126 seem to interact with the substrate hydroxyl group in the eTA-Thr crystal structure (Fig. 6), single mutations of either residues may be functionally compensated by the presence of the remaining histidine residue. With the intent to verify this possibility, a double mutant enzyme was produced and characterised. Tables 4 and 5 show that although kcat for the cleavage of both l-threonine isomers was drastically reduced, the double mutant enzyme still had a measurable aldolase activity. In particular, Km for l-allo-threonine was largely increased (while Km for l-threonine stayed the same), so that the preference of the double mutant for this substrate was lowered about ten times with respect to wild type eTA. In this respect, the effect of the double mutation resembles that of the H83F single mutation.
Table 4.
Kinetic parameters of the retro-aldol cleavage reactions catalysed by wild type and mutant eTAs with the threo and erythro isomers of l-threonine.
| Substrate | WT | H83F/H126F | F87A | F87D | K222A | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| kcata | Kmb | kcat | Km | kcat | Km | kcat | Km | kcat | Km | ||||||||||
| L-allo-Threonine | 213 | 0.24 | 887 | 1.7 | 7.4 | 0.23 | 160 | 0.31 | 516 | 117 | 0.41 | 285 | 80 | 1.2 | 67 | ||||
| L-Threonine | 112 | 19.4 | 5.8 | 0.44 | 21 | 0.02 | 43 | 8.7 | 4.9 | 19.8 | 24.9 | 0.79 | 43 | 72 | 0.6 | ||||
All parameters are the average of three independent determinations, with the range between values being always less than ± 5%.
min−1
mM
Table 5.
Relative kinetic parameters of retro-aldol cleavage reactions catalysed by eTA mutant forms with the threo and erythro isomers of l-threonine.
| Substrate | H83F/H126F | F87A | F87D | K222A | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| kcata | Km | S.Pb | kcat | Km | S.P. | kcat | Km | S.P. | kcat | Km | S.P. | ||||||||
| L-allo-Threonine | 0.008 | 31 | 0.0003 | 11.5 | 0.7 | 1.3 | 0.6 | 105 | 0.5 | 1.7 | 0.3 | 361 | 0.4 | 5.0 | 0.07 | 112 | |||
| L-Threonine | 0.004 | 1.1 | 0.003 | 0.4 | 0.4 | 0.8 | 0.2 | 1.3 | 0.1 | 0.4 | 3.7 | 0.1 | |||||||
Values in the table are the ratio between the parameters determined with wild type and mutant enzymes, and shown in Table 4 (e.g. kcat mutant enzyme/kcat wild type enzyme). In the table, ratios higher than 2 are in bold, while ratios lower than 0.5 are underlined.
Substrate Preference, expressed as the ratio between the specificity constant (kcat/Km) determined with the erythro substrate and the specificity constant determined with the threo substrate. S.P. of the wild type enzyme is 153
Kielkopf et al. (12) attributed to the residue at position 87 a crucial role in TAs stereospecificity. This residue is either a Tyr or a Phe in enzymes showing a marked preference for l-allo-threonine. In P. aerugnosa TA, which shows no substrate preference, an Asp residue occupies this position. We investigated the actual role of residue 87 in eTA through the characterization of F87A and F87D mutants. The F87A mutation had a very mild effect on the kinetic parameters of the aldol cleavage reaction, leaving the substrate preference of the enzyme basically unaffected (Tables 4 and 5). A slightly more pronounced effect may be ascribed to the F87D mutation, which showed marginally perturbed kinetic parameters and doubled substrate preference in favour of l-allo-threonine, although this may not be regarded as a clear switch of stereospecificity.
Another relevant conserved active site residue in TAs is Lys222. This residue is involved in the interaction network that keeps wat C in place (Fig. 6b), a water molecule that might play an important role in catalysis. The effect of the K222A mutation in eTA was to decrease kcat and increase Km of approximately the same extent with both l-threonine isomers. As a result, the catalytic efficiency with both substrates was lowered to the same extent and the S.P. was not affected (Tables 4 and 5).
DISCUSSION
Pyridoxal-5’-phosphate-dependent enzymes are grouped into at least five evolutionarily unrelated families, each having a different protein fold (26). Within the fold-type I family, l-threonine aldolase, serine hydroxymethyltransferase (SHMT) and fungal alanine racemase (AR) form a small subgroup of enzymes that are structurally and mechanistically strictly related (11, 15). The active site structures of these enzymes are very similar and this feature is probably at the base of their overlapping catalytic properties. Although each enzyme shows the highest specificity constant (kcat/KM) for the reaction from which its name derives, all three enzymes are able to catalyse at significant speed racemization, aldol cleavage and half-transamination reactions. Interestingly, also SHMT and fungal AR show a neat preference for l-allo-threonine over l-threonine, further confirming the structural similarity of the active sites (15).
Although the crystal structure of fungal AR is still missing, many three-dimensional structures of SHMT from several different sources have been solved (18, 22, 27–29). In contrast to l-TAs, where each active each site is formed by three monomers (using the active site formed by monomers A, B and D as a typical example; Fig. 3), in SHMT the corresponding active site is formed by monomers A and D. The active site loop from monomer B, which in l-TAs contributes His126B, is missing in SHMT. In this enzyme, another loop contributed by monomer A occupies the same position and is involved in the binding of tetrahydropteroylglutamate, the folate co-substrate of SHMT (11, 28). Obviously, this region of the active site neatly differentiates SHMT from l-TAs and confers reaction specificity. Although the active site pockets of eTA and E. coli SHMT seem to be similar in size, the pocket opening to the bulk solvent is significantly smaller in eSHMT than in eTA. The 4-residue corner between β-strands 11 and 12 (residues 303–306) from monomer A and a 9-residue loop between α-helices 2 and 3 (residues 26 −34) from monomer D, both guarding the mouth of the active site of the eTA structure have become 14- and 20-residue loops in the eSHMT, significantly decreasing the active site opening.
It should be remarked here that the physiological function of eTA is still unknown. The enzyme cleaves l-allo-threonine much more efficiently than l-threonine, however l-allo-threonine is not a recognized metabolite in E. coli. A very interesting investigation on pyridoxal 5’-phosphate synthesis in E. coli serendipitously demonstrated that eTA is able to efficiently catalyse the aldol condensation of glycolaldehyde and glycine to form 4-hydroxy-l-threonine, which serves as a precursor in PLP synthesis (both His83A and H126B may be involved in the binding of the two hydroxyl groups of glycolaldehyde) (30). This observation suggests that eTA may act on several different substrates in E. coli and insinuates that the genuine substrate of this enzyme (and therefore the real function of eTA) has yet to be discovered. Actually, the very large active site mouth of eTA does suggest that a much larger molecule than l-threonine isomers may be easily accommodated. The active site loop contributed by monomer B in eTA, and in particular His126B, may then play an important role in the recognition of this unknown substrate.
Catalytic mechanism and stereospecificity of E. coli l-threonine aldolase
As mentioned above, l-threonine aldolases are structurally and functionally strictly related to serine hydroxymethyltransferase (SHMT), a much more investigated PLP-dependent enzyme that is capable to catalyse the cleavage of a number of different β-hydroxyamino acids (11). The mechanism of SHMT-catalysed (tetrahydrofolate-independent) aldolase reactions has been explored for almost thirty years, yet it is still not fully understood. A recent, theoretical evaluation of possible mechanisms indicated the retro-aldol cleavage (Scheme 1) as a plausible one and also suggested that the cleavage of the Cα-Cβ bond is the rate-limiting step of the catalysed reaction (31). The retro-aldol cleavage mechanism has also been proposed as the mechanism followed by l-TAs (11). The results obtained in the present study with the double H83F/H126F mutant, which is still able to catalyse the cleavage of both l-threonine and l-allo-threonine at a measurable rate, demonstrate that neither of the histidine residues acts as a catalytic base in the retro-aldol cleavage mechanism. Since no other active site amino acid residues are at appropriate distance from the substrate hydroxyl group so as to act as catalytic base, we presume that a water molecule might be involved in the proton abstraction step of the retro-aldol cleavage mechanism. If there is no strict need for a strong catalytic base to remove the proton from the hydroxyl group and initiate the reaction, a weak base such as a polarized water molecule could easily carry out this job. A structural water molecule is in fact placed in a suitable position, interacting with several active site residues (His126, His83, and Lys222), the substrate hydroxyl group and the PLP phosphate (wat C in Fig. 6b) This water molecule may be mediating a proton transfer to the phosphate through hydrogen bonding. At this point, an important stereoelectronic requirement of the retro-aldol cleavage mechanism should be taken into consideration: the bonds to the eliminated substituents (a proton eliminated from the hydroxyl group and the quinonoid intermediate eliminated from Cβ) must lie approximately in the same plane (i.e. they must be periplanar). This is because the sp3 orbital of the oxygen bonded to the proton and the sp3 orbital of the Cβ bonded to the leaving quinonoid become overlapping π orbitals in the product aldehyde. This overlap provides significant stabilization in the transition state of the reaction. The fact that the wat C molecule is placed so as to allow a syn-periplanar conformation of the Oγ-H bond with respect to the Cα-Cβ bond (with a Cα-Cβ-OγH dihedral angle of approximately 0°; Fig. 1b) is strongly in favour of its possible role as catalytic base in the retro-aldol cleavage mechanism. It should be noticed that neither H83 nor H126 might orient the Oγ-H bond so as to allow a periplanar conformation. The symmetric effect of the Lys222A mutation on the kinetic parameters of l-threonine and l-allo-threonine cleavage (slight decrease of kcat and slight increase of Km) and the substantial upholding of substrate preference agrees with the hypothesis of a catalytic water molecule as part of a hydrogen bond network to which Lys222 participates.
Our crystallographic data suggest that the hydroxyl groups of the enzyme-bound l-allo-threonine and l-threonine are very similarly oriented, so that the difference between the two substrates resides in the orientation of the Cβ methyl group (Fig. 6b). Although neither H83 nor H126 act as a catalytic base, both residues clearly play some role in substrate binding and in determining the substrate preference of the enzyme.
The H126N mutation has the effect to increase both kcat and Km to the same extent for l-allo-threonine and l-threonine cleavage, so that the specificity constant and the substrate preference are basically unaltered. One could postulate that the H126N mutation affects the hydrogen bonds network involving H126, the catalytic water molecule and the substrate hydroxyl group, in a way that decreases the stability of enzyme-substrate complexes and at the same time decreases the activation energy of the retro-aldol cleavage rate limiting step. The H126F mutation has on kcat approximately the same effect as that observed with the H126N mutation. However, the presence of a phenyl group at the active site markedly decreases Km for l-threonine, while Km for l-allo-threonine is unaffected. As a consequence, the substrate preference of H126F eTA is greatly reduced with respect to wild type eTA (Table 3). Inspection of the active site shows that the methyl group of l-threonine is expected to be oriented towards F126 in this mutant form of the enzyme. F126 may therefore create a hydrophobic environment for the methyl group of l-threonine that favours binding of this substrate.
The H83 mutations are generally highly detrimental to eTA catalysis, since the stacking interaction between PLP and H83 is compromised. However, the catalytic properties of the H83 mutant clearly testify the role of this residue in substrate binding. While the H83N mutation does not alter substrate specificity, the H83F mutant shows a greatly reduced preference for l-allo-threonine, mainly due to the increase of Km for this substrate with respect of Km for l-threonine. This behaviour may result from the steric hindrance determined by the presence of F83 that stacks to PLP in the H83F mutant and is expected to be in very close proximity to the methyl group of l-allo-threonine.
Kielkopf et al. (12) pointed out that a single variable residue in l-TAs active site might be involved in discriminating l-threonine from l-allo-threonine. According to these authors, side chain bulk at position 87 is correlated with specificity for l-allo-threonine, with larger side chain displaying higher preferences for the allo isomer. However, the results we obtained with the F87A eTA mutant clearly show that reducing the bulk at position 87 does not alter the substrate preference of the enzyme. It is worth noting that in P. aeruginosa TA, which does not show any substrate preference (20), an aspartate residue is present at position 87 (Fig 2). In our study, the F87D mutation doubled the preference of the enzyme for l-allo-threonine, which is somehow different from what expected according to the above-mentioned hypothesis.
In the light of our kinetic and crystallographic results, we believe that the substrate preference of l-TAs is determined by the overall microenvironment that surrounds the substrate bound at the enzyme active site rather than to one or more specific residues. Moreover, this substrate preference, which is based on l-threonine stereoisomers (probably not the actual substrates of the enzymes), may not be related to the physiological role of l-TAs. The l-TAs family may be a group of structurally and mechanistically similar enzymes that act of different l-3-hydroxyamino substrates and therefore play diverse physiological functions in different organisms. As an example, in Candida albicans, the second enzyme of carnitine biosynthesis, namely a hydroxytrimethyllysine aldolase, is encoded by a member of the l-TA family (2).
EXPERIMENTAL PROCEDURES
Materials
Ingredients for bacterial growth were from Sigma-Aldrich. DEAE-Sepharose and Phenyl-Sepharose used for chromatography were from GE Healthcare. l-Lactic dehydrogenase, alcohol dehydrogenase, l-alanine dehydrogenase and d-amino acid oxidase were from Sigma-Aldrich. The oligonucleotide primers used in site directed mutagenesis were from MWG-Biotech (Ebersberg, Germany). The QuickChange site-directed mutagenesis kit was from Stratagene (La Jolla, CA). All other reagents were from Sigma-Aldrich. Wild type and mutant forms of l-threonine aldolase from E. coli (eTA) was expressed and purified to homogeneity as previously described (11).
Crystallization
Freshly dialyzed protein (in 20 mM potassium phosphate, pH 7.0) was used for crystallization. Crystals of eTA in the absence and presence of reactive ligands, L-serine and l-threonine were grown by hanging drop vapor diffusion at 25 °C using the Hampton Research Crystal Screens. Optimized condition for growing native (unliganded) eTA crystals consisted of 603 µM of the enzyme, 0.2 M magnesium chloride hexahydrate, 0.1 M HEPES, pH 7.5, and 30% PEG 400. Crystals of eTA (480 µM) with l-threonine (28 mM) were also grown using the same crystallization buffer/precipitant. The crystallization solution also served as a cryoprotectant. Co-crystals of eTA (274 µM) with L-serine (2 mM) were grown in 0.2 M calcium chloride hexahydrate, 0.1 M HEPES, pH 7.5, and 28% PEG 200, which also worked as a cryoprotectant.
Data Collection
Diffraction data of the flash frozen crystals were collected at 100 K with a Rigaku IV ++ image plate detector using a CuKα X-rays (λ = 1.54 Å) from a MicroMax-007 source fitted with Varimax Confocal optics (Rigaku, The Woodlands, TX). The native and liganded eTA crystalized in an orthorhombic space group with two molecules per asymmetric unit. The datasets were processed with the d*trek software (Rigaku) and the CCP4 suite of programs (32). The X-ray data are summarized in Table 1.
Structure determination
Initial phases for the native eTA structure were obtained from the web-based molecular replacement program, CaspR (33) using the structure of l-threonine aldolase from Thermatoga maritima (PDB ID 1LW5; 24.1% sequence identity) as a search model. The correct amino sequence of eTA was built in Phenix (34), and the model subsequently refined with CNS (35). The electron density maps were well defined except for a loop of five residues (284–288) and the residues 136–138. PLP showed a well-defined density at the active site and was subsequently modelled in. Three magnesium ions, from the crystallization buffer (0.2 M magnesium chloride hexahydrate) were added to the model. COOT (36) was used for model correction. The model was refined to Rwork/Rfree 20.9/27.9 at 2.2 Å resolution. The isomorphous eTA complex with L-serine and l-threonine were refined using the refined native eTA structure. PLP, as well as Mg2+ or Ca2+ ions from the crystallization buffers were added to the l-threonine and l-serine complex structures, respectively. The complexes were refined to Rwork/Rfree of 22.6/28.9 at 2.2 Å resolution and 18.2/24.4 at 2.1 Å resolution, respectively.
Site-directed mutagenesis, expression and purification of the mutant forms
The ltaE gene, encoding E. coli low-specificity eTA (11), inserted into a pET22b(+) plasmid vector (Novagen Inc., Madison, WI, USA) was used as template in site-directed mutagenesis reactions carried out using the QuickChange kit from Stratagene. For each enzyme mutant form, two complementary oligonucleotides containing the mutations were used as primers. E. coli DH5α cells were transformed and used to amplify the mutated plasmid. Both strands of the coding region of the mutated gene were sequenced. The only differences with respect to wild type were those intended. All mutant forms were then expressed in E. coli HMS174(λDE3) strain cells (Novagen Inc., Madison, WI, USA) and were purified to homogeneity and in good yield using the same procedure adopted for wild type eTA (11). When purifying the H83 mutant forms, 0.5 mM PLP and 10 mM L-serine were added to all chromatography and dialysis buffers in order to prevent dissociation of the cofactor from the active site.
Kinetic studies and data analysis
The rate of threonine cleavage was measured by coupling the reaction with reduction of the product acetaldehyde by NADH and alcohol dehydrogenase (37). The rate of the reaction was calculated from the rate of disappearance in absorbance at 340 nm, using a value of ε340 = 6220 cm−1 M−1. Both reactions were carried out in 20 mM potassium phosphate, pH 7.0, at 30 °C. When using H83 mutants as catalysts, 500 µM PLP was included in the reaction mixture. With these mutants, Kd of PLP was calculated from experiments in which the aldolase activity with l-allo-threonine was measured at various cofactor concentrations, by best fitting of initial velocity data to Eq. 1.
The pseudo first-order rate constants of transamination of D- and L-alanine were determined by measuring the disappearance of absorbance at either 498 nm or 420 nm, during the conversion of the enzyme-bound PLP to pyridoxamine 5’-phosphate (PMP) (38). Each reaction was carried out in 50 mM sodium N,N-bis(2-hydroxyethyl)-2-aminoethanesulfonate (NaBES), pH 7.6, at 37 °C and contained 37 µM eTA and 210 mM alanine. With H83 mutants, inclusion of PLP (500 µM) in the reaction established a catalytic cycle. In fact, PMP is bound very loosely to eTA and, once is formed, dissociates from the active site (11) and can easily be replaced by PLP if this is present at 500 µM concentration. In the conditions used in our experiments, the enzyme was constantly saturated with PLP, therefore the course of the reaction could not be followed by measuring the decrease of enzyme-bound PLP (decrease of absorbance at 420 or 498 nm) but had to be tracked by measuring the steady state increase of PMP concentration. The extinction coefficient of PMP (2566 M−1 cm−1) was worked out from single turnover transamination experiments in which the initial concentration of enzyme-bound PLP was known. The apparent Kd for both alanine enantiomers were determined by titrating the enzyme with increasing concentrations of alanine and determining the maximum absorbance at 498 nm due to formation of the quinonoid intermediate. The Kd was calculated from a best fit of ΔA498 values to Eq. 2. With the H83N and H83F mutants, which did not accumulate a quinonoid intermediate, the Kd was calculated from a series of experiments in which the pseudo-first order rate of transamination was measured at various alanine concentrations (Eq. 3).
Racemization reactions of d- and l-alanine were carried out in 50 mM NaBES, pH 7.6, at 37 °C. The reaction mixture contained 30 µM enzyme, 200 mM d- or l-alanine and 1 mM PLP in a volume of 0.5 ml. Control reactions containing no enzyme were carried out in order to take in account a possible contamination of the opposite enantiomer. However, no contaminations could be detected. At various time intervals, 45 µl aliquots of the reaction mixture were removed and the reaction was stopped by addition of 160 mM HClO4. The solution was neutralised adding an equivalent amount of KOH and centrifuged to remove the precipitated protein and KClO4. The sample was then assayed for either D- or L-alanine. The assay for l-alanine consisted of 10 mM NAD+, 0.2 M hydrazine, the sample and 5 units of L-alanine dehydrogenase in 100 mM sodium borate pH 9.5. The change in absorbance at 340 nm due to reduction of NAD+ was used to calculate the concentration of L-alanine produced. For the l → d direction, d-amino acid oxidase and lactate dehydrogenase were used as the coupling enzymes. In the assay, the sample deriving from the reaction mixture was mixed with 0.2 mM NADH and 5 units of lactate dehydrogenase in 20 mM NaBES pH 7.0. In these conditions, the side-product pyruvate, generated from alanine during transamination, was eliminated by conversion into lactate. The following addition of 1.5 units of d-amino acid oxidase then converted the D-alanine produced in the racemization reaction into pyruvate and the latter into lactate with simultaneous consumption of NADH. The concentration of d-alanine in the sample was calculated from the decrease in absorbance at 340 nm observed after addition of d-amino acid oxidase. All enzymes used in the assays were dialysed against either 100 mM sodium borate, pH 9.5, or 20 mM NaBES, pH 7.0, and mixed with 50% glycerol before use. With H83 mutants, 500 µM PLP was included in the reaction mixture.
All spectral and kinetic studies were carried out on a Hewlett-Packard 8452A diode array spectrophotometer. Kinetic data analysis, curve-fitting procedures and statistical analysis were performed using the software GraphPad Prism version 4.01 (GraphPad Software, San Diego California USA). The following equations were used to fit the data:
| Equation 1 |
| Equation 2 |
| Equation 3 |
ACKNOWLEDGEMENTS
This work was supported by grants from the Italian Ministero dell’Istruzione, dell’Università e della Ricerca and from Finanziamento Progetti di Ricerca 2011 of Sapienza University of Rome. Mirella Vivoli and Simona D’Aguanno carried out the described work in partial fulfilment of the requirements for the degree of Doctor of Philosophy in Biochemistry. Structural biology resources were provided in part by NIH award CA16059 to the VCU Massey Cancer Center.
Abbreviations
- TA
threonine aldolase
- eTA
low-specificity Escherichia coli l-threonine aldolase
- tTA
Thermotoga maritima l-threonine aldolase
- SHMT
serine hydroxymethyltransferase
- AR
alanine racemase
- PLP
pyridoxal-5’-phosphate
- PMP
pyridoxamine 5’-phosphate
- eTA-Ser
crystal structure obtained from co-crystallyzation of eTA with l-serine
- eTA-Thr
crystal structure obtained from co-crystallyzation of eTA with l-threonine
Footnotes
RCSB PDB (www.pdb.org): structural data are available in the Protein Data Bank/BioMagResBank databases under the accession numbers 4LNJ, 4LNM and 4LNL for the unliganded, eTA-Thr and eTA-Ser structures.
REFERENCES
- 1.Duckers N, Baer K, Simon S, Groger H, Hummel W. Threonine aldolases-screening, properties and applications in the synthesis of non-proteinogenic beta-hydroxy-alpha-amino acids. Appl Microbiol Biotechnol. 2010;88:409–424. doi: 10.1007/s00253-010-2751-8. [DOI] [PubMed] [Google Scholar]
- 2.Strijbis K, van Roermund CW, Hardy GP, van den Burg J, Bloem K, de Haan J, van Vlies N, Wanders RJ, Vaz FM, Distel B. Identification and characterization of a complete carnitine biosynthesis pathway in Candida albicans. FASEB J. 2009;23:2349–2359. doi: 10.1096/fj.08-127985. [DOI] [PubMed] [Google Scholar]
- 3.Sagui F, Conti P, Roda G, Contestabile R, Riva S. Enzymatic synthesis of omega-carboxy-beta-hydroxy-L-alpha-amino acids. Tetrahedron. 2008;64:5079–5084. [Google Scholar]
- 4.Gwon HJ, Yoshioka H, Song NE, Kim JH, Song YR, Jeong DY, Baik SH. Optimal production of L-threo-2,3-dihydroxyphenylserine (L-threo-DOPS) on a large scale by diastereoselectivity-enhanced variant of L-threonine aldolase expressed in Escherichia coli. Prep Biochem Biotechnol. 2012;42:143–154. doi: 10.1080/10826068.2011.583975. [DOI] [PubMed] [Google Scholar]
- 5.di salvo ML, Budisa N, Contestabile R. PLP- dependent Enzymes: a Powerful Tool for Metabolic Synthesis of Non- canonical Amino Acids in Molecular evolution and control (Molekulare Entwicklung und Kontrolle; Beilstein Symposium Ed; 2013. Vol. in press. [Google Scholar]
- 6.Liu JQ, Dairi T, Kataoka M, Shimizu S, Yama H. Diversity of microbial threonine aldolases and their application. Mol. Catal. B. Enzym. 2000;10:107–115. [Google Scholar]
- 7.Liu JQ, Odani M, Yasuoka T, Dairi T, Itoh N, Kataoka M, Shimizu S, Yamada H. Gene cloning and overproduction of low-specificity D-threonine aldolase from Alcaligenes xylosoxidans and its application for production of a key intermediate for parkinsonism drug. Appl Microbiol Biotechnol. 2000;54:44–51. doi: 10.1007/s002539900301. [DOI] [PubMed] [Google Scholar]
- 8.Liu JQ, Dairi T, Itoh N, Kataoka M, Shimizu S, Yamada H. A novel metal-activated pyridoxal enzyme with a unique primary structure, low specificity D-threonine aldolase from Arthrobacter sp. Strain DK-38. Molecular cloning and cofactor characterization. J Biol Chem. 1998;273:16678–16685. doi: 10.1074/jbc.273.27.16678. [DOI] [PubMed] [Google Scholar]
- 9.Schneider G, Kack H, Lindqvist Y. The manifold of vitamin B6 dependent enzymes. Structure. 2000;8:R1–R6. doi: 10.1016/s0969-2126(00)00085-x. [DOI] [PubMed] [Google Scholar]
- 10.Paiardini A, Contestabile R, D’Aguanno S, Pascarella S, Bossa F. Threonine aldolase and alanine racemase: novel examples of convergent evolution in the superfamily of vitamin B6-dependent enzymes. Biochim Biophys Acta. 2003;1647:214–219. doi: 10.1016/s1570-9639(03)00050-5. [DOI] [PubMed] [Google Scholar]
- 11.Contestabile R, Paiardini A, Pascarella S, di Salvo ML, D’Aguanno S, Bossa F. l-Threonine aldolase, serine hydroxymethyltransferase and fungal alanine racemase. A subgroup of strictly related enzymes specialized for different functions. Eur J Biochem. 2001;268:6508–6525. doi: 10.1046/j.0014-2956.2001.02606.x. [DOI] [PubMed] [Google Scholar]
- 12.Kielkopf CL, Burley SK. X-ray structures of threonine aldolase complexes: structural basis of substrate recognition. Biochemistry. 2002;41:11711–11720. doi: 10.1021/bi020393+. [DOI] [PubMed] [Google Scholar]
- 13.Giger L, Toscano MD, Madeleine B, Marlierre P, Hilvert D. A novel genetic selection system for PLP-dependent threonine aldolases. Tetrahedron. 2012;68:7549–7557. [Google Scholar]
- 14.Gwon HJ, Baik SH. Diastereoselective synthesis of L: -threo-3,4-dihydroxyphenylserine by low-specific L: -threonine aldolase mutants. Biotechnol Lett. 2010;32:143–149. doi: 10.1007/s10529-009-0125-z. [DOI] [PubMed] [Google Scholar]
- 15.di Salvo L, Florio R, Paiardini A, Vivoli M, D’Aguanno S, Contestabile R. Alanine racemase from Tolypocladium inflatum: a key PLP-dependent enzyme in cyclosporin biosynthesis and a model of catalytic promiscuity. Arch Biochem Biophys. 2013;529:55–65. doi: 10.1016/j.abb.2012.11.011. [DOI] [PubMed] [Google Scholar]
- 16.Liu JQ, Dairi T, Itoh N, Kataoka M, Shimizu S, Yamada H. Gene cloning, biochemical characterization and physiological role of a thermostable low-specificity L-threonine aldolase from Escherichia coli. Eur J Biochem. 1998;255:220–226. doi: 10.1046/j.1432-1327.1998.2550220.x. [DOI] [PubMed] [Google Scholar]
- 17.Szebenyi DM, Musayev FN, di Salvo ML, Safo MK, Schirch V. Serine hydroxymethyltransferase: role of glu75 and evidence that serine is cleaved by a retroaldol mechanism. Biochemistry. 2004;43:6865–6876. doi: 10.1021/bi049791y. [DOI] [PubMed] [Google Scholar]
- 18.Renwick SB, Snell K, Baumann U. The crystal structure of human cytosolic serine hydroxymethyltransferase: a target for cancer chemotherapy. Structure. 1998;6:1105–1116. doi: 10.1016/s0969-2126(98)00112-9. [DOI] [PubMed] [Google Scholar]
- 19.Capitani G, De Biase D, Aurizi C, Gut H, Bossa F, Grutter MG. Crystal structure and functional analysis of Escherichia coli glutamate decarboxylase. Embo J. 2003;22:4027–4037. doi: 10.1093/emboj/cdg403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu JQ, Ito S, Dairi T, Itoh N, Kataoka M, Shimizu S, Yamada H. Gene cloning, nucleotide sequencing, and purification and characterization of the low-specificity L-threonine aldolase from Pseudomonas sp. strain NCIMB 10558. Appl Environ Microbiol. 1998;64:549–554. doi: 10.1128/aem.64.2.549-554.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu JQ, Nagata S, Dairi T, Misono H, Shimizu S, Yamada H. The GLY1 gene of Saccharomyces cerevisiae encodes a low-specific L-threonine aldolase that catalyzes cleavage of L-allo-threonine and L-threonine to glycine--expression of the gene in Escherichia coli and purification and characterization of the enzyme. Eur J Biochem. 1997;245:289–293. doi: 10.1111/j.1432-1033.1997.00289.x. [DOI] [PubMed] [Google Scholar]
- 22.Trivedi V, Gupta A, Jala VR, Saravanan P, Rao GS, Rao NA, Savithri HS, Subramanya HS. Crystal structure of binary and ternary complexes of serine hydroxymethyltransferase from Bacillus stearothermophilus: insights into the catalytic mechanism. J Biol Chem. 2002;277:17161–17169. doi: 10.1074/jbc.M111976200. [DOI] [PubMed] [Google Scholar]
- 23.Kirsch JF, Eichele G, Ford GC, Vincent MG, Jansonius JN, Gehring H, Christen P. Mechanism of action of aspartate aminotransferase proposed on the basis of its spatial structure. J Mol Biol. 1984;174:497–525. doi: 10.1016/0022-2836(84)90333-4. [DOI] [PubMed] [Google Scholar]
- 24.Di Salvo ML, Scarsdale JN, Kazanina G, Contestabile R, Schirch V, Wright HT. Structure-Based Mechanism for Early PLP-Mediated Steps of Rabbit Cytosolic Serine Hydroxymethyltransferase Reaction. Biomed Res Int in press. 2013 doi: 10.1155/2013/458571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delle Fratte S, Iurescia S, Angelaccio S, Bossa F, Schirch V. The function of arginine 363 as the substrate carboxyl-binding site in Escherichia coli serine hydroxymethyltransferase. Eur J Biochem. 1994;225:395–401. doi: 10.1111/j.1432-1033.1994.00395.x. [DOI] [PubMed] [Google Scholar]
- 26.Eliot AC, Kirsch JF. Pyridoxal phosphate enzymes: mechanistic, structural, and evolutionary considerations. Annu Rev Biochem. 2004;73:383–415. doi: 10.1146/annurev.biochem.73.011303.074021. [DOI] [PubMed] [Google Scholar]
- 27.Scarsdale JN, Kazanina G, Radaev S, Schirch V, Wright HT. Crystal structure of rabbit cytosolic serine hydroxymethyltransferase at 2.8 A resolution: mechanistic implications. Biochemistry. 1999;38:8347–8358. doi: 10.1021/bi9904151. [DOI] [PubMed] [Google Scholar]
- 28.Scarsdale JN, Radaev S, Kazanina G, Schirch V, Wright HT. Crystal structure at 2.4 A resolution of E. coli serine hydroxymethyltransferase in complex with glycine substrate and 5-formyl tetrahydrofolate. J Mol Biol. 2000;296:155–168. doi: 10.1006/jmbi.1999.3453. [DOI] [PubMed] [Google Scholar]
- 29.Szebenyi DM, Liu X, Kriksunov IA, Stover PJ, Thiel DJ. Structure of a murine cytoplasmic serine hydroxymethyltransferase quinonoid ternary complex: evidence for asymmetric obligate dimers. Biochemistry. 2000;39:13313–13323. doi: 10.1021/bi000635a. [DOI] [PubMed] [Google Scholar]
- 30.Kim J, Kershner JP, Novikov Y, Shoemaker RK, Copley SD. Three serendipitous pathways in E. coli can bypass a block in pyridoxal-5’-phosphate synthesis. Mol Syst Biol. 2010;6:436. doi: 10.1038/msb.2010.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chiba Y, Terada T, Kameya M, Shimizu K, Arai H, Ishii M, Igarashi Y. Mechanism for folate-independent aldolase reaction catalyzed by serine hydroxymethyltransferase. FEBS J. 2012;279:504–514. doi: 10.1111/j.1742-4658.2011.08443.x. [DOI] [PubMed] [Google Scholar]
- 32.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Claude JB, Suhre K, Notredame C, Claverie JM, Abergel C. CaspR: a web server for automated molecular replacement using homology modelling. Nucleic Acids Res. 2004;32:W606–W609. doi: 10.1093/nar/gkh400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 35.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 36.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 37.Schirch L, Peterson D. Purification and properties of mitochondrial serine hydroxymethyltransferase. J Biol Chem. 1980;255:7801–7806. [PubMed] [Google Scholar]
- 38.Shostak K, Schirch V. Serine hydroxymethyltransferase: mechanism of the racemization and transamination of D- and L-alanine. Biochemistry. 1988;27:8007–8014. doi: 10.1021/bi00421a006. [DOI] [PubMed] [Google Scholar]