Abstract
Induced plant defenses against herbivores are modulated by jasmonic acid-, salicylic acid-, and ethylene-signaling pathways. Although there is evidence that some pathogens suppress plant defenses by interfering with the crosstalk between different signaling pathways, such evidence is scarce for herbivores. Here, we demonstrate that the mealybug Phenacoccus solenopsis suppresses the induced defenses in tomato. We found that exogenous JA, but not SA, significantly decreased mealybug feeding time and reduced nymphal performance. In addition, constitutive activation of JA signaling in 35s::prosys plants reduced mealybug survival. These data indicate that the JA signaling pathway plays a key role in mediating the defense responses against P. solenopsis. We also found that mealybug feeding decreased JA production and JA-dependent defense gene expression, but increased SA accumulation and SA-dependent gene expression. In SA-deficient plants, mealybug feeding did not suppress but activated JA accumulation, indicating that the suppression of JA-regulated defenses depends on the SA signaling pathway. Mealybugs benefit from suppression of JA-regulated defenses by exhibiting enhanced nymphal performance. These findings confirm that P. solenopsis manipulates plants for its own benefits by modulating the JA-SA crosstalk and thereby suppressing induced defenses.
Plants possess an array of direct and indirect defenses to protect them against herbivore attack. The role of phytohormones and signaling pathways in the regulation of theses direct and indirect defenses is well established. The octadecanoid pathway, with the central phytohormone jasmonic acid (JA); the shikimate pathway, with the central phytohormone salicylic acid (SA); and the ethylene (ET) pathway are recognized as key signaling pathways in mediating plant defense responses1,2,3,4. According to the model proposed by Reymond and Farmer5, a plant tailors its defense responses to a specific attacker by eliciting signaling molecules from the three pathways to different degrees. Supporting evidence has been accumulating that the crosstalk between JA/ET and SA signaling pathways allows plants to fine-tune the induction of their defense in response to different herbivores or pathogens6,7,8,9. JA-SA crosstalk often results in reciprocal antagonism between these two pathways has been well-demonstrated to be an adaptive strategy that enhances plant fitness10.
Although crosstalk among different signaling pathways tailors plant responses to specific herbivores, the selective advantage of manipulating such crosstalk from the perspective of herbivores must be high. Some recent studies showed that insect herbivores can suppress induced defenses of plants11,12,13,14. In these cases, the suppression of plant defense responses is often associated with changes of phytohormone biosynthesis or signaling pathways. For example, oral secretions of the beet armyworm Spodoptera exigua can suppress JA and ET accumulation but enhance SA accumulation in Nicotiana attenuata8, and the SA induction in Arabidopsis facilitates the growth of S. exigua larvae15. Similarly, the silverleaf whitefly Bemisia tabaci activates SA-dependent responses and represses JA-dependent defense responses to their own advantage in Arabidopsis thaliana11,16,17. Considering the well-demonstrated JA-SA crosstalk, researchers speculated that insects can manipulate plant defenses for their own benefits by modulating the JA-SA crosstalk18. Although this speculation has been supported by a few studies12,13,15,19, the mechanisms involved have received little attention.
Recently, we found that the solenopsis mealybug Phenacoccus solenopsis, which is a newly recognized invasive insect in China20, can suppress the induction of JA-regulated genes and defense metabolites to enhance its nymphal performance on cotton21. However, the underlying mechanism involved in the suppression of JA defense responses by P. solenopsis remains unknown. Considering that P. solenopsis may also activate the induction of SA-dependent responses and show enhanced performance on SA-treated plants21, we hypothesized that suppression of JA defense by P. solenopsis might be mediated by crosstalk with the SA signaling pathway. To test this hypothesis, we used wild-type tomato that is a host plant for P. solenopsis and two transgenic tomato lines including NahG with defective SA biosynthesis and 35S::prosys with constitutive JA signaling. First, we examined the effects of exogenous JA, SA, and herbivory treatments on the performance of P. solenopsis larvae. Second, we used the electronic penetration graph (EPG) technique to record the P. solenopsis feeding behavior on undamaged plants and plants treated with JA, SA, and herbivory. Third, we quantified the accumulation of endogenous JA and SA as well as the transcript levels of JA- and SA-dependent genes in host plants in response to P. solenopsis feeding. Finally, we examined the survival rate of P. solenopsis on NahG, 35S::prosys, and their corresponding wild-type plants, and quantified JA and SA in the two transgenic lines in response to P. solenopsis feeding. Our results demonstrate that P. solenopsis feeding enhances SA accumulation, which suppresses the JA signaling pathway. As a consequence of this herbivore modification of JA-SA crosstalk, the performance of P. solenopsis nymphs is enhanced.
Methods
Plants and insects
Wild-type tomato (Lycopersicon esculentum) cv Moneymaker (MM) is the parental line for the SA-non-accumulating NahG mutant. Wild-type tomato (Solanum lycopersicum) cv Castlemart (CM) is the parental line for the transgenic tomato line 35s::prosys in which JA signaling is constitutive. 35S::prosys seeds were collected from a 35S::prosys homozygote that had been backcrossed five times to its wild-type line cv Castlemart22,34. Tomato seedlings were grown in 500-ml pots containing a commercial potting mix (Fafard Growing Mix 1, Agawam, MA), and were kept in an insect-free greenhouse compartment under natural light and 28/24°C. Plants with four to five fully expanded leaves were used for experiments.
The mealybug, Phenacoccus solenopsis Tinsley (Hemiptera: Pseudococcidae), was originally collected from Hibiscus rosa-sinensis in Hangzhou (30°10′N, 120°15′E), China, and was maintained on wild-type tomato cv Moneymaker in a climate-controlled room (25 ± 3°C, 60–70% RH, 12L: 12D photoperiod).
Plant treatments
Chemical treatment: Jasmonic acid or salicylic acid (Sigma-Aldrich) was dissolved in 0.5 mL of acetone and dispersed in water (containing 0.1% Tween 20) to produce a 1.0 mM JA or SA solution. Each plant was sprayed with 1.0 mL/leaf of the JA or SA solution with a hand-sprayer. Twenty-four hours later, JA- or SA-treated plants were used for experiments.
Mealybug treatment: A mixture of third-instar nymphs and newly emerged adults (40 in total) of P. solenopsis were carefully transferred onto each plant and allowed to feed freely for 6, 12, 24, 72, or 120 h. After that, leaf samples were collected for phytohormone analysis. Plants pre-infested with 40 P. solenopsis adults (2- to 4-days old) for 120 h were used for performance bioassays, the EPG experiment, and gene-expression analysis.
Control treatment: In a preliminary experiment, we found that treatment with a water solution (including 0.5 mL of acetone and 0.1% Tween 20) had no effect on the performance of P. solenopsis reared on tomato plants. Thus, healthy and intact plants that received no treatment were used as control plants.
To avoid the potential interference among plants from different treatments, plants from the same treatment were maintained in a separate climate-controlled room (25 ± 3°C, 60–70% RH, 12L: 12D photoperiod). And each plant was individually kept in a glass cage (25 × 25 × 50 cm).
Performance of mealybugs feeding on tomato plants
We determined the effects of JA, SA, and herbivory treatments on the survival of P. solenopsis. Young nymphs (≤24 h old) from the same cohort were transferred onto the leaves (50 nymphs per plant) of control plants or plants treated with JA, SA, or herbivory (mealybugs). The plants were kept in a climate-controlled room (26 ± 2°C, 60–70% RH, 12L: 12D photoperiod). The mealybugs were assessed twice daily for survival, and survival rates were calculated at 14, 21, and 28 d after the young nymphs had been placed on the plants. Each treatment was represented by four replicate plants.
A similar experiment was performed to determine the effects of control, JA, SA, and herbivory treatments on P. solenopsis developmental duration (from egg to last molt). The procedure was the same as that described in the previous paragraph except that developmental duration was recorded twice daily until adults emerged, and each treatment was represented by five replicate plants.
EPG recording of P. solenopsis feeding behavior on tomato plants
P. solenopsis stylet penetration activities on the leaves of control plants or plants treated with JA, SA, or herbivory (mealybugs) were recorded using a four-channel DC-EPG system (Giga-4; Wageningen, The Netherlands). The method used for recording was the same as that described by Huang et al. (2014)23. In brief, 7- to 8-day-old adult females were carefully collected from tomato plants. The dorsal wax was partially and gently removed from each specimen with a fine brush to enable the attachment of a gold wire electrode (18 μm diameter, 2 cm long). Experiments were conducted in a Faraday cage in the laboratory at 27 ± 2°C. The recordings were started around 08:30 hours and continued for 12 h under florescent light. For each treatment, 20 recordings, each done with a separate mealybug on a separate plant, were used for characterization and analysis of the EPG signals using the EPG analysis worksheet.
Quantification of endogenous JA and SA
Endogenous JA and SA in tomato plants were quantified as described by Wei et al. (2013)24 and Almeida-Trapp et al. (2014)25. In brief, plant material (250–300 mg) was frozen and ground in liquid nitrogen. The resulting powder was mixed with 4 mL of HPLC grade methanol (Sigma-Aldrich) and kept at −20°C for 12 h. For quantification, [9, 10]-dihydro-JA (300 ng) and d6-SA (500 ng) were added as internal standards. JA, SA, and their internal standards were partitioned to an aqueous phase by centrifugation and vaporization. After three rounds of freezing and thawing, the aqueous phase was centrifuged, and the pH of the supernatant was adjusted to 3.0 using 0.1 M HCl. JA, SA, and their internal standards were extracted from the supernatant with an equal volume of ethyl acetate and then dried. The dried extract was re-suspended in 0.1 M acetic acid and loaded onto a C18 column (Waters Company, Milford, MA, USA). The C18 column was sequentially eluted with a series of solvent mixtures [acetic acid/methanol (v/v) at 83/17, 60/40 and 40/60]. After evaporation of the solvent and esterification of the residue using excess ethereal diazomethane, the elution sample volume was adjusted to 50 mL with ethyl acetate. Samples were analyzed using a GC/MS system (6890N/5973 MSD, Agilent Technologies, Inc., Palo Alto, CA, USA) equipped with an HP-5-MS column (30 m × 0.25 mm × 0.25 mm; 19091S-433, J&W Scientific, Agilent Technologies). Endogenous JA, SA, and their internal standards were analyzed in full-scan mode as described by Wei et al. (2013)24.
Quantitative real-time PCR
Total RNA was extracted and purified as described by Zhang et al. (2013)19. First stand cDNA was synthesized from 200 ng RNA using a First-Strand cDNA Synthesis Kit (Bio-Rad, Hercules, CA, USA) according to the manufacturer's instructions. To quantify LoxA, Pin2, PR1, and GluA transcript levels in different samples, real-time quantitative RT-PCR was performed. The real-time PCR was carried out on an ABI 7500 Real Time PCR System (Applied Biosystems, Foster City, CA) with a 96-well rotor. The amplification reactions were performed in a 20-μL final volume containing 10 μl of iQTM SYBR® supermix (BioRad, Hercules, CA), 0.8 μL of forward primer (5 μM) and reverse primer (5 μM) pairs, and 2 μL of cDNA first-strand template. Thermal cycling conditions were 5 min at 95°C followed by 40 cycles of 15 s at 95°C and 15 s at 58°C. The gene-specific primers, which were designed and checked as described previously29, were as follows: for LoxA (U09026; F: 5′- TGGTAGACCACCAACACGAA-3′, R: 5′-GACCAAAACGCTCGTCTCTC-3′); for Pin-2 (AY129402; F: 5′-TGATGCCAAGGCTTGTACTAGAGA-3′, R: 5′-AGCGGACTTCCTTCTGAACGT-3′); for PR-1a (M69247; F: 5′-GAGGGCAGCCGTGCAA-3′, R: 5′-CACATTTTTCCACCAACACATTG-3′); for GluA (M80604; F: 5′-TCA GCAGGGTTGCAAAATCA-3, R: 5′-CTCTAGGTGGGTAGGTGTTGGTTAA-3′); and for GAPDH (U93208; F: 5′- CTCCATCACAGCCACTCAGA-3′, R: 5′-TTCCACCTCTCCAATCCTTG-3′). All reactions were run in duplicate, and average values were used in the analyses. Normalized gene expression was calculated using the 2−ΔCt method with GAPDH as the endogenous control gene, and values were subsequently log2 transformed.
Statistical analysis
Fisher's protected least significant difference (PLSD) test of ANOVA was used to analyze phytohormone data and the data for development time and survival rate. The gene expression data were statistically analyzed by one-way ANOVA. Proportional data were arcsine square root transformed for analysis and back-transformed to percentages for presentation. EPG data were transformed where appropriate (square-root transformation for number of occurrences and natural log transformation for duration) and analyzed by one-way ANOVA.
Results
Performance of P. solenopsis on tomato plants
After 14 d of infestation, the survival rate of P. solenopsis nymphs on control plants was 78.3 ± 3.3%, and survival on JA-, SA-treated, or P. solenopsis-infested plants did not significantly differ from survival on control plants (F3, 12 = 1.14, P = 0.37; Fig. 1A). In contrast, survival at 21 d significantly differed among the four treatments (F3, 12 = 3.73, P = 0.04; Fig. 1A). At 21 d after infestation, survival was significantly lower (P = 0.04) on JA-treated plants than on control plants; survival did not differ on SA-treated, P. solenopsis-infested, and control plants (SA: P = 0.86; mealybug: P = 0.94; Fig. 1A). Survival at 28 d also differed among the four treatments (F3, 12 = 10.39, P = 0.001; Fig. 1A). At 28 d after infestation, survival was significantly lower (P = 0.007) on JA-treated plants than on control plants, but survival did not significantly differ on SA-treated, P. solenopsis-infested, and control plants (SA: P = 0.99; mealybug: P = 0.81; Fig. 1A).
Figure 1. Performance of P. solenopsis on non-infested (Control), P. solenopsis-infested (Mealybug), SA-treated, and JA-treated tomato plants.
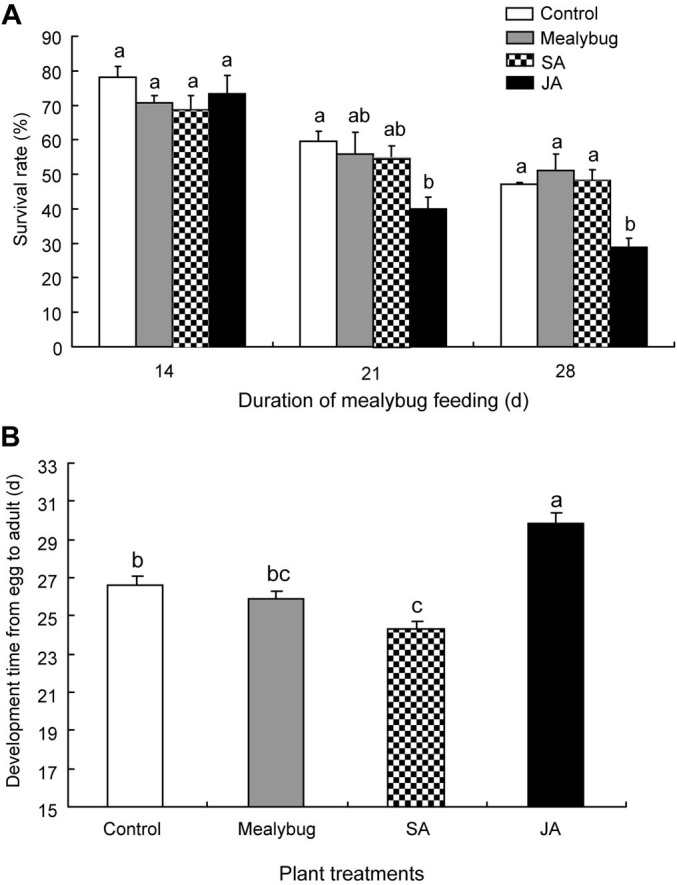
(A) Survival rate of P. solenopsis at different time points, and (B) developmental duration from egg to adult of P. solenopsis. Values are means ± SE, n = 4 in (A) and 25–33 in (B). For each time point in (A) and for the four treatments in (B), bars with different letters are significantly different (Fisher's PLSD test of ANOVA; P < 0.05).
Developmental duration of P. solenopsis from egg to adult significantly differed among the four treatments (F3, 109 = 22.87, P < 0.001; Fig. 1B). Developmental duration was significantly longer on JA-treated plants than on control plants (P < 0.001; Fig. 1B) but was significantly shorter on SA-treated plants than on control plants (P = 0.01; Fig. 1B). Pre-infestation by P. solenopsis did not affect the developmental duration of P. solenopsis (P = 0.74; Fig. 1B).
Feeding behavior of P. solenopsis as indicated by EPG
EPG detects the electrical fluctuations caused by probing behavior of mealybugs during foraging and feeding26,27. Three main phases during stylet penetration have been defined: the pathway phase, the xylem phase, and the phloem (or sieve element) phase28. Given that the probing behavior of P. solenopsis has seldom been recorded in the xylem phase23,27, we mainly analyzed the occurrence of cell puncture (as indicated by the rate at which potential dropped) in the pathway phase and the duration and insertion rate in the phloem (or sieve element) phase. Based on the EPG data, the mealybug treatment did not affect the feeding behavior of P. solenopsis adults (Table 1). The JA treatment significantly reduced the total feeding time in the phloem phase but did not affect the feeding rate in the phloem phase, the number of probes, or the rate of cell puncture (Table 1). The SA treatment significantly increased the rates of feeding in the phloem phase and cell puncture in the pathway phase, but did not affect the total feeding time in the phloem phase or the number of probes (Table 1).
Table 1. Feeding behavior of the mealybug P. solenopsis as indicated by EPG recordings.
| Plant Treatments | Feeding parameters | |||||||
|---|---|---|---|---|---|---|---|---|
| Total duration of SEP feeding (min) | P value | Rate of SEP feeding | P value | No. of probes | P value | Rate of potential drop | P value | |
| Control | 88.5 ± 26.2 | – | 2.9 ± 0.6 | – | 3.3 ± 0.39 | – | 63.2 ± 8.9 | – |
| Mealybug | 96.5 ± 29.1 | 0.26 | 2.4 ± 0.6 | 0.59 | 5.2 ± 1.4 | 0.24 | 60.7 ± 12.8 | 0.61 |
| JA | 14.5 ± 5.9 | 0.004* | 1.4 ± 0.5 | 0.08 | 3.8 ± 0.7 | 0.56 | 41.4 ± 8.5 | 0.09 |
| SA | 111.9 ± 122.6 | 0.53 | 4.4 ± 0.6 | 0.04* | 4.6 ± 0.8 | 0.24 | 84.9 ± 6.3 | 0.03* |
EPGs were recorded for 12 h per insect. Control: undamaged plant (n = 15); Mealybug: plant pre-infested with 40 adults of P. solenopsis for 120 h (n = 10); JA: plant pre-treated with 1.0 mL/leaf of JA solution for 24 h (n = 10); SA: plant pre-treated with 1.0 mL/leaf of SA solution for 24 h (n = 16). Values are means ± SE. Potential drop: caused by stylets puncturing cells; SEP: sieve element phase, observed during feeding in a sieve element. Rate of SEP feeding: the number of probes in sieve element phase during 12-h recording period.
*indicates significant difference (at α = 0.05) from control treatment as determined by one-way ANOVA.
Quantification of endogenous JA and SA in host leaves
The amount of JA was significantly lower in P. solenopsis-infested leaves than in control leaves at 12 h after infestation (P = 0.008) but not at 6, 24, 72, or 120 h after infestation (6 h: P = 0.88; 24 h: P = 0.08; 72 h: P = 0.42; 120 h: P = 0.08) (Fig. 2A). This is consistent with previous findings that the effect of herbivory on endogenous JA level generally occurs within 12 h after infestation8,29.
Figure 2. The accumulation of (A) JA and (B) SA in non-infested tomato leaves (Control) and in P. solenopsis-infested (Mealybug) tomato leaves at different time points.
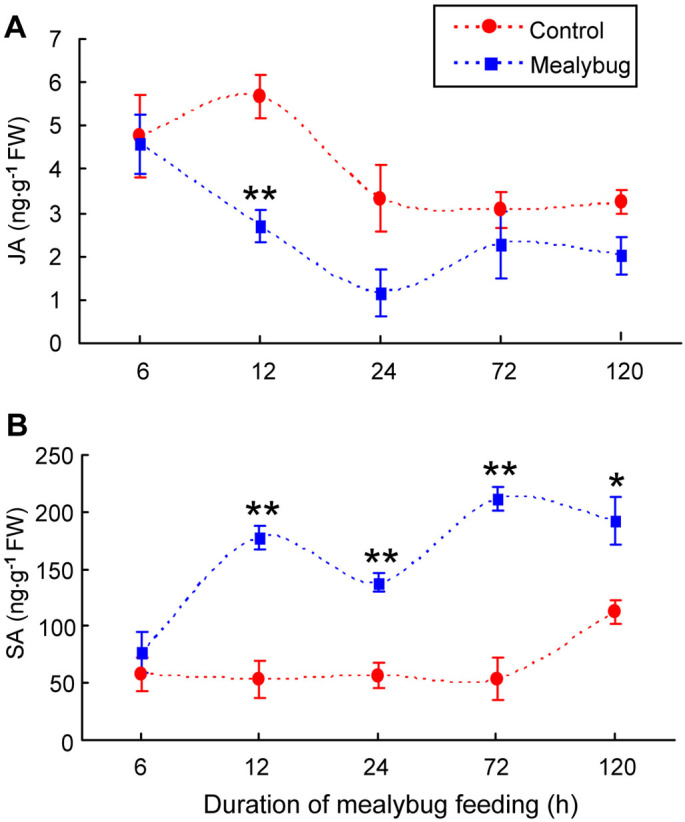
Values are means ± SE of three biological replicates. Asterisks above bars indicated significant differences compared to the control (Fisher's PLSD test of ANOVA; * P < 0.05; ** P < 0.01). FW means fresh weight.
The amount of SA was significantly higher in P. solenopsis-infested leaves than in control leaves at 12, 24, 72, and 120 h after infestation (12 h: P = 0.003; 24 h: P = 0.004; 72 h: P = 0.002; 120 h: P = 0.03), but not at 6 h after infestation (P = 0.45; Fig. 2B).
To test whether SA signaling plays a key role in mediating the suppression of JA defensive responses by P. solenopsis, we further examined the JA and SA levels in NahG (SA-deficient) and 35s::prosys (JA-overexpression) plants infested with P. solenopsis. For NahG plants, the amount of JA was significantly higher in infested leaves than in control leaves at 12, 24, and 72 h after infestation (12 h: P = 0.004; 24 h: P = 0.002; 72 h: P = 0.001) (Fig. 3A). In contrast, the amounts of SA in control NahG plants were consistently low (<9 ng g−1 fresh weight), and P. solenopsis infestation of NahG plants did not increase SA amounts at 12, 24, and 72 h after infestation (12 h: P = 0.12; 24 h: P = 0.20; 72 h: P = 0.91) (Fig. 3B).
Figure 3. Accumulations of (A) JA and (B) SA in non-infested leaves (Control) and in P. solenopsis-infested (Mealybug) leaves of NahG tomato plants.
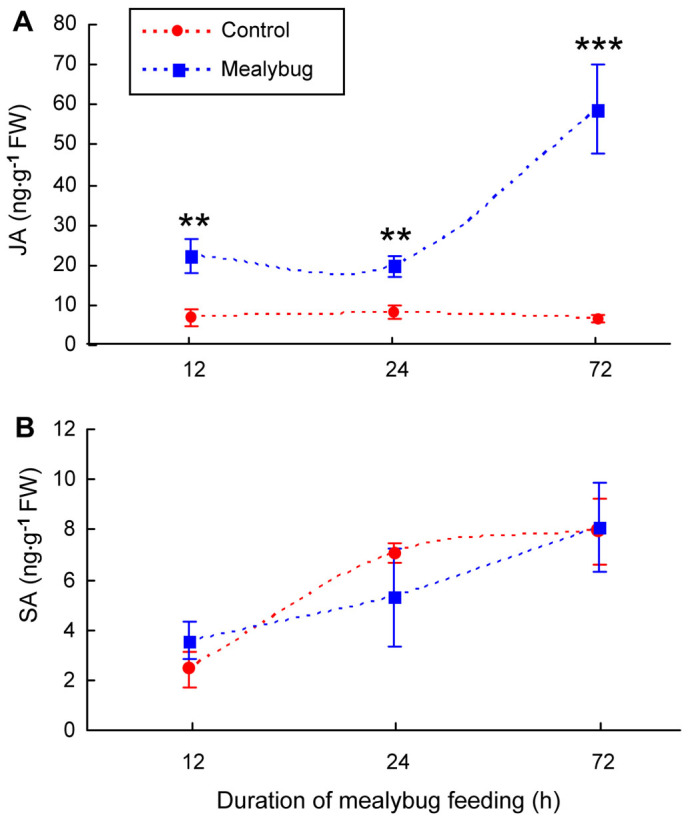
Values are means ± SE of three biological replicates. Asterisks above bars indicated significant differences compared to the control (Fisher's PLSD test of ANOVA; ** P < 0.01; *** P < 0.001). FW means fresh weight.
For 35s::prosys plants, the amounts of JA were significantly higher in infested leaves than in control leaves at 72 and 120 h after infestation (72 h: P = 0.04; 120 h: P = 0.015) (Fig. 4A). Similarly, the amounts of SA were significantly higher in infested leaves than in control leaves at 72 and 120 h after infestation (72 h: P = 0.001; 120 h: P = 0.001) (Fig. 4B).
Figure 4. Accumulations of (A) JA and (B) SA in non-infested leaves (Control) and in P. solenopsis-infested (Mealybug) leaves of 35s::prosys tomato plants.
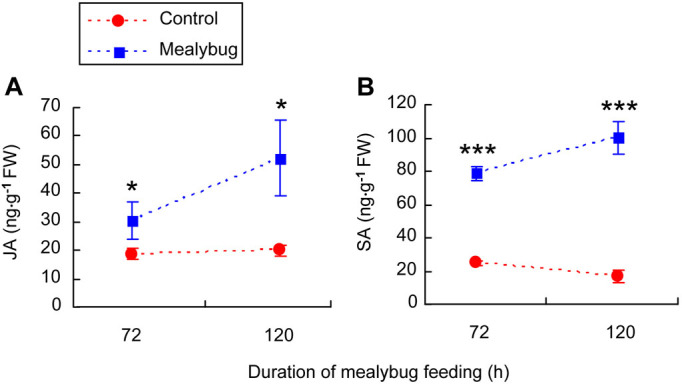
Values are means ± SE of three biological replicates. Asterisks above bars indicated significant differences compared to the control (Fisher's PLSD test of ANOVA; * P < 0.05; *** P < 0.001). FW means fresh weight.
Changes in gene-expression in response to P. solenopsis feeding
To further investigate the effects of P. solenopsis feeding on JA- or SA-dependent defense responses, we examined the transcript levels of two JA-dependent genes (LoxA and Pin-2) and SA-dependent genes (PR-1a and GluA). Lipoxygenase (LOX) is a key enzyme in the biosynthesis of JA along the octadecanoid pathway30. Proteinase inhibitor II (Pin-2), a serine proteinase inhibitor with trypsin and chymotrypsin inhibitory activities, is known to confer insect resistance in many Solanaceae plants31. In tomato, LoxA and Pin-2 are marker genes of the JA signaling pathway33. PR-1a and GluA are known as two pathogenesis-related genes, which are mainly regulated by the SA signaling pathway32,33. Mealybug feeding significantly suppressed the expression of Pin-2 (F1, 5 = 8.47, P = 0.04) but not of LoxA (Fig. 5A). In contrast, mealybug feeding significantly induced the expression of PR-1a and GluA (PR-1a: F1, 5 = 9.17, P = 0.04; GluA: F1, 5 = 12.32, P = 0.02) (Fig. 5B).
Figure 5. Expression of (A) JA-dependent genes and (B) SA-dependent genes in non-infested tomato leaves (Control) and in P. solenopsis-infested (Mealybug) tomato leaves.
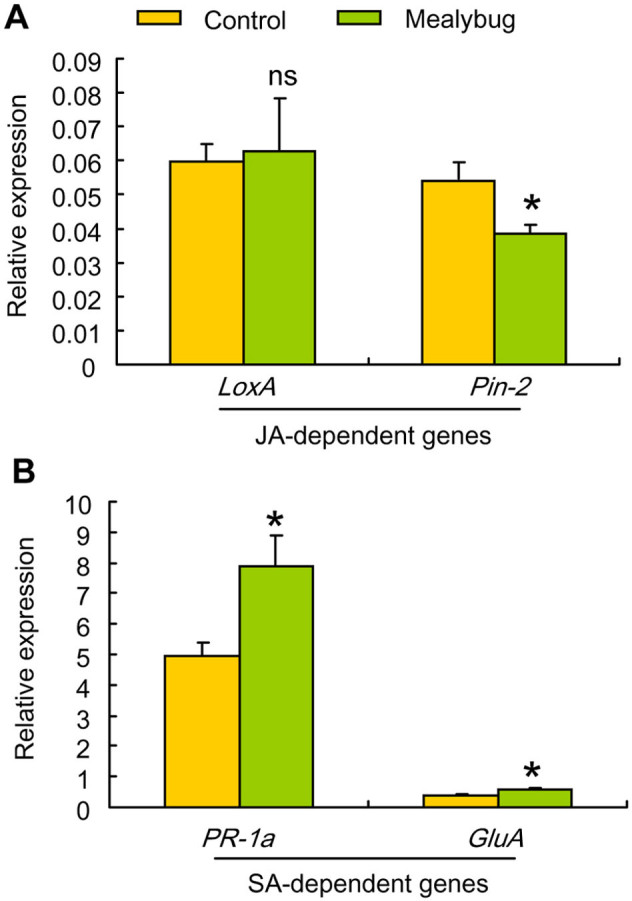
Wild-type tomato (Moneymaker) plants were infested for 120 h with 40 adults of P. solenopsis. Expression was measured by real-time PCR. Values are means ± SE of three biological replicates. Asterisks above bars indicated significant differences compared to the control (one-way ANOVA; ns, not significant; * P < 0.05).
Performance of P. solenopsis on tomato mutants
Survival of P. solenopsis were significantly lower on SA-deficient NahG plants than on wild-type MM plants at 14, 21, and 28 d after infestation (14 d: P < 0.001; 21 d: P < 0.001; 28 d: P = 0.008; Fig. 6A). Likewise, survival of P. solenopsis was significantly lower on JA-overexpression 35s::prosys plants than on wild-type CM plants at 14, 21, and 28 d after infestation (14 d: P < 0.001; 21 d: P < 0.001; 28 d: P < 0.001) (Fig. 6B).
Figure 6. Survival rate of P. solenopsis on (A) MM (wild-type cv Moneymaker) vs. NahG tomato plants and on (B) CM (wild-type cv Castlemart) vs. 35s::prosys tomato plants.
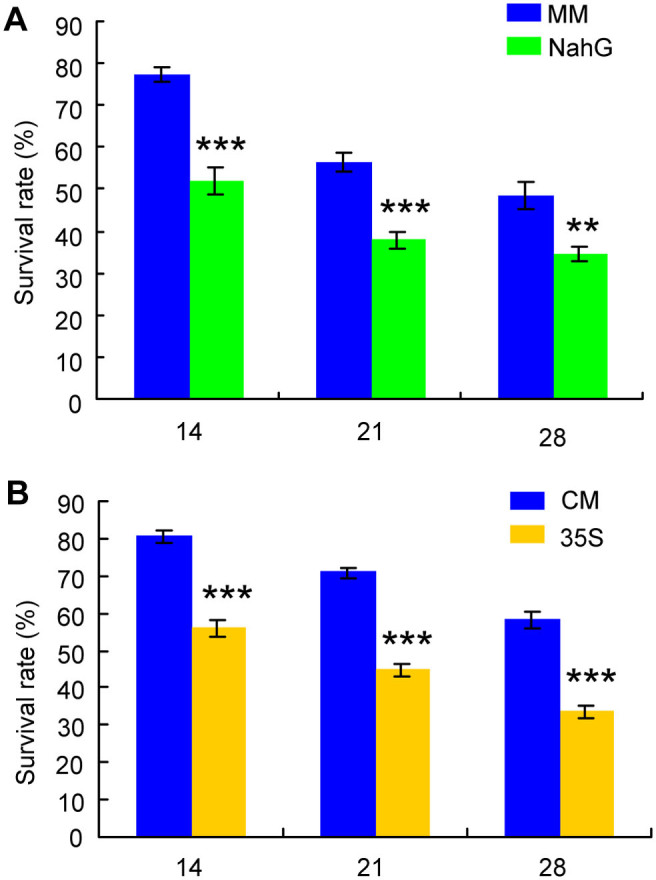
Values are means ± SE (n = 4). Asterisks above bars indicated significant differences compared to the wild type (Fisher's PLSD test of ANOVA; ** P < 0.01; *** P < 0.001).
Discussion
The results of the present study show that application of JA but not SA significantly decreases the feeding time of the mealybug P. solenopsis in the phloem tissue and consequently decreases nymphal performance. In addition, constitutive expression of JA signaling in 35s::prosys plants reduces P. solenopsis survival. These data demonstrate that the JA signaling pathway plays a key role in mediating the defense responses against P. solenopsis in tomato. This result is consistent with a previous finding that the JA-dependent defense pathway may be involved in basal defense against P. solenopsis in cotton21. Our data further show that P. solenopsis can suppress the JA-dependent responses, as evidenced by the decrease in expression of JA-dependent Pin2 and the reduction of JA level in infested plants. Meanwhile, P. solenopsis can induce the SA-dependent responses as indicated by the SA accumulation and increased expression of SA-dependent PR-1a and GluA in infested plants. That the suppression of JA defense responses is mediated by the SA signaling pathway is indicated by the accumulation of JA in infested SA-deficient NahG plants and significantly higher levels of SA in infested WT plants. Moreover, P. solenopsis survival is better on control WT plants than on SA-deficient NahG plants and on 35s::prosys plants that constitutively express JA, which suggests that manipulation of the pathways by nymphs allows them to perform better. Taken together, our results indicate that P. solenopsis can manipulate plants for its own benefit by modulating JA-SA crosstalk in order to suppress induced defenses.
The JA signaling pathway, including the wound hormones JA and JA-Ile, is widely recognized as a key regulator of plant defense against insect herbivores1,34,35. Several recent studies have found that insects can suppress JA-mediated defense responses to enhance their performance on food plant11,12,17,19. These suppression mechanisms often involve some herbivore effectors, including specific enzymes from oral secretions36, egg-derived elicitor12, and vectored microorganisms14,37. For example, caterpillars of the beet armyworm Spodoptera exigua can suppress the accumulation of JA and the induction of JA-dependent genes in Arabidopsis by releasing effectors in oral secretions38. Glucose oxidase in the saliva of Helicoverpa zea caterpillars was the first effector identified that reduces JA-regulated nicotine production in Nicotiana tabacum36. Similarly, phloem-feeding insects like aphids and whiteflies can inhibit the JA defense responses in different plant species although the effectors involved have not been identified11,39,40. However, the mechanism by which plants respond to herbivore effectors and eventually mediate the suppression of JA signaling has not been extensively studied.
Considering that pathogens can reduce plant defenses by manipulating antagonistic crosstalk between JA and SA signaling pathways41, researchers have speculated that insects can circumvent plant defenses in a similar way18. This possibility has been supported by several studies. For instance, oviposition by the cabbage butterfly Pieris brassicae induces SA accumulation and reduces the induction of JA-regulated defense genes in Arabidopsis and leads to the enhanced performance of Spodoptera littoralis; that the suppression of JA-dependent genes and enhancement of S. littoralis performance were not observed in the SA-deficient mutant sid2-1 indicates that SA mediates this phenomenon12. In tomato, larvae of Colorado potato beetle, Leptinotarsa decemlineata, use bacteria in their oral secretions to decrease JA accumulation, decrease JA-responsive anti-herbivore defenses, and increase SA production; these effects were not observed, however, in SA-deficient plants13. In the present study, we found that the expression level of the JA-dependent gene Pin-2 and JA production were significantly lower in P. solenopsis-infested plants than in non-infested plants, which indicates that, unlike herbivores in the two above cases, P. solenopsis does not prevent induction of JA-regulated defenses but reduces the constitutive level of JA defenses. This is consistent with the previous finding that P. solenopsis feeding strongly reduces the constitutive level of JA-regulated gossypol in cotton21. Our data further showed that if SA signaling is blocked, P. solenopsis does activate JA accumulation. Similar phenomena were also observed in the interaction between the whitefly Bemisia tabaci and Arabidopsis. The expression level of the JA-regulated defense genes PDF1.2 and VSP1 is significantly lower in B. tabaci-infested plants than in non-infested plants, indicating that B. tabaci reduces the constitutive levels of defense genes; however, once SA synthesis or its action is impaired, B. tabaci feeding does activate the induction of the two genes11,19. Considering that chewing insects cause extensive tissue damage whereas phloem-feeding insects cause minimal tissue damage18, the above cases indicated that the mode of mechanical damage caused by different herbivores possibly might be an additional factor that influences the strength of the reciprocal antagonism between JA and SA pathways. Collectively, these data indicate that herbivore modulation of the antagonistic crosstalk between the JA and SA signaling pathways might be more intricate than previously thought10.
Our results showed that P. solenopsis feeding did not affect the expression level of LoxA, which encodes a key enzyme involved in JA biosynthesis30, but decreased the expression level of JA-dependent Pin-2 and endogenous JA level. Based on these data we speculated that the suppression of JA defense responses by P. solenopsis is likely located downstream of JA biosynthesis pathway. This speculation is supported by the observation that B. tabaci nymphs induce the upstream JA-responsive genes (LOX2 and OPR3), but suppress the downstream JA-responsive gene (VPS1 and PDF1.2) in Arabidopsis11,19, indicating that nymph feeding can suppress the downstream components of the JA signaling pathway. Future experiments should attempt to explore the target genes manipulated by phloem-feeding insects in the JA signaling pathway.
In addition to the role of SA in the suppression of JA defense responses, the consistent accumulation of SA induced by P. solenopsis feeding might benefit the development of larvae. One reason is that exogenous SA treatment strongly increases the feeding rate of P. solenopsis in the phloem tissue (Table 1), which might directly contribute to rapid larval development (Fig 1B). Alternatively, given that SA pathway plays a central role in plant defense against microbial and fungal pathogens6, the accumulation of SA near the feeding site might protect the leaf tissue from infection and ensure that the tissue retains its full nutrient content (i.e., is not degraded) during larval feeding.
Although considerable SA accumulated in response to P. solenopsis infestation, JA accumulation was also strongly induced by P. solenopsis infestation of the 35s::prosys plant. Because JA signaling is constitutively enhanced in the 35s::prosys plant22, we speculate that if the JA signaling is activated before SA signaling, the tomato plant might become insensitive to SA-mediated suppression of JA signaling by mealybug infestation. Our speculation is supported by a previous study in which simultaneous activation of the JA and ET signaling before induction of the SA response rendered Arabidopsis plants insensitive to SA-mediated suppression of JA-responsive gene expression42. These findings support the view that the sequence of hormone activation affects realization of the antagonism between JA and SA crosstalk7,43. Alternatively, we cannot rule out the possibility that the enhanced SA level induced by mealybug feeding is insufficient to suppress JA accumulation in the 35s::prosys plant.
The mealybug P. solenopsis has been rapidly spreading throughout South China, where it is causing serious economic losses to cotton production20. Our data confirm that, at least in the laboratory, P. solenopsis can circumvent plant defenses by modulating the JA-SA crosstalk. This capability has possibly facilitated the rapid invasion of mealybugs in China and elsewhere21. Future studies should determine whether the modulation of the JA-SA antagonism by P. solenopsis occurs in the field, and if so, whether it enhances P. solenopsis population development.
Author Contributions
P.J.Z. and Y.B.L. conceived and designed the research. P.J.Z., F.H., J.M.Z. and J.N.W. conducted the experiments and analyzed the data. F.H. and P.J.Z. interpreted the results and wrote the paper.
Acknowledgments
This work was financially supported by National Basic Research Program of China (2012CB114105; 2012CB722504), the National Natural Science Foundation of China (31201523; 31270580), and Special Fund for Agro-scientific Research in the Public Interest of China (201103026).
References
- Kessler A. & Baldwin I. T. Plant responses to insect herbivory: the emerging molecular analysis. Annu. Rev. Plant Biol. 53, 299–328 (2002). [DOI] [PubMed] [Google Scholar]
- Dicke M., Van Poecke R. M. P. & De Boer J. G. Inducible indirect defence of plants: from mechanisms to ecological functions. Basic Appl. Ecol. 4, 27–42 (2003). [Google Scholar]
- Ament K., Kant M. R., Sabelis M. W., Haring M. A. & Schuurink R. C. Jasmonic acid is a key regulator of spider mite-induced volatile terpenoid and methyl salicylate emission in tomato. Plant Physiol. 135, 2025–2037 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Dahl C. C. & Baldwin I. T. Deciphering the role of ethylene in plant herbivore interactions. J. Plant Growth Regul. 26, 201–209 (2007). [Google Scholar]
- Reymond P. & Farmer E. E. Jasmonate and salicylate as global signals for defense gene expression. Curr. Opin. Plant Biol. 1, 404–411 (1998). [DOI] [PubMed] [Google Scholar]
- Koornneef A. & Pieterse C. M. J. Cross talk in defense signaling. Plant Physiol. 146, 839–844 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef A. et al. Kinetics of salicylate-mediated suppression of jasmonate signaling reveal a role for redox modulation. Plant Physiol. 147, 1358–1368 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diezel C., Von Dahl C. C., Gaquerel E. & Baldwin I. T. Different lepidopteran elicitors account for crosstalk in herbivory-induced phytohormone signaling. Plant Physiol. 150, 1576–1586 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J. N. et al. Reciprocal crosstalk between jasmonate and salicylate defence-signalling pathways modulates plant volatile emission and herbivore host-selection behaviour. J. Exp. Bot. 65, 3289–3298 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaler J. S., Humphrey P. T. & Whiteman N. K. Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci. 17, 260–270 (2012). [DOI] [PubMed] [Google Scholar]
- Zarate S. I., Kempema L. A. & Walling L. L. Silverleaf whitefly induces salicylic acid defenses and suppresses effectual jasmonic acid defenses. Plant Physiol. 143, 866–875 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruessow F., Gouhier-Darimont C., Buchala A., Metraux J. P. & Reymond P. Insect eggs suppress plant defence against chewing herbivores. Plant J. 62, 876–885 (2010). [DOI] [PubMed] [Google Scholar]
- Chung S. H. et al. Herbivore exploits orally secreted bacteria to suppress plant defenses. Proc. Natl. Acad. Sci. U. S. A. 110, 15728–15733 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X. B. et al. Bemisia tabaci Q carrying tomato yellow leaf curl virus strongly suppresses host plant defenses. Sci. Rep. 4, 5230 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipollini D., Enright S., Traw M. B. & Bergelson J. Salicylic acid inhibits jasmonic acid-induced resistance of Arabidopsis thaliana to Spodoptera exigua. Mol. Ecol. 13, 1643–1653 (2004). [DOI] [PubMed] [Google Scholar]
- Kempema L. A., Cui X. P., Holzer F. M. & Walling L. L. Arabidopsis transcriptome changes in response to phloem-feeding silverleaf whitefly nymphs. Similarities and distinctions in responses to aphids. Plant Physiol. 143, 849–865 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P. J. et al. Phloem-feeding whiteflies can fool their host plants, but not their parasitoids. Funct. Ecol. 27, 1304–1312 (2013). [Google Scholar]
- Walling L. L. Avoiding effective defenses: strategies employed by phloem-feeding insects. Plant Physiol. 146, 859–866 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P. J. et al. Feeding by whiteflies suppresses downstream jasmonic acid signaling by eliciting salicylic acid signaling. J. Chem. Ecol. 39, 612–619 (2013). [DOI] [PubMed] [Google Scholar]
- Wang Y. P., Watson G. W. & Zhang R. Z. The potential distribution of an invasive mealybug Phenacoccus solenopsis and its threat to cotton in Asia. Agr. Forest Entomol. 12, 403–416 (2010). [Google Scholar]
- Zhang P. J. et al. Suppression of jasmonic acid-dependent defense in cotton plant by the mealybug Phenacoccus solenopsis. PLoS ONE 6, e22378 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe G. A. & Ryan C. A. Suppressors of systemin signaling identify genes in the tomato wound response pathway. Genetics 153, 1411–1421 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F. et al. Host Plant probing analysis reveals quick settlement of the solenopsis mealybug during host shift. J. Econ. Entomol. 107, 1419–1425 (2014). [DOI] [PubMed] [Google Scholar]
- Wei J. N. et al. Antagonism between herbivore-induced plant volatiles and trichomes affects tritrophic interactions. Plant Cell Environ. 36, 315–327 (2013). [DOI] [PubMed] [Google Scholar]
- Almeida-Trapp A., De_Souza G. D., Rodrigues-Filho E., Boland W. & Mithöfer A. Validated for phytohormone quantification in plants. Front. Plant Sci. 5, 417 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cid M. & Fereres A. Characterization of the probing and feeding behavior of Planococcus citri (Hemiptera: Pseudococcidae) on grapevine. Ann. Entomol. Soc. Am. 103, 404–417 (2010). [Google Scholar]
- Huang F. et al. EPG waveform characteristics of solenopsis mealybug stylet penetration on cotton. Entomol. Exp. Appl. 143, 47–54 (2012). [Google Scholar]
- Tjallingii W. F. Salivary secretions by aphids interacting with proteins of phloem wound responses. J. Exp. Bot. 57, 739–745 (2006). [DOI] [PubMed] [Google Scholar]
- Engelberth J. et al. Ion channel-forming alamethicin is a potent elicitor of volatile biosynthesis and tendril coiling. Cross talk between jasmonate and salicylate signaling in lima bean. Plant Physiol. 125, 369–377 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell E., Creelman R. A. & Mullet J. E. A chloroplast lipoxygenase is required for wound-induced jasmonic acid accumulation in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 92, 8675–8679 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R., Narvaez J., An G. & Ryan C. Expression of proteinase inhibitors I and II in transgenic tobacco plants: effects on natural defense against Manduca sexta larvae. Proc. Natl. Acad. Sci. U. S. A. 86, 9871–9875 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block A., Schmelz E., O'Donnell P. J., Jones J. B. & Klee H. J. Systemic acquired tolerance to virulent bacterial pathogens in tomato. Plant Physiol. 138, 1481–1490 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milling A., Babujee L. & Allen C. Ralstonia solanacearum extracellular polysaccharide is a specific elicitor of defense responses in wilt-resistant tomato plants. PLoS ONE 6, e15853 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J. N. et al. Ecological trade-offs between jasmonic acid-dependent direct and indirect plant defences in tritrophic interactions. New Phytol. 189, 557–567 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe G. & Jander G. Plant immunity to insect herbivores. Annu. Rev. Plant Biol. 59, 41–66 (2008). [DOI] [PubMed] [Google Scholar]
- Musser R. O. et al. Herbivory: caterpillar saliva beats plant defences. Nature 416, 599–600 (2002). [DOI] [PubMed] [Google Scholar]
- Sugio A., Kingdom H. N., MacLean A. M., Grieve V. M. & Hogenhout S. A. Phytoplasma protein effector SAP11 enhances insect vector reproduction by manipulating plant development and defense hormone biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 108, E1254–E1263 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weech M. H., Chapleau M., Pan L., Ide C. & Bede J. C. Caterpillar saliva interferes with induced Arabidopsis thaliana defence responses via the systemic acquired resistance pathway. J. Exp. Bot. 59, 2437–2448. (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu-Salzman K., Salzman R. A., Ahn J. E. & Koiwa H. Transcriptional regulation of sorghum defense determinants against a phloem-feeding aphid. Plant Physiol. 134, 420–431 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P. J. et al. Whiteflies interfere with indirect plant defense against spider mites in Lima bean. Proc. Natl. Acad. Sci. U. S. A. 106, 21202–21207 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel B. N. & Brooks D. M. Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant Biol. 5, 325–331 (2002). [DOI] [PubMed] [Google Scholar]
- Leon-Reyes A. et al. Ethylene signaling renders the jasmonate response of Arabidopsis insensitive to future suppression by salicylic acid. Mol. Plant Microbe Interact. 23, 187–197 (2010). [DOI] [PubMed] [Google Scholar]
- Mur L. A. J., Kenton P., Atzorn R., Miersch O. & Wasternack C. The outcomes of concentration-specific interactions between salicylate and jasmonate signaling include synergy, antagonism, and oxidative stress leading to cell death. Plant Physiol. 140, 249–262 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]


