Abstract
Hirschsprung disease (HSCR) is a well-known congenital digestive disease that originates due to the developmental disorder of neural crest cells. MiR-206 is kown to have a relationship with digestive malfunctions. Therefore, we investigated whether or not miR-206 was involved in the pathogenesis of HSCR. qRT-PCR and Western blot assays were used to detect the expression levels of miRNA and mRNAs, and proteins in case and control tissue samples and two cell lines (293T and SH-SY5Y). The functions of miR-206 in vitro were measured by transwell assay, CCK8 assay and flow cytometry. Finally, we conducted dual-luciferase reporter assay to verify the connections between miR-206 and the target mRNA SDPR. Down-regulation of miR-206 was found in HSCR case tissue samples compared with controls, which was validated to be connected with the increased level of mRNA and protein of SDPR by qRT-PCR and dual-luciferase reporter assay. Moreover, miR-206 suppressed the cell migration and proliferation and silencing of SDPR could rescue the extent of the suppressing effects by miR-206 inhibitor. The findings suggest that miR-206 may play a significant role in the pathogenesis of HSCR, as well as inhibiting the cell migration and proliferation by targeting SDPR in disease models.
Hirschsprung disease (HSCR) is a common congenital digestive malformation which is characterized by the absence of the ganglion cells in the sub-mucosal and mesenteric plexuses. The incidence of HSCR is two in 10000 live births worldwide with significant differences in various ethnic groups (1.5, 2.1 and 2.8 per 10000 live births in Caucasians, African-Americans and Asians, respectively)1. Still, the mechanism of the pathogenesis of HSCR remains unclear except that during the 5th to 12th week in fetal period, the enteric neural crest cells, strongly associated with the bowel functions, fails to migrate to the hindgut2. HSCR is proven to have a complex genetic etiology involving several genes, including RET, EDNRB, SOX10 and PHOX2B3,4,5,6. However, there are few reports on the roles of non-coding genes, such as miRNA, in the pathogenesis of HSCR.
MicroRNAs (miRNAs) are endogenous 20 ~ 24 nt RNAs that play significant role in regulating gene expression post-transcriptionally in animals and plants by binding to the 3'UTR of the mRNA of the target genes7,8. MiRNAs are reported to have strong association with diverse diseases, including malignancies, such as gastric cancer, breast cancer, colon cancer and lung cancer, by affecting the cell migration, metastasis, proliferation and apoptosis9,10,11,12. Recent studies have demonstrated that miR-206 is responsible for various cancers due to its impact on the cell biological processes, such as cell proliferation, cell differentiation and apoptosis13,14,15. However, until now, there are no reports on the involvement of miR-206 in the early pathogenesis of HSCR.
In this study, we conducted experiments to unravel how miR-206 interacts with its target gene that contributes to the pathogenesis of HSCR in disease models.
Methods
Ethics Statement and subject tissue samples
The study was approved by the Institutional Ethics Committee of Nanjing Medical University. All of the experiments in the research were in compliance with the government policies and defined protocols which are accepted in current practices. In total, 80 HSCR case samples were enrolled into the research, which were earlier diagnosed by barium enema and anorectal manometry evaluation after surgery between 2009 and 2013 (NJMU Birth Cohort). Also, the entire group of control was 80 matched subjects that were confirmed HSCR-free. Written informed consent was obtained from patients' guardians after full explanation of the experiment. All tissue samples were stored at −80°C immediately after surgery.
RNA extraction and quantitative real-time PCR (RT-PCR)
Total RNAs, including miRNAs, were extracted from 80 matched controls, HSCR-stenosed segments (HSCR-S) and 80 HSCR-dilated segments (HSCR-D) colon tissue samples and two cell lines by the method of Trizol reagent (Life Technologies, CA, US). TaqMan® MicroRNA Assays (Applied Biosystems, CA, US) was applied for the detection of expression level of miR-206 in tissue samples with the normal endogenous control. Meanwhile, the mRNA of SDPR was measured by ABI 7900HT with SYBR (Takara, Tokyo, Japan) along with the GAPDH as the internal control. Details of the probes and primers are given in Supplementary Table 1.
Protein extraction and Western blot
The tissue samples and cells were lysed by RIPA buffer (Beyotime, Nantong, China). The incubation of the primary antibody (cat. # AP9935c, Abgent, SanDiego, US) against SDPR proteins with Polyvinylidene Fluoride (PVDF) membranes was performed at 4°C overnight. After rinsing, the secondary antibody (Beyotime, Nantong, China) was incubated with the PVDF membranes for 1 hour at room temperature. During the whole process, GAPDH was regarded as the normal control. Image J software was applied for the detection and quantification of the protein level in Western Blot.
Cell culture and transfection
In this study, due to the limitation of ENS cell model of HSCR, we searched many papers and found two appropriate cell lines, namely, human 293T and SH-SY5Y cells, which were acquired from American Type Culture Collection (ATCC, Manassas VA, US) were employed for the experiments in vitro. Both cell lines were cultured in DMEM medium (Hyclone, UT, US), which contained 10% heat-inactivated fetal bovine serum (FBS), penicillin (100 U/ml) and streptomycin (100 ug/ml) under the condition of 37°C and 5% CO2. The miRNA precursor molecules and SDPR siRNA used in transfection were synthesized and purchased from GenePharma (Shanghai, China). Lipofectamine 2000 Reagent (Invitrogen, CA, US) was used as the vector for transfection reagents during the procedure according to the manufacturers' protocols.
Cell transwell and proliferation assay
Cell transwell assay, which was designed to evaluate the capacity of cell migration, was mainly performed with the application of the Transwell migration chambers (8 μm pore size, Millipore Corporation, Billerica, MA). Firstly, two cell lines were cultured in six-well plates and transfected with miR-206 inhibitor/mimics or SDPR siRNA. After 48 h of transfection, cells were harvested with serum-free medium as single-cell suspension and 100 μl of cell suspension was seeded in the upper chamber (1 × 106 cells/ml), along with the lower chamber filled with the 600 μl DMEM medium with 10% FBS. After incubation from 24 h to 48 h, the cells were stained with crystal violet staining solution (Beyotime, Nantong, China) after cell fixation in 95% methyl aldehyde. The images of migrated cells were captured and counted by Image-pro Plus 6.0 with the amount of normal control cells standardized to 1. Accompanied with the transwell assay, CCK8 assay was conducted to confirm the cell proliferation after 24 h transfection. Transfected cells were planted into 96-well plates and add CCK8 reagent into each well for 1 h incubation at 37°C, which was evaluated by 450 nm absorption measured by TECAN infinite M200 multimodemicroplate reader (Tecan, Mechelen, Belgium). Experiments of cell transwell and proliferation were performed in triplicate independently.
Cell cycle and apoptosis assay
To investigate whether miR-206 had any impact on cell cycle and apoptosis, cells transfected with miR-206 inhibitor were harvested and detected by BD Biasciences FACS Calibur Flow Cytometry (BD Biosciences, NJ, US). For detection of apoptosis, after collection, cells were stained with Annexin V-FITC/Propidium Iodide Kit (KeyGen Biotech, Nanjing, China)and data analyzed with FlowJo V7 software (Tree Star, Oregon, US). Experiments of cell cycle and apoptosis were also performed in triplicate independently.
Dual-luciferase reporter assay
Dual-luciferase reporter assay was used to validate whether or not miR-206 regulated SDPR by binding to the 3'UTR region of SDPR mRNA. Thus, the sequence of 3'UTR region of SDPR predicted to have interaction with miR-206 was inserted into the KpnI and SacI sites of pGL3 promoter vector (Genscript, Nanjing, China). These constructs were named pGL3-SDPR and pGL3-SDPR-mut, respectively. According to the manufacturers' protocols, after transfection with negative control, miR-206 mimics, pGL3-SDPR and pGL3-SDPR-mut, cells were collected to measure fire fly and renilla luciferase activities by Dual Luciferase Assay (Promega, Madison, WI). Experiments of dual-luciferase reporter assay were also performed in triplicate independently.
Statistical analysis
Statistical analyses were performed by using Stata 9.0 statistical software package (Stata Corp. Texas, US) and presented by Graphpad software (GraphPad Software, Inc., CA, US). Data of the relative expression level of miR-206 and SDPR in human tissue samples were presented as a box plot of the median and range of log-transformed expression level accessed by Wilcoxon rank-sum test. The data for the experiments in vitro that were repeated three times, were plotted as mean ± SEM via double-sided Student's t-test. Results were considered to have statistically significant differences if p < 0.05.
Results
Study population
In total, 160 colon tissue specimens were recruited from Department of Pediatric Surgery, Nanjing Children's Hospital affiliated Nanjing Medical University, including 80 HSCR-confirmed cases and 80 matched controls. The clinical characteristics of the study subjects are shown in Table 1,ranging over age, sex and disease classification. As displayed in Table 1, there were no statistically significant differences in terms of age and sex between HSCR cases and controls. Moreover, according to the length of aganglionosis in colon, HSCR was divided into two main types, short-segment HSCR (S-HSCR) and long-segment HSCR (L-HSCR).
Table 1. Clinical characteristics of the study population.
| Variable | HSCR (n = 80) | Control (n = 80) | P |
|---|---|---|---|
| Age(months, mean, SE) | 4.25(0.30) | 4.77(0.83) | 0.57a |
| Sex | |||
| Male | 70 | 55 | 0.17b |
| Female | 10 | 25 | |
| Disease Classifcation | |||
| S-HSCR | 29 | ||
| L-HSCR | 51 |
aStudent t-test
bTwo-sided χ2 test.
S-HSCR: Short-segment HSCR; L-HSCR: Long-segment HSCR.
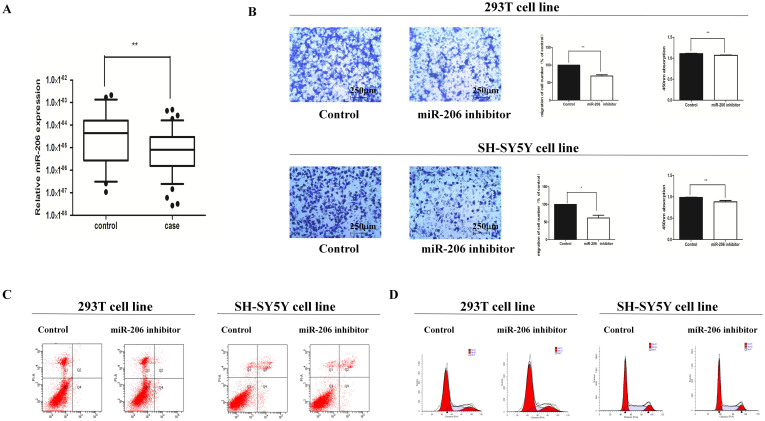
Down-regulation of miR-206 in HSCR
Figure 1A shows that the relative expression level of miR-206 in HSCR cases was significantly lower as compared with matched controls. This implies that miR-206 might have connections with the pathogenesis of HSCR. Another experiment was conducted to examine the miR-206 expression level in 80 matched controls, HSCR-stenosed segments (HSCR-S) and 80 HSCR-dilated segments (HSCR-D). The results showed that the expression level of HSCR-D and HSCR-S were both much lower than controls (Supplement Figure C).
Figure 1. MiR-206 was down-regulated and its cell biological change after treating cell lines with miR-206 inhibitor.
(A): The relative expression levels of miR-206 in human HSCR tissues (n = 80) and control tissues (n = 80) were evaluated by qRT-PCR. Data were presented as box plot of the median and range of log-transformed relative expression levels. The top and bottom of the box represent the seventy-fifth and twenty-fifth percentile. The whiskers indicate the 10th and 90th points. * Significantly different compared with that of control (P < 0.05). (B): Transwell assay was performed as described in Materials and Methods. The representative images of invasive cells at the bottom of the membrane stained with crystal violet were visualized as shown (left). The quantifications of cell migration were presented as percentage migrated cell numbers (right). * indicates significant difference compared with control group (P < 0.05). Absorbance at 450 nm was presented with Mean ± SE. * indicates significant difference compared with control group P < 0.05. (C–D): Flow cytometry assay was performed to evaluate cell apoptosis and cell cycle.
MiR-206 inhibitor suppressed cell migration and proliferation without impacting cell cycle and apoptosis
In order to confirm the functional performance of miR-206 in vitro, we examined how miR-206 impacted cell migration, cell proliferation, cell cycle process and apoptosis. To achieve this, 293t and SH-SY5Y cell lines were transfected with miR-206 inhibitor and then subjected to transwell and CCK8 assays. Both the cell lines showed reduction in number of migrating and proliferating cells suggesting that the down-regulation of miR-206 had a suppressive affection on cell migration and proliferation (Figure 1B). Furthermore, flow cytometry analysis was performed to investigate the impact of miR-206 on cell cycle and apoptosis. The results show no statistical difference in the percentage of apoptotic cells between cells transfected with miR-206 inhibitor and the negative control. Likewise, there were no changes in the cell cycle process (Figure 1C,D).
Bioinformatics prediction of target gene for miR-206
We applied three main databases (DIANA LAB, Targetscan and Pictar) to predict the underlying target genes, which may be regulated by miR-206. Finally, after the prediction and function analysis,the three common target genes, SDPR(serum deprivation response), FN1(fibronectin 1) and PAX3(paired box 3), were selected. SDPR and FN1 are admitted for the confirmed remarkable associations with plasma membrane, which contributes to the dysfunction of caveolae, cell adhesion and migration. PAX3 is generally accepted to have a key role in the fetal development and pathogenesis of colonic aganglionosis.
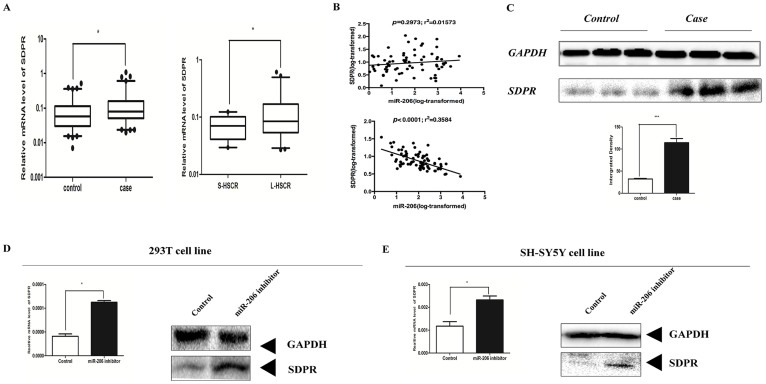
Up-regulation of SDPR in HSCR patients
To determine whether all of three target genes were involved in HSCR, qRT-PCR was used for the examination of the mRNA level in 80 HSCR case and matched control tissue samples. SDPR was the only candidate gene that showed significant up-regulation between HSCR cases and matched controls. Moreover, in order to reveal whether expression level of SDPR had a relationship with the diseases classification, we checked the expression level of SDPR in two main types of HSCR. The results indicate that SDPR expressed much more in L-HSCR than S-HSCR (p = 0.0241) (Figure 2A). Immediately, correlation analysis was conducted between miR-206 and SDPR in match controls and cases, respectively. The findings demonstrated that compared with the poor correlation in controls, there were evident associations between miR-206 and SDPR in HSCR cases (Figure 2B). Simultaneously, via western blot, the protein expression level of SDPR was consistent with the mRNA expression level (Figure 2C).
Figure 2. SDPR was up-regulated in HSCR cases and two cell lines.
(A): The mRNA expression levels of SDPR in human HSCR case and control tissues and the relative expression level of SDPR in S-HSCR and L-HSCR. (B): Correlation analysis between miR-206 and SDPR in controls and cases, respectively. The upper correlation was performed in controls and it showed poor relationships between miR-206 and SDPR, while the lower correlation analysis indicated that miR-206 is connected with SDPR in HSCR cases. (C): The protein expression levels of PTEN in human HSCR tissues and controls (3 representative samples from both groups are shown) (above). Quantization of Western-blotting was done by Image J software (blow). (D–E): Cells were transfected with 100 nM miR-206 inhibitor for 48 h, qRT-PCR was performed to evaluate the mRNA level of SDPR. SDPR protein expression levels were analyzed by western-blotting.
In contrast, FN1 and PAX3 were invariant between HSCR cases and controls (Supplementary Figure A, B). In addition, we evaluated the changes of expression level of SDPR in 293T and SH-SY5Y cell lines after transfection with miR-206 inhibitor. After 48 hours, the expression level of SDPR mRNA and the protein level were detected by qRT-PCR and western blot, respectively. As expected, SDPR expression was remarkably up-regulated at both mRNA and protein levels in 293T and SH-SY5Y cell lines (Figure 2D, E).
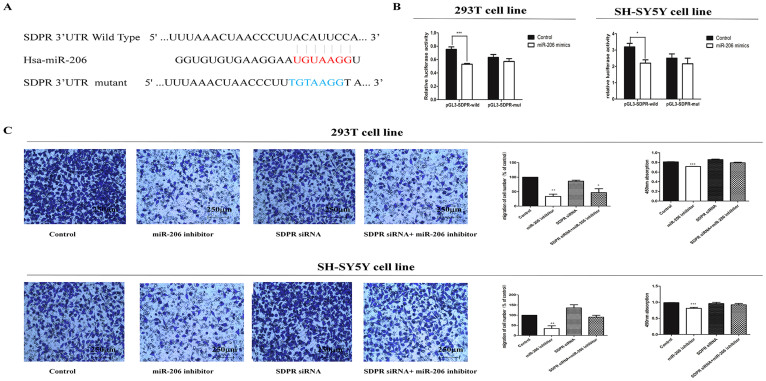
SDPR was target gene for miR-206
To verify the relationship between miR-206 and SDPR, two independent methods were applied to validate the miRNA-target gene interaction. Firstly, we constructed the wild and mutant type luciferase plasmids with the binding area of 3'UTR of SDPR mRNA, which was referred to as pGL3-SDPR and pGL3-SDPR-mut, respectively (Figure 3A). Transfection of miR-206 mimics with pGL3-SDPR into 293T cell line and SH-SY5Y cell lines significantly inhibited the luciferase activity as compared with the control. Meanwhile, there was no significant alteration in luciferase activity for cell lines transfected with negative control, miR-206 mimics and pGL3-SDPR-mut (Figure 3B). The findings demonstrate that miR-206 regulated SDPR by combining the 3'UTR region of its mRNA.
Figure 3. Migration was reversed after co-transfected with miR-206 inhibitor and SDPR siRNA.
(A): Sequence alignment of human miR-206 with 3' UTR of SDPR. Bottom: mutations in the 3'-UTR of SDPR in order to create the mutant luciferase reporter construct. (B): Cells were transfected with miR-206 and control, renilla luciferase vector pRL-SV40 and SDPR3'UTR luciferase reporters for 48 h. Both firefly and Renilla luciferase activities are measured in the same sample. Firefly luciferase signals were normalized with Renilla luciferase signals. All tests were performed in triplicate and presented as mean ± SE. (C): The cell biological changes in migration and proliferation were reversed after co-transfected miR-206 inhibitor and SDPR siRNA. Cells were divided into four groups in two cell lines, respectively. The first group was transfected with control, followed by miR-206 inhibitor, SDPR siRNA and the last one that co-transfected with miR-206 and SDPR siRNA. We compared the co-transfected group with others and found migration and proliferation were partly reversed after co-transfected with miR-206 inhibitor and SDPR siRNA when compared with the cells transfected with miR-206 inhibitor, and were much more restored after co-transfected with miR-206 inhibitor and SDPR siRNA. Data were presented as mean ± SEM from three separate experiments performed in triplicates, and were analyzed by double-sided Student's t-test. (* indicates P < 0.05).
Silencing of SDPR partially rescued the cell migration and proliferation with miR-206 inhibitor mediation
We had validated the fact that miR-206 interacted with SDPR by binding to its 3'UTR region and the suppressed affections of miR-206 on cell migration and proliferation. In order to further verify whether miR-206 had impact on cell migration and proliferation through SDPR directly, we performed a series of rescue experiments. Specific small interfering RNAs (siRNAs) were designed to silence the expression of SDPR in two cell lines. The capacity of cell migration transfected with SDPR siRNAs was familiar with the normal control. Accompanied with the transfection of miR-206 inhibitor, the silencing of SDPR by specific siRNAs could partially draw up the suppression of cell migration, which was still weaker than the normal control. Furthermore, the parallel results from CCK8 assay were displayed after co-transfection with miR-206 inhibitor and SDPR siRNAs were performed (Figure 3C). These results clearly indicated that cell migration and proliferation were impaired by up-regulation of SDPR due to down-regulation of miR-206.
Discussion
In our study, three potential target genes of miR-206 were predicted by DIANA LAB, Pictar and Targetscan, while the results showed SDPR was the only one that had significant differences between HSCR cases and matched controls. In the following step, we detected the expression level of miR-206 and SDPR mRNA in HSCR case and control tissue samples. The results show that miR-206 was down-regulated in HSCR with concomitant up-regulation of SDPR. Therefore, dual-luciferase reporter assay was conducted to reveal the underlying relationships between miR-206 and SDPR. As expected, the results illustrate that miR-206 had an inverse regulatory relationship with SDPR by directly binding to the 3'UTR region of SDPR mRNA, which might have caused degradation or structural changes leading to aberrant expression of proteins.
There are many reports suggesting that miR-206 had strong association with regulation of cellular and biological processes. Especially miR-206 was observed to regulate cell movement during zebrafish gastrulation by regulating mitogen-activated protein kinase (MAPK) JNK signaling16,17. In order to confirm the functional effect of miR-206 on cell biological processes in HSCR, assays such as, cell transwell, cell proliferation, apoptosis and cell cycle were used for validation. The results indicate that the down-regulation of miR-206 suppressed cell migration and proliferation. Moreover, the successful rescue in transwell assay by SDPR siRNA implied that miR-206 performed the suppression by up-regulating SDPR.
SDPR (serum deprivation response) is a key substrate for protein kinase C (PKC) phosphorylation and this interaction determines the compartimentalization of PKC to caveolae18. SDPR was further verified to play a key role in inducing membrane curvature and participate in the formation of caveolae19. Caveolae is calcium channel related to gut electrophysiological pacing function, which has also been identified to have impacts on cell migration and proliferation20,21. Recently, it has been demonstrated that the absence of the caveolae on the membrane surface contributed to the formation of fibroblast-like ICC (Interstitial Cajal Progenitors Cells) in the narrow segment of HSCR compared with the normal adult colon, which was shown to have surface caveolae22. Meantime, fibroblast-like ICC was observed in Igf1r+/CD34+ ICC in Ws/Ws rat colon23. Accordingly, the over-expression of SDPR was validated to play a key role in inducing deformation of caveolae and extensive tubulation of the plasma membrane19. In our study, the expression level of SDPR in HSCR cases was much higher than the normal matched controls. Thus, we speculate that through negative regulation, down-regulation of miR-206 led to the up-regulation of SDPR inducing the deformation of caveolae of ENCCs (enteric neural crest cells) in colon, which would contribute to the pathogenesis of HSCR. Further research needs to be performed to validate the hypothesis, especially the caveolae of ENCCs.
In conclusion, our research reveals that miR-206 plays an important role in the pathogenesis of HSCR and suppresses cell proliferation and migration by regulating SDPR in disease models. Our study provides a new approach for understanding the pathogenesis of HSCR and might contribute to a novel approach to the therapy of HSCR in the future.
Author Contributions
A.S., H.L. and Y.X. designed the experiments; A.S., H.Z. and H.X. performed the experiments; W.T., H.L., J.T. and H.W.Z. contributed essential technical assistance; H.Z. and A.S. wrote the paper.
Supplementary Material
Supplementary Information
Acknowledgments
This study was supported by Natural Science Foundation of China [NSFC 81370473], Natural Science Foundation of Jiangsu Province of China [BK20131388], New Century Excellent Talents of MOE [NCET-13-0870] and Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
References
- Amiel J. & Lyonnet S. Hirschsprung disease, associated syndromes, and genetics: a review. J Med Genet. 38, 729–739 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeown S. J. et al. Hirschsprung disease: a developmental disorder of the enteric nervous system. Wiley Interdiscip Rev Dev Biol. 2, 113–129 (2013). [DOI] [PubMed] [Google Scholar]
- Zhu L. et al. Spatiotemporal regulation of endothelin receptor-B by SOX10 in neural crest-derived enteric neuron precursors. Nat Genet. 36, 732–737 (2004). [DOI] [PubMed] [Google Scholar]
- Emison E. S. et al. A common sex-dependent mutation in a RET enhancer underlies Hirschsprung disease risk. Nature. 434, 857–863 (2005). [DOI] [PubMed] [Google Scholar]
- Fernandez R. M. et al. Contributions of PHOX2B in the pathogenesis of Hirschsprung disease. PLoS One. 8, e54043 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W. et al. Methylation analysis of EDNRB in human colon tissues of Hirschsprung's disease. Pediatr Surg Int. 29, 683–688 (2013). [DOI] [PubMed] [Google Scholar]
- Brodersen P. & Voinnet O. Revisiting the principles of microRNA target recognition and mode of action. Nat Rev Mol Cell Biol. 10, 141–148 (2009). [DOI] [PubMed] [Google Scholar]
- Fabian M. R., Sonenberg N. & Filipowicz, W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 79, 351–379 (2010). [DOI] [PubMed] [Google Scholar]
- Peng Y. et al. MicroRNA-338 Inhibits Growth, Invasion and Metastasis of Gastric Cancer by Targeting NRP1 Expression. PLoS One. 9, e94422 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Q. et al. miR-483-5p promotes invasion and metastasis of lung adenocarcinoma by targeting RhoGDI1 and ALCAM. Cancer Res. 74, 3031–3042 (2014). [DOI] [PubMed] [Google Scholar]
- Wang Y. et al. MicroRNA-375 inhibits colorectal cancer growth by targeting PIK3CA. Biochem Biophys Res Commun. 444, 199–204 (2014). [DOI] [PubMed] [Google Scholar]
- Yu Z. et al. miR-17/20 sensitization of breast cancer cells to chemotherapy-induced apoptosis requires Akt1. Oncotarget. 5, 1083–1090 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R. et al. MiR-206 regulates neural cells proliferation and apoptosis via Otx2. Cell Physiol Biochem. 29, 381–390 (2012). [DOI] [PubMed] [Google Scholar]
- Bao Y. P. et al. Roles of microRNA-206 in osteosarcoma pathogenesis and progression. Asian Pac J Cancer Prev. 14, 3751–3755 (2013). [DOI] [PubMed] [Google Scholar]
- Zhang L. et al. miR-206 inhibits gastric cancer proliferation in part by repressing cyclinD2. Cancer Lett. 332, 94–101 (2013). [DOI] [PubMed] [Google Scholar]
- Liu X. et al. MicroRNA-206 regulates cell movements during zebrafish gastrulation by targeting prickle1a and regulating c-Jun N-terminal kinase 2 phosphorylation. Mol Cell Biol. 32, 2934–2942 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. et al. Nodal promotes mir206 expression to control convergence and extension movements during zebrafish gastrulation. J Genet Genomics. 40, 515–521 (2013). [DOI] [PubMed] [Google Scholar]
- Gustincich S. et al. The human serum deprivation response gene (SDPR) maps to 2q32-q33 and codes for a phosphatidylserine-binding protein. Genomics. 57, 120–129 (1999). [DOI] [PubMed] [Google Scholar]
- Hansen C. G. et al. SDPR induces membrane curvature and functions in the formation of caveolae. Nat Cell Biol. 11, 807–814 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drab M. et al. Loss of caveolae, vascular dysfunction, and pulmonary defects in caveolin-1 gene-disrupted mice. Science. 293, 2449–2452 (2001). [DOI] [PubMed] [Google Scholar]
- Acevedo L. et al. A new role for Nogo as a regulator of vascular remodeling. Nat Med. 10, 382–388 (2004). [DOI] [PubMed] [Google Scholar]
- Chen Z. H. et al. Characterization of interstitial Cajal progenitors cells and their changes in Hirschsprung's disease. PLoS One. 9, e86100 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. Y. et al. Igf1r+/CD34+ immature ICC are putative adult progenitor cells, identified ultrastructurally as fibroblast-like ICC in Ws/Ws rat colon. J Cell Mol Med. 13, 3528–3540 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information