Abstract
Background:
Better biomarkers must be found to develop clinically useful urine tests for bladder cancer. Proteomics can be used to identify the proteins released by cancer cell lines and generate candidate markers for developing such tests.
Methods:
We used shotgun proteomics to identify proteins released into culture media by eight bladder cancer cell lines. These data were compared with protein expression data from the Human Protein Atlas. Epidermal growth factor receptor (EGFR) was identified as a candidate biomarker and measured by ELISA in urine from 60 noncancer control subjects and from 436 patients with bladder cancer and long-term clinical follow-up.
Results:
Bladder cancer cell lines shed soluble EGFR ectodomain. Soluble EGFR is also detectable in urine and is highly elevated in some patients with high-grade bladder cancer. Urinary EGFR is an independent indicator of poor bladder cancer-specific survival with a hazard ratio of 2.89 (95% CI 1.81–4.62, P<0.001). In multivariable models including both urinary EGFR and EpCAM, both biomarkers are predictive of bladder cancer-specific survival and have prognostic value over and above that provided by standard clinical observations.
Conclusions:
Measuring urinary EGFR and EpCAM may represent a simple and useful approach for fast-tracking the investigation and treatment of patients with the most aggressive bladder cancers.
Keywords: EGFR, EpCAM, bladder cancer, urine, prognosis
Urothelial bladder cancer (UBC) is the seventh commonest cancer in Western societies (Burger et al, 2013). At presentation, 75–85% will be non-muscle invasive tumours (NMIBC, stages pTa/pT1/pTis), with the remainder being muscle-invasive (MIBC, stages pT2–4) (Kaufman et al, 2009). Progression to, or presentation with, MIBC represents the critical step in the disease course, necessitating radical therapies, including chemoradiotherapy or neoadjuvant chemotherapy followed by radical cystectomy (Witjes et al, 2014). Despite such approaches, outcomes have changed little for three decades (Kaplan et al, 2014). Reliably identifying patients with the most aggressive UBCs early (both NMIBC and MIBC) and expediting aggressive therapeutic strategies could go some way to improving outcomes (Bryan et al, 2014a), yet this remains a significant challenge despite sophisticated clinicopathological algorithms (Babjuk et al, 2013; Witjes et al, 2014).
As urine directly contacts UBCs, it is an attractive biospecimen for developing noninvasive tests to detect and characterise bladder tumours. Challenges in developing such tests include UBC heterogeneity, such that different tumours may release different biomarkers (thus necessitating multimarker tests), and early-stage and low-grade tumours may only release very small amounts of such markers, potentially impairing test sensitivity. In addition, markers must be highly tumour specific so that haematuria and other nonmalignant conditions do not generate false positives. In the search for better urinary biomarkers genomic, proteomic and metabolomic approaches have all yielded promising results (Orenes-Piñero et al, 2007; Huang et al, 2011; Kandimalla et al, 2013).
Protein ectodomain shedding is becoming increasingly recognised as an important cancer-related phenomenon, and may be a useful source of biomarkers (Zatovicova et al, 2005; Hansen et al, 2013). We have previously reported that the ectodomain of EpCAM is shed into the urine by bladder tumours (Bryan et al, 2014b). Here, we use proteomics to identify the proteins released by a panel of UBC cells in vitro, and compare them with tissue expression data. One of the shortlisted proteins, EGFR, is released into the urine in a soluble form and is a promising biomarker for rapidly identifying UBC patients with the most aggressive disease. This work represents the first description of the prognostic value of soluble urinary EGFR in UBC, alone and in combination with soluble urinary EpCAM (Bryan et al, 2014b).
Materials and methods
Materials and samples
The cell lines used in this study were selected on the basis that they exhibit diverse mutation profiles and may capture some of the heterogeneity of UBC (Supplementary Table S1). Urine samples were prospectively collected for biomarker research between 2006 and 2009 as part of the Bladder Cancer Prognosis Programme (BCPP, ethics approval 06/MRE04/65) (Zeegers et al, 2010). Patients were enrolled on the basis of initial cystoscopic findings suggestive of primary UBC. All UBC patients were newly diagnosed, had not received treatment for UBC before urine collection and were subsequently treated according to current standard practice. Inclusion and exclusion criteria are detailed elsewhere (Zeegers et al, 2010). Samples were placed on ice, centrifuged at 2000 g for 10 min within 8 h of collection, and supernatants stored at −80 °C. As patient recruitment occurred before histopathological confirmation of UBC, a proportion of patients were ultimately diagnosed with nonmalignant conditions and these serve as noncancer ‘controls'. All patients were followed-up to July 2014 with causes of death notified to the BCPP study office.
Proteomic analysis of secretomes
Cells were grown to >80% confluence (apart from RT4 cells that grow as islands) in 4 × T75 flasks. Adherent cells were then washed 4 times with serum-free media and incubated overnight in serum-free media±200 nM phorbol 12-myristate 13-acetate (PMA) (2 flasks for each condition equating to 100–200 μg protein). The low protein concentration in the conditioned media and the fact that >95% of the cells remained adherent indicate that any cell death during secretome generation was minimal. Cells and debris were removed by centrifuging the conditioned media at 2000 g for 20 min prior and the entire supernatant processed using filter-aided tryptic digestion. Proteins were reduced with 20 mM DTT in 8 M urea, 1% CHAPS, 100 mM triethylammonium bicarbonate (1 h at room temperature), alkylated with 50 mM iodoacetamide and then concentrated in 30 kDa centrifugal filters. Following 4 washes with 100 mM triethylammonium bicarbonate, 10 μg of sequencing grade trypsin was added to the filters (<50 : 1 protein/trypsin) and incubated at 37 °C overnight. The peptides were centrifuged through the filters and their primary amines dimethylated by incubating with 25 mM sodium cyanoborohydride and 0.2% formaldehyde (control secretome) or 0.2% deuterated formaldehyde (+PMA secretome) for 30 min at room temperature. Following quenching with 100 mM ammonium bicarbonate, the samples were separated into 24 fractions by mixed-mode HPLC (Phillips et al, 2010) and each fraction analysed by LC-MS/MS. For each LC-MS/MS run, peptides were separated using a linear gradient of 1–37% acetonitrile in 0.1% formic acid over 1 h at a flow rate of 350 nl min−1 (25 cm × 75 μm Acclaim PepMap C18 (Thermo Scientific, Hemel Hempstead, UK) 3 μm 100 Å column, Ultimate 3000 RS HPLC) and sprayed into a maXis impact (Bruker Daltonics, Coventry, UK) via a captive source with the instrument operating in data-dependent mode (8 MS/MS per cycle). The data were searched against a database containing Swissprot human and bovine protein sequences and randomised versions thereof with MASCOT (Matrix Science Ltd, London, UK). Search parameters were: enzyme=trypsin, up to 2 missed cleavages, precursor ion tolerance=20 p.p.m., fragment ion tolerance=0.02 Da, mowse score >25, fixed modifications=cysteine carbamidomethyl, variable modifications=methionine oxidation and light/heavy dimethylation on N-termini and lysines. Multiple data sets were combined using Proteinscape 3 (Bruker Daltonics) and filtered at 1% protein false discovery. Relative quantitation (±PMA) was performed using WARP-LC software (Bruker Daltonics) with expression ratios normalised to the median ratio in each experiment. Cell compartment analysis was performed using http://string-db.org/.
EGFR ELISA and urine testing
Epidermal growth factor receptor (EGFR) was measured with a sandwich ELISA utilising goat polyclonal antibodies against the extracellular domain of EGFR (R&D Systems Europe Ltd, Abingdon, UK, Cat no. DYC1854-5). All urine samples were measured using 50 μl of urine plus 50 μl of 1% BSA in PBST per well. Urinary EGFR is expressed as pg EGFR per mg creatinine (determined by the Jaffe method). Cell lysates were prepared in PBS containing 1% Igepal CA-630 and Complete Protease Inhibitor Cocktail (Roche Diagnostics Ltd, Burgess Hill, UK) and the protein concentrations determined using the bicinchoninic acid assay calibrated with bovine serum albumin. Secretome and lysate EGFR concentrations were determined using appropriate volumes of sample (2–100 μl) for the concentration range of the assay. Secretome and lysate EGFR concentrations are expressed as ng EGFR per mg of total cell protein. For western blotting, proteins were separated with SDS–PAGE on 4–12% gels using MOPS running buffer (Life Technologies, Paisley, UK), blotted onto PVDF and probed with 0.2 μg ml−1 biotinylated anti-EGFR antibody (R&D Systems Europe Ltd, Cat no. BAF231) followed by streptavidin-HRP.
Human Proteome Atlas analysis
Data were downloaded from http://www.proteinatlas.org (Uhlen et al, 2010). For a protein to be a candidate biomarker it should be absent or expressed at low levels in normal urothelium and highly expressed in UBC; however, because of the heterogeneity of UBC, clinically useful biomarkers may be overexpressed in a subset of tumours rather than in all cases. We arbitrarily set our selection criteria for candidate biomarkers as high or medium staining in at least 4 cases of UBC (out of 11 or 12 cases depending on the protein) and low or undetectable staining in normal urothelium, generating a list of 54 proteins.
Statistical analyses
Urinary EGFR levels are presented as medians and statistical significance was calculated using Mann–Whitney tests. The UBC-specific survival was defined as the time from registration into the BCPP study to date of death from UBC. Patients were censored at the date last known to be alive or date of UBC-related death. Univariable Cox proportional hazards models were employed (α 0.1) to identify factors to be included in a Cox multivariable model; significance was set at 0.05. Univariable factors included age (years), tumour number (1 vs 2+), grade (1, 2 vs 3), size of largest tumour (<3 cm vs >3 cm), CIS (present vs absent), stage (pTa or pT1 vs pT2+), sex (male vs female), urinary EGFR (normal vs elevated) and urinary EpCAM (normal vs elevated). Analysis was done in Stata 12.1 (StataCorp, College Station, TX, USA).
Results
Proteomic analysis of secretomes
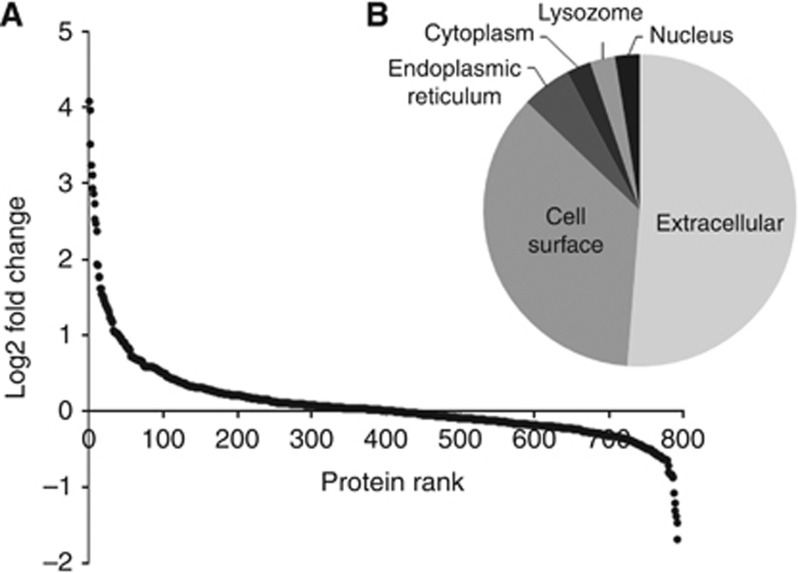
In total, 2078 unique proteins were identified in the conditioned media of one or more of the UBC cell lines with 1338, 1025, 794, 593, 443, 312 and 188 proteins identified in 2, 3, 4, 5, 6, 7 or 8 UBC cell lines respectively (Table 1 and Supplementary Tables S2 and S3). All secretomes were analysed±PMA to stimulate secretion and shedding. Relative quantitation was achieved using stable isotope labelling and is shown for all proteins quantitated±PMA in at least four UBC cell lines in Figure 1A and Supplementary Table S4. The cellular compartmentalisation of the 39 proteins with a >2-fold median upregulation in the secretomes due to PMA is shown in Figure 1B. The upregulated proteins are almost exclusively secreted and cell surface proteins (34 out of 39=87%), whereas the corresponding percentage for all proteins found in at least 4 UBC cell lines was 69% and for all proteins found in at least 2 UBC cell lines it was 63%. We found reports of 11 of these proteins previously being investigated as urinary biomarkers for UBC, with 8 being significantly elevated in patients with UBC (Supplementary Table S4).
Table 1. The UBC cell line secretome analysis.
| Cell line | Grade | No. of human peptides | No. of human proteins | No. of EGFR peptides | Secretome (EGFR) ng mg−1 | Cellular (EGFR) ng mg−1 | % Sec |
|---|---|---|---|---|---|---|---|
| UROtsa | NA | 5499 | 763 | 0 | 0.40 (0.03), 0.72 (0.08) | 43.7 (1.3), 61.4 (3.7) | 1.0 |
| MGH-U3 | 1 | 10 954 | 1041 | 5 | 4.29 (0.92), 4.46 (1.19) | 90.8 (4.7), 94.7 (8.9) | 4.7 |
| R-T4 | 1 | 8145 | 1160 | 1 | 1.30 (0.36), 2.62 (0.29) | 34.0 (18.0), 22.1 (11.8) | 9.6 |
| SW-780 | 1 | 4181 | 627 | 1 | 2.81 (0.32), 4.11 (0.62) | 49.3 (4.50), 62.2 (16.7) | 5.9 |
| RT112 | 2 | 5900 | 787 | 4 | 0.66 (0.07), 1.18 (0.17) | 82.4 (3.3), 62.0 (5.6) | 1.3 |
| VM-CUB-1 | 2 | 5915 | 765 | 16 | 10.8 (2.39), 9.40 (1.18) | 227 (3.79), 151 (41.7) | 5.5 |
| 5637 | 2 | 4188 | 628 | 3 | 0.41 (0.03), 0.68 (0.04) | 102 (3.81), 72.8 (0.87) | 0.7 |
| T-24 | 3 | 7087 | 916 | 0 | 1.19 (0.32), 2.37 (0.57) | 27.1 (1.36), 29.0 (3.18) | 6.3 |
| HB-CLS-2 | 3 | 7135 | 847 | 53 | 252 (92.8), 298 (22.3) | 2720 (161), 2646 (927) | 10.3 |
Abbreviations: EGFR=epidermal growth factor receptor; NA=not applicable; % Sec=percentage of secreted EGFR; UBC=urothelial bladder cancer.
The number of unique peptides identified by liquid chromatography-tandem mass spectrometry (LC-MS/MS) in each secretome and the resulting protein identifications are shown alongside the number of EGFR peptides identified. Proteomic analysis was performed once per cell line. The concentration of EGFR (determined by ELISA on biological triplicates) in paired secretomes and cell lysates is shown as ng EGFR per mg of cellular protein (determined by bicinchoninic acid assay); left value= −PMA, right value= +PMA. The percentage of secreted EGFR (% Sec) is relative to cellular EGFR.
Figure 1.
The effect of PMA on UBC cell line secretomes. (A) The log2 of median fold change for proteins quantitated in ≥4 UBC cell line secretomes±PMA (each data point is for a single protein and the proteins have been sorted from largest increase to largest decrease): 39 proteins show a >2-fold increase and 6 proteins a >2-fold decrease. The cell compartment for the 39 upregulated proteins is shown in (B).
Selection of EGFR as a candidate biomarker
Although PMA helps to identify proteins that are genuinely secreted or shed from UBC cells, it does not necessarily indicate that they are UBC specific, especially as release of the same proteins was also stimulated in the immortalised ‘normal' urothelial UROtsa cell line (Supplementary Table S4). To address this issue we compared the secretome data with immunohistochemistry data in the Human Protein Atlas. This approach, requiring proteins to be overexpressed in UBC and released from UBC cell lines, generated a shortlist of 5 candidates (EGFR, glucose-6-phosphate dehydrogenase, peroxiredoxin 6, LY6/PLAUR domain containing 3 and fibulin 1); EGFR was unique in that it was the only one of these five proteins that was identified in the cancer cell line secretomes and not in the UROtsa secretome. Tryptic peptides from EGFR were detected in seven of the eight UBC cell lines with a tendency for more peptides (suggestive of a higher EGFR protein concentration) in the cell lines derived from higher-grade tumours (Table 1). Both ‘light' and ‘heavy' EGFR peptides were identified at similar levels in the secretomes, indicating that EGFR is released into the conditioned media in the absence and presence of PMA. These findings were confirmed by ELISA: all of the cell lines express and release some EGFR, although levels range from very low in UROtsa and several of the UBC lines through to extremely high in HB-CLS-2. The PMA has a modest effect (on average <2-fold increase) on secretome EGFR levels and the amount of EGFR released into the conditioned media varies from cell line to cell line, between ∼1% and 10% of the total cellular EGFR.
Urinary EGFR as a diagnostic marker
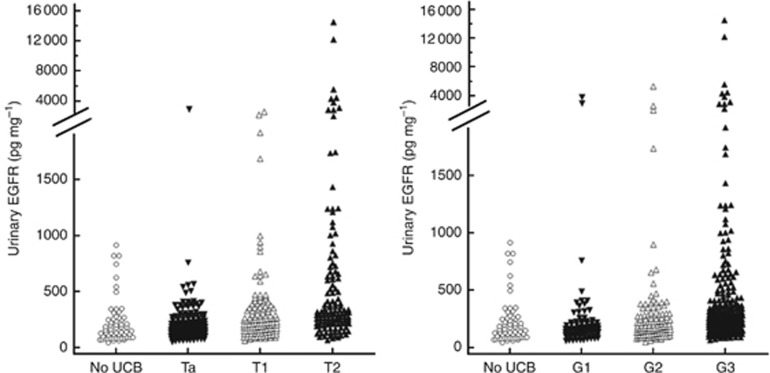
Epidermal growth factor receptor was measured in the urine of 436 UBC patients and 60 noncancer controls. The data are summarised in Table 2. The median urinary EGFR concentration was 153 pg per mg creatinine in noncancer controls and was not significantly increased in patients with pTa UBC. There were significantly higher levels of EGFR in the urine of patients with pT1 or pT2+ UBC (224 and 317 pg per mg creatinine, respectively). The data for individual patients stratified according to stage and grade are shown in Figure 2. High levels of urinary EGFR were observed in a proportion of patients, but not all patients, with high-grade and high-stage UBC. Defining the upper limit of the reference range as the mean+2 s.d. in the noncancer control group gives a threshold of 630 pg EGFR per mg creatinine, and the percentage of positive cases is 7, 4, 13 and 27 in noncancer controls, pTa, pT1 and pT2+ UBC, respectively.
Table 2. Patient data and urinary EGFR summary.
| Stage | N | Male/female | Grade (G1/G2/G3) | Age | EGFR (pg per mg creatinine) | P-value | ROC | No. of positive cases |
|---|---|---|---|---|---|---|---|---|
| Noncancer | 60 | 45/15 | NA | 75 (66–79) | 153 (91–261) | NA | NA | 4 (7%) |
| pTa | 184 | 140/44 | 79/84/21 | 74 (65–81) | 167 (124–236) | 0.395 | 0.536 | 3 (2%) |
| pT1 | 130 | 112/18 | 2/37/91 | 75 (67–80) | 224 (151–353) | 0.0003 | 0.650 | 13 (10%) |
| pT2+ | 122 | 97/25 | 0/6/116 | 78 (68–83) | 317 (215–658) | 0.0001 | 0.766 | 33 (27%) |
Abbreviations: EGFR=epidermal growth factor receptor; ROC=receiver operating characteristic.
The total numbers of patients in each stage group and the numbers of males and females and number of patients with grade 1, 2 or 3 urothelial bladder cancer (UBC) are shown. Age and urinary EGFR for each stage group are shown as median (interquartile range (IQR)). P-values and ROC areas are provided for each stage of UBC vs noncancer controls. The number of positive cases in each group is calculated using a threshold of 630 pg EGFR per mg creatinine (mean+2 s.d. of the urinary EGFR concentrations in the noncancer group).
Figure 2.
Urinary EGFR in UBC patients. The left-hand panel shows urinary EGFR data for patients stratified according to stage and the right-hand panel according to grade.
Characterisation of ‘soluble' EGFR
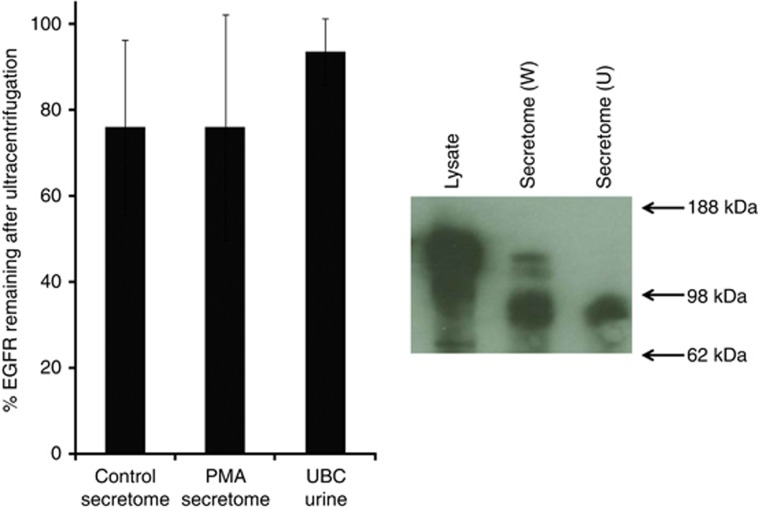
Peptides from the extracellular domain, but not the intracellular domain, of EGFR were identified in secretomes from MGH-U3, RT112, 5637, RT-4 and SW-780 cell lines (EGFR sequence coverage shown in Supplementary Figure S1). VM-CUB-1 and HB-CLS-2 secretomes contained the highest levels of EGFR, and peptides from both the intracellular and extracellular domains were identified; however, both spectral counting and peptide intensity suggested a higher concentration of the extracellular domain than the intracellular domain; that is, the extracellular domain of EGFR is shed into the conditioned media but full-length membrane-bound EGFR is also present. To test this hypothesis, we ultracentrifuged the secretomes at 136 000 g for 90 min to pellet membranes and membrane-bound proteins and measured the EGFR remaining in the supernatants. On average, across the secretomes, 73% of the EGFR remained in the supernatant, confirming that most of the EGFR is in a soluble rather than membrane-bound form (Figure 3). Western blotting of the proteins released by HB-CLS-2 cells indicated that the predominant form of EGFR in the secretome has a molecular weight of ∼100 kDa. A small amount of full-length EGFR (∼170 kDa) is also present but is removed by ultracentrifugation (Figure 3).
Figure 3.
The effect of ultracentrifugation on EGFR. The histogram shows the percentage of EGFR remaining in the supernatant following ultracentrifugation. Secretome data are shown as the mean±s.d. across all 9 cell lines (±PMA) and urine data as the mean±s.d. for urine from 12 patients with UBC and elevated EGFR. The western blot shows EGFR in HB-CLS-2 cell lysate and whole (W) and ultracentrifuged (U) HB-CLS-2 conditioned media.
Ultracentrifugation was also used to test whether urinary EGFR was shed extracellular domain or membrane-bound EGFR: 12 urine samples with a range of elevated EGFR levels were ultracentrifuged and EGFR measured in the supernatants. On average, 93.5% (s.d. 7.8%) remained in the supernatant, indicating that the predominant form of EGFR in urine is the soluble extracellular domain.
Urinary EGFR as a prognostic marker
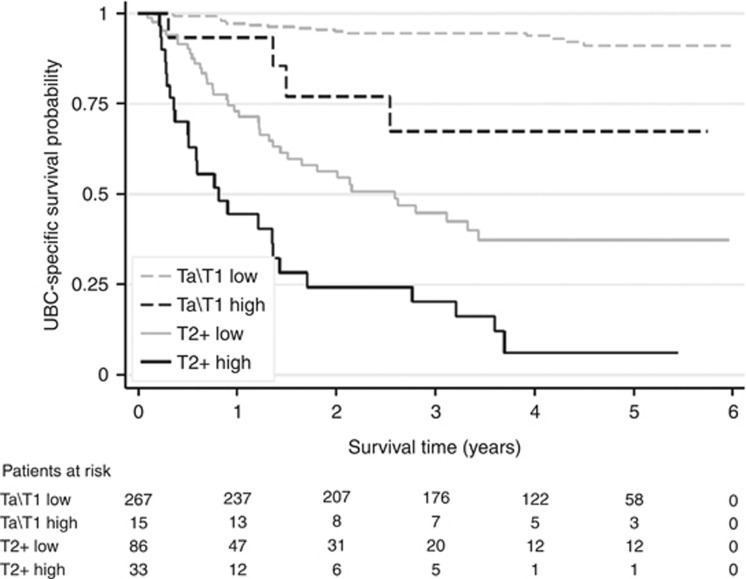
Potential prognostic factors (age, number of tumours, grade, size of largest tumour, presence of CIS, stage, gender and urinary EGFR) were initially analysed in univariable models. Epidermal growth factor receptor was found to be highly significant (HR=6.9, 90% CI 4.7–10.1, P<0.001) and this can be seen in the Kaplan–Meier curves for UBC-specific survival in both NMIBC and MIBC (Figure 4). In the univariable analyses grade, stage, age, tumour size, CIS and urinary EGFR all reached statistical significance and were included in a multivariable analysis. The multivariable analysis showed that elevated urinary EGFR has prognostic value (HR=2.9 95% CI 1.1–4.6: P<0.001) over and above that provided by grade and stage for UBC-specific survival (Table 3a).
Figure 4.
The prognostic value of urinary EGFR. Kaplan–Meier curves are shown for UBC-specific survival with patients stratified according to low/high urinary EGFR (>630 pg per mg creatinine).
Table 3a. Cox multivariable analysis of prognostic indicators.
| Variable | β | s.e. (β) | HR | P-value | HR 95% CI |
|---|---|---|---|---|---|
| Grade 3 (reference grade 1 or 2) | 0.982 | 0.395 | 2.670 | 0.013 | 1.231–5.792 |
| Stage pT2+ (reference pTa or pT1) | 1.997 | 0.306 | 7.369 | <0.001 | 4.043–13.429 |
| High EGFR (reference low EGFR) | 1.062 | 0.239 | 2.891 | <0.001 | 1.809–4.620 |
Abbreviations: CI=confidence interval; EGFR=epidermal growth factor receptor; HR=hazard ratio.
Combination of urinary EpCAM and EGFR for prognostication
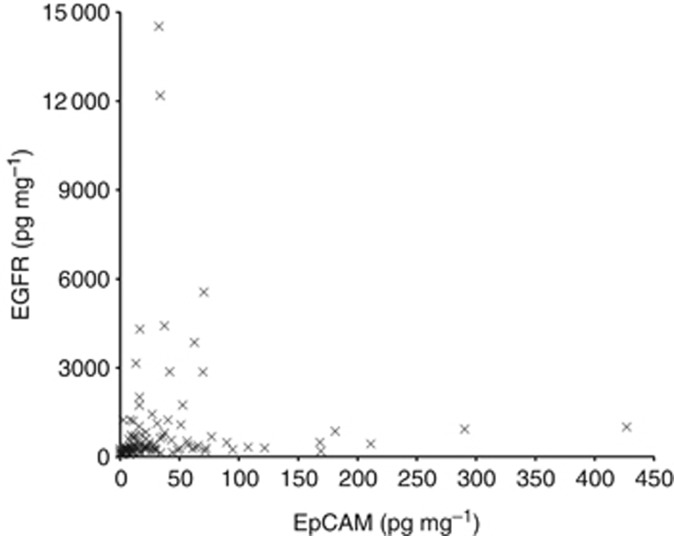
We have previously reported that the urinary concentration of the extracellular domain of EpCAM is associated with a shorter UBC-specific survival time (Bryan et al, 2014b). The urinary concentrations of EGFR and EpCAM have quite different distributions in our cohort of UBC patients: many patients have high levels of either EpCAM or EGFR (Figure 5), suggesting that they are not redundant as biomarkers and might be used in combination to improve prognostication. Multivariable modelling including both biomarkers and all univariable factors was performed; grade and stage were both highly prognostic (P=0.011 and P<0.001 respectively), as were EGFR (HR=6.69, 95% CI 3.41–13.12, P<0.001) and EpCAM (HR=2.13, 95% CI 1.19–3.82, P=0.011). In addition, the interaction between EGFR and EPCAM was statistically significant (P=0.001). It might be predicted that being ‘double-positive' (high levels of both biomarkers) would carry an extremely poor prognosis, but we found that this was not the case: compared with being ‘double-negative' (low levels of both biomarkers), the risk of death from UBC doubles if a patient is EpCAM high and EGFR low, increases 6.7-fold with high EGFR and low EpCAM and for ‘double-positive' the risk is increased 2.6-fold. Therefore, although the risk associated with being double positive is greater than only being EpCAM positive, it is less than being only EGFR positive (P=0.001) (Table 3b and Supplementary Figure S2). Utilising the two biomarkers enables the identification of a double-negative group of patients who have the best prognosis and those patients who are EGFR high and EpCAM low who have the worst prognosis.
Figure 5.
Relationship between urinary EGFR and EpCAM in UBC: the creatinine-normalised concentrations of EGFR and EpCAM in the urine of patients with MIBC.
Table 3b. Cox multivariable analysis of prognostic indicators including EpCAM.
| Variable | β | s.e. (β) | HR | P-value | HR 95% CI |
|---|---|---|---|---|---|
| Grade 3 (reference grade 1 or 2) | 1.005 | 0.394 | 2.732 | 0.011 | 1.262–5.912 |
| Stage pT2+ (reference pTa or pT1) | 1.892 | 0.301 | 6.632 | <0.001 | 3.680–11.953 |
| High EGFR (reference low EGFR) | 1.900 | 0.344 | 6.691 | <0.001 | 3.411–13.124 |
| High EpCAM (reference low EpCAM) | 0.756 | 0.298 | 2.131 | 0.011 | 1.189–3.818 |
| EGFR and EpCAM interaction (high EGFR and EpCAM) | −1.689 | 0.487 | 0.185 | 0.001 | 0.071–0.480 |
Abbreviations: CI=confidence interval; EGFR=epidermal growth factor receptor; EpCAM=epithelial cell adhesion molecule; HR=hazard ratio.
Discussion
Combining proteomic analysis of UBC cell line secretomes and immunohistochemistry data available in the Human Protein Atlas identified EGFR as a potential urinary biomarker for UBC. We then tested the diagnostic and prognostic utility of urinary EGFR in a cohort of 496 patients with long-term clinical follow-up. Urinary EGFR is elevated in 27% of MIBC cases and 17% of grade 3 UBCs but normal levels are observed in pTa and low-grade disease; thus, overall, urinary EGFR is not a good diagnostic marker for UBC. However, in patients with confirmed UBC, urinary EGFR is a strong independent prognostic indicator of UBC-specific survival. Urinary EGFR could have utility for rapidly identifying patients with the most aggressive disease, and expediting their subsequent investigation and management.
Shedding of EGFR from cancer cell lines (although not UBC cell lines) has been reported to be related to overexpression, to be stimulated by PMA (and hence may be under the control of protein kinase C) and to be blocked by metalloprotease inhibitors (Perez-Torres et al, 2008). Adamczyk et al (2011) reported that pancreatic cancer cell lines release both exosomal (full length, 170 kDa) and ectodomain (110 kDa) forms of EGFR. Evidence suggests that EGFR ectodomain is shed as the result of proteolytic cleavage and that it is probably cleavage of an alternatively spliced 3-kb transcript of EGFR that generates the soluble ectodomain (Wilken et al, 2013). Although this splice variant lacks both the transmembrane and intracellular domains, it seems that it remains membrane associated until cleaved (Wilken et al, 2013). Our UBC cell line data are consistent with the body of evidence suggesting that both full-length and ectodomain EGFR are released from cancer cells by a regulated mechanism involving proteolytic cleavage.
Soluble EGFR has been detected in plasma and investigated in several cancers (Baron et al, 2003; Hudelist et al, 2006; Müller et al, 2006; Lemos-González et al, 2007; Zampino et al, 2008). However, plasma levels appear to be lower in cancer patients than healthy controls, are not related to EGFR expression in tumours and there is little evidence of diagnostic, predictive or prognostic value. Although EGF has been measured in the urine of UBC patients (Chow et al, 1997), the only case of EGFR being measured in urine that we have encountered looks at a number of distal tumour sites and not UBC (Witters et al, 1995). Thus, we believe that the current work is the first exploration of soluble urinary EGFR as a biomarker for UBC, with a much clearer relationship between urinary EGFR and UBC than in studies of plasma EGFR in other solid malignancies.
Epidermal growth factor receptor has been reported to be overexpressed in many epithelial cancers and was reported as a poor prognostic indicator in UBC as early as 1990 (Neal et al, 1990), representing a late event in the progression of the disease (Lipponen and Eskelinen, 1994). More recently, EGFR overexpression has been shown to be a characteristic of basal-like aggressive MIBC, and in mouse models these tumours respond to anti-EGFR therapy (Rebouissou et al, 2014). Several clinical trials using anti-EGFR therapies in combination with chemotherapy or radiotherapy in MIBC are currently underway. Although further work is required to determine whether urinary levels of EGFR are entirely determined by the level of expression in tumour cells, or whether regulation of shedding is also important, the fact that many bladder tumours release detectable levels of EGFR into the urine suggests that urinary EGFR should be considered as a possible predictive marker in clinical trials evaluating anti-EGFR therapy in UBC.
Conclusions
We present the first description of the prognostic value of soluble urinary EGFR in UBC, both alone and in combination with soluble urinary EpCAM (Bryan et al, 2014b). These ELISA-based tests have significant potential clinical utility and we intend to evaluate this potential prospectively in UBC, as well as determining whether other urological malignancies also increase urine EGFR levels. Mechanisms of ectodomain shedding may add an extra layer of complexity to the molecular pathology of UBC that may not be uncovered by genomic approaches and appear to unmask potentially important prognostic markers. Better understanding of these phenomena may also reveal new therapeutic targets, targets that are desperately needed for UBC (Bryan et al, 2014a).
Acknowledgments
We thank all the West Midlands Consultant Urologists and their units involved with BCPP, as well as the BCPP research nurses and Margaret Grant, Deborah Bird, Jennifer Barnwell, Duncan Nekeman and Eline van Roekel. We also thank Ashley Martin, Neil Shimwell and Wenbin Wei for help with mass spectrometry and bioinformatics. BCPP is funded by Cancer Research UK, the University of Birmingham and the Birmingham and The Black Country and West Midlands North and South Comprehensive Local Research Networks, and sponsored by the University of Birmingham. The BCPP biospecimen collection is supported by funding from the Birmingham ECMC. Douglas Ward is funded by a philanthropic donation to the University of Birmingham in support of bladder cancer research.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Supplementary Material
References
- Adamczyk K, Tehrani M, Warnken U, Schmiegel W, Schnölzer M, Schwarte-Waldhoff I. Characterization of soluble and exosomal forms of the EGFR released from pancreatic cancer cells. Life Sci. 2011;89:304–312. doi: 10.1016/j.lfs.2011.06.020. [DOI] [PubMed] [Google Scholar]
- Babjuk M, Burger M, Zigeuner R, Shariat S, van Rhijn B, Compérat E, Sylvester R, Kaasinen E, Böhle A, Palou Redorta J, Rouprêt M. EAU guidelines on non-muscle-invasive urothelial carcinoma of the bladder: update 2013. Eur Urol. 2013;64:639–653. doi: 10.1016/j.eururo.2013.06.003. [DOI] [PubMed] [Google Scholar]
- Baron A, Cora E, Lafky J, Boardman C, Buenafe M, Rademaker A, Liu D, Fishman D, Podratz K, Maihle N. Soluble epidermal growth factor receptor (sEGFR/sErbB1) as a potential risk, screening, and diagnostic serum biomarker of epithelial ovarian cancer. Cancer Epidemiol Biomarkers Prev. 2003;12:103–113. [PubMed] [Google Scholar]
- Bryan R, Kirby R, O'Brien T, Mostafid H. So much cost, such little progress. Eur Urol. 2014;66:253–262. doi: 10.1016/j.eururo.2014.02.031. [DOI] [PubMed] [Google Scholar]
- Bryan R, Shimwell N, Wei W, Devall A, Pirrie S, James N, Zeegers M, Cheng K, Martin A, Ward D. Urinary EpCAM in urothelial bladder cancer patients: characterisation and evaluation of biomarker potential. Br J Cancer. 2014;110:679–685. doi: 10.1038/bjc.2013.744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger M, Catto J, Dalbagni G, Grossman H, Herr H, Karakiewicz P, Kassouf W, Kiemeney L, La Vecchia C, Shariat S, Lotan Y. Epidemiology and risk factors of urothelial bladder cancer. Eur Urol. 2013;63:234–241. doi: 10.1016/j.eururo.2012.07.033. [DOI] [PubMed] [Google Scholar]
- Chow N, Liu H, Lee E, Chang C, Chan S, Cheng H, Tzai T, Lin J. Significance of urinary epidermal growth factor and its receptor expression in human bladder cancer. Anticancer Res. 1997;17:1293–1296. [PubMed] [Google Scholar]
- Hansen A, Freeman T, Arnold S, Starchenko A, Jones-Paris C, Gilger M, Washington M, Fan K, Shyr Y, Beauchamp R, Zijlstra A. Elevated ALCAM shedding in colorectal cancer correlates with poor patient outcome. Cancer Res. 2013;15:2955–2964. doi: 10.1158/0008-5472.CAN-12-2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Lin L, Gao Y, Chen Y, Yan X, Xing J, Hang W. Bladder cancer determination via two urinary metabolites: a biomarker pattern approach. Mol Cell Proteomics. 2011;10:1–10. doi: 10.1074/mcp.M111.007922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudelist G, Köstler W, Gschwantler-Kaulich D, Czerwenka K, Kubista E, Müller R, Helmy S, Manavi M, Zielinski C, Singer C. Serum EGFR levels and efficacy of trastuzumab-based therapy in patients with metastatic breast cancer. Eur J Cancer. 2006;42:186–192. doi: 10.1016/j.ejca.2005.08.036. [DOI] [PubMed] [Google Scholar]
- Kandimalla R, Masius R, Beukers W, Bangma C, Orntoft T, Dyrskjot L, van Leeuwen N, Lingsma H, van Tilborg A, Zwarthoff E. A 3-plex methylation assay combined with the FGFR3 mutation assay sensitively detects recurrent bladder cancer in voided urine. Clin Cancer Res. 2013;19:4760–4769. doi: 10.1158/1078-0432.CCR-12-3276. [DOI] [PubMed] [Google Scholar]
- Kaplan A, Litwin M, Chamie K. The future of bladder cancer care in the USA. Nat Rev Urol. 2014;11:59–62. doi: 10.1038/nrurol.2013.180. [DOI] [PubMed] [Google Scholar]
- Kaufman D, Shipley W, Feldman A. Bladder cancer. Lancet. 2009;374:239–249. doi: 10.1016/S0140-6736(09)60491-8. [DOI] [PubMed] [Google Scholar]
- Lemos-González Y, Rodríguez-Berrocal F, Cordero O, Gómez C, Páez de la Cadena M. Alteration of the serum levels of the epidermal growth factor receptor and its ligands in patients with non-small cell lung cancer and head and neck carcinoma. Br J Cancer. 2007;96:1569–1578. doi: 10.1038/sj.bjc.6603770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipponen P, Eskelinen M. Expression of epidermal growth factor receptor in bladder cancer as related to established prognostic factors, oncoprotein (c-erbB-2, p53) expression and long-term prognosis. Br J Cancer. 1994;69:1120–1125. doi: 10.1038/bjc.1994.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller V, Witzel I, Pantel K, Krenkel S, Lück H, Neumann R, Keller T, Dittmer J, Jänicke F, Thomssen C. Prognostic and predictive impact of soluble epidermal growth factor receptor (sEGFR) protein in the serum of patients treated with chemotherapy for metastatic breast cancer. Anticancer Res. 2006;26:1479–1487. [PubMed] [Google Scholar]
- Neal D, Sharples L, Smith K, Fennelly J, Hall R, Harris A. The epidermal growth factor receptor and the prognosis of bladder cancer. Cancer. 1990;65:1619–1625. doi: 10.1002/1097-0142(19900401)65:7<1619::aid-cncr2820650728>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Orenes-Piñero E, Cortón M, González-Peramato P, Algaba F, Casal I, Serrano A, Sánchez-Carbayo M. Searching urinary tumor markers for bladder cancer using a two-dimensional differential gel electrophoresis (2D-DIGE) approach. J Proteome Res. 2007;11:4440–4448. doi: 10.1021/pr070368w. [DOI] [PubMed] [Google Scholar]
- Perez-Torres M, Valle B, Maihle N, Negron-Vega L, Nieves-Alicea R, Cora E. Shedding of epidermal growth factor receptor is a regulated process that occurs with overexpression in malignant cells. Exp Cell Res. 2008;314:2907–2918. doi: 10.1016/j.yexcr.2008.07.013. [DOI] [PubMed] [Google Scholar]
- Phillips H, Williamson J, van Elburg K, Snijders A, Wright P, Dickman M. Shotgun proteome analysis utilising mixed mode (reversed phase-anion exchange chromatography) in conjunction with reversed phase liquid chromatography mass spectrometry analysis. Proteomics. 2010;10:2950–2960. doi: 10.1002/pmic.200900669. [DOI] [PubMed] [Google Scholar]
- Rebouissou S, Bernard-Pierrot I, de Reyniès A, Lepage M, Krucker C, Chapeaublanc E, Hérault A, Kamoun A, Caillault A, Letouzé E, Elarouci N, Neuzillet Y, Molinié V, Vordos D, Laplanche A, Maillé P, Soyeux P, Ofualuka K, Reyal F, Biton A, Sibony M, Paoletti X, Southgate J, Benhamou S, Lebret T, Allory Y, Radvanyi F. EGFR as a potential therapeutic target for a subset of muscle-invasive bladder cancers presenting a basal-like phenotype. Sci Transl Med. 2014;6:244ra91. doi: 10.1126/scitranslmed.3008970. [DOI] [PubMed] [Google Scholar]
- Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S, Wernerus H, Björling L, Ponten F. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol. 2010;28:1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- Wilken J, Perez-Torres M, Nieves-Alicea R, Cora E, Christensen T, Baron A, Maihle N. Shedding of soluble epidermal growth factor receptor (sEGFR) is mediated by a metalloprotease/fibronectin/integrin axis and inhibited by cetuximab. Biochemistry. 2013;52:4531–4540. doi: 10.1021/bi400437d. [DOI] [PubMed] [Google Scholar]
- Witjes J, Compérat E, Cowan N, De Santis M, Gakis G, Lebret T, Ribal M, Van der Heijden A, Sherif A. EAU guidelines on muscle-invasive and metastatic bladder cancer: summary of the 2013 guidelines. Eur Urol. 2014;65:778–792. doi: 10.1016/j.eururo.2013.11.046. [DOI] [PubMed] [Google Scholar]
- Witters L, Curley E, Kumar R, Chinchilli V, Harvey J, Crebbin V, Harvey H, Lipton A. Epidermal growth factor receptor ectodomain in the urine of patients with squamous cell carcinoma. Clin Cancer Res. 1995;1:551–557. [PubMed] [Google Scholar]
- Zampino M, Magni E, Santoro L, Zorzino L, Dell'Orto P, Sonzogni A, Fazio N, Monfardini L, Chiappa A, Biffi R, de Braud F. Epidermal growth factor receptor serum (sEGFR) level may predict response in patients with EGFR-positive advanced colorectal cancer treated with gefitinib. Cancer Chemother Pharmacol. 2008;63:139–148. doi: 10.1007/s00280-008-0722-x. [DOI] [PubMed] [Google Scholar]
- Zatovicova M, Sedlakova O, Svastova E, Ohradanova A, Ciampor F, Arribas J, Pastorek J, Pastorekova S. Ectodomain shedding of the hypoxia-induced carbonic anhydrase IX is a metalloprotease-dependent process regulated by TACE/ADAM17. Br J Cancer. 2005;93:1267–1276. doi: 10.1038/sj.bjc.6602861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeegers M, Bryan R, Langford C, Billingham L, Murray P, Deshmukh N, Hussain S, James N, Wallace D, Cheng K. The West Midlands Bladder Cancer Prognosis Programme: rationale and design. BJU Int. 2010;105:784–788. doi: 10.1111/j.1464-410X.2009.08849.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.