Abstract
Background:
hERG1 channels are aberrantly expressed in human cancers. The expression, functional role and clinical significance of hERG1 channels in pancreatic ductal adenocarcinoma (PDAC) is lacking.
Methods:
hERG1 expression was tested in PDAC primary samples assembled as tissue microarray by immunohistochemistry using an anti-hERG1 monoclonal antibody (α-hERG1-MoAb). The functional role of hERG1 was studied in PDAC cell lines and primary cultures. ERG1 expression during PDAC progression was studied in Pdx-1-Cre,LSL-KrasG12D/+,LSL-Trp53R175H/+ transgenic (KPC) mice. ERG1 expression in vivo was determined by optical imaging using Alexa-680-labelled α-hERG1-MoAb.
Results:
(i) hERG1 was expressed at high levels in 59% of primary PDAC; (ii) hERG1 blockade decreased PDAC cell growth and migration; (iii) hERG1 was physically and functionally linked to the Epidermal Growth Factor-Receptor pathway; (iv) in transgenic mice, ERG1 was expressed in PanIN lesions, reaching high expression levels in PDAC; (v) PDAC patients whose primary tumour showed high hERG1 expression had a worse prognosis; (vi) the α-hERG1-MoAb could detect PDAC in vivo.
Conclusions:
hERG1 regulates PDAC malignancy and its expression, once validated in a larger cohort also comprising of late-stage, non-surgically resected cases, may be exploited for diagnostic and prognostic purposes in PDAC either ex vivo or in vivo.
Keywords: pancreatic ductal adenocarcinoma (PDAC), hERG1 potassium channels, EGF-R, prognosis, molecular-imaging
Pancreatic ductal adenocarcinoma (PDAC) is the most commonly diagnosed pancreatic neoplasm, and represents the tenth most common cause of death from cancer in both sexes combined (Ferlay et al, 2013). Although surgery remains the cornerstone of cure, pharmacological treatments which are currently being applied, either as an adjuvant to surgery or as a definitive treatment for unresectable disease, have not substantially improved the patients' outcome (Partensky, 2013). Indeed, PDAC 5-year survival rate is still below 6% (Ferlay et al, 2013; Partensky, 2013). The main reasons of PDAC poor prognosis include aggressive growth and a pro-invasive behaviour, which account for rapid development of distant metastases, as well as the rapid onset of chemoresistance. PDAC malignancy is frequently correlated with the activation of EGF-R signalling pathways, which in turn mediate cell proliferation and invasiveness, hence sustaining the tumour malignancy (Boeck et al, 2013).Indeed, a high percentage of PDAC show EGF-R overexpression, as well as Kras mutations (Mulcahy et al, 1998; Sivecke and Crawford, 2012). Notably, and at difference from what occurs in colorectal cancer (Krasinskas, 2011), in PDAC a mutated Kras does not totally abrogate (or bypass) EGF-R-triggered signalling (Sivecke and Crawford, 2012).
Traditional PDAC prognostic factors include tumour size and grade, lymph node status, resection margins and vascular or neural invasion. In the last few years, many studies have been performed to identify novel biomarkers to predict the clinical and therapeutic outcomes accurately, as well as to design a multimodal therapeutic strategy (Apte et al, 2004). However, none of the molecular markers described so far can be recommended for routine clinical use (Ansari et al, 2011). The identification of novel biomarkers that accurately predict disease recurrence and patients' survival would substantially improve the identification of individual risk assessment and treatment selection.
In the last few years, a new class of proteins has acquired increasing relevance in oncological research: the ion channels and transporters (ICTs). Indeed, ICTs control many ‘cancer hallmarks' in different types of human cancers (Prevarskaya et al, 2010) and the blockage of their activity impairs the growth of different types of tumours, both in vitro and in vivo (Becchetti, 2011). A detailed characterization of ICTs in different cancer types is also allowing to exploit these proteins for diagnostic and patients' stratification purposes (Pedersen and Stock, 2013).
Our group contributed to this topic, focusing on channels encoded by the human ether à-go-go-related gene 1 (hERG1). hERG1 channels are over- and mis-expressed in a wide variety of human cancers and their activity is involved in the regulation of neoplastic cell growth and progression (Arcangeli, 2005). hERG1 has a clinical significance in colorectal cancer patients, where it contributes to identify at-risk subjects (Lastraioli et al, 2012), as well as in gastric cancer where it displays a negative prognostic impact in T1-stage patients (Crociani et al, 2014). In both cancer types, hERG1 is directly linked to VEGF-A and the angiogenic pathway (Crociani et al, 2013).
Some specific ICTs have been detected and characterised in PDAC cells: TRP cationic channels of either the ‘melastatin-related' (TRPM) or ‘capsaicin' (TRPV1) type (Hartel et al, 2006), Cl− channels, in particular CLIC3 (Dozynkiewicz et al, 2012), the sodium hydrogen exchanger 1 (HNE1) (Provost and Wallert, 2013). The latter has been recently shown to interact with the Epidermal Growth Factor Receptor (EGF-R) in PDAC (Olszewski et al, 2010). However none of them has been exploited for diagnostic or prognostic purposes so far.
Only very few studies examining the role of hERG1 in human PDAC have been produced: it was recently shown that hERG1 is expressed in pancreatic cancer (Zhou et al, 2012), and its expression is negatively regulated by miR-96 (Feng et al, 2014). hERG1 silencing, as well as miR-96 over-expression inhibits the PDAC cell growth and invasiveness in vitro, and reduces tumorigenicity in vivo (Feng et al, 2014).
In the present paper we determined hERG1 expression and function in PDAC and evaluated its potential as a diagnostic and prognostic biomarker.
Materials and Methods
Cell culture: PDAC cell lines
Cells were cultured in either RPMI-1640 medium supplemented with 2 mM L-glutamine (MIAPaCa-2 and BxPC-3) or Dulbecco's Modified Eagle Medium (DMEM, Euroclone, Milan, Italy) supplemented with 4 mM L-glutamine (PANC-1) and 10% foetal bovine serum (FBS), in a humidified 5% CO2 atmosphere at 37 °C.
Cell culture: primary PDAC surgical samples
PDAC surgical samples were collected during surgical procedures, and a small piece of the tumour was put in DMEM supplemented with 4 mM L-glutamine and kept refrigerated until use. The collected tissues were washed three times with PBS-Gentamicin (50 mg ml−1) and Penicillin/Streptomycin (100 μg ml−1) and transferred into a petri dish containing the ‘complete medium' (CM) consisting of DMEM supplemented with 4 mM L-glutamine, 15 mM Hepes buffer, 10% heat inactivated FBS, Gentamicin (50 mg ml−1), Penicillin/Streptomycin (100 μg ml−1) and Amphotericin B (Fungizone, 5 μg ml−1). The tumour was finely minced with a sterile scalpel and transferred into a sterile tube containing CM, supplemented with Liver Digest Medium (Life Technologies, Carlsbad, CA, USA) following the manufacturer's instructions. The tubes were incubated under moderate stirring at 37 °C for 45 min. After digestion, gross debris were left to sediment, and the supernatant containing dispersed cells was centrifuged, washed once in CM and then resuspended in a small volume (depending on the size of the tumour sample) of CM to have a final cell concentration of 50 000–100 000 cells per ml. Cells were seeded onto a ‘Basement membrane' consisting of RPMI 5X, HEPES (50 mmol l−1), NaOH(1 mol l−1), laminin (0.1 mg ml−1) fibronectin (1 mg ml−1), Collagen IV (1 mg ml−1) and incubated in a humidified 5% CO2 atmosphere at 37 °C. After 24 h of incubation the medium was changed and fresh CM was added. After further 36 h of incubation, EGF was added at 20 ng ml−1 final concentration. When needed, E4031 was added at 40 μM final concentration. After 6 h of incubation in the presence of EGF and/or E4031, cells were photographed under an inverted microscope, and cell vitality was assessed through the WST-1 assay (see Supplementary Information).
RNA extraction and real-time quantitative PCR (RQ-PCR)
RNA extraction and RQ-PCR were performed as reported by Pillozzi et al (2007). The β-actin gene was used as a standard reference. The relative quantification of hERG1 gene expression was performed by the ΔCt method. Primer sequences are reported in Supplementary Information (Material and Methods of molecular biology experiments).
Protein extraction, immunoprecipitation (IP) and Western blotting
Whole-cell proteins were extracted from cultured cells as reported by Crociani et al (2013). The protocol is detailed in Supplementary Information.
Immunofluorescence (IF) laser-confocal microscopy
MIAPaCa-2, BxPC-3 and PANC-1 were plated and on the following day were fixed for 15 min in PBS plus 4% paraformaldehyde at room temperature. The protocol is fully detailed in Supplementary Information.
Study on PDAC patients and TMA construction
Once optimised the IHC protocol, 44 carcinoma samples (23 males, 21 females; mean age of 62.7 years, range 27–85 years) from consecutive, surgically resected tumours were retrieved from the archives of the Pathology Unit, Campus Bio-Medico University in Rome. Data on clinical variables, including sex and age were gathered retrospectively from patients' records. In our series of surgically resected patients there was not a selection criteria. Tumour-node-metastasis status classification was reassessed according to the Union for International Cancer Control (Sobin et al, 2009). Follow-up data were available for 42 included patients; none of the patients received chemotherapy or radiation therapy as a part of a neoadjuvant program. As the patients were all consecutive patients submitted to surgery, the majority of them were finally classified as TNM-stage I or II patients. Patients (3 out of the 42 patients with available follow-up data) which resulted to belong to TNM-stage III or IV were excluded from our survival analysis to eliminate a potential confounding factor on evaluating the survival. The study was carried out with approval of the Campus Bio-Medico institutional research board. Clinicopathological data are reported in Supplementary Table S1 and TMA construction is reported in Supplementary Information (Materials and Methods of experiments on primary samples).
Immunohistochemistry (IHC) and scoring assessment
IHC for hERG1 and EGF-R detection was performed as previously reported (Lastraioli et al, 2012) using two different preparations (either unpurified or affinity purified through a protein A column) of the α-hERG1-MoAb directed against the S5-pore region (DIVAL Toscana Srl). Protocols and data were compared to determine the best experimental conditions. Proliferation index was evaluated by performing IHC with anti-Ki67 antibody (clone MIB-1, DAKO Cytomation, Glostrup, Denmark), as previously reported (Lastraioli et al, 2006). IHC protocol is detailed in Supplementary Information.
hERG1 and EGF-R expression were evaluated by an immunohistochemical score calculated by combining the estimate of the percentage of immunoreactive cells (quantity score) with the estimate of staining intensity (staining-intensity score). This method is frequently applied in similar conditions when working with cytoplasmic/membrane proteins (Perrone et al, 2005). The details of the scoring system are reported in Supplementary Information (Materials and Methods of experiments on primary samples).
In vivo experiments
All experiments involving mice were approved by the Italian Ministry of Health. In vivo experiments were performed at the Laboratory of Genetic Engineering for the Production of Animal Models (LIGeMA) at the Animal House of the University of Florence.
Six-week old, female immunodeficient nu/nu mice were used for tumour MIA PaCa-luc2 cell implantation, as detailed in Supplementary Information.
Pdx-1-Cre,LSL-KrasG12D/+,LSL- Trp53R175H/+ (KPC) transgenic mice (Hingorani et al, 2003, 2005) were kindly provided from Dr DA Tuveson (Cancer Research UK, Cambridge Research Institute, Cambridge, UK).
Fluorescent imaging was performed using the α-hERG1-MoAb. The detailed protocol is reported in Supplementary Information.
Statistical analysis
Descriptive statistic was used to identify associations among age, clinical-pathological characteristics and presence of hERG1 and EGF-R using Mann–Whitney's U-test for continuous parameters and χ2 or Fisher's exact test for categorical parameters. Survival analysis (disease-free survival and overall survival) was aimed to estimate the prognostic value of these parameters. Cox-regression model and Kaplan–Meier curves were used.
Results
Analysis of hERG1 expression in PDAC
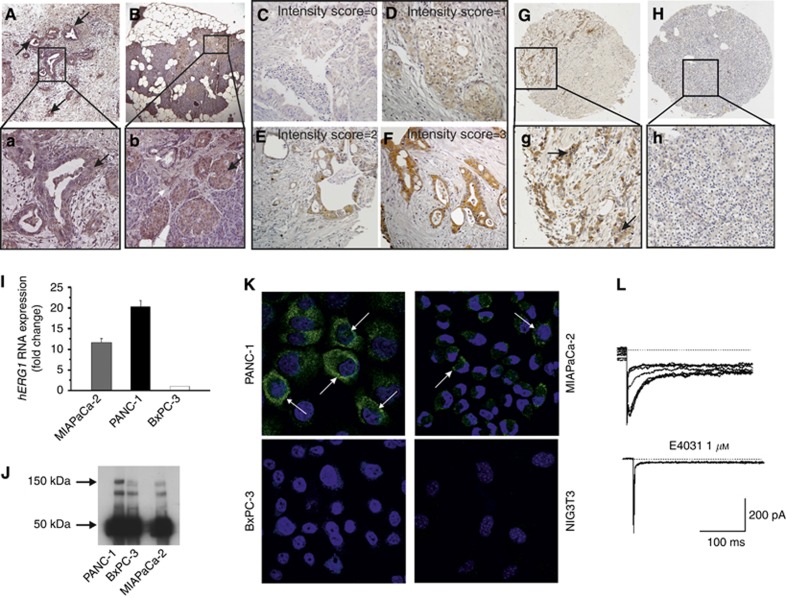
The expression of hERG1 was first evaluated in PDAC primary samples. A plasma membrane and cytoplasmic positivity for hERG1, with a granular pattern was detected in malignant neoplastic cells (Figure 1A and a, see black arrows). Stromal tissue and normal ductal epithelium were negative for hERG1 expression (Figure 1B and b, see white arrows). On the other hand, in agreement with functional data (Rosati et al, 2000), beta cells in Langerhans islets expressed hERG1 (Figure 1B and b, see black arrows).
Figure 1.
hERG1 expression in PDAC primary samples. (A) IHC for hERG1 in a PDAC sample. Magnification: 50x; a: higher magnification (200x) of the area in the black box in panel A. Arrow indicate hERG1 immunostaining at the cytoplasmic level. (B) IHC for hERG1 in a healthy pancreas. Magnification: 50x; b: higher magnification (200x) of the area in the black box in panel B. The black arrow indicates the hERG1 immunostaining in Langerhans islets, mainly attributable to the expression in beta cells; the white arrows indicate the lack of expression in the pancreatic stroma, as well as in normal ductal epithelial cells. (C–F) Representative microphotographs of samples showing different hERG1 intensity are reported; E: intensity score=0; F: intensity score=1; G: intensity score=2; H: intensity score=3. Magnification: 400x. (G) IHC for hERG1 in a PDAC sample assembled in TMA. Magnification: 50x; g: higher magnification of the area in the black box in panel G. (H) IHC for hERG1 protein in the corresponding healthy pancreatic tissue of panel G. Magnification: 50x. (I) Expression of hERG1 RNA. hERG1 transcript levels were normalised to the β-actin gene. (J) Expression of hERG1 protein: immunoprecipitation data. Two miligrams of total cell lysates were immunoprecipitated with the α-hERG1-MoAb and the corresponding blots were probed with a polyclonal antibody directed against the hERG1 C-terminus (C54 antibody). The 150 kDa band (arrow) corresponds to the mature, fully glycosylated hERG1 protein; the lower band (135 kDa) represents the partially glycosylated, immature hERG1 protein; the 50 kDa band (arrow) corresponds to the Ig used for the immunoprecipitation. (K) Expression of hERG1 protein: immunofluorescence data. In the merged images here reported, the green signal indicates the presence of hERG1 protein at the plasma membrane and cytoplasmic levels, whereas nuclei are stained in blue by DAPI. Magnification: 60x. (L) hERG1 current in PDAC (MIAPaCa-2) cells. Current traces (left panel) are sensitive to the specific hERG1 blocker E4031 (right panel).
For the purposes of the present study, 44 PDAC primary samples assembled as TMA were analysed, and a scoring system for hERG1 quantification was determined. Beta cells in Langerhans islets in the corresponding healthy pancreas sample were used as a positive internal control. hERG1 expression varied among the whole spectrum of PDAC primary samples, from null or very low expression to a very high expression. We applied a scoring system (detailed in Supplementary Information), evaluating the percentage of positive cells and the staining intensity in each tumour sample in comparison with its normal counterpart (i.e. beta cells in Langerhans islets). Figure 1C–F shows representative samples displaying different degrees of hERG1 staining intensity. When setting a threshold value at 6 (as detailed in Supplementary Information), and considering (i) samples with a score value between 0 and 5 as negative, (ii) samples between 6 and 12 as positive, 59.1% of the samples turned out to be positive for hERG1 expression. An example of a ‘hERG1 positive' PDAC, as well as of the corresponding healthy pancreas, is shown in Figure 1G and H, and relative higher magnification pictures (panels g and h, respectively).
hERG1 expression was then evaluated in three PDAC cell lines: PANC-1, MIAPaCa-2 and BxPC-3. We analysed hERG1 mRNA (by RQ-PCR) and hERG1 protein (by IP and IF) levels, as well as the functionality of the channel through the patch clamp technique. All the PDAC cell lines expressed the hERG1 mRNA (Figure 1I) and the hERG1 protein (Figure 1J) although at different levels: PANC-1 cells have the highest, MIAPaCa-2 an intermediate, whereas BxPC-3 cells the lowest level of expression. The proper expression of the hERG1 protein at the plasma membrane level of PDAC cells was confirmed by IF (Figure 1K), Both PANC-1 and MIAPaCa-2 cells were positive for hERG1 immunostaining, whereas BxPC-3 cells showed only a faint hERG1 IF signal. Setting a threshold equal to the IF displayed by NIH-3T3 cells (taken as negative control), 85% of PANC-1, 76% of MIAPaCa-2 and 5% of BxPC-3 cells turned out to be positive. A high hERG1 expression, with a dotty pattern (see arrows in Figure 1K), was evident on the plasma membrane of PANC-1 cells and, although to a lower extent, of MIAPaCa-2 cells. Setting a threshold equal to that shown by the four cells indicated by the arrow in Figure 1K, 15% of PANC-1, 9% of MIAPaCa-2 and almost none of the BxPC-3 cells expressed the channel at high levels onto the plasma membrane.
hERG1 channels expressed in PDAC cells were functional, as witnessed by patch clamp experiments. Figure 1L shows a representative example of membrane currents recorded in MIAPaCa-2 cells: traces show the typical biophysical and pharmacological features (e.g. the blockade by the hERG1-specific inhibitor E4031) of hERG1 currents in tumour cells (Faravelli et al, 1996; Schönherr et al, 1999). Collectively, proper hERG1 currents were detected, in steady-state culture conditions, in 13% of PANC-1 cells and 11% of MIAPaCa-2 cells, whereas none of the BxPC-3 cells expressed significant hERG1 current.
On the whole, hERG1 is expressed and is functional in the PDAC cells, although with different expression levels in the different cell lines examined. PANC-1 and, to a lower extent, MIAPaCa-2 cells, express the channel at significant levels.
hERG1 regulates malignancy in PDAC cells
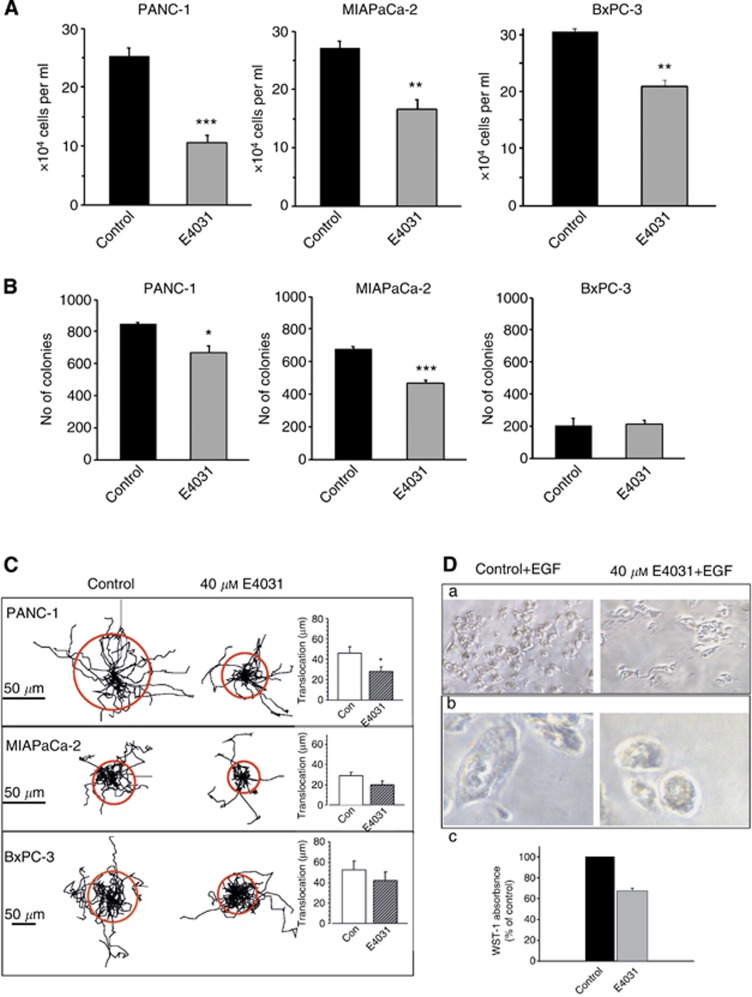
Then, we investigated whether hERG1 exerted any cancer-related function in the PDAC cells where it is expressed. To this purpose, hERG1 channel activity was inhibited with the hERG1-specific blocker E4031 at 40 μM final concentration as reported by Crociani et al (2013). We found that blocking hERG1 activity inhibited PDAC cell proliferation, measured as the number of viable cells after 72 h of incubation (Figure 2A). The strongest inhibitory effect was evident in PANC-1 cells (P<0.01, Student's t-test).
Figure 2.
Functional role of hERG1 channels in PDAC cells. (A) Effect of E4031 on PDAC cell proliferation. PANC-1, MIAPaCa-2 and BxPC-3 cells were seeded at 10 000 cells per well and treated at time zero with E4031 (40 μM, final concentration). The number of viable cells were estimated through the trypan blue exclusion method at different times of incubation. (B) Effect of E4031 on PDAC cell growth in soft agar. When PDAC cell lines were cultured in soft agar, clonal growth was inhibited significantly by hERG1-specific inhibitor E4031. Colonies were morphologically similar and reached the same size. For PANC-1 and MIAPaCa-2 cell lines 3000 cells per plate were seeded, whereas for BxPC-3 cell line 5000 cells per plate were seeded. (C) Effect of E4031 on migration of PDAC cells. The cell trajectories are normalised using a common starting point. Each track represents the movement of one cell during the 5 h period. The radii of the circles represent the mean distances covered within ∼5 h. The method is fully described in Supplementary Information (Materials and Methods of in vitro experiments). *P<0.05; **P<0.02; ***P<0.01 (Student's t-test). (D) E4031 effects on primary PDAC cells. a, b: representative microphotographs of a primary PDAC cell culture. Left panels: control cells; right panels: E4031-treated cells. Magnification: 200x (a) and 400x (b). In panel c, a histogram showing cell viability (evaluated by WST-1 assay, as described in Supplementary Information) in control (black bar) and E4031-treated cells (grey bar) is reported.
E4031 also decreased the ability of PANC-1 and in MIAPaca-2 cells to form colonies in soft agar (P<0.05 and P<0.02, respectively (Student's t-test)), whereas was not effective in BxPC-3 cells, which also displayed the lowest capacity to give rise to colonies in soft agar (Figure 2B).
Finally, we tested whether hERG1 activity affected PDAC cell random migration on a reconstituted ‘basement' membrane. Figure 2C shows that PANC-1 cells, in control condition, were motile, whereas E4031-treated cells covered a shorter distance and migrated more slowly. These effects, well documented and statistically significant in PANC-1 cells (P<0.05; Student's t-test), were less evident in MIAPaCa-2 and almost absent in BXPC-3 cells. The latter displayed a lower basal-migratory capacity.
hERG1 function was also assessed in short-term in vitro cultures obtained from PDAC clinical samples. Figure 2D shows the results of a representative experiment: in control culture conditions numerous cells are present (panel a, photograph on the left), and appear adherent to the substrate and vital (panel a, photograph on the left). On the contrary, the same culture treated with E4031 for 6 h shows less adherent cells (panel a, photograph on the right), which start to round up and to detach from the substrate, showing evident signs of cytotoxicity (panel b, photograph on the right). The E4031-induced decrease in cell vitality is further supported by the WST-1 data (panel c).
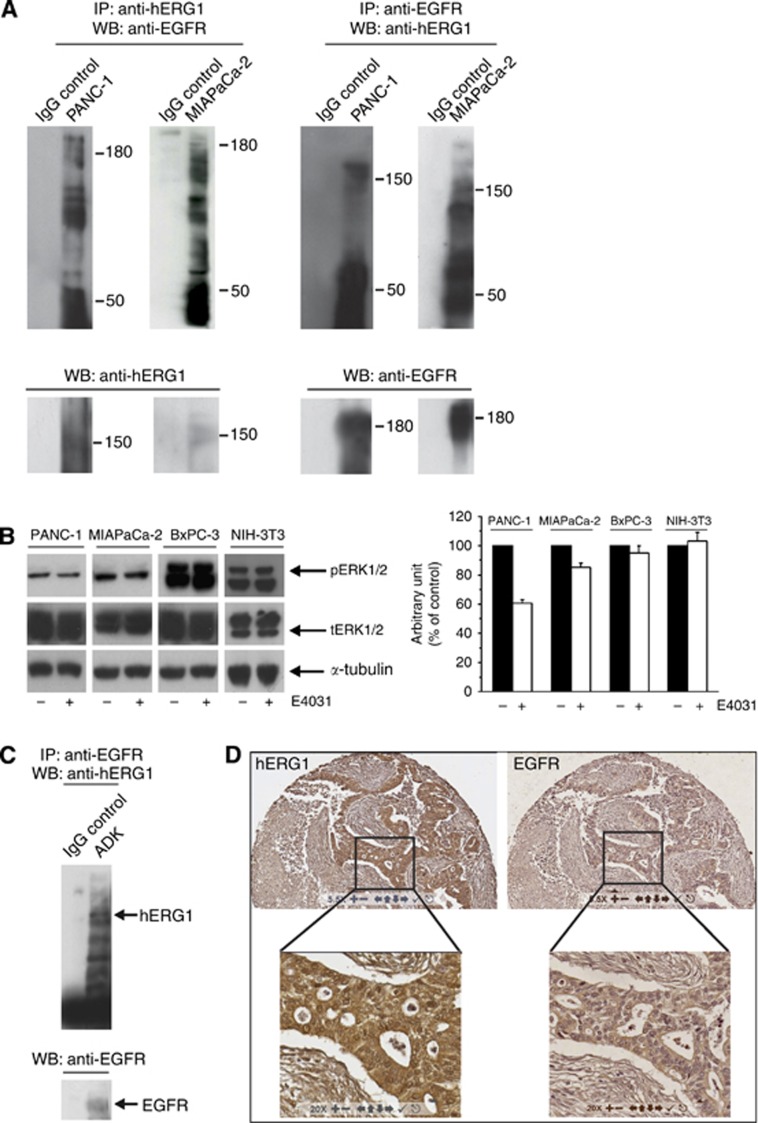
In tumour cells hERG1 function is related to its ability to trigger intracellular signalling pathways through the interaction with plasma membrane proteins (Pillozzi and Arcangeli, 2010). In search of hERG1 plasma membrane partner proteins in PDAC cells, we found that hERG1 co-immunoprecipitated with the EGF-R in PANC-1 and, although at a lower extent, in MIAPaCa-2 cells (Figure 3A). In PANC-1 cells, E4031 decreased the phosphorylation of ERK1 and ERK2, key signalling proteins downstream to EGF-R (Neuzillet et al, 2014). This inhibitory effect of E4031 was evident and statistically significant after 180 min of incubation in standard culture conditions in PANC-1 cells (P<0.01; Student's t-test), whereas less evident in the MIAPaCa-2 cells (Figure 3B). Consistently with the very low or null expression of hERG1, E4031 had no effect on the phosphorylation of ERK1 and ERK2 in BxPC-3 cells and NIH-3T3 (Figure 3B).
Figure 3.
Relationships between hERG1 and EGF-R in PDAC cell lines and primary samples. (A) Co-immunoprecipitation (co-IP) of hERG1 and EGF-R in PANC-1 and MIAPaCa-2 cells (expressing EGF-R at high levels (Supplementary Information, Supplementary Figure S1). Co-IP experiments were performed using either a α-hERG1-MoAb (panels on the right) or the anti-EGF-R (sc-03, Santa Cruz, Dallas, TX, USA; panels on the left) to immunoprecipitate and the same EGF-R antibody or the polyclonal antibody directed against the hERG1 C-terminus (C54 antibody) for membrane decoration. (B) Effect of E4031 on ERK1/2 phosphorylation after 3 h treatment. NIH-3T3 cells were used as negative control. For membrane decoration, phospho-p44/42 MAP kinase (Cell Signaling, Danvers, MA, USA) and ERK1 antibodies (C-16, Santa Cruz), that are able to recognize both ERK1 and ERK2 total protein were used. Monoclonal anti-α-Tubulin antibody (Sigma-Aldrich) was used as a probe for α-Tubulin as an internal loading control. ***P<0.01 (Student's t-test). (C) Co-IP of hERG1 and EGF-R in PDAC primary samples. Co-IP experiments were performed on a total protein extract (1 mg) pooled from three different primary PDAC samples, using the same protocol as in panel A. (D) IHC for hERG1 and EGF-R. A representative picture of IHC staining with the two antibodies in the same tumour sample is shown. Magnification: 55x (upper panels) and 200x (lower panels).
The co-IP of hERG1 and EGF-R also occurred in PDAC primary samples (Figure 3C). IHC experiments further confirmed the co-expression of the two proteins in PDAC. Figure 3D shows two consecutive sections of a representative sample: it is evident that hERG1 positive cells match with EGF-R positive cells.
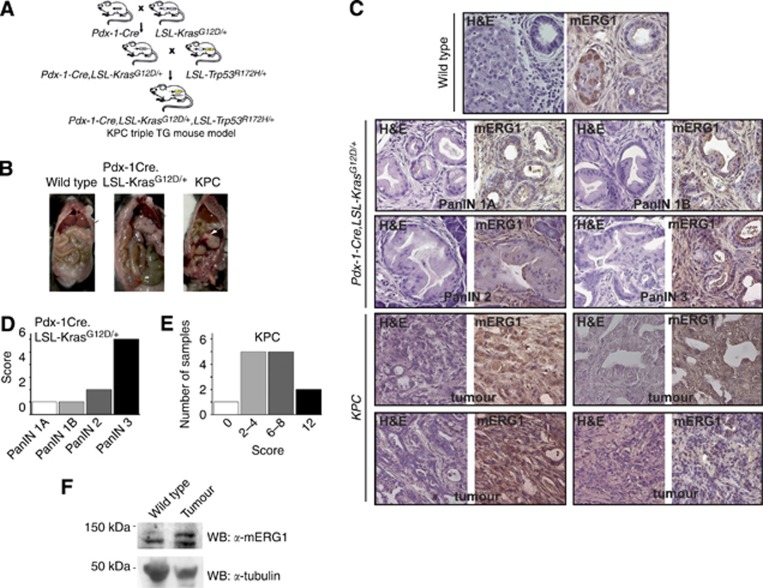
mERG1 expression during PDAC progression: in vivo studies on KPC mice
We studied the pattern of expression of the murine homologue of hERG1, mERG1, during PDAC progression in vivo by IHC. A widely used mouse model of PDAC, that is, KPC transgenic mice (Hingorani et al, 2003, 2005) was used. The model is detailed in Supplementary Information (Materials and Methods of in vivo experiments and Supplementary Figure S2) (Figure 4A). Once mice showed clear signs of cachexia and abdominal distension (around 5 months of age), they were killed and the autoptic examination revealed the presence of tumours (Figure 4B), whose histopathological characteristics are shown in Figure 4C and in Supplementary Information (Supplementary Figure S3). Some of the tumours showed a high degree of tissue organization (panels a–c) whereas others were made of islet of tumour cells (panel d) or small groups of neoplastic cells with extended areas of necrosis (panel e). Double-transgenic Pdx-1-Cre,LSL- KrasG12D/+ mice were used to detect and analyse PDAC pre-cancerous lesions, in particular PanIN. Figure 4C shows Haematoxylin and Eosin staining of the four different grades of PanIN. Notably, PanIN lesions which occurred in transgenic mice showed the same histological features as those observed in human patients.
Figure 4.
mERG1 expression in KPC transgenic mouse model. (A) Breeding strategy for the generation of Pdx-1-Cre,LSL-KrasG12D/+,LSL-Trp53R175H/+ (KPC) triple-transgenic mice. (B) Representative photographs of macroscopic examination of organs and tumours in control (wild type, WT), double-transgenic (Pdx-1-Cre,LSL-KrasG12D/+) and KPC triple-transgenic mice. Once the abdominal cavity is opened, primary PDAC (white arrow) in KPC mice was easily detected, as compared with the normal appearing pancreas of the wild-type and double-transgenic mice (black arrows). (C) Histological appearance of tumour masses in control (WT), double-transgenic (Pdx-1-Cre,LSL-KrasG12D/+) and KPC triple-transgenic mice. Haematoxylin and Eosin (H&E; left panel of each couple of microphotographs), magnification: 400x. IHC of mERG1 in control, double-transgenic and KPC mice (right panel of each couple of images). Representative pictures of samples showing a different score are reported (PanIN 1A and PanIN 1B: score 1; PanIN 2: score 2; PanIN 3: score 6; KPC tumours: upper panels score 12 and 8, lower panels score 12 and 2). Magnification: 400x. (D) Histogram summarising mERG1 score distribution in PanIN lesions developed by Pdx-1-Cre,LSL-KrasG12D/+ mice. (E) Histogram showing mERG1 score in KPC tumour samples. (F) WBs on total lysates from normal pancreas of WT mice and tumours of KPC mice. Protein extracts were probed using the polyclonal antibody α-C54. mERG1 specific bands showed a lower molecular weight than the expected 155 kDa and 135 kDa bands: this difference is probably owing to an alternative glycosylation pattern present in primary samples. The membrane was further decorated with α–tubulin antibody.
mERG1 was expressed in beta cells of the Langerhans islets of healthy pancreas, whereas no mERG1 immunoreaction was detectable either in ductal or acinar cells (Figure 4C, left panel ‘Wild Type'). mERG1 was expressed at quite low levels and with a focal pattern in PanIN (Figure 4C upper panels) whereas a more diffuse and intense staining was observed in tumours of KPC mice (Figure 4C, lower panels). A certain degree of variability could be observed in terms of intensity and diffusion (compare the panels at the bottom of Figure 4C).
We applied the same scoring system described above to analyse the Pdx-1-Cre,LSL- KrasG12D/+ and KPC samples. Figure 4D shows that in PanIN lesions which developed in Pdx-1-Cre,LSL- KrasG12D/+ mice, mERG1 score increases (from 1 to 6) with the severity of the lesion, being higher in PanIN3 with respect to PanIN2 and PanIN1 lesions. In the tumours which developed in KPC mice, the majority of the samples showed an intermediate scoring (Figure 4E). The different score values of the single samples shown in Figure 4C are reported in the figure legend. The expression of the mERG1 protein in the tumours of KPC mice was also confirmed by Western Blot analysis (Figure 4F).
Diagnostic and prognostic significance of hERG1 in PDAC: IHC data
Given the functional role of hERG1 in PDAC, its potential prognostic value was then established using IHC data from the PDAC tissue samples assembled in TMA, and taking into account the scoring groups (see MATERIALS AND METHODS).
As stated above, 26/44 (59.1%) of the samples were positive for hERG1 expression. hERG1 positive PDAC tumours showed a positivity to Ki67 (i.e., a high proliferative index). When applying a 20% cut-off for Ki67 expression, a statistically significant association with hERG1 emerged (P=0.035, Fisher's exact test) (Table 1). A representative sample, showing two consecutive sections, clearly shows that hERG1 positive cells match with Ki67 positive cells (Supplementary Figure S4). Moreover, hERG1 expression significantly correlated with the tumour grading (G; P=0.050, Fisher's exact test) and with EGF-R expression (P=0.027, Fisher's exact test; Table 1).
Table 1. Associations between hERG1 expression and clinicopathological parameters.
| Variable | hERG1 negative (%) | hERG1 positive (%) | Fisher's exact test (P value) |
|---|---|---|---|
|
Category | |||
| Gender | |||
| Female | 7 (33.33) | 14 (66.67) | 0.373 |
| Male | 11 (47.83) | 12 (52.17) | |
| hERG1 test | |||
| Negative | |||
| Positive | |||
| EGF-R test | |||
| Negative | 9 (52.94) | 9 (33.33) | 0.027* |
| Positive | 8 (47.06) | 18 (66.67) | |
| hERG1 and EGF-R test | |||
| Both negative | 17 (41.46) | 24 (58.54) | |
| Both positive | 1 (33.33) | 2 (66.67) | |
| One positive | 9 (52.94) | 8 (47.06) | |
| T stage | |||
| T2 | 1 (14.29) | 6 (85.71) | 0.211 |
| T3 | 17 (45.95) | 20 (54.05) | |
| N stage | |||
| N0 | 9 (52.94) | 9 (33.33) | 0.225 |
| N1 | 8 (47.06) | 18 (66.67) | |
| M stage | |||
| M0 | 17 (41.46) | 24 (58.54) | 1.000 |
| M1 | 1 (33.33) | 2 (66.67) | |
| Staging | |||
| I | 9 (52.94) | 8 (47.06) | 0.561 |
| II | 8 (33.33) | 16 (66.67) | |
| IV | 1 (33.33) | 2 (66.67) | |
| Grading | |||
| Grade 1 | 3 (100.00) | 0 (0.00) | 0.050* |
| Grade 2 | 11 (44.00) | 14 (56.00) | |
| Grade 3 | 4 (25.00) | 12 (75.00) | |
| Ki67 | |||
| <20% | 14 (87.50) | 10 (52.63) | 0.035* |
| ⩾20% | 2 (12.50) | 9 (47.37) | |
Abbreviations: EGF-R=Epidermal Growth Factor Receptor; hERG1=human ether a go-go related gene 1. Significant correlations (P<0.05) are indicated in bold and marked with *.
Furthermore, survival analysis was conducted in 39 patients, whose complete follow-up data were available. The median follow-up was 9.3 years (IQR: 8.4–11.2). At the univariate analysis the following variables turned out to have a significant impact on overall survival (OS): TNM stage (II vs I: HR: 2–11 (0.98–4.56); P=0.049) and hERG1 expression (positive vs negative: HR=2.23 (1.07–4.66); P=0.029; Table 2). These results were confirmed at the multivariate analysis (Table 2).
Table 2. Univariate and multivariate overall survival analyses and cumulative 1-year overall survival in stage I and II patients.
|
Univariate analysis |
Final multivariate model |
||||||
|---|---|---|---|---|---|---|---|
| Variable | n | n failed | Cumulative proportion 1-year survival | Hazard ratio (95% CI) | LR test (P value) | Hazard ratio (95% CI) | LR test (P value) |
|
Category | |||||||
| Gender | |||||||
| Female | 19 | 15 | 0.684 | 1 (ref.) | 0.869 | ||
| Male | 20 | 16 | 0.700 | 0.94 (0.46–1.92) | |||
| hERG1 test | |||||||
| Negative | 17 | 12 | 0.882 | 1 (ref.) | 0.029* | 1 (ref.) | 0.049* |
| Positive | 22 | 19 | 0.545 | 2.23 (1.07–4.66) | 2.12 (1.01–4.48) | ||
| EGF-R test | |||||||
| Negative | 14 | 8 | 0.677 | 1 (ref.) | 0.195 | ||
| Positive | 25 | 16 | 0.45 | 1.75 (0.74–4.14) | |||
| hERG1 and EGF-R test | |||||||
| Both negative | 9 | 6 | 0.889 | 1 (ref.) | 0.067 | ||
| Both positive | 17 | 15 | 0.529 | 2.85 (1.09–7.49) | |||
| One positive | 13 | 10 | 0.769 | 1.47 (0.53–4.07) | |||
| TNM stage | |||||||
| I | 16 | 10 | 0.75 | 1 (ref.) | 0.049* | 1 (ref.) | 0.085 |
| II | 23 | 21 | 0.652 | 2.11 (0.98–4.56) | 2.00 (0.91–4.41) | ||
| Grading | |||||||
| Grade 1 | 3 | 2 | 0.667 | 1 (ref.) | 0.461 | ||
| Grade 2 | 22 | 17 | 0.682 | 1.59 (0.36–6.92) | |||
| Grade 3 | 14 | 12 | 0.714 | 2.24 (0.49–10.18) | |||
| Ki67 | |||||||
| <20% | 22 | 16 | 0.727 | 1 (ref.) | 0.346 | ||
| ⩾20% | 9 | 8 | 0.667 | 1.53 (0.65–3.63) | |||
Abbreviations: CI=confidence interval; EGF-R=Epidermal Growth Factor Receptor; hERG1=human ether a go-go related gene 1; LR=log rank test. Significant associations (P<0.05) are indicated in bold and marked with *.
We also tested whether hERG1 expression correlated with Kras mutation status. The cases, reported in Table 1, for which Kras mutational status was available (24 out of 44) were all Kras mutated (see raw data in Supplementary Table S2). Therefore, we analysed a small subset of patients, not included in those, whose samples were assembled in TMA, either Kras wild type (n=5) or Kras mutated (n=4). No association emerged between Kras mutation status and hERG1 expression (P=0.444, Fisher's exact test; Supplementary Table S3).
Diagnostic and prognostic significance of hERG1 in PDAC: in vivo optical imaging
We then determined whether hERG1 expression could be also exploited as a molecular marker for in vivo diagnosis. For this purpose we used α-hERG1-MoAb labelled it with Alexa-680, and tested its ability to bind hERG1 expressed in PDAC, using either (i) immunodeficient nu/nu mice, injected into the pancreas with luciferase-transfected MIAPaCa-2 cells (MIAPaCa-2-luc-nu/nu mice) or (ii) the KPC transgenic mice described above. The details of the two models are described in Supplementary Information (Materials and Methods of in vivo experiments and Supplementary Figure S5–S7).
MIAPaCa-2-luc-nu/nu mice were treated, 21 days after tumour-cell injection (e.g., when the counts per minute reached significant levels, but mice did not show gross signs of illness), with 100 μg of Alexa-680-labelled α-hERG1-MoAb injected intravenously (i.v.) and NIR images collected 3 days afterwards. As shown in Figure 5A, a fluorescent signal was detected from the abdominal area suggesting the distribution of the Alexa-680-labelled α-hERG1-MoAb at tumour site level. The fluorescence signal of the tumour masses was more clearly evident in excised organs. The fluorescence intensities measured as cpm cm−2 in the tumour and in normal tissues are depicted in Figure 5B.
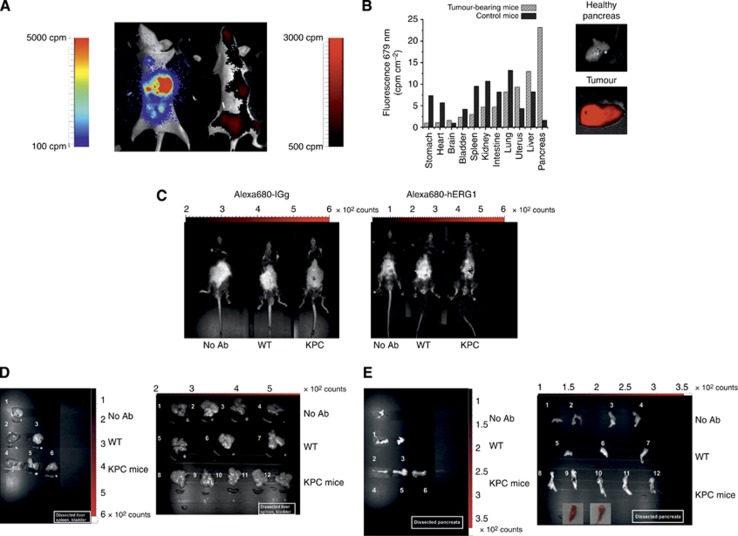
Figure 5.
(A) In vivo detection of Alexa-680-labelled α-hERG1-MoAb derived fluorescence. Whole-body fluorescence compared with bioluminescence. (B) The uptake and retention of Alexa-680-labelled-hERG1-MoAb in a surgically dissected tumour (lower panel) compared with the healthy pancreas (upper panel). Tumour tissue displayed strong signals both in vivo and ex vivo imaging. (C) Representative pictures of whole-body in vivo imaging of Alexa-680-labelled α-IgG nonspecific control antibody (left panel) and the Alexa-680-labelled-hERG1-MoAb fluorescence. Imaging was performed in untreated WT and KPC mice and in Ab-injected WT and KPC mice aged 5 months. Autofluorescence was subtracted based on the signal relative to WT non-injected mouse (on the left). (D–E) In vivo imaging of the Alexa-680-labelled-hERG1-MoAb fluorescence within dissected organs (D) and pancreas (E). Left panels: 1: organs and pancreas of untreated WT mice; 2–3: organs and pancreas of control Ab-injected WT mice; 4–6: organs and pancreas of control Ab-injected KPC mice. Right panels: 1–2: organs and pancreas of untreated WT mice; 3–4: organs and pancreas of untreated KPC mice; 5–7: organs and pancreas of MoAb-injected WT mice; 8–12: organs and pancreas of MoAb-injected KPC mice; PDAC tumours are shown in the insets (9–10).
Molecular optical imaging was also performed in KPC mice, 5-month old, after i.v. injection of Alexa-680-labelled α-hERG1-MoAb or Alexa-680-labelled α-IGg nonspecific control antibody. As shown in Figure 5C (right panel), a significant fluorescent signal was detected only from the abdominal area of Alexa-680-labelled α-hERG1-MoAb treated KPC mice, whereas only a faint signal was present in wild-type or KPC mice treated with the control antibody. This suggested the accumulation of the Alexa-680-labelled α-hERG1-MoAb in the tumour site. This was confirmed by the ex vivo imaging on different excised organs (Figure 5D and E, right panel). Only in pancreata carrying a tumour mass a high fluorescent signal was detected (Figure 5E, right panel, samples 9 and 10), after the subtraction of the fluorescence level of the values obtained using the control antibody (left panels), to remove the nonspecific fluorescence intensity. No appreciable fluorescent signal was revealed in the other organs, such as liver, spleen or bladder (Figure 5D).
Discussion
Despite many efforts, pancreatic cancer, and its most frequent form, the PDAC, still carries a poor prognosis. In search of molecular markers to improve the early diagnosis and accurate prognosis, we studied hERG1 channels in PDAC. We provide evidence that hERG1 channels are functionally expressed in PDAC and may be exploited, once validated, for diagnostic and prognostic purposes in PDAC.
The expression of hERG1 in PDAC cells was analysed at different levels with different techniques, reaching good consistency in the whole set of data. Roughly half of the PDAC cell lines expressed functional hERG1 channels at very high levels. These results are consistent with those recently reported by Feng et al (2014).
Interestingly, a high hERG1 expression was detected in Kras mutated cell lines (PANC-1 and MIAPaCa-2; Deer et al, 2010). However, no correlation between hERG1 expression and Kras mutational status emerged in clinical samples. As activating the mutations of Kras can concomitantly upregulate EGF-R and its downstream signalling pathways (Bera et al, 2013), we studied such pathway, providing evidence that hERG1 interacted with EGF-R. Overall, hERG1 turned out to contribute to the control of EGF-R-triggered intracellular signalling pathways, and, by this way, to the regulation of PDAC cell proliferation and migratory activity. In cancer cells, hERG1 is known to interact with plasma membrane-adhesion receptors, in particular integrins (Arcangeli and Becchetti, 2006), as well as with growth factor receptors, such as the VEGF-R1 in leukaemia (Pillozzi et al, 2007). We demonstrate for the first time that a physical and functional interaction of hERG1 with EGF-R occurs in PDAC. This finding could have relevance in PDAC where anti-EGF-R therapy, alone or in combination with gemcitabine, did not give relevant benefits in terms of OS in PDAC in the Canadian NCI trial (Moore et al, 2007). Hence, the understanding of EGF-R-associated molecules could help in identifying alternative approaches. In this light, the targeting of NHE1, which also interacts with EGF-R, with specific inhibitors has been proven to be an exciting novel pharmacological approach (Miraglia et al, 2005).
We also provided evidence that hERG1 is switched on during pancreatic cancer progression as occurs in transgenic mice. In fact, mERG1 expression turned out to be limited to the beta cells of the Langerhans islets in wild-type mice, as in humans (Rosati et al, 2000), and no expression was detected in normal ductal cells. However, mERG1 started to be aberrantly is expressed in ductal cells of Pancreatic Intraepithelial Neoplasia (PanIN) lesions, fo example, the precursors lesions of PDAC, which arise in the pancreas of Pdx-1-Cre,LSL- KrasG12D/+mice. As PDAC carcinogenesis progresses, as it occurs in KPC mice, mERG1 expression increases to reach high levels in adenocarcinomas of KPC mice. This not-obvious result is similar to what occurs in APC mice (Fiore et al, 2013) which over-express mERG1 in the tumours they develop in the small and large intestine, and further stresses the relevance of hERG1 in tumour progression.
hERG1 could be proposed as a therapeutic target in oncology (Arcangeli et al, 2009). Indeed in the present paper we provide evidence that hERG1 blockers could inhibit PDAC cell growth and migration in vitro. Consistently, hERG1 silencing, or miRNA-96 over-expression, has been shown to affect PDAC tumorigenicity either in vitro or in vivo (Feng et al, 2014). However, considering hERG1 as a therapeutic target often encounters hindrances, because of the cardiotoxicity that many hERG1 blockers exert. Besides designing novel therapeutic strategies or new non cardiotoxic hERG1 blockers (Gasparoli et al, 2015), we favoured to test the potential usefulness of hERG1 as a prognostic marker in PDAC. To this purpose, we analysed hERG1 expression in a cohort of 44 PDAC samples from surgically resected patients, reaching the demonstration that: (i) roughly half of the primary PDAC samples over-express hERG1; (ii) hERG1 expression correlates with EGF-R expression, and with (iii) the proliferative index (measured through the Ki67 staining); (iv) in TNM-stage I and II patients, hERG1 positive PDAC had a worse prognosis compared with the hERG1 negative. It is worth noting that in our series of surgically resected patients there was no selection criteria. They were all consecutive patients submitted to surgery and the majority of them were finally classified as TNM-stage I or II patients. This is not surprising, as surgical resection is indicated only in this category of patients, whereas stage III patients are generally submitted, when possible, to a preoperative neoadiuvant treatment based on chemotherapy or radiochemotherapy. In our study we planned to compare the hERG1 expression in PDAC cells with the normal cells in the same specimen (e.g., the same patients). This is feasible and easy only in resected surgical specimens; on the other hand, in the majority of not operated patients we can have only cancer cells coming from needle biopsy or fine needle aspiration. It can be therefore difficult to gather normal pancreatic cells. For all these reasons, the few (3 out of 39) patients belonging to more advanced TNM stages were excluded from our survival analysis to eliminate a potential confounding factor on evaluating the survival. Overall, these results suggest the possibility that hERG1 expression might have a prognostic impact in PDAC. It will be necessary to validate these results in a larger cohort of patients, comprising also TNM-stages III and IV, in which hERG1 expression will be assessed in short-term cultures, applying the method we have developed in the present study.
All these results were obtained determining hERG1 expression by IHC, the gold standard diagnostic tool used to assess the presence or absence of molecular markers on tumour tissues (Taylor, 1994). Moreover, we determined appropriate methodologies and cut-off values, to facilitate the employment of a categorical scoring system by pathologists for an easier interpretation of the results, as well as to validate the results in order to further propose hERG1 as a potential prognostic marker. In fact, the real prognostic impact of hERG1, and in general of ion channels, on cancer patients' survival is still under study and merits great attention. For example, in acute myeloid leukaemia, hERG1 identifies a subgroup of patients with worse prognosis (Pillozzi et al, 2007), whereas in colorectal cancer, hERG1 positivity identifies a high risk group in stage I and II patients, only when associated to Glut-1 negativity (Lastraioli et al, 2012). We recently showed in a huge cohort of gastric cancer patients that hERG1 expression displays a negative prognostic impact in T1-stage patients (Crociani et al, 2014). Hence, the impact of hERG1 in cancer progression may depend on the characteristics of the tumour sample.
Finally, the translatability of our data to the clinical setting is further sustained by the possibility of detecting hERG1 in vivo, using the α-hERG1-MoAb as a tracer for in vivo imaging. In fact, the antibody, once labelled with Alexa-680, was able to detect hERG1-expressing tumours either when PDAC cells were injected into the pancreas of immunodeficient mice, or in transgenic KPC mice that express mutated Kras and Trp53 in the pancreatic ductal cells (e.g., under the control of the Pdx-1 promoter). The possibility to exploit the α-hERG1-MoAb to target hERG1 in vivo could have an immediate clinical fallout for the diagnosis and clinical follow-up of PDAC. In view of the detected mERG1 expression in PanIN lesions in transgenic mice, the possibility exists of exploiting hERG1 for an early diagnosis of PDAC pre-cancerous lesions (such as Intraductal papillary mucinous neoplasm and PanIN). Finally the antibody, whose capacity to drive nanoparticles into hERG1-expressing cells in vitro has already been proven (Sette et al, 2013), could be used to drive nanoparticles to the tumour site in vivo, hence opening new therapeutic perspectives (Sette et al, 2013). A similar approach has been successfully utilised by Soroceanu et al (1999) who exploited the expression of Cl− channels in glioblastoma to utilise chlorotoxin either as a diagnostic agent or as a vehicle for drug delivery (Sontheimer, 2008).
Overall data here provided could qualify hERG1 as a candidate for diagnosis, prognosis and patients' stratification in PDAC.
Acknowledgments
We thank Dr I Marzi for confocal microscopy, Dr F Zanieri for performing some preliminary experiments, Dr D Righi for experiments on IHC and ISH Leica BOND-MAX system and Dr D Tomcikova for helpful suggestions on the statistical analysis. This work was supported by Associazione Italiana per la Ricerca sul Cancro (AIRC, Grant No 1662) and Regione Toscana-POR CREO FESR 2007-2013 (Attività 1.1-Linea di intervento 1.1.c), to AA, Ion TraC-Marie Curie. SM is an IonTraC-Marie Curie fellow.
The authors declare no conflict of interests.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Supplementary Material
References
- Ansari D, Rosendahl A, Elebro J, Andersson R. Systematic review of immunohistochemical biomarkers to identify prognostic subgroups of patients with pancreatic cancer. Br J Surg. 2011;98:1041–1055. doi: 10.1002/bjs.7574. [DOI] [PubMed] [Google Scholar]
- Apte MV, Park S, Phillips PA, Santucci N, Goldstein D, Kumar RK, Ramm GA, Buchler M, Friess H, McCarroll JA, Keogh G, Merrett N, Pirola R, Wilson JS. Desmoplastic reaction in pancreatic cancer: role of pancreatic stellate cells. Pancreas. 2004;29:179–187. doi: 10.1097/00006676-200410000-00002. [DOI] [PubMed] [Google Scholar]
- Arcangeli A.2005Expression and role of hERG channels in cancer cells Novartis Found Symp 266225–232.discussion 232-4. [PubMed] [Google Scholar]
- Arcangeli A, Becchetti A. Complex functional interaction between integrin receptors and ion channels. Trends Cell Biol. 2006;16:631–639. doi: 10.1016/j.tcb.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Arcangeli A, Crociani O, Lastraioli E, Masi A, Pillozzi S, Becchetti A. Targeting ion channels in cancer: a novel frontier in antineoplastic therapy. Curr Med Chem. 2009;16:66–93. doi: 10.2174/092986709787002835. [DOI] [PubMed] [Google Scholar]
- Becchetti A. Ion channels and transporters in cancer. 1. Ion channels and cell proliferation in cancer. Am J Physiol Cell Physiol. 2011;301:C255–C265. doi: 10.1152/ajpcell.00047.2011. [DOI] [PubMed] [Google Scholar]
- Bera A, Zhao S, Cao L, Chiao PJ, Freeman JW. Oncogenic K-Ras and loss of Smad4 mediate invasion by activating an EGFR/NF-κB axis that induces expression of MMP9 and uPA in human pancreas progenitor cells. PLoS One. 2013;8:e82282. doi: 10.1371/journal.pone.0082282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeck S, Jung A, Laubender RP, Neumann J, Egg R, Goritschan C, Vehling-Kaiser U, Winkelmann C, Fischer von Weikersthal L, Clemens MR, Gauler TC, Märten A, Klein S, Kojouharoff G, Barner M, Geissler M, Greten TF, Mansmann U, Kirchner T, Heinemann V. EGFR pathway biomarkers in erlotinib-treated patients with advanced pancreatic cancer: translational results from the randomised, crossover phase 3 trial AIO-PK0104. Br J Cancer. 2013;108:469–476. doi: 10.1038/bjc.2012.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crociani O, Zanieri F, Pillozzi S, Lastraioli E, Stefanini M, Fiore A, Fortunato A, D'Amico M, Masselli M, De Lorenzo E, Gasparoli L, Chiu M, Bussolati O, Becchetti A, Arcangeli A. hERG1 channels modulate integrin signaling to trigger angiogenesis and tumor progression in colorectal cancer. Sci Rep. 2013;3:3308. doi: 10.1038/srep03308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crociani O, Lastraioli E, Boni L, Pillozzi S, Romoli MR, D'Amico M, Stefanini M, Crescioli S, Taddei A, Bencini L, Bernini M, Farsi M, Beghelli S, Scarpa A, Messerini L, Tomezzoli A, Vindigni C, Morgagni P, Saragoni L, Giommoni E, Gasperoni S, Di Costanzo F, Roviello F, De Manzoni G, Bechi P, Arcangeli A. hERG1 channels regulate VEGF-A secretion in human gastric cancer: clinicopathological correlations and therapeutical implications. Clin Cancer Res. 2014;20:1502–1512. doi: 10.1158/1078-0432.CCR-13-2633. [DOI] [PubMed] [Google Scholar]
- Deer EL, González-Hernández J, Coursen JD, Shea JE, Ngatia J, Scaife CL, Firpo MA, Mulvihill SJ. Phenotype and genotype of pancreatic cancer cell lines. Pancreas. 2010;39:425–435. doi: 10.1097/MPA.0b013e3181c15963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dozynkiewicz MA, Jamieson NB, Macpherson I, Grindlay J, van den Berghe PV, von Thun A, Morton JP, Gourley C, Timpson P, Nixon C, McKay CJ, Carter R, Strachan D, Anderson K, Sansom OJ, Caswell PT, Norman JC. Rab25 and CLIC3 collaborate to promote integrin recycling from late endosomes/lysosomes and drive cancer progression. Dev Cell. 2012;22:131–145. doi: 10.1016/j.devcel.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faravelli L, Arcangeli A, Olivotto M, Wanke E. A HERG-like K+ channel in rat F-11 DRG cell line: pharmacological identification and biophysical characterization. J Physiol. 1996;496 (Pt 1:13–23. doi: 10.1113/jphysiol.1996.sp021661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Yu J, Pan X, Li Z, Chen Z, Zhang W, Wang B, Yang L, Zhang G, Xu Z. HERG1 functions as an oncogene in pancreatic cancer and is downregulated by miR-96. Oncotarget. 2014;5:5832–5844. doi: 10.18632/oncotarget.2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F.2013. IARC CancerBase No. 11. Available from http://globocan.iarc.fr accessed on 26 February 2014.
- Fiore A, Carraresi L, Morabito A, Polvani S, Fortunato A, Lastraioli E, Femia AP, De Lorenzo E, Caderni G, Arcangeli A. Characterization of hERG1 channel role in mouse colorectal carcinogenesis. Cancer Med. 2013;2:583–594. doi: 10.1002/cam4.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasparoli L, D'Amico M, Masselli M, Pillozzi S, Caves R, Khuwaileh R, Tiedke W, Mugridge K, Pratesi A, Mitcheson JS, Basso G, Becchetti A, Arcangeli A. The new pyrimido-indole compound CD-160130 preferentially inhibits the KV11.1B isoform and produces antileukemic effects without cardio-toxicity. Mol Pharmacol. 2015;87:183–196. doi: 10.1124/mol.114.094920. [DOI] [PubMed] [Google Scholar]
- Hartel M, di Mola FF, Selvaggi F, Mascetta G, Wente MN, Felix K, Giese NA, Hinz U, Di Sebastiano P, Büchler MW, Friess H. Vanilloids in pancreatic cancer: potential for chemotherapy and pain management. Gut. 2006;55:519–528. doi: 10.1136/gut.2005.073205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, Ross S, Conrads TP, Veenstra TD, Hitt BA, Kawaguchi Y, Johann D, Liotta LA, Crawford HC, Putt ME, Jacks T, Wright CV, Hruban RH, Lowy AM, Tuveson DA. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, Rustgi AK, Chang S, Tuveson DA. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7:469–483. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- Krasinskas AM. EGFR signaling in colorectal carcinoma. Patholog Res Int. 2011;2011:932932. doi: 10.4061/2011/932932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lastraioli E, Taddei A, Messerini L, Comin CE, Festini M, Giannelli M, Tomezzoli A, Paglierani M, Mugnai G, De Manzoni G, Bechi P, Arcangeli A. hERG1 channels in human esophagus: evidence for their aberrant expression in the malignant progression of Barrett's esophagus. J Cell Physiol. 2006;209:398–404. doi: 10.1002/jcp.20748. [DOI] [PubMed] [Google Scholar]
- Lastraioli E, Bencini L, Bianchini E, Romoli MR, Crociani O, Giommoni E, Messerini L, Gasperoni S, Moretti R, Di Costanzo F, Boni L, Arcangeli A. hERG1 Channels and Glut-1 as independent prognostic indicators of worse outcome in stage I and II colorectal cancer: a pilot study. Transl Oncol. 2012;5:105–112. doi: 10.1593/tlo.11250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miraglia E, Viarisio D, Riganti C, Costamagna C, Ghigo D, Bosia A. Na+/H+ exchanger activity is increased in doxorubicin-resistant human colon cancer cells and its modulation modifies the sensitivity of the cells to doxorubicin. Int J Cancer. 2005;115:924–929. doi: 10.1002/ijc.20959. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Goldstein D, Hamm J, Figer A, Hecht JR, Gallinger S, Au HJ, Murawa P, Walde D, Wolff RA, Campos D, Lim R, Ding K, Clark G, Voskoglou-Nomikos T, Ptasynski M, Parulekar W, National Cancer Institute of Canada Clinical Trials Group Erlotinib plus gemcitabine compared with gemcitabine alone in patients with advanced pancreatic cancer: a phase III trial of the National Cancer Institute of Canada Clinical Trials Group. J Clin Oncol. 2007;25:1960–1966. doi: 10.1200/JCO.2006.07.9525. [DOI] [PubMed] [Google Scholar]
- Mulcahy HE, Lyautey J, Lederrey C, qi Chen X, Anker P, Alstead EM, Ballinger A, Farthing MJ, Stroun M. A prospective study of K-ras mutations in the plasma of pancreatic cancer patients. Clin Cancer Res. 1998;4:271–275. [PubMed] [Google Scholar]
- Neuzillet C, de Gramont A, Tijeras-Raballand A, de Mestier L, Cros J, Faivre S, Raymond E. Perspectives of TGF-β inhibition in pancreatic and hepatocellular carcinomas. Oncotarget. 2014;5:78–94. doi: 10.18632/oncotarget.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski U, Hlozek M, Hamilton G. Activation of Na+/H+ exchanger 1 by neurotensin signaling in pancreatic cancer cell lines. Biochem Biophys Res Commun. 2010;393:414–419. doi: 10.1016/j.bbrc.2010.02.009. [DOI] [PubMed] [Google Scholar]
- Partensky C. Toward a better understanding of pancreatic ductal adenocarcinoma: glimmers of hope. Pancreas. 2013;42:729–739. doi: 10.1097/MPA.0b013e318288107a. [DOI] [PubMed] [Google Scholar]
- Pedersen SF, Stock C. Ion channels and transporters in cancer: pathophysiology, regulation, and clinical potential. Cancer Res. 2013;73:1658–1661. doi: 10.1158/0008-5472.CAN-12-4188. [DOI] [PubMed] [Google Scholar]
- Perrone G, Santini D, Vincenzi B, Zagami M, La Cesa A, Bianchi A, Altomare V, Primavera A, Battista C, Vetrani A, Tonini G, Rabitti C. COX-2 expression in DCIS: correlation with VEGF, HER-2/neu, prognostic molecular markers and clinicopathological features. Histopathology. 2005;46:561–568. doi: 10.1111/j.1365-2559.2005.02132.x. [DOI] [PubMed] [Google Scholar]
- Pillozzi S, Brizzi MF, Bernabei PA, Bartolozzi B, Caporale R, Basile V, Boddi V, Pegoraro L, Becchetti A, Arcangeli A. VEGFR-1 (FLT-1), beta1 integrin, and hERG K+ channel for a macromolecular signaling complex in acute myeloid leukemia: role in cell migration and clinical outcome. Blood. 2007;110:1238–1250. doi: 10.1182/blood-2006-02-003772. [DOI] [PubMed] [Google Scholar]
- Pillozzi S, Arcangeli A. Physical and functional interaction between integrins and hERG1 channels in cancer cells. Adv Exp Med Biol. 2010;674:55–67. doi: 10.1007/978-1-4419-6066-5_6. [DOI] [PubMed] [Google Scholar]
- Prevarskaya N, Skryma R, Shuba Y. Ion channels and the hallmarks of cancer. Trends Mol Med. 2010;16:107–121. doi: 10.1016/j.molmed.2010.01.005. [DOI] [PubMed] [Google Scholar]
- Provost JJ, Wallert MA. Inside out: targeting NHE1 as an intracellular and extracellular regulator of cancer progression. Chem Biol Drug Des. 2013;81:85–101. doi: 10.1111/cbdd.12035. [DOI] [PubMed] [Google Scholar]
- Rosati B, Marchetti P, Crociani O, Lecchi M, Lupi R, Arcangeli A, Olivotto M, Wanke E. Glucose- and arginine-induced insulin secretion by human pancreatic beta-cells: the role of HERG K(+) channels in firing and release. FASEB J. 2000;14:2601–2610. doi: 10.1096/fj.00-0077com. [DOI] [PubMed] [Google Scholar]
- Schönherr R, Rosati B, Hehl S, Rao VG, Arcangeli A, Olivotto M, Heinemann SH, Wanke E. Functional role of the slow activation property of ERG K+ channels. Eur J Neurosci. 1999;11:753–760. doi: 10.1046/j.1460-9568.1999.00493.x. [DOI] [PubMed] [Google Scholar]
- Sette A, Spadavecchia J, Landoulsi J, Casale S, Haye B, Crociani O, Arcangeli A. Development of novel anti-Kv 11.1 antibody-conjugated PEG-TiO2 nanoparticles for targeting pancreatic ductal adenocarcinoma cells. J Nanopart Res. 2013;15:2111. doi: 10.1007/s11051-013-2111-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siveke JT, Crawford HC. KRAS above and beyond–EGFR in pancreatic cancer. Oncotarget. 2012;3:1262–1263. doi: 10.18632/oncotarget.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobin LH, Gospodarowicz M, Wittekind Ch.2009TNM Classification of Malignant Tumors. UICC International Union Against Cancer7th edWiley-Blackwell: New York [Google Scholar]
- Sontheimer H. An unexpected role for ion channels in brain tumor metastasis. Exp Biol Med. 2008;233:779–791. doi: 10.3181/0711-MR-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soroceanu L, Manning TJ, Jr, Sontheimer H. Modulation of glioma cell migration and invasion using Cl(−) and K(+) ion channel blockers. J Neurosci. 1999;19:5942–5954. doi: 10.1523/JNEUROSCI.19-14-05942.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor CR. An exaltation of experts: concerted efforts in the standardization of immunohistochemistry. Hum Pathol. 1994;25:2–11. doi: 10.1016/0046-8177(94)90164-3. [DOI] [PubMed] [Google Scholar]
- Zhou B, Irwanto A, Guo YM, Bei JX, Wu Q, Chen G, Zhang TP, Lei JJ, Feng QS, Chen LZ, Liu J, Zhao YP. Exome sequencing and digital PCR analyses reveal novel mutated genes related to the metastasis of pancreatic ductal adenocarcinoma. Cancer Biol Ther. 2012;13:871–879. doi: 10.4161/cbt.20839. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.