Abstract
Background:
Mitochondrial mutations are commonly reported in tumours, but it is unclear whether impaired mitochondrial function per se is a cause or consequence of cancer. To elucidate this, we examined the risk of cancer in a nationwide cohort of patients with mitochondrial dysfunction.
Methods:
We used nationwide results on genetic testing for mitochondrial disease and the Danish Civil Registration System, to construct a cohort of 311 patients with mitochondrial dysfunction. A total of 177 cohort members were identified from genetic testing and 134 genetically untested cohort members were matrilineal relatives to a cohort member with a genetically confirmed maternally inherited mDNA mutation. Information on cancer was obtained by linkage to the Danish Cancer Register. Standardised incidence ratios (SIRs) were used to assess the relative risk of cancer.
Results:
During 7334 person-years of follow-up, 19 subjects developed a primary cancer. The corresponding SIR for any primary cancer was 1.06 (95% confidence interval 0.68–1.63). Subgroup analyses according to mutational subtype yielded similar results, for example, a SIR of 0.94 (95% CI 0.53 to 1.67) for the m.3243A>G maternally inherited mDNA mutation, cases=13.
Conclusions:
Patients with mitochondrial dysfunction do not appear to be at increased risk of cancer compared with the general population.
Keywords: mitochondrial dysfunction, epidemiology, cohort study
During the first part of the twentieth century, Otto Warburg introduced the concept of aerobic glycolysis in cancer (Warburg, 1956). The role of mitochondria in carcinogenesis has been further enlightened by the subsequent demonstration of increased production of reactive oxygen species by many tumours (Petros et al, 2005), which can cause damage to both mitochondrial DNA (mDNA) and nuclear DNA (nDNA) (Lee and Wei, 2005; Hegde et al, 2012).
Considerable debate has taken place over whether mitochondrial mutations, which are frequently reported in cancers, are a cause or a consequence of the cancer (Brandon et al, 2006). With respect to mutations in nDNA encoding mitochondrial function, mutations in the fumarase gene have been shown to predispose to the distinct cancer susceptibility syndrome, Hereditary Leiomyomatosis and Renal Cell Cancer (OMIM: 150800) (Kiuru et al, 2001; Launonen et al, 2001; Tomlinson et al, 2002), and mutations in the succinate dehydrogenase gene to predispose to various presentations of hereditary paragangliomas (Baysal et al, 2000; Astuti et al, 2001). Linkage of these nDNA mutations to cancer raises the possibility that aberrations in mDNA could also contribute to carcinogenesis. Following this, a link between mitochondrial dysfunction and carcinogenesis would be particularly supported by demonstration of germline vs somatic mDNA mutations from patients with cancer. However, the bulk of mDNA aberrations reported in cancer patients have been somatic, whereas reports of germline mDNA mutations are rare (Brandon et al, 2006). For instance, in a study of the mDNA gene encoding the mitochondrial complex 1 (MT-CO1) in European-American prostate cancer patients, 11% of the prostectomy specimens harboured MT-CO1 mutations, of which four different mutations were thought to be germline (Petros et al, 2005). In other studies of Afro-American women, the m.10398A allele in the ND3 gene was linked to an increased risk of breast cancer in some studies (Canter et al, 2005; Bai et al, 2007), but not in others (Mosquera-Miguel et al, 2008; Setiawan et al, 2008).
Although the mentioned mDNA variants may be associated with an increased risk of cancer in some organs, it is important to note that until now no pathogenic mDNA mutation has been unambiguously associated with cancer (Schon et al, 2012). Also distinct from the hypothesis that mDNA copy number is associated with cancer (Yu, 2011), the enigma as to whether impaired mitochondrial function per se predisposes to development of cancer therefore persists. To elucidate this further, we set up a nationwide cohort of more than 300 patients with mitochondrial dysfunction and assessed prospective cancer development among cohort members compared with the general population.
Materials and methods
Study design and setting
A nationwide cohort of patients with mitochondrial dysfunction was established. By use of the unique personal identification number assigned to all Danish citizens, we linked individual-level information on mitochondrial dysfunction with information on incident cancer from the Danish Cancer Register (Gjerstorff, 2011) and with demographic information from the Danish Civil Registration System (CRS) (Pedersen, 2011). The CRS was established on 2 April 1968, and contains among others, information on sex, date of birth and updated information on vital status and emigration, thus minimising loss to follow-up.
Mitochondrial dysfunction
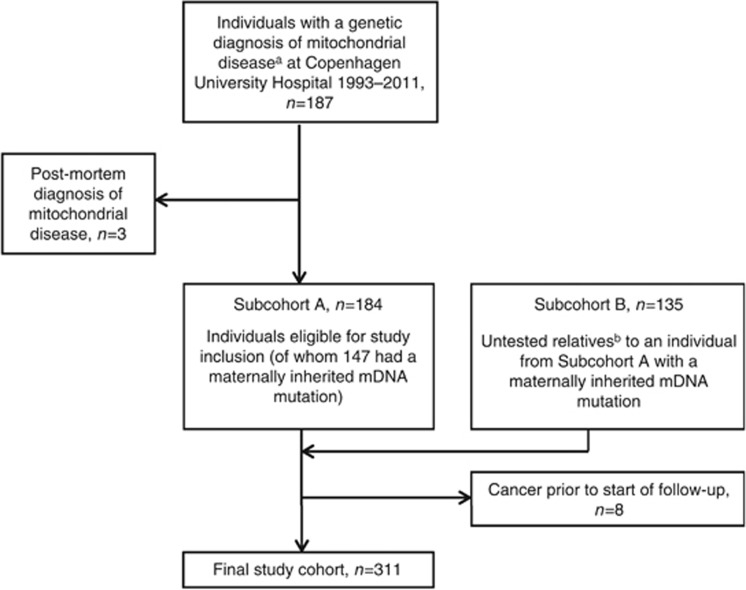
The cohort of patients with mitochondrial dysfunction was based on two subcohorts: A) a clinical cohort of individuals with genetically verified mitochondrial dysfunction and B) a register-based cohort (identified through a family relations database) consisting of untested maternal relatives to members of subcohort A with maternally inherited mDNA mutations. Subcohort A consisted of all mutation-positive individuals that had undergone genetic testing for a suspected mitochondrial disorder at the Laboratory of Molecular Genetics, Rigshospitalet, which is the only laboratory in Denmark where analyses for mitochondrial mutations are carried out officially. Individuals with the following types of mitochondrial mutations were included in the study: maternally inherited mDNA mutations, de novo mDNA mutations or nDNA mutations affecting mitochondrial function. Individuals with multiple mDNA deletions were included as cases with mitochondrial dysfunction only if the activity of respiratory enzymes was found to be abnormal in skeletal muscle, if the individual was <40 years of age at time of diagnosis or if the multiple mDNA deletions were explained by the presence of pathogenic nDNA mutation(s) known to cause mDNA deletions. Older persons with multiple mDNA deletions and normal respiratory chain function were excluded as low levels of multiple mDNA deletions develop in many with age (Fayet et al, 2002). Furthermore, individuals in whom single large-scale deletions of mDNA had been detected were not included in the cohort, because these cases are usually sporadic and in most cases only carry deleted mDNA molecules in skeletal muscle. DNA for genetic analysis was isolated from a standard EDTA blood sample or a muscle biopsy depending on the analysis. All genetic analyses were carried out as part of a diagnostic workup. Allele-specific high-sensitive PCR analysis was used to assess the m.3243A>G mutation, whereas the remaining mDNA mutations were assayed by PCR and direct sequencing. Nuclear genes were PCR-amplified and sequenced directly for the coding and exon flanking sequences of the requested genes. DNA from muscle biopsies, initially sent for assessment of the respiratory chain activity, was isolated and assessed for single large-scale or multiple mDNA deletions using a long PCR-based analysis (Kleinle et al, 1997). Members of subcohort B were identified by use of the Danish Family Relations Database (DFRD), established at Statens Serum Institut, Copenhagen. Subcohort B included genetically untested matrilineal relatives to members of subcohort A, in whom a maternally inherited mDNA mutation had been detected. The following types of matrilineal relatives were identified: first degree (mothers and full siblings), second degree (grandmother through mother, uncles/aunts through mother, nephews/nieces through sisters, half siblings through mother, half-uncles and half-aunts through mother and grandmother, and for female mutation carriers, grandchildren through daughter) and third degree (cousins through mothers sister). Thus, irrespective of the degree of family relation, all identified subjects had a direct maternal relationship, and thus should have inherited the mDNA aberration observed in the proband. The Danish Family Relations Database is based on parent–child links registered in the Danish Civil Registration System and allows for identification of first degree relatives (parents, children and siblings) and half-siblings for nearly all individuals born in 1950 or later, whereas second-degree relatives (grandparents, grandchildren, aunts/uncles, nieces/nephews) and third-degree relatives (full cousins) can be identified for the majority of individuals born after 1985. A flow chart of the study cohort is presented in Figure 1.
Figure 1.
Selection of patients with mitochondrial dysfunction for study inclusion. aWe included patients with functional maternally inherited mDNA mutations, de novo mDNA mutations or nDNA mutations in genes encoding mitochondrial functions (as defined in the methods section). bIdentified through the Danish Family Relations Database, established at Statens Serum Institut, Copenhagen. We included only relatives who under the assumption of matrilineal inheritance of mDNA mutations would be expected to carry the mutation (defined in the methods section). Abbreviations: mDNA=mitochondrial DNA; nDNA=nuclear DNA.
Cancer diagnosis
The Danish Cancer Register is a high-quality population-based register with mandatory recording of all diagnosed cancers in Denmark. The register was established in 1943, and diagnostic groups of individual cancers have been coded according to the International Classification of Diseases (ICD) version 7 until 1977. Thereafter, ICD10 has been used (Gjerstorff, 2011). To allow for comparability between cancers coded using ICD7 and ICD10, we used the NORDCAN codes provided by the Association of the Nordic Cancer Registers (ANCR) (Engholm et al, 2010).
Statistical methods
We evaluated risk of cancer (all cancers with a NORDCAN code, groups 1 to 88) among individuals with a mitochondrial disease, using standardised incidence ratios (SIRs). Each SIR was calculated by dividing the number of observed events of cancer in the study cohort during follow-up by the expected number of cancers. The expected number of cancers was calculated by applying age-, sex- and calendar time-specific national cancer incidence rates to the person-years of follow-up observed in the study cohort. Confidence intervals were based on a sandwich estimator assuming working independence and all tests of statistical association were score tests based on this assumption (Liang and Zeger, 1986). Confidence intervals were also estimated assuming exchangeable correlation for family members in the cohort to account for familial correlation, but results were similar (not shown) and therefore working independence was used for all results presented. For mutation groups where less than 10 cancers were observed, exact confidence intervals were estimated using the Poisson distribution. To avoid survivor bias, start of follow-up depended on mode of identification of cohort members: (i) for probands: date of withdrawal of blood used for diagnosis of mitochondrial disease; (ii) for cohort members identified by genetic testing and who were identifiable in the DFRD: date of birth or 2 April 1968, whichever came last; (iii) for cohort members identified by genetic testing, but with no registration in the DFRD: date of withdrawal of blood used for diagnosis of mitochondrial disease; (iv) for cohort members without genetic testing but identifiable in the DFRD (except for mothers of probands): date of birth or 2 April 1968, whichever came last; and v) for mothers of probands where the mother had no genetic testing, but was identifiable in the DFRD: date of birth of the proband or 2 April 1968, whichever came last. Follow-up ended at the first of the following: cancer, death, emigration, designated missing as defined by the CRS or at 31 December 2011. Accordingly, cohort members with cancer prior to follow-up were excluded from the analyses. When considering any cancer except non-melanoma skin cancer (all cancer with a NORDCAN code except groups 30 and 88: common skin cancer and basal cell carcinoma, respectively), non-melanoma skin cancer cases were disregarded when calculating follow-up time and number of cases. In an extra analysis, we assessed the risk of multiple cancers (i.e., not just primary cancers) and the analysis was performed as described above with the exception that follow-up did not end at time of first cancer when calculating follow-up time in the cohort and when calculating national rates. Analyses were performed using SAS software (version 9.4).
Ethics
The study was approved by the Danish Data Protection Agency (journal numbers 2008-54-0472 and 2007-58-0015). According to Danish legislation, research based exclusively on register data is exempt from approval by a biomedical ethics committee.
Results
The cohort included 311 individuals (56% female) with mitochondrial dysfunction of whom almost 90% had a maternally inherited mDNA mutation. Almost 60% were identified from genetic testing and among tested cohort members, 62% were female. Descriptive characteristics of the study cohort are presented in Table 1 and information on subtype of mitochondrial mutations in Table 2.
Table 1. Characteristics of a nationwide cohort of patients with mitochondrial dysfunction by mode of ascertainment.
|
Subcohort A |
Subcohort B | |||
|---|---|---|---|---|
| Probands (n=61) | Tested relatives, clinic based (n=116) | Untested relatives, register based (n=134) | All (n=311) | |
| n (%) | n (%) | n (%) | n (%) | |
|
Type of mutation | ||||
| Maternally inherited mDNA | 32 (52.5) | 112 (96.6) | 134 (100) | 278 (89.4) |
| De novo mDNA | 8 (13.1) | 0 (0) | 0 (0) | 8 (2.57) |
| nDNA | 21 (34.4) | 4 (3.45) | 0 (0) | 25 (8.04) |
|
Sex | ||||
| Male | 29 (47.5) | 39 (33.6) | 69 (51.5) | 137 (44.1) |
| Female | 32 (52.5) | 77 (66.4) | 65 (48.5) | 174 (56.0) |
|
Year of birth | ||||
| Before 1950 | 12 (19.7) | 29 (25.0) | 22 (16.4) | 63 (20.3) |
| 1950–1969 | 23 (37.7) | 49 (42.2) | 21 (15.7) | 93 (29.9) |
| 1970–1989 | 13 (21.3) | 27 (23.3) | 44 (32.8) | 84 (27.0) |
| 1990–2011 | 13 (21.3) | 11 (9.48) | 47 (35.1) | 71 (22.8) |
|
Year of study entrya | ||||
| 1968–1972 | 0 (0) | 64 (55.2) | 45 (33.6) | 109 (35.1) |
| 1973–1982 | 0 (0) | 13 (11.2) | 22 (16.4) | 35 (11.3) |
| 1983–1992 | 0 (0) | 16 (13.8) | 28 (20.9) | 44 (14.2) |
| 1993–2002 | 19 (31.2) | 10 (8.62) | 27 (20.2) | 56 (18.0) |
| 2003–2011 | 42 (68.9) | 13 (11.2) | 12 (8.96) | 67 (21.5) |
Abbreviations: mDNA=mitochondrial DNA; nDNA=nuclear DNA.
Probands were classified as the first individual diagnosed in a family, tested relatives as relatives that had tested positive for the mutation found in the family proband and untested relatives as relatives with a direct maternal relationship identified through the family relations database (as defined in the methods section).
The majority of the cohort members who had undergone genetic testing were older at study entry, whereas the untested relatives were generally younger: median (inter quartile interval) age at study entry, probands=36.1 (18.7–50.9) years, tested relatives= 6.63 (0.0–26.5) years, untested relatives=0.0 (0.0–6.61) years. The differences reflect that time for entry into the study differed for the three groups in order to avoid survivor bias (see methods section).
Table 2. Specific types of mDNA and nDNA mutations in a nationwide cohort of 311 patientsa with mitochondrial dysfunction.
| Subcohort | n | Affected nucleotide and position | Protein | Gene |
|---|---|---|---|---|
|
nDNA mutations affecting mitochondrial function | ||||
| A | 1 | c.659C>T +/+ | p.A220V +/+ | ACAD9 |
| A | 1 | c.248delT +/+ | p.Val83Glyfs*2 +/+ | C12orf65 |
| A | 2 | c.137A>G +/+ | p.N46S +/+ | DGUOK |
| A | 1 | c.178C>T +/+ | p. R60* +/+ | NDUFA12 |
| A | 1 | del ex 2-4 +/+ | - | NDUFAF2 |
| A | 2 | c.2827C>T +/− | p.R943C +/- | POLG |
| A | 1 | c.752C>T/c.1399G>A | p.T251I/p.A467T | POLG |
| A | 1 | c.[752C>T;1760C>T]/c.2591A>G | p.[T251I;p587L]/p.N864S | POLG |
| A | 1 | c.911T>G +/+ | p.L304R +/+ | POLG |
| A | 1 | c.2529G>C/c.2828G>A | p.W748S/p.G848S | POLG |
| A | 2 | c.752C>T/c.1760C>T | p.T251I/p.P587L | POLG |
| A | 1 | c.133G>A +/+ | p.G45R +/+ | PRF1 |
| A | 1 | c.312del10insAT/c.901_902delTG | — | SURF1 |
| A | 3 | c.1110C>A +/+ | p.F370L +/+ | C10orf2 |
| A | 6 | multiple mDNA deletions | — | — |
|
De novo mDNA | ||||
| A | 1 | m.14453G>A | — | — |
| A | 1 | m.15579A>G | — | — |
| A | 1 | m.3256C>T | — | — |
| A | 1 | m.4409T>C | — | — |
| A | 1 | m.4450G>A | — | — |
| A | 1 | m.8156dupG | — | — |
| A | 1 | m.8340G>A | — | — |
| A | 1 | m.8989G>C | — | — |
|
Maternally inherited mDNA, clinic-based | ||||
| A | 110 | m.3243A>G | — | — |
| A | 4 | m.4078A>G | — | — |
| A | 21 | m.8344A>G | — | — |
| A | 7 | m.8993T>C | — | — |
| A | 2 | m.9176T>C | — | — |
|
Maternally inherited mDNA, register-basedb | ||||
| B | 108 | m.3243A>G | — | — |
| B | 5 | m.4078A>G | — | — |
| B | 10 | m.8344A>G | — | — |
| B | 8 | m.8993T>C | — | — |
| B | 3 | m.9176T>C | — | — |
Abbreviations: mDNA=mitochondrial DNA; nDNA=nuclear DNA.
In the present table, the 311 cohort members are grouped according to four mutation subgroups and membership of Subcohort A or Subcohort B as defined in the methods section. The number of cohort members in the four categories of mutation subgroups thus sum to the total number of cohort members in the main cohort (25+8+144+134=311).
For the subcohort of untested cohort members identified through the family relations database, the indicated mDNA point mutation is the mutation of the family proband.
Main analysis
Nineteen cohort members out of 311 developed a primary cancer during 7334 person-years of follow-up; median age at cancer diagnosis (inter quartile interval) was 57.4 (49.0–69.7) years. The corresponding SIR for any cancer was 1.06 (95% CI 0.68–1.63) (Table 3). Stratification according to sex and current age indicated no heterogeneity. Observed and expected numbers of organ-specific cancers during follow-up are presented in Supplementary Table 1.
Table 3. Standardised incidence ratios of cancer in a nationwide cohort of patients with mitochondrial dysfunction, overall and according to proband status, subgroup of mitochondrial mutation, sex and current age.
| Mitochondrial mutation | Cohort members | obs | exp | SIR | 95% CIa |
|---|---|---|---|---|---|
| Main cohort (all mutations) | 311 | 19 | 18.0 | 1.06 | 0.68 to 1.63 |
| All genetically certain casesb | 162 | 10 | 9.51 | 1.05 | 0.60 to 1.85 |
|
Stratification by proband status | |||||
| Probands | 61 | 0 | 1.76 | — | — |
| Non-probands | 250 | 19 | 16.2 | 1.17 | 0.76 to 1.81 |
|
Stratification by mutational subgroup | |||||
| Maternally inherited mDNA mutation | 278 | 19 | 17.4 | 1.09 | 0.71 to 1.69 |
| Register based | 134 | 8c | 6.52 | 1.23 | 0.70 to 2.17 |
| Clinic based | 144 | 11 | 10.9 | 1.01 | 0.58 to 1.76 |
| m.3243A>G mutation | 218 | 13 | 13.8 | 0.94 | 0.53 to 1.67 |
| Non-m.3243A>G mutationd | 60 | 6c | 3.61 | 1.66 | 0.98 to 2.83 |
| Other mutations | 33 | 0 | 0.60 | — | — |
| De novo mDNA mutation | 8 | 0 | 0.13 | — | — |
| Nuclear DNA mutation | 25 | 0 | 0.47 | — | — |
|
Stratification by sex and current age | |||||
| Female | 174 | 15 | 14.0 | 1.07 | 0.66 to 1.74 |
| Male | 137 | 4c | 4.03 | 0.99 | 0.39 to 2.51 |
| Current age<50 | 282e | 5c | 5.72 | 0.87 | 0.38 to 1.98 |
| Current age≥50 | 103e | 14 | 12.3 | 1.14 | 0.72 to 1.80 |
Abbreviations: CI=confidence interval; exp=expected number of primary cancers during follow-up; mDNA=mitochondrial DNA; nDNA=nuclear DNA; obs=observed number of primary cancers during follow-up; SIR=standardised incidence ratio.
P-homogeneity: proband vs non-proband (P-value not evaluated), maternally inherited mDNA mutation vs other mutations (P-value not evaluated), female vs male= 0.88 and current age less than 50 vs current age 50 or older =0.52, register-based vs clinic-based=0.60 and m.3243A>G vs non-m.3243A>G =0.19.
Genetically, certain cases denotes cohort members with a genetically confirmed diagnosis, that is, in this subanalysis, the following cohort members were excluded: (i) cohort members identified only through the Danish Family Relations Database, (ii) 12 tested cohort members from families with the m.3243A>G-mutation in whom the degree of mDNA heteroplasmy was very low, that is, below the detection limit of the assay of 1% and (iii) 3 tested cohort members from a family where the m.8344A>G mutation was detected in the proband but neither in the mother nor in the tested siblings. This proband was diagnosed at the beginning of the study period, where the assay (direct sequencing) had an estimated sensitivity of approximately 15%, that is, a low level of heteroplasmy for the tested relatives cannot be ruled out.
As asymptotic and exact confidence intervals may differ when having few cancer cases, we also estimated exact confidence intervals for mutation groups with less than 10 cancer cases: register-based (0.53 to 2.42), non-m.3243A>G mutation (0.61 to 3.61), male (0.27 to 2.54) and current age<50 (0.28 to 2.04). Use of exact confidence intervals did not change the study conclusions.
The non-m.3243A>G-group (register-based and clinic-based cohort members) consisted of the following mutations (cohort members eligible for follow-up, obs/exp): m.8344A>G (31, 4/1.99), m.4078A>G (9, 0/0.07), m.9176T>C (5, 0/0.35), m.8993T>C (15, 2/1.20).
As current age is a time-dependent variable, the same cohort member can contribute to both categories as exposed and explaining why the number of cohort members in the two categories exceeds 311.
Additional analyses
To investigate the possibility that an association might be seen for a particular subgroup of the main cohort, we estimated SIRs of cancer according to selected subdivisions of mitochondrial dysfunction with similar results observed among the different subgroups (Table 3).
We further investigated the risk of both primary and subsequent cancers. Among the 311 cohort members followed for cancer, there were a total of 23 cancers in 19 cohort members during 7452 person-years of follow-up, corresponding to a SIR of 1.15 (95% CI 0.77–1.74). Descriptive characteristics of cancer cases among cohort members are presented in Supplementary Table 2.
Finally, we explored the risk of any cancer, except non-melanoma skin cancer (hypothesising that non-melanoma skin cancer would be more prone to surveillance bias). In this analysis, 313 cohort members were followed for cancer. During 7379 person-years of follow-up there were 13 cancer cases corresponding to a SIR of 0.87 (95% CI 0.49–1.52). Similar results were observed in sub-analyses with stratification according to selected subdivisions of mitochondrial dysfunction (Supplementary Table 3).
Discussion
In this nationwide cohort study, patients with mitochondrial dysfunction were not at increased risk of developing cancer compared with the general population. The same result was observed for specific subtypes of mitochondrial mutations.
The notion that a causal association between mitochondrial dysfunction and cancer exists is inferred by studies of specific nDNA mutations in genes affecting mitochondrial function that associate with particular cancer susceptibility syndromes, for example, Hereditary Leiomyomatosis and Renal Cell Cancer (Kiuru et al, 2001; Launonen et al, 2001; Tomlinson et al, 2002). However, in studies of the association between mDNA variants of presumed germline origin and cancer, results are heterogeneous and based on patients with cancer rather than patients with mitochondrial disease (Canter et al, 2005; Petros et al, 2005; Bai et al, 2007; Mosquera-Miguel et al, 2008; Setiawan et al, 2008). Increased cancer prevalence has been observed in a group of patients with not clearly defined mitochondrial disease in adults compared with the total population of Austria (including children). However, the authors did not adjust for the age difference between the study group and the general population and therefore this observation cannot be used to elucidate the causal question (Finsterer and Krexner, 2013). Thus, it remains to be shown whether impaired mitochondrial function per se is a cause or a consequence of cancer. In contrast to the existing literature, the present study used a cohort approach to address this issue. We selected patients on the basis of genetically confirmed mitochondrial dysfunction and absence of cancer at the start of follow-up. This approach ensured that information on the exposure (mitochondrial dysfunction) was obtained prior to and independent from the outcome (cancer) and thereby we avoided issues of reverse causality.
We found no overall association between mitochondrial dysfunction and subsequent cancer development arguing against impaired mitochondrial function as a cause of cancer development. One could speculate that associations may exist for organ-specific cancers or for specific mitochondrial mutations. We note that cancers such as biliary cystadenocarcinoma (Ohno et al, 2010), renal cell carcinoma (Sangkhathat et al, 2005) and renal oncocytoma (Piccoli et al, 2012) have been reported in patients with mitochondrial diseases such as mitochondrial myopathy, encephalomyopathy, lactic acidosis and stroke-like syndrome (MELAS) due to the m.3243A>G mDNA mutation. In the present study, we observed cancers covering a wide spectrum of different cancer types; however, given the size of the study, inferences about the risk of organ-specific cancers were not feasible. The main cohort included individuals with various mitochondrial mutations: maternally inherited mDNA mutations, de novo mDNA mutations and mutations in nDNA affecting mitochondrial function. For the majority of the specific mutations, stratification according to mutational subtype was not feasible owing to small numbers, however, for the maternally inherited m.3243A>G mutation, we investigated the risk of cancer among cohort members with a maternally inherited m.3243A>G mutation vs those with a maternally inherited mDNA mutation other than m.3243A>G and found no indication of heterogeneity.
Strengths of the study include the nationwide cohort design applied to a unique combination of a large cohort of patients with mitochondrial dysfunction and a national follow-up on cancer allowing for both sufficient precision and minimal bias. Thus, with an upper limit of the confidence interval for the main estimate of 63%, this study does not support an increased risk of cancer of clinical relevance in patients with mitochondrial disease. Moreover, we consider potential selection and information biases in the study minimal. First, by combining demographic information from the Danish Civil Registration System and information on patients with a clinical diagnosis of mitochondrial disease, we were able to perform a prospective cohort study with minimal loss to follow-up. Second, with respect to the outcome of cancer, the Danish Cancer Register is considered close to complete (Gjerstorff, 2011). A limitation of our study is the lack of information on the degree of mDNA heteroplasmy. However, there are various arguments against this influencing the conclusions of the study. First, the genetically tested part of the cohort was initially selected on the basis of symptomatic disease having led to referral for genetic testing of the proband. Probands likely represent cohort members with the most severe clinical manifestations, and thus most severely perturbed mitochondrial dysfunction. One could thus speculate that the degree of mDNA heteroplasmy would thus be lower in genetically tested non-probands compared with probands and even lower in members of subcohort B (genetically untested cohort members) vs members of subcohort A. However, we observed no increased cancer risk in probands, when restricting the main cohort to only cohort members with a genetically confirmed diagnosis nor when comparing cohort members identified from the clinic vs those identified from the registers. Second, one could speculate that differences in the detection limits of the assays used to identify the various mDNA mutations may have biased the conclusions of the study (apart from the m.3243A>G mutations where a specific high-sensitive PCR assay was used, mDNA mutations were identified and assessed by PCR and direct sequencing, which does not allow for detection of low level heteroplasmy). However, cancer risk was similar among cohort members with the m.3243A>G mutation and those with a non-m.3243A>G mutation. Subsequently, neither selection on severity of mitochondrial disease nor on degrees of mDNA heteroplasmy, are likely to have biased the conclusion of our study.
The present study was not designed to evaluate the effect of screening for cancer in patients with mitochondrial dysfunction. Still, with respect to future decisions on clinical management of patients with mitochondrial dysfunction, the findings provide the most extensive information about risk of cancer in these patients that is available to date.
In conclusion, the results of the present study do not support the existence of an increased risk of cancer in patients with perturbed mitochondrial function. Although the opposite scenario is not contradicted by our results, that is, that cancer-induced alterations of cellular metabolism can cause secondary mDNA aberrations that subsequently start a vicious circle of further malignant transformation (Gogvadze et al, 2008), impaired mitochondrial function per se does not appear to cause cancer.
Acknowledgments
This work was supported by a PhD-study grant from the University of Copenhagen to Dr. Lund and a research grant from the Danish Cancer Society (Kræftens Bekæmpelse) to Dr. Melbye. The funding agency had no role in the design and conduct of the study; collection, management, analysis and interpretation of the data, and preparation, review, or approval of the manuscript.
Author contributions
Marie Lund: draft of manuscript, study concept or design, analysis or interpretation of data, acquisition of data; Mads Melbye: critical revision of the manuscript for content, study concept or design, analysis or interpretation of data, study supervision or coordination, proof of final version; Lars Diaz: critical revision of the manuscript for content, analysis or interpretation of data, statistical analysis, proof of final version; Morten Duno: critical revision of the manuscript for content, acquisition of data, proof of final version; Jan Wohlfahrt: critical revision of the manuscript for content, study concept or design, analysis or interpretation of data, study supervision or coordination, proof of final version; John Vissing: critical revision of the manuscript for content, study concept or design, analysis or interpretation of data, acquisition of data, study supervision or coordination, proof of final version.
Dr. Lund reports grants from University of Copenhagen, grants from Danish Cancer Society (Kræftens Bekæmpelse), during the conduct of the study; Dr. Melbye reports grants from Danish Cancer Society (Kræftens Bekæmpelse), during the conduct of the study; Mr. Diaz reports grants from Danish Cancer Society (Kræftens Bekæmpelse), during the conduct of the study; Dr. Vissing reports personal fees from Genzyme Incorporation, outside the submitted work; the remaining co-authors declare no conflicts of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Supplementary Material
References
- Astuti D, Latif F, Dallol A, Dahia PL, Douglas F, George E, Skoldberg F, Husebye ES, Eng C, Maher ER. Gene mutations in the succinate dehydrogenase subunit SDHB cause susceptibility to familial pheochromocytoma and to familial paraganglioma. Am J Hum Genet. 2001;69:49–54. doi: 10.1086/321282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai R-K, Leal SM, Covarrubias D, Liu A, Wong L-JC. Mitochondrial genetic background modifies breast cancer risk. Cancer Res. 2007;67:4687–4694. doi: 10.1158/0008-5472.CAN-06-3554. [DOI] [PubMed] [Google Scholar]
- Baysal BE, Ferrell RE, Willett-Brozick JE, Lawrence EC, Myssiorek D, Bosch A, van der Mey A, Taschner PE, Rubinstein WS, Myers EN, Richard CW, III, Cornelisse CJ, Devilee P, Devlin B. Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science. 2000;287:848–851. doi: 10.1126/science.287.5454.848. [DOI] [PubMed] [Google Scholar]
- Brandon M, Baldi P, Wallace DC. Mitochondrial mutations in cancer. Oncogene. 2006;25:4647–4662. doi: 10.1038/sj.onc.1209607. [DOI] [PubMed] [Google Scholar]
- Canter JA, Kallianpur AR, Parl FF, Millikan RC. Mitochondrial DNA G10398A polymorphism and invasive breast cancer in African-American women. Cancer Res. 2005;65:8028–8033. doi: 10.1158/0008-5472.CAN-05-1428. [DOI] [PubMed] [Google Scholar]
- Engholm G, Ferlay J, Christensen N, Bray F, Gjerstorff ML, Klint A, Kotlum JE, Olafsdottir E, Pukkala E, Storm HH. NORDCAN—a Nordic tool for cancer information, planning, quality control and research. Acta Oncol. 2010;49:725–736. doi: 10.3109/02841861003782017. [DOI] [PubMed] [Google Scholar]
- Fayet G, Jansson M, Sternberg D, Moslemi A-R, Blondy P, Lombès A, Fardeau M, Oldfors A. Ageing muscle: clonal expansions of mitochondrial DNA point mutations and deletions cause focal impairment of mitochondrial function. Neuromuscul Disord. 2002;12:484–493. doi: 10.1016/s0960-8966(01)00332-7. [DOI] [PubMed] [Google Scholar]
- Finsterer J, Krexner E. Increased prevalence of malignancy in adult mitochondrial disorders. J Med Life. 2013;6:477–481. [PMC free article] [PubMed] [Google Scholar]
- Gjerstorff ML. The Danish Cancer Registry. Scand J Public Heal. 2011;39:42–45. doi: 10.1177/1403494810393562. [DOI] [PubMed] [Google Scholar]
- Gogvadze V, Orrenius S, Zhivotovsky B. Mitochondria in cancer cells: what is so special about them. Trends Cell Biol. 2008;18:165–173. doi: 10.1016/j.tcb.2008.01.006. [DOI] [PubMed] [Google Scholar]
- Hegde ML, Mantha AK, Hazra TK, Bhakat KK, Mitra S, Szczesny B. Oxidative genome damage and its repair: implications in aging and neurodegenerative diseases. Mech Ageing Dev. 2012;133:157–168. doi: 10.1016/j.mad.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiuru M, Launonen V, Hietala M, Aittomaki K, Vierimaa O, Salovaara R, Arola J, Pukkala E, Sistonen P, Herva R, Aaltonen LA. Familial cutaneous leiomyomatosis is a two-hit condition associated with renal cell cancer of characteristic histopathology. Am J Pathol. 2001;159:825–829. doi: 10.1016/S0002-9440(10)61757-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinle S, Wiesmann U, Superti-Furga A, Krähenbühl S, Boltshauser E, Reichen J, Liechti-Gallati S. Detection and characterization of mitochondrial DNA rearrangements in Pearson and Kearns-Sayre syndromes by long PCR. Hum Genet. 1997;100:643–650. doi: 10.1007/s004390050567. [DOI] [PubMed] [Google Scholar]
- Launonen V, Vierimaa O, Kiuru M, Isola J, Roth S, Pukkala E, Sistonen P, Herva R, Aaltonen LA. Inherited susceptibility to uterine leiomyomas and renal cell cancer. Proc Natl Acad Sci USA. 2001;98:3387–3392. doi: 10.1073/pnas.051633798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H-C, Wei Y-H. Mitochondrial biogenesis and mitochondrial DNA maintenance of mammalian cells under oxidative stress. Int J Biochem Cell Biol. 2005;37:822–834. doi: 10.1016/j.biocel.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Liang KY, Zeger SL. Longitudinal data analysis using generalized linear models. Biometrika. 1986;73:13–22. [Google Scholar]
- Mosquera-Miguel A, Alvarez-Iglesias V, Carracedo A, Salas A, Vega A, Milne R, de Leon AC, Benitez J. Is mitochondrial DNA variation associated with sporadic breast cancer risk. Cancer Res. 2008;68:623–625. doi: 10.1158/0008-5472.CAN-07-2385. [DOI] [PubMed] [Google Scholar]
- Ohno A, Mori A, Doi R, Yonenaga Y, Asano N, Uemoto S. Successful left hemihepatectomy and perioperative management of a patient with biliary cystadenocarcinoma, complicated with MELAS syndrome: report of a case. Surg Today. 2010;40:878–882. doi: 10.1007/s00595-009-4145-z. [DOI] [PubMed] [Google Scholar]
- Pedersen CB. The Danish Civil Registration System. Scand J Public Heal. 2011;39:22–25. doi: 10.1177/1403494810387965. [DOI] [PubMed] [Google Scholar]
- Petros JA, Baumann AK, Ruiz-Pesini E, Amin MB, Sun CQ, Hall J, Lim S, Issa MM, Flanders WD, Hosseini SH, Marshall FF, Wallace DC. mtDNA mutations increase tumorigenicity in prostate cancer. Proc Natl Acad Sci USA. 2005;102:719–724. doi: 10.1073/pnas.0408894102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccoli GB, Bonino LD, Campisi P, Vigotti FN, Ferraresi M, Fassio F, Brocheriou I, Porpiglia F, Restagno G. Chronic kidney disease, severe arterial and arteriolar sclerosis and kidney neoplasia: on the spectrum of kidney involvement in MELAS syndrome. BMC Nephrol. 2012;13:9. doi: 10.1186/1471-2369-13-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangkhathat S, Kusafuka T, Yoneda A, Kuroda S, Tanaka Y, Sakai N, Fukuzawa M. Renal cell carcinoma in a pediatric patient with an inherited mitochondrial mutation. Pediatr Surg Int. 2005;21:745–748. doi: 10.1007/s00383-005-1471-0. [DOI] [PubMed] [Google Scholar]
- Schon EA, Dimauro S, Hirano M. Human mitochondrial DNA: roles of inherited and somatic mutations. Nat Rev Genet. 2012;13:878–890. doi: 10.1038/nrg3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setiawan VW, Chu L-H, John EM, Ding YC, Ingles SA, Bernstein L, Press MF, Ursin G, Haiman CA, Neuhausen SL. Mitochondrial DNA G10398A variant is not associated with breast cancer in African-American women. Cancer Genet Cytogenet. 2008;181:16–19. doi: 10.1016/j.cancergencyto.2007.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson IP, Alam NA, Rowan AJ, Barclay E, Jaeger EE, Kelsell D, Leigh I, Gorman P, Lamlum H, Rahman S, Roylance RR, Olpin S, Bevan S, Barker K, Hearle N, Houlston RS, Kiuru M, Lehtonen R, Karhu A, Vilkki S, Laiho P, Eklund C, Vierimaa O, Aittomaki K, Hietala M, Sistonen P, Paetau A, Salovaara R, Herva R, Launonen V, Aaltonen LA. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat Genet. 2002;30:406–410. doi: 10.1038/ng849. [DOI] [PubMed] [Google Scholar]
- Warburg O. Origin of cancer cells. Oncologia. 1956;9:75–83. [PubMed] [Google Scholar]
- Yu M. Generation, function and diagnostic value of mitochondrial DNA copy number alternations in human cancers. Life Sci. 2011;89:65–71. doi: 10.1016/j.lfs.2011.05.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.