Abstract
The hSuv3 (human Suv3) helicase has been shown to be a major player in mitochondrial RNA surveillance and decay, but its physiological role might go beyond this functional niche. hSuv3 has been found to interact with BLM (Bloom’s syndrome protein) and WRN (Werner’s syndrome protein), members of the RecQ helicase family involved in multiple DNA metabolic processes, and in protection and stabilization of the genome. In the present study, we have addressed the possible role of hSuv3 in genome maintenance by examining its potential association with key interaction partners of the RecQ helicases. By analysis of hSuv3 co-IP (co-immunoprecipitation) complexes, we identify two new interaction partners of hSuv3: the RPA (replication protein A) and FEN1 (flap endonuclease 1). Utilizing an in vitro biochemical assay we find that low amounts of RPA inhibit helicase activity of hSuv3 on a forked substrate. Another single-strand-binding protein, mtSSB (mitochondrial single-strand-binding protein), fails to affect hSuv3 activity, indicating that the functional interaction is specific for hSuv3 and RPA. Further in vitro studies demonstrate that the flap endonuclease activity of FEN1 is stimulated by hSuv3 independently of flap length. hSuv3 is generally thought to be a mitochondrial helicase, but the physical and functional interactions between hSuv3 and known RecQ helicase-associated proteins strengthen the hypothesis that hSuv3 may play a significant role in nuclear DNA metabolism as well.
Keywords: DNA repair, flap endonuclease 1, genome stability, hSuv3 helicase, RecQ helicase family, replication protein A
INTRODUCTION
The Suv3 helicases (SUV3, hSUV3p and SUPV3L1) were originally identified in a study on mitochondrial RNA metabolism in Saccharomyces cerevisiae [1]. Together with the nuclease Dss1p, ScSuv3p (S. cerevisiae Suv3) forms the mitochondrial degradosome complex that plays a pivotal role in mitochondrial RNA metabolism, and consequently in mitochondrial and cellular homoeostasis [2,3]. Suv3 is ubiquitously present in all eukaryotes [4]. The human SUV3 gene was identified based on a high level of sequence conservation [4], and characterized as a DExH-box RNA helicase of the Ski2 superfamily capable of unwinding RNA/RNA, RNA/DNA and DNA/DNA duplexes in vitro [4,5]. Originally, hSuv3 (human Suv3) was found to primarily unwind substrates in the 5′→3′ direction [5]; however, recently the directionality was reported to be 3′→5′ [6].
Expression analysis in human tissues found the gene to be expressed in all tissues, with the highest levels in the liver [4]. In mice, Suv3 expression begins at the blastocyst stage and becomes extensive in all cell types throughout life, with expression in the mature animal being highest in the brain, sensory organs and testis [7]. Complete loss of the SUV3 gene in mice results in embryonic lethality [8], whereas conditional post-natal loss of Suv3 function in mice leads to an accelerated aging-like phenotype [9].
hSuv3 localizes mainly to the mitochondrial matrix [10,11]. It appears to have a central role in mitochondrial RNA metabolism, and its knockdown is associated with accumulation of truncated mitochondrial RNA species, and decrease in mtDNA (mitochondrial DNA) copy number and mitochondrial protein expression, eventually resulting in cell death [12–14]. To date, no orthologue of the Dss1p nuclease has been found in humans, and results proposing the PNPase (polynucleotide phosphorylase) as a possible alternative [6,15] remain a matter of controversy due to the lack of PNPase in the mitochondrial matrix [16].
In addition to the mitochondrial localization, a small fraction of hSuv3 has been detected in the nucleus [13]. Knockdown of hSuv3 in HeLa cells led to an increase in homologous recombination during mitosis as measured by SCE (sister chromatid exchange) [8], indicating a potential nuclear role of hSuv3. However, no nuclear function of hSuv3 has yet been reported. Nonetheless, a high-throughput interaction screen in S. cerevisiae found that ScSuv3p interacts with Sgs1, the sole yeast RecQ helicase [17]. Importantly, the interaction seems to be conserved as hSuv3 was found to interact with two of the human RecQ helicases, WRN (Werner’s syndrome protein) and BLM (Bloom’s syndrome protein) [8]. The RecQ helicase family is well known for their crucial role in genome maintenance, with deficiencies leading to severe DNA instability associated with premature aging syndromes. These helicases participate in an intricate interplay with a variety of DNA metabolic proteins to achieve an optimal activity for each of their numerous roles [18]. One such partner is RPA (replication protein A), a heterotrimeric ssDNA (single-stranded DNA)-binding protein complex, consisting of 70, 32 and 14 kDa subunits, which was identified for its role in SV40 (simian virus 40) DNA replication [19]. RPA has been shown to stimulate helicase activities of the RecQ helicases RECQ1 [20], BLM [21], WRN [22], RECQL4 [23] and RECQ5β [24]. Through its binding to DNA, RPA appears to actively coordinate assembly and disassembly of DNA-processing proteins on ssDNA, thereby serving multiple roles in DNA metabolism ranging from DNA replication and telomere maintenance to homologous recombination and DNA repair [25].
FEN1 (flap endonuclease 1) is another well-documented partner of the RecQ helicases [18]. It is a multifunctional structure-specific nuclease, displaying an endonuclease activity on 5′-flap ssDNA or RNA substrates, a low-efficiency 5′→3′ exonuclease activity on DNA structures [26] and a GEN (gap endonuclease) activity [27]. FEN1 plays a critical role in DNA repair, homologous recombination, telomere maintenance, RNA primer removal and resolution of stalled replication forks [27–30]. Although at present a potential relationship between FEN1 and RECQ1 has not been characterized [31], stimulation of FEN1 in the presence of BLM [32], WRN [33], RECQL4 [34] and RECQ5β [35] has been reported.
We speculate that hSuv3 might participate in genome maintenance via a RecQ helicase-like complex through interactions with members of the RecQ family and other proteins known to be key partners of the RecQ helicases. In the present study we present evidence that hSuv3 interacts with FEN1 and RPA, and examine the functional consequences of these interactions.
MATERIALS AND METHODS
Cell culture
HeLa cells were grown in DMEM (Dulbecco’s modified Eagle’s medium) supplemented with 10% FBS (fetal bovine serum), 50 units/ml of penicillin and 50 µg/ml of streptomycin in a humidified 5% CO2 incubator at 37°C. GM38 primary fibroblasts were grown in DMEM supplemented with 15% FBS, nonessential amino acids, essential amino acids, 50 units/ml penicillin and 50 µg/ml streptomycin in a humidified 5% CO2 incubator at 37°C.
hSuv3 plasmids and purification
Plasmid expressing His6-MBP-Δ46NhSuv3, a fusion of His-tagged MBP (maltose-binding protein) and amino acids 47–786 of hSuv3 was generated as previously described [13]. The above plasmid was modified by PCR-mediated C-terminal deletion with the primers: hSUV3-722Cdel-F2, 5′-TAGACCCAGCTTTCTTGTACAAAGTGGTG-3′, and hSUV3-722Cdel-R, 5′-CTCAGTAGCTTTACTCCCTAGAGCTTTGGT-3′, resulting in a construct pEXPHisMBP-hSuv3-(47–722)-WT (wild-type) encoding residues 47–722 of the hSuv3 with N-terminal His6-MBP tag. Furthermore, point mutation of the conserved Lys213 (in the Walker A motif)was introduced into construct pEXPHisMBP-hSuv3-(47–722)-WT with the primers: hSUV3-K213A-F, 5′-GCGACTTATCACGCAATCCAGAAATAC-3′ and hSUV-3-K213A-R, 5′-TCCACTGTTTGTGGGGCCTGAATG-3′, generating pE-XPHisMBP-hSuv3-(47–722)-K213A. Both plasmids, pEXP-HisMBP-hSuv3-(47–722)-WT and pEXPHisMBP-hSuv3-(47–722)-K213A, were verified by direct sequencing. Protein expression and purification was carried out as previously described [13], with the following modifications. Protein expression was induced with 0.2 mM IPTG (isopropyl β-d-thiogalactoside) for 18 h at 20°C. Metal affinity chromatography steps were performed on Ni2+ -charged HisTrap FF columns (GE Healthcare) equilibrated with Buffer A [50 mM Hepes NaOH, pH 7.5, 300 mM KCl, 10 mM 2-mercaptoethanol, 10% (v/v) glycerol and 25 mM imidazole] and developed with Buffer A supplemented with 250 mM imidazole. Size-exclusion chromatography was carried on a Superdex 200 16/60 column (GE Healthcare) equilibrated with Buffer A.
Recombinant proteins
The recombinant RPA was purified as a complex from Escherichia coli by methods described previously [36]. Recombinant mtSSB (mitochondrial single-strand-binding protein) was purchased from Abnova. Purified recombinant FEN1 was a gift from Dr David M. Wilson, III (National Institute of Aging, Baltimore, MD, U.S.A.).
Preparation of cell extracts
Whole cell extracts were prepared by incubating harvested HeLa cells with the CelLytic™ M reagent (Sigma) for 30 min at 4°C, followed by centrifugation at 18000 g for 20 min. The supernatant was used directly for further analysis. Nuclear extracts were prepared as described previously [37].
Antibodies
Polyclonal anti-hSuv3 antibodies were raised against the recombinant purified hSuv3-(47–722) in rabbits by Covance. The antibodies were purified from whole serum using an AminoLink Plus Immobilization Kit (Pierce) according to manufacturer instructions. The following antibodies were purchased: mouse monoclonal anti-RPA (Santa Cruz Biotechnology, sc-81372), mouse monoclonal anti-FEN1 (Abcam, Ab462), mouse monoclonal anti-Cdc47 (Thermo Scientific, MS862-P1), rabbit polyclonal anti-calregulin (Santa Cruz Biotechnology, sc11398) and rabbit polyclonal anti-VDAC (voltage-dependent anion channel; Abcam, ab15895).
Co-IP (co-immunoprecipitation)
The cell extracts were pre-cleared with recombinant Protein A–agarose beads for 2 h at 4°C. The pre-cleared extracts were incubated with 4 µg of antibody overnight at 4°C. Protein A–agarose beads were pre-blocked with 0.1 µg/ml BSA, before incubation with the lysate–antibody complexes, for 2 h at 4°C. The beads were washed three times in the indicated wash buffer: hSuv3-RPA, 50 mM Tris/HCl, pH 7.4, 150 mM NaCl and 0.01% Triton X-100; hSuv3-FEN1, 20 mM Hepes, pH 7.9, 20% glycerol, 300 mM KCl and 0.1% Tween 20. The final complexes were eluted by boiling the beads in reducing SDS/PAGE loading buffer for 5 min at 95°C. The immunoprecipitates were analysed by Western blotting. In the case of treatment, the nuclear extracts were incubated with 0.2 unit/(µg of protein) DNase I (Ambion) for 20 min at 37°C or 50 µg/ml EtBr (ethidium bromide) prior to immunoprecipitation.
Western blotting
Extracts were separated on an 8–16% Tris/glycine gradient SDS gel followed by transfer to a PVDF membrane. The membrane was blocked for 1 h at 37°C with TBST [TBS (Tris-buffered saline; 150 mM NaCl and 20 mM Tris/HCl, pH 7.4) with 0.1% Tween 20] containing 3% (w/v) non-fat dried skimmed milk powder, and incubated with the primary antibodies for 1 h at room temperature (25°C). The membrane was further incubated with HRP (horseradish peroxidase)-linked secondary antibodies (Sigma) and visualized using an ECL® Plus chemiluminescent detection kit.
Oligonucleotide substrates
PAGE-purified oligonucleotides (Integrated DNA Technologies) were used for preparation of substrates: D50, 5′-GGGACGCGTCGGCCTGGCACGTCGGCCGCTGCGGCCAGGCACCCGATGGC-3′; D49, 5′-TTTGTTTGTTTGTTTGTTTGTTTGCCGACGTGCCAGGCCGACGCGTCCC-3′; D50-2, 5′-GGGACGCGTCGGCCTGGCACGTCGGCTTTGTTTGTTTGTTTGTTTGTTTT-3′; D25F, 5′- CCGACGTGCCAGGCCGACGCGTCCC-3′; D25-2, 5′-GGGACGCGTCGGCCTGGCACGTCGG-3′; FLAP1, 5′-AGTAAAACGACGGCCAGTGC-3′; FLAP15, 5′-TTTTTTTTTTTCCAAGTAAAACGACGGCCAGTGC-3′; and FLAP template (T44), 5′-GCACTGGCCGTCGTTTTACGGTCGTGACTGGGAAAACCCTGGCG-3′; and FLAP upstream (F25): 5′-CGCCAGGGTTTTCCCAGTCACGACC-3′.
5′-End labelling was performed by incubating the oligonucleotide in the presence of [γ-32P]ATP and T4 polynucleotide kinase at 37°C for 1 h. For annealing, 5′-32P-labelled oligonucleotides were mixed with a 2-fold excess of the unlabelled complementary strand in 40 mM Tris/HCl, pH 8.0, and 50 mM NaCl and incubated at 70°C for 10 min, followed by slow-cooling to 30°C. Finally, the resulting substrates were purified using G-25 spin columns.
Helicase activity assay
The reactions contained 0.5 nM substrate and the indicated concentration of protein in 20 mM Hepes/NaOH, pH 7.5, 1 mM ATP, 3 mM MgCl2, 1 mM DTT (dithiothreitol) and 5% glycerol. Reactions for the interaction studies with RPA and mtSSB were performed in 20 mM Tris/HCl, pH 7.4, 1 mM ATP, 3 mM MgCl2, 1 mM DTT, 5% glycerol and 0.1 mg/ml BSA. The reactions were started by addition of the 5′-end-labelled substrate, incubated at 37°C for 30 min, terminated by addition of helicase stop buffer to a final concentration of 10 mM Tris/HCl, pH 8.0, 10 mM EDTA, 10% glycerol, 0.3% SDS, 0.01% Bromophenol Blue, 15 nM unlabelled oligonucleotide and 10 ng/µl proteinase K (New England Biolabs), and incubated for 10 min at 37°C. The products were resolved on a non-denaturing 8% polyacrylamide gel and detected using a PhosphorImager followed by analysis with ImageQuant software.
Incision activity assay
Reactions were prepared as described for the helicase activity assay in 20 mM Hepes/NaOH, pH 7.5, 1 mM ATP, 3 mM MgCl2, 1 mM DTT and 5% glycerol. After incubation with the substrate for 15 min at 37°C, the reactions were terminated by addition of form amide loading dye to a final concentration of 30% (v/v) form amide, 3.3 mM EDTA, 0.003% Bromophenol Blue. Samples were heated at 95°C for 5 min and loaded on to a 7 M urea, 15% polyacrylamide gel. Results were analysed as for the helicase activity assay.
RESULTS
Substrate specificity and directionality of hSuv3
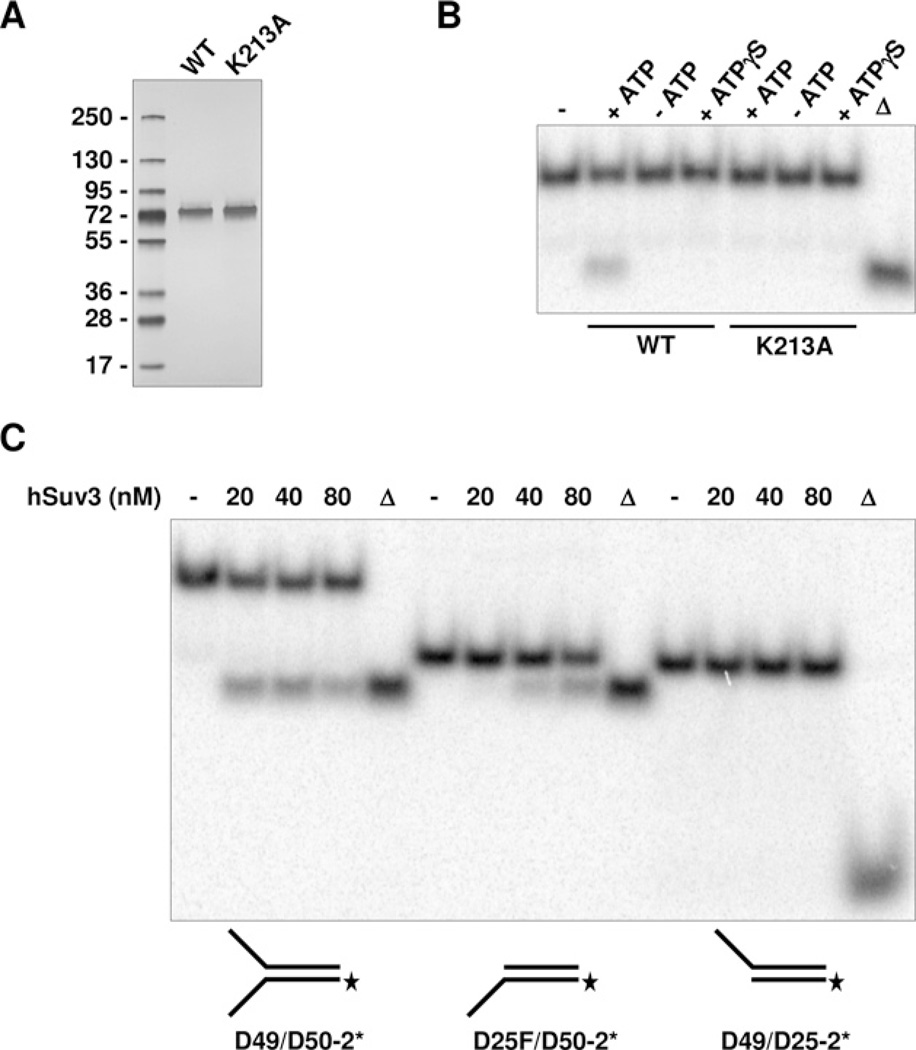
Initial biochemical characterization of hSuv3 helicase activity conducted by Shu et al. [5] revealed that the enzyme possesses 5′→3′ directionality. Although a slight increase in the product indicating the reverse 3′→5′ direction was shown, it was concluded that hSuv3 preferentially unwinds tailed substrates in the 5′→3′ direction. More recently, the same group reported an opposite finding demonstrating 3′→5′ directionality of hSuv3 [6]. In the second study the tailed substrates used were exclusively unwound in the 3′→5′ fashion with no evidence for activity in the reverse direction. In light of the reported discrepancy in directionality of hSuv3, we decided to analyse the helicase activity of hSuv3 on multiple DNA duplex substrates. To do so, we expressed and purified to apparent homogeneity hSuv3-(47–722)-WT and its corresponding hSuv3-(47–722)-K213A point mutant lacking ATPase activity (Figure 1A). To verify that the helicase activity was solely from hSuv3 and not from a co-purifying E. coli helicase, both the WT and the mutant protein were tested on a forked DNA substrate D49/D50-2*. The WT protein unwound DNA only in the presence of ATP and the activity was abolished when the energy co-factor was substituted by a non-hydrolysable analogue ATPγ S. No helicase activity was observed when the hSuv3 (47–722)-K213A point mutant was used (Figure 1B). To test the directionality of hSuv3 helicase we used the D49/D50-2* forked substrate with a 26 bp duplex region and 24–23 nt single-stranded arms, and corresponding tailed substrates in which one of the single-stranded regions was removed. We found that hSuv3 translocates in the 3′→5′ direction during unwinding of DNA duplexes, as demonstrated by strand separation of the 3′-tailed D25F/D50-2* substrate (Figure 1C). No helicase activity was observed with the 5′-tailed D49*/D25-2 substrate. Additionally, our results demonstrate that the enzyme preferentially unwind the forked DNA/DNA duplex (D49/D50-2*) over the corresponding tailed substrate (D25F/D50-2) (Figure 1C).
Figure 1. Substrate specificity and directionality of hSuv3.
(A) SDS/PAGE of purified hSuv3-(47–722)-WT and -K213A mutant, 500 ng per lane, 4–15% gel, silver stained. (B) Helicase assay performed as described in the Materials and methods section with D49/D50-2* forked substrate. WT and K213A hSuv3-(47–722) were used at 20 nM concentration. Energy cofactors used are indicated. Triangle denotes heat-denatured substrate. (C) Helicase assays with indicated DNA substrates. hSuv3-(47–722)-WT was used at the indicated concentrations.
Physical interaction between hSuv3 and RPA in vivo
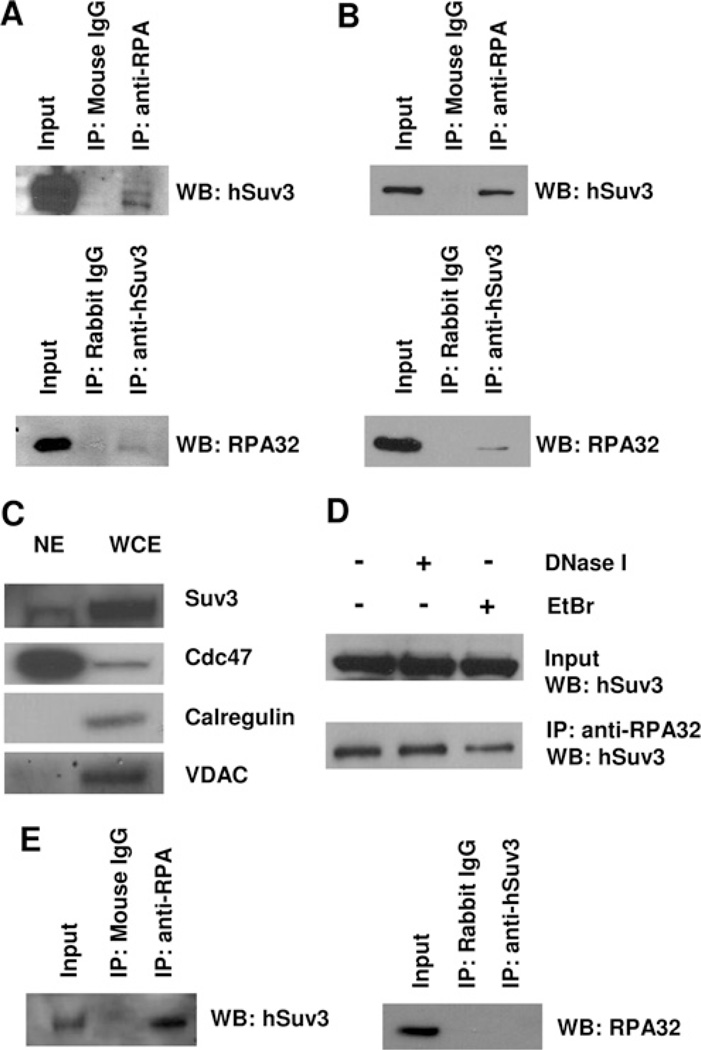
Studies have shown that Suv3 interacts with several RecQ helicases, WRN and BLM in human cells [8] and Sgs1 in yeast [17]. In order to explore the possibility of the involvement of hSuv3 in DNA metabolism through a RecQ helicase-like complex, we decided to search for interactions between hSuv3 and well-documented protein-binding partners of RecQ family members. To investigate a potential association of endogenous hSuv3 and RecQ helicase partners, we performed co-IP using HeLa cell lysates. RPA was one of the proteins we were able to detect by Western blotting in the anti-hSuv3 immunoprecipitates (Figure 2A). The result was further supported by the reciprocal experiment, in which hSuv3 was immunoprecipitated by the anti-RPA antibody. As controls, normal IgGs (from mouse or rabbit) were used under the same assay conditions, and neither hSuv3 nor RPA were detected.
Figure 2. Interaction of RPA with hSuv3.
(A) Interaction of RPA with hSuv3 in HeLa whole cell extract. Total cell lysates were used for co-IP assays with anti-RPA, anti-hSuv3 or normal IgG antibody. (B) RPA and hSuv3 interact in the nuclear fraction. HeLa nuclear extracts (1 mg) were used for co-IP as in (A). (C) Western blot analysis (WB) of nuclear extract (NE). The nuclear extract used in (B) and whole cell extract (WCE) were analysed for the presence of subcellular markers calregulin (cytoplasm), VDAC (mitochondria) and Cdc47 (nucleus). (D) The hSuv3–RPA interaction is independent of DNA. HeLa nuclear extracts (1 mg) were either untreated or treated with 0.2 unit/(µg of protein) DNase I for 20 min at 37°C or 50 µg/ml EtBr for 30 min on ice, before co-IP as in (A). (E) RPA and hSuv3 interact in the nuclear fraction of GM38 primary human fibroblasts. Nuclear extracts were used for co-IP as in (B).
To date, RPA has been described as an exclusively nuclear protein [38], whereas hSuv3 localizes to both the mitochondrial matrix [10] and the nucleus [13]. On this basis, we hypothesized that it is the nuclear fraction of hSuv3 that is binding RPA. To test this, we repeated the co-IP experiment using HeLa nuclear extract (Figure 2B). Indeed, the result was confirmed and an even stronger interaction was seen in the nuclear fraction. The quality of the nuclear extract was analysed using subcellular markers (Figure 2C). The nuclear extract was negative for calregulin and VDAC, used as cytoplasmic and mitochondrial markers, respectively, while enriched for the nuclear protein Cdc47.
Since both hSuv3 and RPA are able to bind DNA, we wanted to rule out the possibility that the observed interaction was mediated through DNA. Pretreatment of the nuclear extract with either DNase I or EtBr did not decrease the signal compared with the non-treated control, supporting the notion that the interaction is protein–protein-mediated (Figure 2D).
To investigate if the finding could be extended to non-transformed cells, additional nuclear extracts were prepared from GM38 primary fibroblasts and used for co-IP. Again, precipitation of Suv3 with RPA was achieved (Figure 2E). The reciprocal co-IP was unsuccessful. Still, these results support the notion that the interaction may have a biological relevance in normal cell function.
RPA inhibits hSuv3 helicase activity
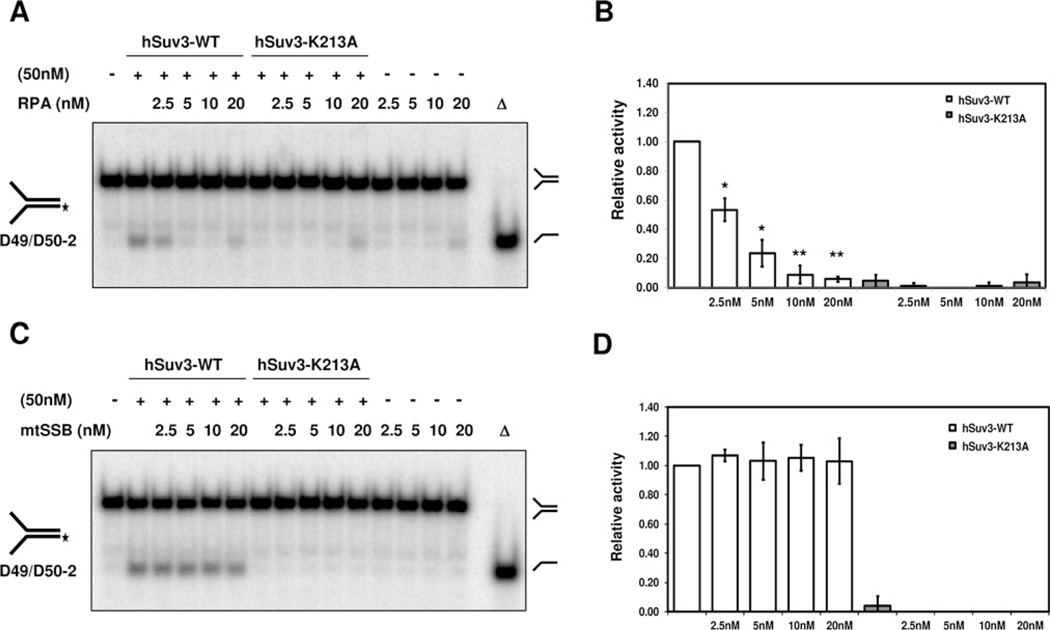
Previous studies have shown that RPA can stimulate DNA-unwinding activity of several RecQ helicases, including RECQ1 [20], BLM [21], WRN [22], RECQL4 [23] and RECQ5β [24]. In light of the observed interaction, we considered that the same could be the case for hSuv3. To test potential functional consequence of the physical interaction between hSuv3 and RPA, we performed the helicase activity assay on a forked duplex substrate in the presence of hSuv3 and increasing concentrations of RPA. The presence of RPA led to inhibition of the unwinding of a 50 bp forked substrate by hSuv3 (Figures 3A and 3B). The highest amounts of RPA alone resulted in a small amount of product as well, which can be explained by the double-strand destabilization by RPA due to its inherent polarity binding [19].
Figure 3. Inhibition of hSuv3 helicase activity by RPA.
(A) Unwinding activity of hSuv3 on a forked substrate in the presence of RPA. Purified recombinant proteins were mixed (50 nM hSuv3 and RPA as indicated) and the reaction was started by addition of the radioactively labelled DNA substrate (0.5 nM), followed by incubation at 37°C for 30 min. (B) Quantification from (A). hSuv3 helicase activity in the presence of RPA relative to helicase activity by hSuv3 alone. Products from RPA alone were subtracted from helicase activity values. Error bars represent S.D. (mean value for three experiments). *P < 0.001 in Student’s t test for values different from hSuv3 alone; **P < 0.0001. (C) Unwinding activity of hSuv3 in the presence of mtSSB as in (A). (D) Quantification of fold stimulation from (C). Error bars represent S.D. (mean value for three experiments).
To determine whether this functional effect was specific to RPA, the human mtSSB was used in a parallel hSuv3 unwinding assay. The mtSSB had no significant effect on the helicase activity of hSuv3 on the forked substrate at all concentrations tested (Figures 3C and 3D). The capability of both RPA and mtSSB to bind the substrate was verified by EMSA (electrophoretic mobility-shift assay) (results not shown). Consequently, the lack of effect by mtSSB supports the idea that a specific protein–protein interaction between hSuv3 and RPA is responsible for the decreased unwinding of the DNA substrate.
Physical interaction between hSuv3 and FEN1 in vivo
To learn more about a potential hSuv3 multiprotein complex, we examined the co-immunoprecipitate from HeLa nuclear extracts for other robust RecQ helicase partners.
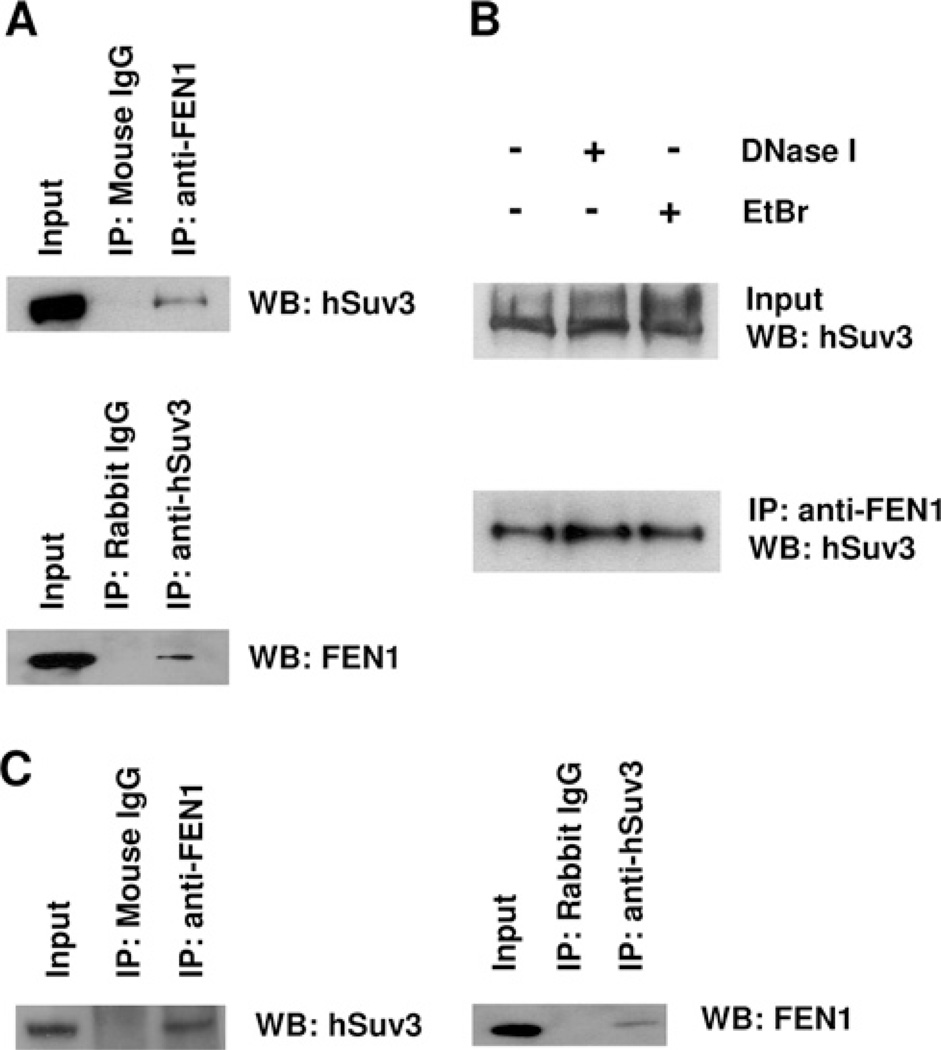
The FEN1 protein was detected in the anti-hSuv3 immunoprecipitates from nuclear extracts (Figure 4A). Again, neither normal mouse nor rabbit IgG were able to co-immunoprecipitate either one of the above proteins.
Figure 4. Interaction of FEN1 with hSuv3.
(A) Interaction of FEN1 with hSuv3 in the nuclear fraction of HeLa cells. HeLa nuclear extracts were used for co-IP assays with anti-FEN1, anti-hSuv3 or normal IgG antibody. (B) The hSuv3-FEN1 interaction is independent of DNA. Nuclear extracts were either untreated or treated with DNase I or EtBr, before co-IP as in (A). (C) FEN1 and hSuv3 interact in the nuclear fraction of GM38 primary human fibroblasts. Nuclear extracts were used for co-IP as in (A). WB, Western blot.
Also, we could demonstrate the presence of hSuv3 protein in the anti-FEN1 immunoprecipitate. Together these results support the notion that nuclear hSuv3 interacts with FEN1. The interaction was not affected by the presence of either EtBr or DNase I, demonstrating that it is not DNA-mediated, but rather a direct protein–protein interaction (Figure 4B). These results were also reproduced using GM38 primary fibroblasts (Figure 4C), again supporting a biological role of the interaction.
Stimulation of flap incision in the presence of hSuv3
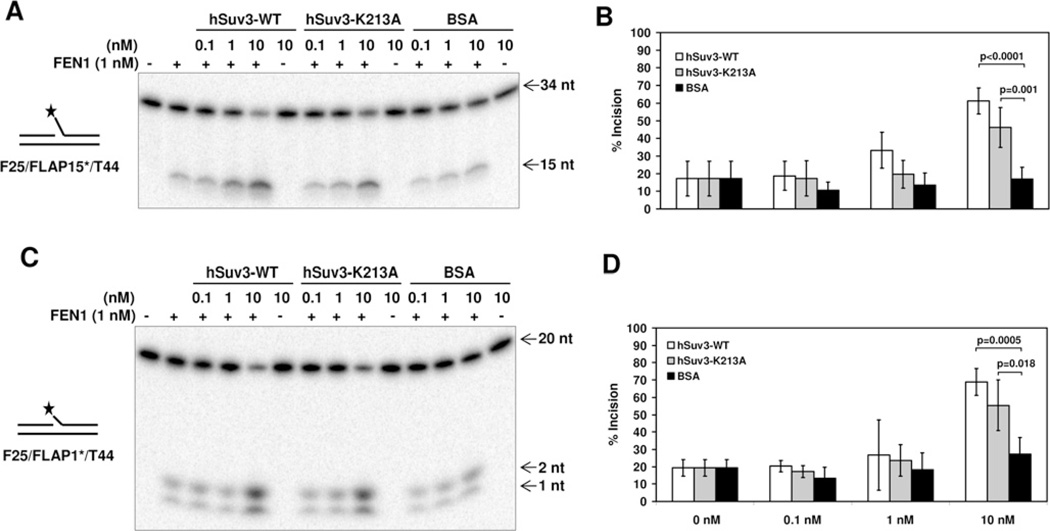
Several of the RecQ helicases, including BLM [32], WRN [33], RECQL4 [34] and RECQ5β [35], have been found to stimulate the 5′-flap incision of FEN1. Hence, based on the physical interaction observed between hSuv3 and FEN1, we tested whether the interaction could modulate the catalytic activity of one or both of the proteins. In order to characterize the effect of hSuv3 on the rate of FEN1 incision, we performed an incision assay utilizing a 44 bp DNA substrate with a 15 nucleotide flap, adjacent to an upstream 19 bp duplex. In the presence of FEN1 alone, 17% of the substrate was incised. When adding increasing amounts of hSuv3-WT to the reaction, an approx. 3-fold stimulation was observed using a 10-fold excess of hSuv3-WT compared with the effect of BSA (Figure 5A). hSuv3-K213A did also cause significant stimulation at similar conditions, although the effect was less pronounced compared with hSuv3-WT.
Figure 5. Stimulation of FEN1 incision activity by hSuv3.
(A) FEN1 incision on a long (15 nt) flap in the presence of hSuv3-WT, hSuv3-K213A or BSA. Purified recombinant proteins were mixed (1 nM FEN1) and the reaction started by addition of the radioactively labelled DNA substrate (0.5 nM), followed by incubation at 37°C for 15 min. (B) Quantification of fold stimulation from (A). Error bars represent S.D. (mean value for five experiments). (C) FEN1 incision on a short (1 nt) flap in the presence of hSuv3-WT, hSuv3-K213A or BSA. (D) Quantification of fold stimulation from (C). Error bars represent S.D. (mean value for four experiments). P values were determined using Student’s t test.
Considering that the flap structure could be important for the stimulatory effect, we performed the assay on a similar substrate with a shorter 1 nucleotide flap (Figures 5C and 5D). Again, 2–3-fold stimulation was achieved by adding the hSuv3 proteins, indicating that hSuv3 stimulates FEN1 incision independently of flap length. Importantly, hSuv3 alone did not catalyse any significant cleavage of the 5′-flap DNA substrates.
DISCUSSION
The importance of the mitochondrial function of conserved Suv3 helicase has been demonstrated in several organisms, ranging from the protozoan Trypanosoma brucei to S. cerevisiae and mammals [2,12,39]. However, evidence of the nuclear role is limited. Interactions with the RecQ helicases demonstrated in S. cerevisiae (Sgs1) [17], and in humans (WRN and BLM) [8] may provide clues to the elucidation of Suv3′s nuclear function. Deficiencies in either BLM or WRN are associated with premature aging, sharing striking similarity to the phenotype caused by disruption of Suv3 in mice that includes sarcopenia, kyphosis, skin defects and premature death [9,18,40,41]. However, only BLM deficiency is associated with elevated SCE, also seen in the hSuv3 knockdown [8,42]. It was previously suggested that hSuv3 could work together with WRN and BLM in resolving replication fork crises. This could explain the elevated SCE levels in cells with knockdown of hSuv3, possibly as a result of alternative repair by illegitimate recombination [8]. Identification of interacting protein partners is crucial to a further characterization of the potential function of hSuv3 in DNA metabolism. The present paper describes, for the first time, non-mitochondrial interactions of hSuv3 in an in vivo experimental setup. Previous data have been based either on yeast two-hybrid systems [43] or in vitro interactions [8].
In the present study, we find that hSuv3 directly interacts with the nuclear protein RPA. RPA is considered to be the primary eukaryotic ssDNA binding protein, serving functional roles in replication, recombination and repair [38]. Hence, the results suggest that hSuv3 cooperates with RPA in one of these essential processes. Interestingly, RPA inhibits hSuv3 unwinding of a forked substrate. This differs from the results previously obtained with RecQ helicases [20–22,24], where RPA was shown to stimulate DNA unwinding activity in a dose-dependent manner. Another single-stranded-binding protein, mtSSB, does not cause the same effect. Therefore the effect of hSuv3′s helicase activity by RPA cannot be ascribed to the mere presence of ssDNA binding protein on the exposed single strands blocking hSuv3 entry.
One possible explanation for the decrease in hSuv3 helicase activity caused by RPA could be that post-translational modifications of one or both proteins, dictated by a specific cellular event, might be needed to stabilize and cause stimulation. Alternatively, the role of the hSuv3–RPA interaction might be independent of helicase activity, and rather be limited to coordinate loading of other proteins facilitating a specific functional outcome. As such, the BLM helicase promotes homologous recombination between diverged sequences through a novel ATPase-independent mechanism [44].
In S. cerevisiae ScSuv3p was found to interact with Ddc1 and MEC3 (mucosae-associated epithelial chemokine 3) [17]. Ddc1 and MEC3 are DNA-damage checkpoint proteins recruited to unrepaired recombination intermediates, and are homologues of human Rad9 and Hus1 of the 9-1-1 checkpoint complex (Saccharomyces Genome Database: http://www.yeastgenome.org). The 9-1-1 complex is a multifunctional protein complex, loaded on to DNA in response to various types of genotoxic stresses, which participates in checkpoint signalling at stalled replication forks [45]. RPA is known to bind to the Rad9 subunit, hence the three proteins in complex could be involved in the resolving of blocked replication forks.
Our present investigation also uncovered an interaction between hSuv3 and FEN1 in the nuclear fraction of human cells. hSuv3 could potentially modulate the activity of FEN1 in vivo in one of its many functions in DNA repair, telomere maintenance, Okazaki fragment maturation or resolving stalled replication forks [46,47]. In this case, hSuv3 was shown to stimulate the incision activity of FEN1, independently of flap length and only in part on helicase activity. The K213A amino acid substitution is located in a Walker A consensus motif in which the lysine residue is believed to be important for ATP binding [48]. Therefore hSuv3-K213A would be expected to lack any conformational change associated with ATP binding, which could explain the difference between the effects of hSuv3-WT and hSuv3-K213A on FEN1 incision. On the other hand, the stimulatory effect of hSuv3-K213A might still be sufficient to fulfil the nuclear role of hSuv3. A potential ATP-independent role of hSuv3 could explain why expression of a nuclear dominant-negative version of hSuv3 did not affect the morphology or growth rate of the cell [15].
Helicase-independent stimulation of FEN1 activity has also been shown for WRN, BLM, RecQ4 and RecQ5 [32–35]. The interaction between WRN and FEN1 has previously been proposed to be involved in initiation of recombination pathways at stalled replication forks by employing FEN1’s GEN activity [27]. The FEN1 GEN activity was also stimulated in the presence of pRPA (phosphorylated RPA). Replication fork arrest is one event that has been reported to lead to phosphorylation of RPA [49], and consequently it has been speculated that FEN1 may coordinate with RPA in a protein complex located at the replication fork [47]. Our findings could suggest the involvement of hSuv3 in this putative complex. Meanwhile, high amounts of RPA were found to inhibit FEN1 flap cleavage, an effect believed to be important in regulation of endonuclease switching in Okazaki fragment maturation [50]. This could implicate hSuv3 in proper lagging-strand synthesis during Okazaki fragment processing.
To summarize, we present evidence of physical and functional interactions of hSuv3 with the RPA and FEN1 proteins. These findings support our hypothesis that hSuv3 participates in a multiprotein complex involved in nuclear DNA metabolism, making hSuv3 a potential new player in the field of genome maintenance.
ACKNOWLEDGEMENTS
We thank David M. Wilson, III for providing purified FEN1, Lale Dawut and Deborah L. Croteau for technical assistance, and Robert Jedrzejczak, Scott Maynard and Martin Borch Jensen for critical reading of the paper prior to submission.
FUNDING
This work was supported by the Intramural Research Program of the National Institutes of Health, National Institute on Aging [grant number Z01 AG000733-15], The Danish Research Council [grant number 271-08-0697] and the Polish National Centre for Research and Development [grant number NR13004704].
Abbreviations used
- BLM
Bloom’s syndrome protein
- Cdc47
cell division cycle 47
- co-IP
co-immunoprecipitation
- DMEM
Dulbecco’s modified Eagle’s medium
- DTT
dithiothreitol
- EtBr
ethidium bromide
- FBS
fetal bovine serum
- FEN1
flap endonuclease 1
- GEN
gap endonuclease
- hSuv3
human Suv3
- MBP
maltose-binding protein
- MEC3
mucosae-associated epithelial chemokine 3
- mtSSB
mitochondrial single strand binding protein
- PNPase
polynucleotide phosphorylase
- RPA
replication protein A
- SCE
sister chromatid exchange
- ssDNA
single-stranded DNA
- ScSuv3p
Saccharomyces cerevisiae Suv3
- VDAC
voltage-dependent anion channel
- WRN
Werner’s syndrome protein
- WT
wild-type
Footnotes
AUTHOR CONTRIBUTION
Susanne Venø performed most of the co-IP, Western blotting and biochemical work. Tomasz Kulikowicz purified the recombinant hSuv3 protein and performed biochemical work. Cezar Pestana performed co-IP. Susanne Venø and Tomasz Kulikowicz prepared the Figures and wrote the paper. Vilhelm Bohr, Tinna Stevnsner and Piotr Stepien designed and supervised the research and wrote the manuscript. All of the authors contributed to and approved the final manuscript.
REFERENCES
- 1.Conrad-Webb H, Perlman PS, Zhu H, Butow RA. The nuclear SUV3-1 mutation affects a variety of post-transcriptional processes in yeast mitochondria. Nucleic Acids Res. 1990;18:1369–1376. doi: 10.1093/nar/18.6.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dziembowski A, Piwowarski J, Hoser R, Minczuk M, Dmochowska A, Siep M, van der Spek H, Grivell L, Stepien PP. The yeast mitochondrial degradosome. Its composition, interplay between RNA helicase and RNase activities and the role in mitochondrial RNA metabolism. J. Biol. Chem. 2003;278:1603–1611. doi: 10.1074/jbc.M208287200. [DOI] [PubMed] [Google Scholar]
- 3.Dziembowski A, Malewicz M, Minczuk M, Golik P, Dmochowska A, Stepien PP. The yeast nuclear gene DSS1, which codes for a putative RNase II, is necessary for the function of the mitochondrial degradosome in processing and turnover of RNA. Mol. Gen. Genet. 1998;260:108–114. doi: 10.1007/s004380050876. [DOI] [PubMed] [Google Scholar]
- 4.Dmochowska A, Kalita K, Krawczyk M, Golik P, Mroczek K, Lazowska J, Stepien PP, Bartnik E. A human putative Suv3-like RNA helicase is conserved between Rhodobacter and all eukaryotes. Acta Biochim. Pol. 1999;46:155–162. [PubMed] [Google Scholar]
- 5.Shu Z, Vijayakumar S, Chen CF, Chen PL, Lee WH. Purified human SUV3p exhibits multiple-substrate unwinding activity upon conformational change. Biochemistry. 2004;43:4781–4790. doi: 10.1021/bi0356449. [DOI] [PubMed] [Google Scholar]
- 6.Wang DD, Shu Z, Lieser SA, Chen PL, Lee WH. Human mitochondrial SUV3 and polynucleotide phosphorylase form a 330-kDa heteropentamer to cooperatively degrade double-stranded RNA with a 3′-to-5′ directionality. J. Biol. Chem. 2009;284:20812–20821. doi: 10.1074/jbc.M109.009605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paul E, Kielbasinski M, Sedivy JM, Murga-Zamalloa C, Khanna H, Klysik JE. Widespread expression of the Supv3L1 mitochondrial RNA helicase in the mouse. Transgenic Res. 2010;19:691–701. doi: 10.1007/s11248-009-9346-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pereira M, Mason P, Szczesny RJ, Maddukuri L, Dziwura S, Jedrzejczak R, Paul E, Wojcik A, Dybczynska L, Tudek B, et al. Interaction of human SUV3 RNA/DNA helicase with BLM helicase; loss of the SUV3 gene results in mouse embryonic lethality. Mech. Ageing Dev. 2007;128:609–617. doi: 10.1016/j.mad.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 9.Paul E, Cronan R, Weston PJ, Boekelheide K, Sedivy JM, Lee SY, Wiest DL, Resnick MB, Klysik JE. Disruption of Supv3L1 damages the skin and causes sarcopenia, loss of fat, death. Mamm. Genome. 2009;20:92–108. doi: 10.1007/s00335-008-9168-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Minczuk M, Piwowarski J, Papworth MA, Awiszus K, Schalinski S, Dziembowski A, Dmochowska A, Bartnik E, Tokatlidis K, Stepien PP, et al. Localisation of the human hSuv3p helicase in the mitochondrial matrix and its preferential unwinding of dsDNA. Nucleic Acids Res. 2002;30:5074–5086. doi: 10.1093/nar/gkf647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bogenhagen DF, Rousseau D, Burke S. The layered structure of human mitochondrial DNA nucleoids. J. Biol. Chem. 2008;283:3665–3675. doi: 10.1074/jbc.M708444200. [DOI] [PubMed] [Google Scholar]
- 12.Khidr L, Wu G, Davila A, Procaccio V, Wallace D, Lee WH. Role of SUV3 helicase in maintaining mitochondrial homeostasis in human cells. J. Biol. Chem. 2008;283:27064–27073. doi: 10.1074/jbc.M802991200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szczesny RJ, Obriot H, Paczkowska A, Jedrzejczak R, Dmochowska A, Bartnik E, Formstecher P, Polakowska R, Stepien PP. Down-regulation of human RNA/DNA helicase SUV3 induces apoptosis by a caspase- and AIF-dependent pathway. Biol. Cell. 2007;99:323–332. doi: 10.1042/BC20060108. [DOI] [PubMed] [Google Scholar]
- 14.Borowski LS, Szczesny RJ, Brzezniak LK, Stepien PP. RNA turnover in human mitochondria: more questions than answers? Biochim. Biophys. Acta. 2010;1797:1066–1070. doi: 10.1016/j.bbabio.2010.01.028. [DOI] [PubMed] [Google Scholar]
- 15.Szczesny RJ, Borowski LS, Brzezniak LK, Dmochowska A, Gewartowski K, Bartnik E, Stepien PP. Human mitochondrial RNA turnover caught in flagranti: involvement of hSuv3p helicase in RNA surveillance. Nucleic Acids Res. 38:279–298. doi: 10.1093/nar/gkp903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bobrowicz AJ, Lightowlers RN, Chrzanowska-Lightowlers Z. Polyadenylation and degradation of mRNA in mammalian mitochondria: a missing link? Biochem. Soc. Trans. 2008;36:517–519. doi: 10.1042/BST0360517. [DOI] [PubMed] [Google Scholar]
- 17.Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 18.Bohr VA. Rising from the RecQ-age: the role of human RecQ helicases in genome maintenance. Trends Biochem. Sci. 2008;33:609–620. doi: 10.1016/j.tibs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wold MS. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 20.Cui S, Arosio D, Doherty KM, Brosh RM, Jr, Falaschi A, Vindigni A. Analysis of the unwinding activity of the dimeric RECQ1 helicase in the presence of human replication protein A. Nucleic Acids Res. 2004;32:2158–2170. doi: 10.1093/nar/gkh540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brosh RM, Jr, Li JL, Kenny MK, Karow JK, Cooper MP, Kureekattil RP, Hickson ID, Bohr VA. Replication protein A physically interacts with the Bloom’s syndrome protein and stimulates its helicase activity. J. Biol. Chem. 2000;275:23500–23508. doi: 10.1074/jbc.M001557200. [DOI] [PubMed] [Google Scholar]
- 22.Brosh RM, Jr, Orren DK, Nehlin JO, Ravn PH, Kenny MK, Machwe A, Bohr VA. Functional and physical interaction between WRN helicase and human replication protein A. J. Biol. Chem. 1999;274:18341–18350. doi: 10.1074/jbc.274.26.18341. [DOI] [PubMed] [Google Scholar]
- 23.Rossi ML, Ghosh AK, Kulikowicz T, Croteau DL, Bohr VA. Conserved helicase domain of human RecQ4 is required for strand annealing-independent DNA unwinding. DNA Repair. 2010;9:796–804. doi: 10.1016/j.dnarep.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia PL, Liu Y, Jiricny J, West SC, Janscak P. Human RECQ5 β, a protein with DNA helicase and strand-annealing activities in a single polypeptide. EMBO J. 2004;23:2882–2891. doi: 10.1038/sj.emboj.7600301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zou Y, Liu Y, Wu X, Shell SM. Functions of human replication protein A (RPA): from DNA replication to DNA damage and stress responses. J. Cell Physiol. 2006;208:267–273. doi: 10.1002/jcp.20622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harrington JJ, Lieber MR. The characterization of a mammalian DNA structure-specific endonuclease. EMBO J. 1994;13:1235–1246. doi: 10.1002/j.1460-2075.1994.tb06373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng L, Zhou M, Chai Q, Parrish J, Xue D, Patrick SM, Turchi JJ, Yannone SM, Chen D, Shen B. Novel function of the flap endonuclease 1 complex in processing stalled DNA replication forks. EMBO Rep. 2005;6:83–89. doi: 10.1038/sj.embor.7400313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen B, Singh P, Liu R, Qiu J, Zheng L, Finger LD, Alas S. Multiple but dissectible functions of FEN-1 nucleases in nucleic acid processing, genome stability and diseases. BioEssays. 2005;27:717–729. doi: 10.1002/bies.20255. [DOI] [PubMed] [Google Scholar]
- 29.Bambara RA, Murante RS, Henricksen LA. Enzymes and reactions at the eukaryotic DNA replication fork. J. Biol. Chem. 1997;272:4647–4650. doi: 10.1074/jbc.272.8.4647. [DOI] [PubMed] [Google Scholar]
- 30.Saharia A, Guittat L, Crocker S, Lim A, Steffen M, Kulkarni S, Stewart SA. Flap endonuclease 1 contributes to telomere stability. Curr. Biol. 2008;18:496–500. doi: 10.1016/j.cub.2008.02.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu Y, Brosh RM., Jr Distinct roles of RECQ1 in the maintenance of genomic stability. DNA Repair. 2010;9:315–324. doi: 10.1016/j.dnarep.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharma S, Sommers JA, Wu L, Bohr VA, Hickson ID, Brosh RM., Jr Stimulation of flap endonuclease-1 by the Bloom’s syndrome protein. J. Biol. Chem. 2004;279:9847–9856. doi: 10.1074/jbc.M309898200. [DOI] [PubMed] [Google Scholar]
- 33.Brosh RM, Jr, von Kobbe C, Sommers JA, Karmakar P, Opresko PL, Piotrowski J, Dianova I, Dianov GL, Bohr VA. Werner syndrome protein interacts with human flap endonuclease 1 and stimulates its cleavage activity. EMBO J. 2001;20:5791–5801. doi: 10.1093/emboj/20.20.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schurman SH, Hedayati M, Wang Z, Singh DK, Speina E, Zhang Y, Becker K, Macris M, Sung P, Wilson DM, III, et al. Direct and indirect roles of RECQL4 in modulating base excision repair capacity. Hum. Mol. Genet. 2009;18:3470–3483. doi: 10.1093/hmg/ddp291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Speina E, Dawut L, Hedayati M, Wang Z, May A, Schwendener S, Janscak P, Croteau DL, Bohr VA. Human RECQL5β stimulates flap endonuclease 1. Nucleic Acids Res. 2010;38:2904–2916. doi: 10.1093/nar/gkp1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Binz SK, Dickson AM, Haring SJ, Wold MS. Functional assays for replication protein A (RPA) Methods Enzymol. 2006;409:11–38. doi: 10.1016/S0076-6879(05)09002-6. [DOI] [PubMed] [Google Scholar]
- 37.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iftode C, Daniely Y, Borowiec JA. Replication protein A (RPA): the eukaryotic SSB. Crit. Rev. Biochem. Mol. Biol. 1999;34:141–180. doi: 10.1080/10409239991209255. [DOI] [PubMed] [Google Scholar]
- 39.Mattiacio JL, Read LK. Evidence for a degradosome-like complex in the mitochondria of Trypanosoma brucei. FEBS Lett. 2009;583:2333–2338. doi: 10.1016/j.febslet.2009.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ellis NA, Lennon DJ, Proytcheva M, Alhadeff B, Henderson EE, German J. Somatic intragenic recombination within the mutated locus BLM can correct the high sister-chromatid exchange phenotype of Bloom syndrome cells. Am. J. Hum. Genet. 1995;57:1019–1027. [PMC free article] [PubMed] [Google Scholar]
- 41.Yu CE, Oshima J, Hisama FM, Matthews S, Trask BJ, Schellenberg GD. A YAC, P1, and cosmid contig and 17 new polymorphic markers for the Werner syndrome region at 8p12–p21. Genomics. 1996;35:431–440. doi: 10.1006/geno.1996.0382. [DOI] [PubMed] [Google Scholar]
- 42.German J. Bloom syndrome: a Mendelian prototype of somatic mutational disease. Medicine. 1993;72:393–406. [PubMed] [Google Scholar]
- 43.Minczuk M, Mroczek S, Pawlak SD, Stepien PP. Human ATP-dependent RNA/DNA helicase hSuv3p interacts with the cofactor of survivin HBXIP. FEBS J. 2005;272:5008–5019. doi: 10.1111/j.1742-4658.2005.04910.x. [DOI] [PubMed] [Google Scholar]
- 44.Kikuchi K, Abdel-Aziz HI, Taniguchi Y, Yamazoe M, Takeda S, Hirota K. Bloom DNA helicase facilitates homologous recombination between diverged homologous sequences. J. Biol. Chem. 2009;284:26360–26367. doi: 10.1074/jbc.M109.029348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parrilla-Castellar ER, Arlander SJ, Karnitz L. Dial 9-1-1 for DNA damage: the Rad9-Hus1-Rad1 (9-1-1) clamp complex. DNA Repair. 2004;3:1009–1014. doi: 10.1016/j.dnarep.2004.03.032. [DOI] [PubMed] [Google Scholar]
- 46.Sampathi S, Bhusari A, Shen B, Chai W. Human flap endonuclease I is in complex with telomerase and is required for telomerase-mediated telomere maintenance. J. Biol. Chem. 2009;284:3682–3690. doi: 10.1074/jbc.M805362200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu Y, Kao HI, Bambara RA. Flap endonuclease 1: a central component of DNA metabolism. Annu. Rev. Biochem. 2004;73:589–615. doi: 10.1146/annurev.biochem.73.012803.092453. [DOI] [PubMed] [Google Scholar]
- 48.Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shao RG, Cao CX, Zhang H, Kohn KW, Wold MS, Pommier Y. Replication-mediated DNA damage by camptothecin induces phosphorylation of RPA by DNA-dependent protein kinase and dissociates RPA:DNA-PK complexes. EMBO J. 1999;18:1397–1406. doi: 10.1093/emboj/18.5.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bae SH, Bae KH, Kim JA, Seo YS. RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature. 2001;412:456–461. doi: 10.1038/35086609. [DOI] [PubMed] [Google Scholar]