Abstract
We conducted a meta-analysis to assess the association between patatin-like phospholipase domain-containing 3 (PNPLA3) rs738409 polymorphism and nonalcoholic fatty liver disease (NAFLD) and its subtypes simple steatosis(SS) and nonalcoholic steatohepatitis (NASH). The study-specific odds ratios (ORs) and 95% confidence intervals (CIs) were calculated using fixed-effects or random-effects models, with assessment for heterogeneity and publication bias. Twenty-three case-control studies involving 6071 NAFLD patients and 10366 controls were identified. The combined results showed a significant association between NAFLD risk and the rs738409 polymorphism in all genetic models (additive model: OR = 3.41, 95% CI = 2.57–4.52; P < 0.00001). In addition, evidence indicated that the rs738409 polymorphism was significantly associated with NASH in all genetic models (additive model: OR = 4.44, 95% CI = 3.39–5.82; P < 0.00001). The subgroup and sensitivity analyses showed that these changes were not influenced by the ethnicities and ages of subjects or by the source of controls. The rs738409 polymorphism was only significantly associated with risk of simple steatosis in the allele contrast and had no effect in the other genetic models. These findings suggest that the rs738409 polymorphism in PNPLA3 gene confers high cross-ethnicity risk for NAFLD and NASH development.
Nonalcoholic fatty liver disease (NAFLD) is the most common cause of liver disease in western countries, affecting up to 20–35% of the general population1, and has emerged as a major public health issue worldwide2,3. NAFLD has a broad spectrum of manifestations and can be histologically subdivided into simple steatosis and non-alcoholic steatohepatitis (NASH), which include steatosis, lobular inflammation and hepatocyte ballooning with or without fibrosis4. Although simple steatosis is generally considered to have a benign hepatological prognosis, NASH much more frequently progresses to fibrosis, cirrhosis and hepatocellular carcinoma in later years5,6 and will be the leading cause of liver transplantation in the United States by 20207. The precise mechanism responsible for the development and progression of NAFLD has not been elucidated. Some NAFLD patients will progress into NASH with cirrhosis, whereas others do not develop beyond simple steatosis. Currently, there is increasing evidence that genetic8,9,10 as well as environmental factors11 play important roles in the progression of NAFLD.
The human patatin-like phospholipase-3 (PNPLA3) gene is localized on human chromosome 22. The PNPLA3 protein, which is also known as adiponutrin, is expressed in both adipocytes and hepatocytes12. PNPLA3 exhibits lipase activity against triglycerides and acylglycerol transacetylase activity, and its expression is highly responsive in energy mobilization and the storage of lipid droplets13. The PNPLA3 gene is one of the potential candidate genes currently related to NAFLD susceptibility. In 1998, Romeo et al. noted that a single nucleotide polymorphism in residue 148 (I148 M, rs738409), which exhibits a C-to-G transition resulting in an amino acid substitution of isoleucine to methionine, was a strong genetic determinant of NAFLD10. Consistent with this result, some following studies also demonstrated an association between the rs738409 polymorphism and NAFLD risk14,15,16. However, it is unclear whether this polymorphism is associated with simple steatosis only or also associated with NASH. Further studies have also attempted to analyze the association between the rs738409 polymorphism and histological parameters of NAFLD17,18,19, but the results are not consistent, partially because only few studies with a limited number of subjects analyzed the association between the rs738409 polymorphism and NASH or simple steatosis.
There is no approved therapy for NAFLD, and the diagnosis of NASH can only be proven by liver biopsy. In addition, it is important to establish whether the associations differ between different subgroups of NAFLD. To clarify the association between the rs738409 polymorphism and risk of NAFLD, we conducted a systematic review and meta-analysis of the available prospective studies with the specific aims of analyzing NAFLD subgroups, including simple steatosis or NASH, to clarify whether the association differed by histological parameters.
Methods
Search strategy
We conducted an electronic search of the PubMed, EMBASE and Web of Science databases from their inception until December 2014 to identify the association between the rs738409 polymorphism and NAFLD risk using the following search terms: PNPLA3 and (polymorphism or variant or variation) and (NAFLD or NASH or (non-alcoholic fatty liver disease) or (fatty liver) or steatohepatitis). Additional studies not captured by our database search were identified by surveying the references of the originally identified reviews and research reports and by using the MEDLINE option “Related Articles”. The search was confined to human studies without country restrictions. In addition, the publication language was restricted to English.
Inclusion and exclusion criteria
Potentially relevant studies were selected based on the following inclusion criteria: (1) studies concerning the association between the PNPLA3 rs738409 polymorphism and risk of NAFLD; (2) case-control studies based on unrelated individuals; (3) studies in which the diagnosis of NAFLD was clear; (4) studies that provide the number of NAFLD cases and controls and the frequency of the rs738409 genotypes; and (5) studies published in English. The major reasons for study exclusion were the following: (1) case-only study or overlapping data; (2) studies with a sample size less than one hundred; (3) studies with abstracts only and reports published as comment and review papers; and (4) studies with secondary causes of steatosis, including alcohol abuse, the use of drugs, surgical procedures and hepatitis B and hepatitis C virus infection.
Data extraction
Two investigators independently selected the trials and extracted the data, and disagreements or uncertainties were resolved by consensus. The following data were extracted: first author, publication year, country of origin, ethnicity of studied population, sex ratio, mean age, diagnostic criteria for NAFLD, number of individuals in the case and control groups, frequency of PNPLA3 genotypes in the cases and controls; and consistency with the Hardy-Weinberg equilibrium(HWEs).
Study quality assessment
The quality of the studies was assessed independently by two investigators according to the quality assessment scores developed from the genetic association studies conducted by Thakkinstian et al. The total scores ranged from 0 (worst) to 13 (best)20. The criteria of the quality assessment used to analyze the studies in this meta-analysis are available in Table S1.
Statistical analysis
The strength of the association between the PNPLA3 polymorphism and NAFLD risk was assessed by the odds ratios (ORs) and 95% confidence interval (CI). The Chi-square test was used to assess the Hardy-Weinberg equilibrium (HWE) in order to analyze the genotype distribution in the control groups. Meta-analyses were performed for four genotype contrasts per outcome: allele contrast (G versus C), dominant model (GG+CG versus CC), recessive model (GG versus CG+CC), and additive model (GG versus CC)21,22. The Cochrane Q statistic and the inconsistency index (I2) were used to calculate the heterogeneity among the studies, and a P value < 0.10 or I2 > 50% was considered to be significant23. If heterogeneity existed among the studies, the random-effect model (the Dersimonian and Laird method) was used to calculate the pooled OR. Otherwise, a fixed-effect model (the Mantel-Haenszel method) was used for outcomes without obvious heterogeneity24. Sensitivity analyses were performed to assess the stability of the results by excluding one study at a time in order to analyze the influence of each study on the overall OR. The publication bias was assessed using funnel plots and Egger's test25.
Three subgroup analyses were additionally carried out by ethnicity (Caucasian, Asian or Hispanics), mean age (pediatric or adult) and source of the controls (hospital based or population based). The statistical analysis was performed with RevMan software version 5 (Cochrane Collaboration) and STATA software version 10.0 (Stata Corporation). A P value < 0.05 was considered to be statistically significant in this trial unless otherwise specified.
Results
Literature search
The search strategy initially identified 419 potentially relevant articles, and 363 articles were determined to be irrelevant after a review of the titles and abstracts. Thus, 56 trials proceeded to a full-text review, and an additional 33 studies were excluded. Finally, 23 articles were ultimately selected for inclusion in the meta-analysis17,18,19,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45. A flow describing the article selection process for this meta-analysis is shown in Figure 1. Of all of the studies included, 10 studies involved Caucasians17,19,26,27,28,29,30,31,32,42, 12 studies investigated Asians18,33,34,35,36,37,38,39,40,43,44,45, and 1 study researched Hispanics41. All of the studies followed a case-control design, 8 studies used population-based controls16,18,28,33,40,41,42,43, and 15 studies used hospital-based controls17,19,26,29,30,31,32,34,35,36,37,38,39,44,45. In addition, 19 studies were conducted in adult patients16,17,18,19,26,28,29,30,31,32,34,35,36,37,38,39,42,44,45, and 4 investigated pediatric patients33,40,41,43. The distribution of genotypes in the controls was consistent with HWE in 21 studies17,18,19,26,27,29,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45 and insufficient in the 2 other studies28,30. The quality score of the included studies ranged from 7 to 11 (Table S1). The characteristics of the included studies are presented in Table 1.
Figure 1. Flowchart of the study selection.
Table 1. Characteristics of Studies Included in this meta-analysis.
| Author | Year | Country or Region | Ethnicity | Source of Control | Genotyping method | Age | Female n (%) | NAFLD diagnosis | Liver Biopsy(n) | Cases | Controls | Quality Score |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Kantartzis et al. | 2009 | Germany | Caucasian | H-B | TaqMan | Adult | 200(60.6) | H-MRS | NA | 105 | 225 | 10 |
| Sookoian et al. | 2009 | Argentina | Caucasian | H-B | Allele specific PCR | Adult | 186(69.9) | US and LB | 103 | 172 | 94 | 10 |
| Valenti et al. | 2010 | Italy/United Kingdom | Caucasian | P-B | TaqMan | Adult | Italy: 114(26.3) UK: 123 (38) | LB | 574 | 574 | 179 | 11 |
| Rotman et al. | 2010 | USA | Caucasian | P-B | MassARRAY Sequenome | Adult | NA | LB | 766 | 120 | 766 | 7 |
| Speliotes et al. | 2010 | USA | Caucasian | H-B | MassARRAY Sequenome | Adult | NA | LB | 678 | 678 | 1405 | 10 |
| Goran et al. | 2010 | USA | Hispanic | P-B | TaqMan | Pediatric | 129 (68.6) | DEXA | NA | 71 | 188 | 9 |
| Lin et al. | 2010 | Taiwan | Asian | P-B | TaqMan | Pediatric | 174 (33.5) | US | NA | 102 | 418 | 11 |
| Hotta et al. | 2010 | Japan | Asian | H-B | TaqMan | Adult | 527 (63.4) | LB | 253 | 253 | 575 | 8 |
| Wang et al. | 2011 | Taiwan | Asian | H-B | TaqMan | Adult | 472 (53.7) | US | NA | 156 | 723 | 10 |
| Petit et al. | 2011 | France | Caucasian | H-B | Real-time PCR | Adult | 120 (51.3) | H-MRS | NA | 149 | 85 | 8 |
| Zain et al. | 2012 | Malaysia | Asian | H-B | TaqMan | Adult | 180 (52.6) | LB | 144 | 144 | 198 | 10 |
| Kawaguchi et al. | 2012 | Japan | Asian | H-B | BeadChip | Adult | 741 (50.7) | LB | 529 | 529 | 932 | 10 |
| Valenti et al. | 2012 | Italian | Caucasian | H-B | Real-time PCR | Adult | 87 (21.7) | LB | 144 | 144 | 257 | 9 |
| Li et al. | 2012 | China | Asian | H-B | TaqMan | Adult | NA | US | NA | 203 | 202 | 10 |
| Peng et al. | 2012 | China | Asian | H-B | MassARRAY Sequenome | Adult | 308 (27.8) | US | NA | 553 | 553 | 11 |
| Lin et al. | 2013 | Taiwan | Asian | P-B | TaqMan | Pediatric | 237 (30.3) | US | NA | 182 | 599 | 9 |
| Guichelaar et al. | 2013 | USA | Caucasian | H-B | TaqMan | Adult | 122 (84.7) | LB | 144 | 132 | 12 | 8 |
| Verrijken et al. | 2013 | Belgium | Caucasian | H-B | TaqMan | Adult | 331 (70.4) | LB | 287 | 208 | 79 | 10 |
| Kitamoto et al. | 2013 | Japan | Asian | P-B | BeadChip | Adult | 782 (49.6) | LB | 564 | 564 | 1946 | 11 |
| Musso et al. | 2013 | Italy | Caucasian | P-B | TaqMan | Adult | 78 (36.8) | US | NA | 51 | 161 | 11 |
| Lin et al. | 2014 | Taiwan | Asian | P-B | TaqMan | Pediatric | 242 (30.4) | US | NA | 191 | 606 | 11 |
| Niu et al. | 2014 | China | Asian | H-B | ABI Sequencer | Adult | 426 (53.3) | US | NA | 390 | 409 | 10 |
| Lee et al. | 2014 | Korea | Asian | H-B | TaqMan | Adult | 178 (52.5) | US | NA | 155 | 184 | 11 |
P-B, population-based study; H-B, hospital-based study; H-MRS: hydrogen magnetic resonance (H-MR) spectroscopy, US: liver ultrasonographic examination, LB: liver biopsy, DEXA: dual energy-ray absorptiometry, NA: not available.
Association between rs738409 and risk for NAFLD
All studies
A total of 23 studies with 6071 cases and 10366 controls reported an association between the rs738409 polymorphism and NAFLD risk17,18,19,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41. Overall, the frequency of the G allele was 49.5% in NAFLD and 34.8% in the controls. The Hispanic population bears the highest frequency of the G allele (69.0% cases vs. 41.9% controls), followed by the Asian (54.2% cases vs. 39.9% controls) and Caucasian (42.2% cases vs. 22.7% controls) populations. The distribution of the rs738409 genotypes and alleles is presented in Table 2. Strong evidence of an association between the rs738409 polymorphism and NAFLD risk was found in all genetic models: allele contrast (OR = 2.10, 95% CI = 1.78–2.48, P < 0.00001; heterogeneity test: I2 = 89%, P < 0.00001); dominant model (OR = 2.06, 95% CI = 1.75–2.43, P < 0.00001; heterogeneity test: I2 = 67%, P < 0.00001); recessive model (OR = 2.49, 95% CI = 2.01–3.08, P < 0.00001; heterogeneity test: I2 = 72%, P < 0.0001); and additive model (OR = 3.41, 95% CI = 2.57–4.52, P < 0.00001; heterogeneity test: I2 = 77%, P < 0.00001) (Figure 2). After exclusion of the two articles deviating from HWE in the cases and controls, the results of the relationship was not influenced significantly in all genetic models (Table 3).
Table 2. The distribution of alleles and genotypes of PNPLA3 in NAFLD studies.
| Sample size | Genotype in cases | Genotype in controls | Case | Control | G allele (%) | C allele (%) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| First Author | Cases | Controls | GG | CG | CC | GG | CG | CC | G | C | G | C | Cases | Controls | Cases | Controls | HWE P value |
| Kantartzis | 105 | 225 | 13 | 41 | 51 | 18 | 70 | 137 | 67 | 143 | 106 | 344 | 31.9 | 23.6 | 68.1 | 76.4 | YES |
| Sookoian | 103 | 94 | NA | NA | NA | NA | NA | NA | 130 | 76 | 63 | 125 | 63.1 | 33.6 | 36.9 | 66.4 | YES |
| Valenti 2010 | 574 | 179 | 75 | 254 | 245 | 5 | 56 | 118 | 404 | 744 | 66 | 292 | 35.2 | 18.4 | 64.8 | 81.6 | 0.59 |
| Rotman | 520 | 336 | NA | NA | NA | NA | NA | NA | 516 | 524 | 153 | 519 | 49.6 | 22.8 | 50.4 | 77.2 | NA |
| Speliotes | 592 | 1405 | NA | NA | NA | NA | NA | NA | 592 | 592 | 618 | 2192 | 50 | 22.0 | 50 | 78.0 | YES |
| Goran | 71 | 188 | 34 | 30 | 7 | 19 | 60 | 38 | 98 | 44 | 98 | 136 | 69 | 26 | 31 | 74 | 0.56 |
| Lin 2011 | 102 | 418 | 26 | 52 | 24 | 59 | 192 | 167 | 104 | 100 | 310 | 526 | 51.0 | 37.1 | 49.0 | 62.9 | 0.75 |
| Hotta | 253 | 575 | 175 | 97 | 111 | 104 | 296 | 175 | 305 | 201 | 504 | 646 | 88.3 | 43.8 | 11.7 | 56.2 | 0.28 |
| Wang | 156 | 723 | 40 | 80 | 36 | 269 | 335 | 119 | 152 | 160 | 573 | 873 | 51.3 | 60.4 | 48.7 | 39.6 | 0.40 |
| Petit | 149 | 85 | NA | NA | 68 | NA | NA | 51 | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Zain | 144 | 198 | NA | NA | NA | NA | NA | NA | 130 | 158 | 95 | 301 | 45.1 | 24.0 | 54.9 | 76.0 | YES |
| Kawaguchi | 529 | 932 | 217 | 468 | 247 | 203 | 236 | 88 | 642 | 412 | 902 | 962 | 85.2 | 34.4 | 14.8 | 65.6 | 0.17 |
| Valenti 2012 | 144 | 257 | 21 | 68 | 55 | 16 | 95 | 146 | 110 | 178 | 127 | 387 | 38.2 | 24.7 | 61.8 | 75.3 | 0.92 |
| Li | 203 | 202 | 49 | 84 | 70 | 18 | 90 | 94 | 182 | 224 | 126 | 278 | 44.8 | 31.0 | 55.2 | 69.0 | 0.59 |
| Peng | 553 | 553 | 93 | 276 | 183 | 59 | 259 | 235 | 462 | 642 | 377 | 729 | 41.8 | 34.1 | 58.2 | 65.9 | 0.32 |
| Lin 2013 | 182 | 599 | 35 | 93 | 54 | 74 | 288 | 237 | 163 | 201 | 436 | 762 | 44.8 | 36.4 | 55.2 | 63.6 | 0.35 |
| Guichelaar | 132 | 12 | 12 | 41 | 79 | 0 | 3 | 9 | 65 | 199 | 3 | 21 | 24.6 | 12.5 | 75.4 | 87.5 | 0.62 |
| Verrijken | 208 | 79 | 17 | 83 | 108 | 0 | 23 | 56 | 117 | 299 | 140 | 434 | 20.4 | 5.5 | 79.6 | 94.5 | 0.13 |
| Kitamoto | 564 | 1946 | 227 | 241 | 96 | 199 | 513 | 300 | 695 | 433 | 911 | 1113 | 61.6 | 23.4 | 38.4 | 76.6 | 0.44 |
| Musso | 51 | 161 | 14 | 23 | 14 | 21 | 49 | 91 | 51 | 51 | 91 | 231 | 50 | 28.3 | 50 | 71.7 | YES |
| Lin | 191 | 606 | 38 | 95 | 58 | 75 | 293 | 238 | 171 | 211 | 443 | 769 | 44.8 | 36.6 | 55.2 | 63.4 | 0.30 |
| Niu | 390 | 409 | 189 | 153 | 48 | 50 | 176 | 183 | 531 | 249 | 276 | 542 | 68.1 | 33.7 | 31.9 | 66.3 | 0.45 |
| Lee | 155 | 184 | 49 | 75 | 31 | 37 | 92 | 55 | 173 | 137 | 166 | 202 | 55.8 | 45.1 | 44.2 | 54.9 | 0.90 |
NA: not applicable YES: studies have already pointed out that the data was HWE, but the data was not applicable.
Figure 2. Forest plot of NAFLD susceptibility associated with rs738409 polymorphism at additive model (GG vs CC).
Table 3. Association between PNPLA3 polymorphism and NAFLD risk.
| Subgroup | Inherited model | Study number | NO. of cases/controls(n/n) | Pheterogeneity | I2 (%) | Pooled OR (95%CI) | P valuea |
|---|---|---|---|---|---|---|---|
| Total studies | Allele contrast | 22 | 11838/18552 | P < 0.00001 | 89 | 2.10 (1.78, 2.48) | P < 0.00001 |
| Dominant model | 19 | 4709/7328 | P < 0.0001 | 67 | 2.06 (1.75, 2.43) | P < 0.00001 | |
| Recessive model | 18 | 4560/7243 | P < 0.0001 | 72 | 2.49 (2.01, 3.08) | P < 0.00001 | |
| Additive model | 18 | 2523/3886 | P < 0.00001 | 77 | 3.41 (2.57,4.52) | P < 0.00001 | |
| Studies excluded for DHWE | |||||||
| Allele contrast | 21 | 10798/17880 | P < 0.00001 | 89 | 2.05 (1.74, 2.42) | P < 0.00001 | |
| Dominant model | 18 | 4560/7243 | P < 0.00001 | 69 | 2.02 (1.84, 2.20) | P < 0.00001 | |
| Recessive model | 18 | 4560/7243 | P < 0.00001 | 72 | 2.51 (2.28, 2.77) | P < 0.00001 | |
| Additive model | 18 | 2523/3886 | P < 0.00001 | 77 | 3.32 (2.94, 3.74) | P < 0.00001 | |
| Ethnicity | |||||||
| Caucasian | Allele contrast | 9 | 4858/5496 | P < 0.0001 | 75 | 2.56 (2.06, 3.18) | P < 0.00001 |
| Dominant model | 7 | 1363/998 | P = 0.60 | 0 | 2.21 (1.83, 2.67) | P < 0.00001 | |
| Recessive model | 6 | 1214/913 | P = 0.35 | 10 | 2.68 (1.78, 4.05) | P < 0.00001 | |
| Additive model | 6 | 704/617 | P = 0.27 | 22 | 3.79 (2.35, 6.13) | P < 0.00001 | |
| Asian | Allele contrast | 12 | 6838/12822 | P < 0.00001 | 88 | 1.82 (1.52,2.18) | P < 0.00001 |
| Dominant model | 11 | 3275/6213 | P < 0.00001 | 78 | 1.95 (1.56, 2.43) | P < 0.00001 | |
| Recessive model | 11 | 3275/6213 | P < 0.00001 | 81 | 2.33 (1.81, 2.99) | P < 0.00001 | |
| Additive model | 11 | 1778/3212 | P < 0.00001 | 84 | 3.08 (2.21, 4.31) | P < 0.00001 | |
| Hispanics | Allele contrast | 1 | 142/234 | NA | NA | 3.09 (1.99, 4.80) | P < 0.00001 |
| Dominant model | 1 | 71/117 | NA | NA | 4.40 (1.84, 10.51) | P = 0.0009 | |
| Recessive model | 1 | 71/117 | NA | NA | 4.74 (2.41, 9.33) | P < 0.00001 | |
| Additive model | 1 | 41/57 | NA | NA | 9.71 (3.64, 25.94) | P < 0.00001 | |
| Control source | |||||||
| Population based | Allele contrast | 7 | 4076/5836 | P < 0.00001 | 90 | 2.17 (1.60, 2.95) | P < 0.00001 |
| Dominant model | 7 | 2038/2918 | P < 0.00001 | 82 | 2.47 (2.14, 2.85) | P < 0.00001 | |
| Recessive model | 7 | 2038/2918 | P < 0.00001 | 83 | 3.04 (2.00,4.62) | P < 0.00001 | |
| Additive model | 7 | 1139/1542 | P < 0.00001 | 87 | 4.61 (2.58, 8.23) | P < 0.00001 | |
| Hospital based | Allele contrast | 15 | 7762/12716 | P < 0.00001 | 89 | 2.07 (1.68, 2.54) | P < 0.00001 |
| Dominant model | 12 | 2671/4410 | P = 0.65 | 0 | 1.76 (1.57,1.97) | P < 0.00001 | |
| Recessive model | 11 | 2522/4325 | P = 0.26 | 19 | 2.10 (1.78, 2.47) | P < 0.00001 | |
| Additive model | 11 | 1384/2344 | P = 0.34 | 11 | 2.62 (2.20, 3.13) | P < 0.00001 | |
| Age of participants | |||||||
| Adult | Allele contrast | 18 | 10746/15072 | P < 0.00001 | 90 | 2.19 (1.82, 2.62) | P < 0.00001 |
| Dominant model | 15 | 4163/5588 | P < 0.0001 | 70 | 2.10 (1.74, 2.54) | P < 0.00001 | |
| Recessive model | 14 | 4014/5503 | P < 0.00001 | 75 | 2.59 (2.01, 3.34) | P < 0.00001 | |
| Additive model | 14 | 2248/2978 | P < 0.00001 | 79 | 3.54 (2.54, 4.94) | P < 0.00001 | |
| Pediatric | Allele contrast | 4 | 1092/3480 | P = 0.01 | 73 | 1.73 (1.31, 2.29) | P = 0.0001 |
| Dominant model | 4 | 546/1740 | P = 0.09 | 54 | 1.89 (1.34, 2.66) | P < 0.00001 | |
| Recessive model | 4 | 546/1740 | P = 0.07 | 58 | 2.18 (1.47, 3.22) | P < 0.0001 | |
| Additive model | 4 | 275/908 | P = 0.03 | 66 | 2.97 (1.75, 5.02) | P < 0.0001 |
a: Test for overall effect. NA: Not applicable.
Subgroup analyses
Subgroup analyses were conducted to explore the differences between ethnicity, mean age and sources of the controls. In the subgroup analysis by ethnicity, significant association was found between the rs738409 polymorphism and NAFLD risk among the Caucasian, Asian and Hispanic populations. The association between rs738409 polymorphism and NAFLD was most significant in Hispanic population, which followed by Caucasian population, and the association was weakest in Asian population. The analyses also showed that the risk of NAFLD was significantly increased in both adult participants and pediatric subjects. In addition, the G allele was strongly associated with NAFLD susceptibility in hospital-based controls and population-based controls. The results were consistent in all genetic models. More details are presented in Table 3.
Histological Severity of NAFLD
Five eligible studies were used to investigate the association between the rs738409 polymorphism and lobular necroinflammation, including 1978 patients. A statistically significant association was seen between carrying GG genotype and higher inflammation scores (OR = 3.13, 95% CI = 2.76–3.56, P < 0.00001; heterogeneity test: I2 = 0%, P = 0.674) with obvious publication bias (Egger test: P = 0.980) (Figure 3). The six eligible studies with 2552 patients analyze the relationship between rs738409 polymorphism and fibrosis. The analysis pointed out that the GG genotype was significantly associated with fibrosis score (OR = 3.11, 95% CI = 2.66–3.65, P < 0.00001; heterogeneity test: I2 = 18.3%, P = 0.295) and the publication bias was not significant (Egger test: P = 0.457) (Figure 3).
Figure 3.
(A)The forest plot for the association between rs738409 polymorphism and the risk of necroinflammation (additive model: GG vs CC). (B)The forest plot for the association between rs738409 polymorphism and the risk of fibrosis (additive model: GG vs CC).
Association between rs738409 and risk for simple steatosis
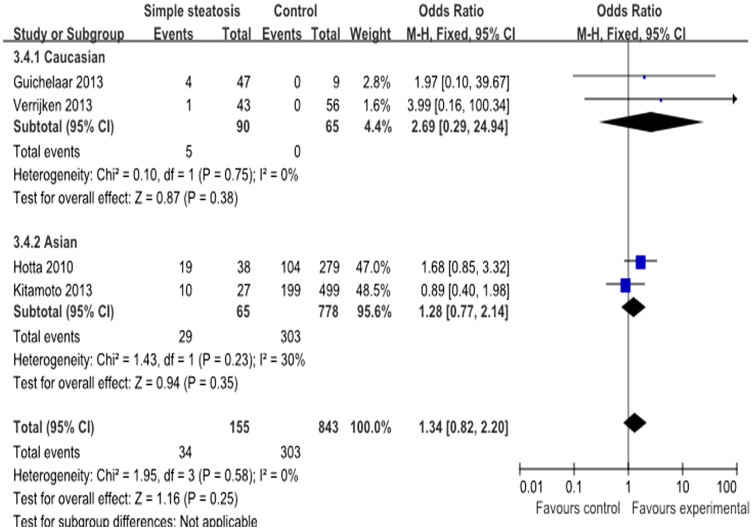
Overall, 7 studies with 387 cases and 2306 controls analyzed the rs738409 polymorphism and risk of simple steatosis17,18,19,28,32,34,36. Interestingly, the frequency of the risk G allele was very close between the cases (38.1%) and controls (38.0%). In Caucasian subjects, the frequency of the G allele was 34.3% in cases and 23.2% in controls, and these values are lower than those found in the Asian population (44.3% cases vs. 42.3% controls) (Table 4). We analyzed the relationship between the G allele and the risk of simple steatosis. No significant association was observed between rs738409 polymorphism and simple steatosis under additive model (OR = 1.34, 95% CI = 0.82–2.20, P = 0.25; heterogeneity test: I2 = 0%, P = 0.58), dominant model and recessive model (Figure 4). However, a significant association was found in allele contrast. A further subgroup analysis based on ethnicity showed no obvious association between the rs738409 polymorphism and simple steatosis in Asian subjects, while a strong association was found in the Caucasian population under the allele contrast instead of the other three genetic models. (Table 5).
Table 4. The distribution of alleles and genotypes of PNPLA3 in SS studies and NASH studies.
| Sample size | Genotype in cases | Genotype in controls | Case | Control | G allele (%) | C allele (%) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| First Author | Cases | Controls | GG | CG | CC | GG | CG | CC | G | C | G | C | cases | controls | cases | controls |
| SS | ||||||||||||||||
| Sookoian | 40 | 94 | NA | NA | NA | NA | NA | NA | 42 | 38 | 63 | 125 | 52.5 | 33.6 | 48.5 | 66.4 |
| Rotman | 82 | 336 | NA | NA | NA | NA | NA | NA | 85 | 79 | 153 | 519 | 51.8 | 22.8 | 48.2 | 77.2 |
| Hotta | 64 | 575 | 19 | 26 | 19 | 104 | 296 | 175 | 64 | 64 | 504 | 646 | 50 | 43.8 | 50 | 56.2 |
| Zain | 33 | 198 | NA | NA | NA | NA | NA | NA | 23 | 43 | 95 | 301 | 35.0 | 24.0 | 65.0 | 76.0 |
| Guichelaar | 60 | 12 | 4 | 13 | 43 | 0 | 3 | 9 | 21 | 99 | 3 | 21 | 17.5 | 12.5 | 82.5 | 87.5 |
| Verrijken | 57 | 79 | 1 | 14 | 42 | 0 | 23 | 56 | 16 | 98 | 23 | 135 | 14.0 | 14.6 | 86.0 | 85.4 |
| Kitamoto | 51 | 1012 | 10 | 24 | 17 | 199 | 513 | 300 | 44 | 58 | 911 | 1113 | 43.1 | 45.0 | 56.9 | 55.0 |
| Total | 387 | 2306 | 34 | 77 | 121 | 303 | 835 | 540 | 295 | 479 | 1752 | 2860 | 38.1 | 38.0 | 61.9 | 62.0 |
| NASH | ||||||||||||||||
| Sookoian | 63 | 94 | NA | NA | NA | NA | NA | NA | 88 | 38 | 63 | 125 | 69.8 | 33.6 | 30.2 | 66.4 |
| Rotman | 438 | 336 | NA | NA | NA | NA | NA | NA | 431 | 445 | 153 | 519 | 49.2 | 22.8 | 45.8 | 77.2 |
| Hotta | 189 | 575 | 78 | 85 | 26 | 104 | 296 | 175 | 241 | 137 | 504 | 646 | 63.8 | 43.8 | 36.2 | 56.2 |
| Zain | 111 | 198 | NA | NA | NA | NA | NA | NA | 106 | 116 | 95 | 301 | 48.0 | 24.0 | 52.0 | 76.0 |
| Guichelaar | 72 | 12 | 8 | 28 | 36 | 0 | 3 | 9 | 44 | 100 | 3 | 21 | 30.6 | 12.5 | 69.4 | 87.5 |
| Verrijken | 151 | 79 | 16 | 69 | 66 | 0 | 23 | 56 | 101 | 201 | 23 | 135 | 33.4 | 14.6 | 66.6 | 85.4 |
| Kitamoto | 442 | 1012 | 187 | 183 | 72 | 199 | 513 | 300 | 557 | 327 | 911 | 1113 | 63.0 | 45.0 | 37.0 | 55.0 |
| Total | 1466 | 2306 | 289 | 365 | 200 | 303 | 835 | 540 | 1568 | 1364 | 1752 | 2860 | 53.5 | 38.0 | 46.5 | 62.0 |
SS: simple steatosis; NASH: nonalcoholic steatohepatitis.
Figure 4. Forest plot of simple steatosis susceptibility associated with rs738409 polymorphism at additive model (GG vs CC).
Table 5. Association between PNPLA3 polymorphism and simple steatosis risk.
| Group | Study number(n) | NO. of cases/controls(n/n) | Pheterogeneity | I2 (%) | Pooled OR (95%CI) | P valuea |
|---|---|---|---|---|---|---|
| SS | ||||||
| Allele contrast | 7 | 774/4612 | P < 0.0001 | 81 | 1.59 (1.02, 2.49) | P = 0.04 |
| Dominant model | 4 | 232/1678 | P = 0.94 | 0 | 0.94 (0.66, 1.33) | P = 0.73 |
| Recessive model | 4 | 232/1678 | P = 0.49 | 0 | 1.49 (0.97, 2.30) | P = 0.07 |
| Additive model | 4 | 155/843 | P = 0.58 | 0 | 1.34 (0.82, 2.20) | P = 0.25 |
| Caucasian | ||||||
| Allele contrast | 4 | 478/1042 | P = 0.005 | 76 | 1.98 (1.05, 3.75) | P = 0.04 |
| Dominant model | 2 | 117/91 | P = 0.71 | 0 | 0.93 (0.48, 1.82) | P = 0.84 |
| Recessive model | 2 | 117/91 | P = 0.74 | 0 | 2.77 (0.30, 25.51) | P = 0.37 |
| Additive model | 2 | 90/65 | P = 0.75 | 0 | 2.69 (0.29, 24.94) | P = 0.38 |
| Asian | ||||||
| Allele contrast | 3 | 296/3570 | P = 0.20 | 37 | 1.22 (0.89, 1.67) | P = 0.22 |
| Dominant model | 2 | 115/1587 | P = 0.62 | 0 | 0.94 (0.62, 1.42) | P = 0.77 |
| Recessive model | 2 | 115/1587 | P = 0.16 | 49 | 1.44 (0.93, 2.25) | P = 0.10 |
| Additive model | 2 | 65/778 | P = 0.23 | 30 | 1.28 (0.77, 2.14) | P = 0.35 |
SS: simple steatosis.
a: Test for overall effect.
Association between rs738409 and NASH risk
Overall, seven studies with 1466 cases and 2306 controls reported the rs738409 polymorphism and risk of NASH17,18,19,28,32,34,36: four studies conducted in Caucasians and three studies performed in Asian populations. The pooled overall frequency of the risk G allele was 53.5% in the cases and 38.0% in the controls. The G allele varied widely between the different populations: high in the Asian populations (60.9% cases vs.42.3% controls) and lower in the Caucasian subjects (45.9% cases vs. 23.2% controls) (Table 4). Strong evidence of an association was detected between the rs738409 polymorphism and NASH risk under the additive model (OR = 4.44, 95% CI = 3.39–5.82, P < 0.00001; heterogeneity test: I2 = 0%, P = 0.49) (Figure 5). The association was also significant in the other three genetic models, and no evidence of heterogeneity was observed between the studies. Evidence of a strong association between the rs738409 polymorphism and NASH susceptibility was also found in both Asian and Caucasian populations with all genetic models. In addition, Caucasian populations with rs738409 polymorphism are more easily develop into NASH than Asian populations. The results are described in Table 6.
Figure 5. Forest plot of NASH susceptibility associated with rs738409 polymorphism at additive model (GG vs CC).
Table 6. Association between PNPLA3 Polymorphism and NASH risk.
| Group | Study number(n) | NO. of cases/controls(n/n) | Pheterogeneity | I2 (%) | Pooled OR (95%CI) | P valuea |
|---|---|---|---|---|---|---|
| NASH | ||||||
| Allele contrast | 7 | 2932/4612 | P = 0.005 | 68 | 2.78 (2.24, 3.44) | P < 0.00001 |
| Dominant model | 4 | 854/1678 | P = 0.63 | 0 | 2.44 (1.95, 3.04) | P < 0.00001 |
| Recessive model | 4 | 854/1678 | P = 0.63 | 0 | 3.15 (2.58, 3.85) | P < 0.00001 |
| Additive model | 4 | 489/843 | P = 0.49 | 0 | 4.44 (3.39, 5.82) | P < 0.00001 |
| Caucasian | ||||||
| Allele contrast | 4 | 1448/1042 | P = 0.59 | 0 | 3.40 (2.82, 4.09) | P < 0.00001 |
| Dominant model | 2 | 223/91 | P = 0.95 | 0 | 3.11 (1.82, 5.33) | P < 0.0001 |
| Recessive model | 2 | 223/91 | P = 0.38 | 0 | 10.33 (1.42, 75.06) | P = 0.02 |
| Additive model | 2 | 126/65 | P = 0.36 | 0 | 14.28 (1.96, 103.92) | P = 0.009 |
| Asian | ||||||
| Allele contrast | 3 | 1484/3570 | P = 0.24 | 30 | 2.26 (1.93, 2.65) | P < 0.00001 |
| Dominant model | 2 | 631/1587 | P = 0.38 | 0 | 2.33 (1.83, 2.96) | P < 0.00001 |
| Recessive model | 2 | 631/1587 | P = 0.79 | 0 | 3.05 (2.49, 3.74) | P < 0.00001 |
| Additive model | 2 | 363/778 | P = 0.41 | 0 | 4.22 (3.21, 5.55) | P < 0.00001 |
a: Test for overall effect.
Sensitivity and Publication Bias
Sensitivity analysis was performed under additive model to evaluate the influence of a specific study on the overall estimate. The corresponding pooled ORs with 95% CIs produced similarly before and after omitting each study at a time, indicating that our results were stable and reliable (Table S2). The funnel plots of the studies were symmetric in the current meta-analysis (Figure 6). Furthermore, the results of Egger's test did not support the existence of publication bias (additive model: NAFLD: P = 0.467; SS: P = 0.611; NASH: P = 0.282).
Figure 6. Publication bias on rs738409 polymorphism under additive model.
(A) Funnel plot of studies of the rs738409 variant and NAFLD. (B) Funnel plot of studies of the rs738409 variant and simple steatosis. (C) Funnel plot of studies of the rs738409 variant and NASH.
Discussion
The current meta-analysis provided a systematic assessment of the association between the PNPLA3 rs738409 polymorphism and susceptibility to NAFLD, including its subtypes simple steatosis and NASH. Our results suggested that rs738409 polymorphism exerted a significant influence not only on NAFLD risk, but also on histological severity of NAFLD. In addition, a further analysis showed that individuals with the rs738409 polymorphism experienced a significantly increased risk for NASH. However, our meta-analysis did not show a definite association of rs738409 polymorphism with simple steatosis.
Our results are consistent with those from a previous meta-analysis conducted by Sookoian et al.14, which showed a significant association between the rs738409 polymorphism and NAFLD (OR = 3.26, 95% CI = 2.73–3.89, P < 0.00001) and a significant association between the rs738409 polymorphism and NASH (OR = 3.26, 95% CI = 2.14–4.95, P < 0.00001), similar to the results reported in this manuscript. In the present meta-analysis, analysis of the rs738409 polymorphism revealed a significantly increased NAFLD risk in all genetic models. When the data were stratified by subject ethnicity, a significant correlation was found in all three populations, suggesting that the susceptibility genes may be a strong indicator across different races. In the population-based and hospital-based control studies, a significant correlation was also observed in all genetic models, suggesting that our results were not influenced by the source of controls. In addition, the association between the rs738409 polymorphism and NAFLD risk was also significant in both adult and children populations, indicating that the results are highly stable and not influenced by ethnicity, source of the controls and age of participants.
A large population-based study that involved 9229 multiethnic population, including African-Americans, Hispanics and European-Americans, revealed that patients with the rs738409 polymorphism are associated with a higher risk of NAFLD compared with normal controls10. These findings are generally consistent with individual published reports because 70–90% of the trials showed an association between the rs738409 polymorphism and NAFLD risk27,38,41. The underlying mechanism for how PNPLA3 genotype increases NAFLD susceptibility remains to be elucidated. The questions that have been raised are whether the I148M polymorphism increases liver damage favoring the accumulation of fatty acids in lipid droplets or increases the susceptibility to progress into NASH and fibrogenesis.
It should be noted that rs738409 polymorphism was only significantly associated with increased simple steatosis risk under allelic model, but not under the other three genetic models. When stratified by ethnicity, we only detect a significant association in the Asian subgroup under allele contrast, but failed to detect a significant association in the Caucasion population under all genetic models. This meta-analysis of the associations of the rs738409 polymorphism with NASH showed a significant relation. In the subgroup analysis stratified by ethnicity, similar correlations were observed in both Caucasian and Asian populations. The results from the allele contrast were consistent with those from the other genetic models. The sensitivity analysis revealed that no single study qualitatively changed the pooled odds ratios. These findings suggested that rs738409 polymorphism was strongly associated with NASH.
In our meta-analysis, it appears that the rs738409 polymorphism is more likely to increase the NASH risk instead of simple steatosis. Consistent with our results, animal studies have revealed that, although PNPLA3 has triglyceride lipase activity and is responsible for the transalkylation of acylglycerol, knockout of PNPLA3 has no effect on liver steatosis or insulin resistance46. Further epidemiological studies have also noted that this G allele variation did not affect the main risk factors for steatosis, including insulin resistance, LDL, HDL, total cholesterol and glucose levels29. Other polymorphisms, such as CD14 rs2569190 and GCLC rs4140528, are also regarded to increase the risk of NASH instead of simple steatosis47. There are some possible reasons to explain this phenomenon. First, the effect of the rs738409 G allele may be involved in the differential expression and function of variant PNPLA3 instead of resulting in a loss of function of the wild-type protein. Second, there may be some gene-gene interactions. It is possible that the difference in phenotypes may be caused by some other genetic variant that is strongly linked to rs738409. Third, although NASH and simple steatosis are currently regarded as two histological subtypes along the unique spectrum of NAFLD, evidence suggests that these two conditions may be not only different from the histological syndrome but also varied from pathophysiological standpoints. The results that the association between NASH risk and the rs738409 genotype is independent of simple steatosis might suggest that simple steatosis may not be the essential condition for the progressive damage. Simple steatosis and NASH are likely to be two independent conditions in the NAFLD spectrum.
Despite the inevitable limitations of this meta-analysis, we believe that our research provides useful information. First, the individual sample size of each study included in our meta-analysis was too small to obtain a definite association between rs738409 polymorphisms and NAFLD risk, but the pooled odds ratios generated from the 23 studies significantly increased the statistical power of the analysis compared to that obtained with a single study. Moreover, the protocol of this meta-analysis has been well-designed with explicit criteria and methods for study selection, data extraction and data analysis, which allowed reliable inferences about causality. Third, there was no significant publication bias in this meta-analysis, and the results of the sensitivity analysis support the stability of the results.
However, some limitations of this meta-analysis should be addressed. First, the retrieved literature may not be sufficiently comprehensive. Only published case-control studies were included in this meta-analysis. Second, most of the study subjects were of Caucasian and Asian ancestry, and the Hispanic subgroup was very limited in this meta-analysis. Thus, potential selective bias and publication bias may have occurred. Third, because NAFLD was a multifactor disease, the potential effects of gene-gene and gene-environment interactions should be considered. Fourth, the sample size of NASH in this meta-analysis was so small that the statistical power for making a definitive conclusion regarding the possible risk of the rs738409 polymorphism was limited.
In conclusion, results from this meta-analysis showed that the G allele at PNPLA3 gene was a risk factor for NAFLD and its subtype NASH, especially in Asian, Caucasian and Hispanic populations. However, no association was observed between the rs738409 polymorphism and simple steatosis risk. Further studies with higher quality, more participants and various ethnicities are needed to obtain a more precise estimate of the genetic effects.
Author Contributions
R.-F.X. and G.-Z.C. conceived the study design, and wrote the manuscript; A.-Y.T., S.-S.Z. and Y.-B.D. performed the analyses. All authors read and approved the final manuscript.
Supplementary Material
Supplementary information
Acknowledgments
The present study was supported by the National Natural Science Foundation of China (No. 81400369).
References
- Angulo P. Nonalcoholic fatty liver disease. N Engl J Med 346, 1221–1231 (2002). [DOI] [PubMed] [Google Scholar]
- Browning J. D. et al. Prevalence of hepatic steatosis in an urban population in the United States: impact of ethnicity. Hepatology 40, 1387–1395 (2004). [DOI] [PubMed] [Google Scholar]
- Farrell G. C. & Larter C. Z. Nonalcoholic fatty liver disease: from steatosis to cirrhosis. Hepatology 43, S99–S112 (2006). [DOI] [PubMed] [Google Scholar]
- Ratziu V., Bellentani S., Cortez-Pinto H., Day C. & Marchesini G. A position statement on NAFLD/NASH based on the EASL 2009 special conference. J Hepatol 53, 372–384 (2010). [DOI] [PubMed] [Google Scholar]
- Powell E. E. et al. The natural history of nonalcoholic steatohepatitis: a follow-up study of forty-two patients for up to 21 years. Hepatology 11, 74–80 (1990). [DOI] [PubMed] [Google Scholar]
- Vernon G., Baranova A. & Younossi Z. M. Systematic review: the epidemiology and natural history of non-alcoholic fatty liver disease and non-alcoholic steatohepatitis in adults. Aliment Pharmacol Ther 34, 274–285 (2011). [DOI] [PubMed] [Google Scholar]
- Musso G., Gambino R., Cassader M. & Pagano G. A meta-analysis of randomized trials for the treatment of nonalcoholic fatty liver disease. Hepatology 52, 79–104 (2010). [DOI] [PubMed] [Google Scholar]
- Wilfred de Alwis N. M. & Day C. P. Genetics of alcoholic liver disease and nonalcoholic fatty liver disease. Semin Liver Dis 27, 44–54 (2007). [DOI] [PubMed] [Google Scholar]
- Miele L. et al. The Kruppel-like factor 6 genotype is associated with fibrosis in nonalcoholic fatty liver disease. Gastroenterology 135, 282–291 e281 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo S. et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet 40, 1461–1465 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day C. P. & James O. F. Steatohepatitis: a tale of two “hits”? Gastroenterology 114, 842–845 (1998). [DOI] [PubMed] [Google Scholar]
- Kotronen A. et al. A common variant in PNPLA3, which encodes adiponutrin, is associated with liver fat content in humans. Diabetologia 52, 1056–1060 (2009). [DOI] [PubMed] [Google Scholar]
- Sookoian S. & Pirola C. J. PNPLA3, the triacylglycerol synthesis/hydrolysis/storage dilemma, and nonalcoholic fatty liver disease. World J Gastroenterol 18, 6018–6026 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sookoian S. & Pirola C. J. Meta-analysis of the influence of I148M variant of patatin-like phospholipase domain containing 3 gene (PNPLA3) on the susceptibility and histological severity of nonalcoholic fatty liver disease. Hepatology 53, 1883–1894 (2011). [DOI] [PubMed] [Google Scholar]
- Santoro N. et al. A common variant in the patatin-like phospholipase 3 gene (PNPLA3) is associated with fatty liver disease in obese children and adolescents. Hepatology 52, 1281–1290 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenti L. et al. I148M patatin-like phospholipase domain-containing 3 gene variant and severity of pediatric nonalcoholic fatty liver disease. Hepatology 52, 1274–1280 (2010). [DOI] [PubMed] [Google Scholar]
- Verrijken A. et al. A gene variant of PNPLA3, but not of APOC3, is associated with histological parameters of NAFLD in an obese population. Obesity (Silver Spring) 21, 2138–2145 (2013). [DOI] [PubMed] [Google Scholar]
- Kitamoto T. et al. Genome-wide scan revealed that polymorphisms in the PNPLA3, SAMM50, and PARVB genes are associated with development and progression of nonalcoholic fatty liver disease in Japan. Hum Genet 132, 783–792 (2013). [DOI] [PubMed] [Google Scholar]
- Sookoian S. et al. A nonsynonymous gene variant in the adiponutrin gene is associated with nonalcoholic fatty liver disease severity. J Lipid Res 50, 2111–2116 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakkinstian A. et al. Systematic review and meta-analysis of the association between {beta}2-adrenoceptor polymorphisms and asthma: a HuGE review. Am J Epidemiol 162, 201–211 (2005). [DOI] [PubMed] [Google Scholar]
- Minelli C., Thompson J. R., Abrams K. R., Thakkinstian A. & Attia J. The choice of a genetic model in the meta-analysis of molecular association studies. International journal of epidemiology 34, 1319–1328 (2005). [DOI] [PubMed] [Google Scholar]
- Zintzaras E. & Lau J. Synthesis of genetic association studies for pertinent gene-disease associations requires appropriate methodological and statistical approaches. Journal of clinical epidemiology 61, 634–645 (2008). [DOI] [PubMed] [Google Scholar]
- Higgins J. P. & Thompson S. G. Quantifying heterogeneity in a meta-analysis. Stat Med 21, 1539–1558 (2002). [DOI] [PubMed] [Google Scholar]
- DerSimonian R. & Laird N. Meta-analysis in clinical trials. Control Clin Trials 7, 177–188 (1986). [DOI] [PubMed] [Google Scholar]
- Egger M., Davey Smith G., Schneider M. & Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ 315, 629–634 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantartzis K. et al. Dissociation between fatty liver and insulin resistance in humans carrying a variant of the patatin-like phospholipase 3 gene. Diabetes 58, 2616–2623 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenti L. et al. Homozygosity for the patatin-like phospholipase-3/adiponutrin I148M polymorphism influences liver fibrosis in patients with nonalcoholic fatty liver disease. Hepatology 51, 1209–1217 (2010). [DOI] [PubMed] [Google Scholar]
- Rotman Y. et al. The association of genetic variability in patatin-like phospholipase domain-containing protein 3 (PNPLA3) with histological severity of nonalcoholic fatty liver disease. Hepatology 52, 894–903 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speliotes E. K. et al. PNPLA3 variants specifically confer increased risk for histologic nonalcoholic fatty liver disease but not metabolic disease. Hepatology 52, 904–912 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit J. M. et al. PNPLA3 polymorphism influences liver fibrosis in unselected patients with type 2 diabetes. Liver Int 31, 1332–1336 (2011). [DOI] [PubMed] [Google Scholar]
- Valenti L. et al. The I148M PNPLA3 polymorphism influences serum adiponectin in patients with fatty liver and healthy controls. BMC Gastroenterol 12, 111 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guichelaar M. M. et al. Interactions of allelic variance of PNPLA3 with nongenetic factors in predicting nonalcoholic steatohepatitis and nonhepatic complications of severe obesity. Obesity (Silver Spring) 21, 1935–1941 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. C. et al. A common variant in the PNPLA3 gene is a risk factor for non-alcoholic fatty liver disease in obese Taiwanese children. J Pediatr 158, 740–744 (2011). [DOI] [PubMed] [Google Scholar]
- Hotta K. et al. Association of the rs738409 polymorphism in PNPLA3 with liver damage and the development of nonalcoholic fatty liver disease. BMC Med Genet 11, 172 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. W. et al. The PNPLA3 I148M polymorphism is associated with insulin resistance and nonalcoholic fatty liver disease in a normoglycaemic population. Liver Int 31, 1326–1331 (2011). [DOI] [PubMed] [Google Scholar]
- Zain S. M. et al. A multi-ethnic study of a PNPLA3 gene variant and its association with disease severity in non-alcoholic fatty liver disease. Hum Genet 131, 1145–1152 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi T. et al. Genetic polymorphisms of the human PNPLA3 gene are strongly associated with severity of non-alcoholic fatty liver disease in Japanese. PLoS One 7, e38322 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Xing C., Tian Z. & Ku H. C. Genetic variant I148M in PNPLA3 is associated with the ultrasonography-determined steatosis degree in a Chinese population. BMC Med Genet 13, 113 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X. E. et al. Genetic variants in PNPLA3 and risk of non-alcoholic fatty liver disease in a Han Chinese population. PLoS One 7, e50256 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. C., Chang P. F., Chang M. H. & Ni Y. H. A common variant in the peroxisome proliferator-activated receptor-gamma coactivator-1alpha gene is associated with nonalcoholic fatty liver disease in obese children. The American journal of clinical nutrition 97, 326–331 (2013). [DOI] [PubMed] [Google Scholar]
- Goran M. I. et al. Effects of PNPLA3 on liver fat and metabolic profile in Hispanic children and adolescents. Diabetes 59, 3127–3130 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musso G., Bo S., Cassader M., De Michieli F. & Gambino R. Impact of sterol regulatory element-binding factor-1c polymorphism on incidence of nonalcoholic fatty liver disease and on the severity of liver disease and of glucose and lipid dysmetabolism. The American journal of clinical nutrition 98, 895–906 (2013). [DOI] [PubMed] [Google Scholar]
- Lin Y. C., Chang P. F., Chang M. H. & Ni Y. H. Genetic variants in GCKR and PNPLA3 confer susceptibility to nonalcoholic fatty liver disease in obese individuals. The American journal of clinical nutrition 99, 869–874 (2014). [DOI] [PubMed] [Google Scholar]
- Niu T. H. et al. Lack of association between apolipoprotein C3 gene polymorphisms and risk of nonalcoholic fatty liver disease in a Chinese Han population. World J Gastroenterol 20, 3655–3662 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. S. et al. Role of the PNPLA3 I148M polymorphism in nonalcoholic fatty liver disease and fibrosis in Korea. Digestive diseases and sciences 59, 2967–2974 (2014). [DOI] [PubMed] [Google Scholar]
- Chen W., Chang B., Li L. & Chan L. Patatin-like phospholipase domain-containing 3/adiponutrin deficiency in mice is not associated with fatty liver disease. Hepatology 52, 1134–1142 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly A. K., Ballestri S., Carulli L., Loria P. & Day C. P. Genetic determinants of susceptibility and severity in nonalcoholic fatty liver disease. Expert Rev Gastroenterol Hepatol 5, 253–263 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information