Abstract
Primary ciliary dyskinesia (PCD) is a genetically heterogeneous, autosomal recessive disorder that results from functional and ultrastructural abnormalities of motile cilia. Patients with PCD have diverse clinical phenotypes that include chronic upper and lower respiratory tract infections, situs inversus, heterotaxy with or without congenital heart disease, and male infertility, among others. In this report, the carrier frequencies for eleven mutations in eight PCD-associated genes (DNAI1, DNAI2, DNAH5, DNAH11, CCDC114, CCDC40, CCDC65, and C21orf59) that had been found in individuals of Ashkenazi Jewish descent were investigated in order to advise on including them in existing clinical mutation panels for this population. Results showed relatively high carrier frequencies for the DNAH5 c.7502G>C mutation (0.58%), the DNAI2 c.1304G>A mutation (0.50%), and the C21orf59 c.735C>G mutation (0.48%), as well as lower frequencies for mutations in DNAI1, CCDC65, CCDC114, and DNAH11 (0.10–0.29%). These results suggest that several of these genes should be considered for inclusion in carrier screening panels in the Ashkenazi Jewish population.
Keywords: Ashkenazi, carrier frequency, Kartagener syndrome, primary ciliary dyskinesia, situs inversus
Introduction
Primary ciliary dyskinesia (PCD, MIM: 244400) is a genetically heterogeneous, autosomal recessive disorder that results from functional and ultrastructural abnormalities of motile cilia. Phenotypes of PCD can be diverse, with irregularities in motile respiratory cilia causing chronic upper and lower respiratory tract infections, in embryonic nodal cilia causing situs inversus and heterotaxy with or without congenital heart disease, and in spermatozoa flagella causing male infertility, among others clinical features (Zariwala et al. 2007; Escudier et al. 2009; Leigh et al. 2009; Knowles et al. 2013a). PCD can be diagnosed in various manners including the detection of ultrastructural defects by transmission electron microscopy, abnormal ciliary beat frequencies or patterns, low levels of nasal nitric oxide, the presentation of typical clinical features, and mutation analysis in ciliary genes (Zariwala et al. 2007; Escudier et al. 2009; Leigh et al. 2009, 2013; Knowles et al. 2013a; Svobodova et al. 2013).
Primary ciliary dyskinesia has extensive locus and allelic heterogeneity that makes it challenging for genetic diagnoses. Due to the high number of causative PCD mutations, tiered mutation screening methods have been suggested for PCD patients (Hornef et al. 2006; Zariwala et al. 2006; Djakow et al. 2012). In tiered screenings, mutations are ranked based on their prevalence in the PCD population, with the most common mutations screened first in an effort to increase efficiency. Founder effects have resulted in certain recessive mutations to appear more frequently in specific ethnic populations as well, which allows for a different ranking system to be made based on the frequency of mutations in a specific ethnic group or geographic isolates. An example of such effects has been seen in PCD where a splice-site mutation in RSPH4A (MIM 612649, 612647) is found in people of Hispanic origin from Puerto Rico (Daniels et al. 2013). Since the Ashkenazi Jewish population has an increased prevalence of recessive mutations due to past founder effects (Bray et al. 2010), carrier frequencies were performed for eleven mutations in eight PCD genes that had been observed in affected individuals of Ashkenazi Jewish descent in order to advise on screening for them.
Of the 11 mutations evaluated, six were in four genes known to cause defective ciliary outer dynein arm (ODA) complexes (Hornef et al. 2006; Zariwala et al. 2006; Knowles et al. 2013b). These included mutations in DNAI1 (OMIM# 604366) (NM_012144.2) c.1490G>A (r.1402_1569del) (p.Arg468_Lys523del) (Zariwala et al. 2006), DNAI2 (OMIM# 605483) (NM_023036.4) c.1304G>A (p.Trp435*) (Knowles et al. 2013b), CCDC114 (OMIM# 615038) (NM_144557.3) c.939delT (p.His313Glnfs*14) (Knowles et al. 2013b), and three in DNAH5 (OMIM# 603335) (NM_001369.2): c.7502G>C (p.Arg2501Pro) (Hornef et al. 2006), novel c.5545G>A (p.Ala1849Thr) and novel c.6988+2T>C (g.IVS42+2T>C) (p. splice?). Two of the remaining mutations were in CCDC40 (OMIM# 613799) (NM_017950.3) c.248delC (p.Ala83Valfs*84) (Becker-Heck et al. 2011; Antony et al. 2012) which causes defective inner dynein arm (IDA) complexes together with microtubular disorganization in a subset of cilia, and C21orf59 (OMIM# 615494) (NM_021254.2) c.735C>G (p.Tyr245*) (Austin-Tse et al. 2013) which causes the absence of both ODAs and IDAs. The remaining three mutations were from two genes that do not cause any detectable ciliary ultrastructural defects (Schwabe et al. 2008; Knowles et al. 2012; Austin-Tse et al. 2013; Horani et al. 2013) and included the c.877_878delAT (p.Ile293Profs*2) in CCDC65 (OMIM# 611088) (NM_033124.4) (Austin-Tse et al. 2013; Horani et al. 2013) and two mutations in DNAH11(OMIM# 603339) (NM_001277115.1): c.6244C>T (p.Arg2082*) and c.11929G>T (p.Glu3977*) (Knowles et al. 2012).
Materials and Methods
Ethics statement
The samples used in this study were obtained with written patient consent from self-identified Ashkenazi Jews enrolled in the carrier testing Dor Yeshorim program (Ekstein and Katzenstein 2001) to be used for research purposes. Consent form information included that patient material would be used for clinical testing and that excess material would be de-identified and used for research purposes to characterize single-gene disorders in the Ashkenazi Jewish population. The positive control samples came from the Institutional review board approved research cohort for the protection of the rights of human subjects that were recruited under the auspices of the Genetic Disorders of Mucociliary Clearance consortium. Institutional review board permission was not required for the control samples used in the carrier frequency study because all sample identifiers were removed prior to receipt by the laboratory where the TaqMan assays were carried out (45 CFR part 46.101(b)(4)).
Patients
The positive control samples came from the individuals harboring mutations who self-identified as being from an Ashkenazi Jewish descent. The genotypes for the majority of the positive controls for the mutations in DNAH11 (NM_001277115.1), DNAI1 (NM_012144.2), DNAI2 (NM_023036.4), CCDC114 (NM_144577.3), CCDC40 (NM_017950.3), C21orf59 (NM_021254.2), and CCDC65 (NM_033124.4) have previously been published (Zariwala et al. 2006; Antony et al. 2012; Knowles et al. 2012; Austin-Tse et al. 2013; Knowles et al. 2013b). In addition, for the previously known c.7502G>C missense mutation (Hornef et al. 2006)and a novel c.6988+2T>C splice-site mutation in DNAH5 (NM_001369.2), we used gDNA from the affected individual (#826), who harbored both mutations. Individual #826 had an affected sibling with the identical DNAH5 genotypes and both unaffected parents were carriers; thus, indicating both mutations in the affected individuals were inherited in trans. For the c.5545G>A mutation in DNAH5, the gDNA from an unaffected carrier mother of Ashkenazi descent was used. This mutation was observed in a PCD affected individual who harbored a splice-site mutation on the trans allele that was inherited from a non-Ashkenazi unaffected father; thus the splice-site mutation was not evaluated.
Assay design and validation
To design the genotyping assays, the full sequence of each gene was obtained from the National Center for Biotechnology Information (NCBI) (U.S. National Library of Medicine, Bethesda, MD). Roughly 200 base pairs upstream and downstream of the mutation site were selected and repetitive sequences and SNPs were masked using Repeat Masker (Institute for Systems Biology, Seattle, WA) and NCBI specialized Basic Local Alignment Search Tool (BLAST) using the SNP Flanks option, respectively. The assays were then made in File Builder software (Life Technologies [LTI], Carlsbad, CA) with sequence-specific forward and reverse primers to amplify the polymorphic sequences, and VIC and FAM fluorescent-labeled probes to detect the normal and mutant alleles, respectively (Table1). A no template control consisting of water, three wild-type samples, and one known heterozygous carrier sample were used to validate all of the assays except for the CCDC40 assay which was validated on homozygous affected gDNA. The genotypes for all of the control samples were confirmed using Sanger Sequencing. The samples and assays were plated in duplicate in a 384 well plate along with TaqMan Genotyping Master Mix (LTI) (final volume 5 μL) and run in duplex real-time PCR reactions followed by allelic discrimination on the ABI PRISM® 7900 HT Sequence Detection System using SDS 2.3 software (LTI).
Table 1.
TaqMan assay design including primer and probe sequences
| Genes | Mutations | Forward primer | Reverse primer | VIC probe | FAM probe |
|---|---|---|---|---|---|
| DNAI1 | c.1490G>A | CCTCTTGGAACTGGGCTAAGC | CATGTAGTCAATCTCTTTGTGGAAGTCA | TCTTTTCCCAAGGTTGTG | TTTTCCCAAGATTGTG |
| DNAI2 | c.1304G>A | CGTTTTCTTTACCACCAGGATGGA | GGATCGCACTGCTCGAACAT | CTGGATATCTGGGACTTC | CCTGGATATCTAGGACTTC |
| DNAH11 | c.6244C>T | GTGCTATTAAGTCTGTCTTGGTTGTG | TCATATTAGCATTGCAGTACCTGATCTTC | ATTTTTATCTCCTCGTTTCAG | TTTTTATCTCCTCATTTCAG |
| DNAH11 | c.11929G>T | CAGGAGACGGTGGCAGAAG | CCCAGTGTCCTCCTTTGGAA | TGGCCCTGGAGAAAG | TGGCCCTGTAGAAAG |
| DNAH5 | c.6988+2T>C | GGGATATTTTCTACGCTTTGGAGGAA | GTATAGCCTCCAAGGATTCTATCTAGAAGT | TTGTAATTTATCTTCTACCTTTCT | AATTTATCTTCTGCCTTTCT |
| DNAH5 | c.7502G>C | TCGCGCTGCTGTGGAG | CGCAGCCAGAGCTCCAG | ACGGACGGCGCCGC | ACGGACCGCGCCGC |
| DNAH5 | c.5545G>A | TGATATGGACACGGGATTCAGAAGA | CTCCAGGAAAGCCTGATTAGTTTTC | AGCCCTTAGAAATGCCAAGT | AGCCCTTAGAAATACCAAGT |
| CCDC40 | c.248delC | AAGCGGAAGCTGCAATTGA | TCCTCTTCGCTTTCAGCATCTC | AGGAGGCTGTGTCCTA | AGGAGGTGTGTCCTATG |
| CCDC65 | c.877_878delAT | GCCATAACTATTTCAAAAGGCAAGATCA | CGCAGTTGTACAAGGACCAATTC | ATGAGAACCGGTATATCCGTA | ATGAGAACCGGTATCCGTA |
| CCDC114 | c.939delT | TCATCAACGAGCAGAACTTGGA | AGGCCTCACTCACCTCCTTGA | CTGGAGCATGTGCAGGA | AGCAGTGCAGGAAGA |
| C21orf59 | c.735C>G | GAGGAGCAGAAGCAGCTGAT | GCACTGCAGAAAGCCCATCT | TGTCTTCTGTGATAGTACAGC | TGTCTTCTGTGATACTACAGC |
GenBank reference sequence and version number for the genes studied: DNAI1 (NM_012144.2), DNAI2 (NM_023036.4), DNAH11 (NM_001277115.1), DNAH5 (NM_001369.2), CCDC40 (NM_017950.3), CCDC65 (NM_033124.4), CCDC114 (NM_144577.3), and C21orf59 (NM_021254.2).
Carrier frequency study
For the carrier frequency study, ∽1000 samples were analyzed for each mutation. The gDNA samples were not normalized prior to plating, but almost all samples fell within the suggested range of 1–20 ng (LTI). The plates were run on the GeneAmp® PCR System 9700 (LTI) at the following setting: holds at 50°C for 2 min and 95°C for 10 min, and then 40 cycles at 95°C for 15 sec and 60°C for 1 min. Allelic discrimination was then performed on the ABI PRISM® 7900 HT Sequence Detection System using SDS 2.3 software and the data were analyzed using TaqMan Genotyper v1.1 software (LTI). Any samples that did not amplify were not included in the carrier frequency calculations. The samples from the initial validation were used as controls. The Wilson score interval (Wilson 1927) was used to calculate the confidence intervals (CI) for carrier frequencies.
Results
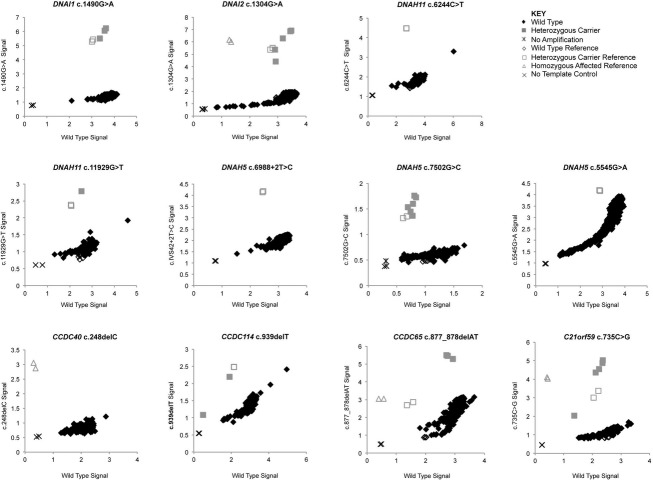
The initial validation of the assays on a small scale yielded 100% genotyping accuracy. The carrier frequency experiments were then performed on ∽1000 samples from individuals of Ashkenazi Jewish descent. The carrier frequency results were as follows: 0.28% (CI 0.09–0.83%) for the DNAI1 c.1490G>A mutation, 0.50% (CI 0.21–1.16%) for the DNAI2 c.1304G>A mutation, 0.10% (CI 0.02–0.54%) for the DNAH11 c.11929G>T mutation, 0.58% (CI 0.27–1.26%) for the DNAH5 c.7502G>C mutation, 0.19% (CI 0.05–0.69%) for the CCDC114 c.939delT mutation, 0.29% (CI 0.10–0.85%) for the CCDC65 c.877_878delAT mutation, and 0.48% (CI 0.20–1.12%) for the C21orf59 c.735C>G mutation. No carriers were detected for the DNAH5 c.6988+2T>C and c.5545G>A, the DNAH11 c.6244C>T, or the CCDC40 c.248delC mutations. TaqMan allelic discrimination result plots can be seen in Figure1 for all of the mutations, and a summary of the carrier frequencies can be seen in Table2. Samples identified as being heterozygous carriers were confirmed by Sanger Sequencing (if gDNA was available), and all genotypes were concordant between the two methods.
Figure 1.
Carrier frequency allelic discrimination plots. For all plots, the VIC probe (wild-type allele) is represented by the X-axis, and the FAM probe (mutant allele) is represented by the Y-axis. Sterile water was used as the no template control. The GenBank reference sequence and version number for the genes studied are as follows: DNAI1 (NM_012144.2), DNAI2 (NM_023036.4), DNAH11 (NM_001277115.1), DNAH5 (NM_001369.2), CCDC40 (NM_017950.3), CCDC65 (NM_033124.4), CCDC114 (NM_144577.3), and C21orf59 (NM_021254.2).
Table 2.
Carrier frequency of 11 mutations in eight primary ciliary dyskinesia-associated genes
| Genes | Mutations | No. of individuals wild type | No. of. individuals heterozygous carrier1 | Carrier frequency (%) and confidence interval (%) |
|---|---|---|---|---|
| DNAI1 | c.1490G>A | 1052 | 3 | 0.28 (0.09–0.83) |
| DNAI2 | c.1304G>A | 1000 | 5 | 0.50 (0.21–1.16) |
| DNAH11 | c.6244C>T | 1052 | 0 | 0.00 |
| DNAH11 | c.11929G>T | 1051 | 1 | 0.10 (0.02–0.54) |
| DNAH5 | c.6988+2T>C | 1050 | 0 | 0.00 |
| DNAH5 | c.7502G>C | 1036 | 6 | 0.58 (0.27–1.26) |
| DNAH5 | c.5545G>A | 1051 | 0 | 0.00 |
| CCDC40 | c.248delC | 1052 | 0 | 0.00 |
| CCDC65 | c.877_878delAT | 1032 | 3 | 0.29 (0.10–0.85) |
| CCDC114 | c.939delT | 1054 | 2 | 0.19 (0.05–0.69) |
| C21orf59 | c.735C>G | 1031 | 5 | 0.48 (0.20–1.12) |
None of the individuals were identified as being homozygous for the mutations.
Discussion
Because of the extensive heterogeneous nature of PCD, knowing if certain mutations are more prevalent in a specific ethnic population can help prioritize variants for expeditious screening. In this study, 11 different mutations represented by eight genes that had all been previously found in individuals of Ashkenazi Jewish descent were examined. Of these, seven mutations representing seven genes were found to have carrier frequencies ranging from 0.1% to 0.58% in the population.
Of all the mutations screened, the highest frequency (0.58%) was found for the c.7502G>C mutation in the DNAH5 gene. Mutations in this gene are the most common in PCD patients in the general population (Hornef et al. 2006; Failly et al. 2009), accounting for 28% of all PCD families and 49% of PCD families that have ODA defects (Zariwala et al. 2007). The 0.48% frequency of the c.735C>G mutation in C21orf59 and the 0.29% for the c.877_878delAT mutation in CCDC65 are believed to be founder mutations, which explains their relatively high frequencies (Austin-Tse et al. 2013). The 0.29% carrier frequency for the c.877_878delAT CCDC65 was slightly lower than the 0.41% previously detected in this population (Horani et al. 2013), which may reflect that this study was conducted on the orthodox as opposed to the general Ashkenazi population. Other mutations that had been previously reported to be at mutation hotspots for PCD patients had no carriers detected, such as the c.248delC mutation in CCDC40 (Antony et al. 2012).
Based on the various frequencies found, we suggest that the seven mutations that had carriers detected be recommended for inclusion in mutation screening panels specific for the Ashkenazi population since there is a chance of compound heterozygosity based on the prevalence of the various mutations. While none of the mutations meet the inclusion criteria of a 1% frequency set by the American College of Medical Genetics (ACMG) (Gross et al. 2008) the advances made in high-throughput screening programs (Fedick et al. 2013) have made screening for additional and/or rare mutations possible and affordable, with many commercial laboratories already offering ethnic screening panels that extend beyond the scope of mutations specifically recommended by the ACMG and other such organizations (Lazarin et al. 2013) Since the ACMG, along with the American College of Obstetricians and Gynecologists, has acknowledged that individuals may want to be screened for additional disorders, we suggest that information for all of the PCD genes and mutations studied here be made available when receiving genetic counseling to aid patients in making informed decisions (Monaghan et al. 2008; ACOG Committee on Genetics 2009).
Acknowledgments
The authors thank the PCD subjects, family members, and the US PCD Foundation. The authors thank Dor Yeshorim for providing the samples used in the carrier frequency studies and Bonei Olam for funding this project. They acknowledge investigators and coordinators of the Genetic Disorders of Mucociliary Clearance consortium (http://rarediseasesnetwork.epi.usf.edu/gdmcc/), Dr. Margaret Leigh, Mr. Daniel Stewart and Ms. Whitney Wolf from the University of North Carolina, Chapel Hill. The Genetic Disorders of Mucociliary Clearance Consortium (U54HL096458) is a part of the National Institutes of Health (NIH) Rare Disease Clinical Research Network (RDCRN), supported through collaboration between the NIH Office of Rare Diseases Research (ORDR) at the National Center for Advancing Translational Science (NCATS), and the National Heart, Lung & Blood Institute (NHLBI). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of Interest
None declared.
References
- ACOG Committee on Genetics. ACOG Committee Opinion No. 442: preconception and prenatal carrier screening for genetic diseases in individuals of Eastern European Jewish descent. Obstet. Gynecol. 2009;114:950–953. doi: 10.1097/AOG.0b013e3181bd12f4. [DOI] [PubMed] [Google Scholar]
- Antony D, Becker-Heck A, Zariwala MA, Schmidts M, Onoufriadis A, Forouhan M, et al. Mutations in CCDC39 and CCDC40 are the major cause of primary ciliary dyskinesia with axonemal disorganization and absent inner dynein arms. Hum. Mutat. 2012;34:462–472. doi: 10.1002/humu.22261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin-Tse C, Halbritter J, Zariwala MA, Gilberti RM, Gee HY, Hellman N, et al. Zebrafish ciliopathy screen plus human mutational analysis identifies C21orf59 and CCDC65 defects as causing primary ciliary dyskinesia. Am. J. Hum. Genet. 2013;93:672–686. doi: 10.1016/j.ajhg.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker-Heck A, Zohn IE, Okabe N, Pollock A, Lenhart KB, Sullivan-Brown J, et al. The coiled-coil domain containing protein CCDC40 is essential for motile cilia function and left-right axis formation. Nat. Genet. 2011;43:79–84. doi: 10.1038/ng.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray SM, Mulle JG, Dodd AF, Pulver AE, Wooding S. Warren ST. Signatures of founder effects, admixture, and selection in the Ashkenazi Jewish population. Proc. Natl Acad. Sci. USA. 2010;107:16222–16227. doi: 10.1073/pnas.1004381107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels ML, Leigh MW, Davis SD, Armstrong MC, Carson JL, Hazucha M, et al. Founder mutation in RSPH4A identified in patients of Hispanic descent with primary ciliary dyskinesia. Hum. Mutat. 2013;34:1352–1356. doi: 10.1002/humu.22371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djakow J, Svobodova T, Hrach K, Uhlik J, Cinek O. Pohunek P. Effectiveness of sequencing selected exons of DNAH5 and DNAI1 in diagnosis of primary ciliary dyskinesia. Pediatr. Pulmonol. 2012;47:864–875. doi: 10.1002/ppul.22520. [DOI] [PubMed] [Google Scholar]
- Ekstein J. Katzenstein H. The Dor Yeshorim story: community-based carrier screening for Tay-Sachs disease. Adv. Genet. 2001;44:297–310. doi: 10.1016/s0065-2660(01)44087-9. [DOI] [PubMed] [Google Scholar]
- Escudier E, Duquesnoy P, Papon JF. Amselem S. Ciliary defects and genetics of primary ciliary dyskinesia. Paediatr. Respir. Rev. 2009;10:51–54. doi: 10.1016/j.prrv.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Failly M, Bartoloni L, Letourneau A, Munoz A, Falconnet E, Rossier C, et al. Mutations in DNAH5 account for only 15% of a non-preselected cohort of patients with primary ciliary dyskinesia. J. Med. Genet. 2009;46:281–286. doi: 10.1136/jmg.2008.061176. [DOI] [PubMed] [Google Scholar]
- Fedick A, Su J, Jalas C, Northrop L, Devkota B, Ekstein J, et al. High-throughput carrier screening using TaqMan allelic discrimination. PLoS One. 2013;8:1–9. doi: 10.1371/journal.pone.0059722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross SJ, Pletcher BA. Monaghan KG. Carrier screening in individuals of Ashkenazi Jewish descent. Genet. Med. 2008;10:54–56. doi: 10.1097/GIM.0b013e31815f247c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horani A, Brody SL, Ferkol TW, Shoseyov D, Wasserman MG, Ta-shma A, et al. CCDC65 mutation causes primary ciliary dyskinesia with normal ultrastructure and hyperkinetic cilia. PLoS One. 2013;8:e72299. doi: 10.1371/journal.pone.0072299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornef N, Olbrich H, Horvath J, Zariwala MA, Fliegauf M, Loges NT, et al. DNAH5 mutations are a common cause of primary ciliary dyskinesia with outer dynein arm defects. Am. J. Respir. Crit. Care Med. 2006;174:120–126. doi: 10.1164/rccm.200601-084OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles MR, Leigh MW, Carson JL, Davis SD, Dell SD, Ferkol TW, et al. Mutations of DNAH11 in patients with primary ciliary dyskinesia with normal ciliary ultrastructure. Thorax. 2012;67:433–441. doi: 10.1136/thoraxjnl-2011-200301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles MR, Daniels LA, Davis SD, Zariwala MA. Leigh MW. Primary ciliary dyskinesia. Recent advances in diagnostics, genetics, and characterization of clinical disease. Am. J. Respir. Crit. Care Med. 2013a;188:913–922. doi: 10.1164/rccm.201301-0059CI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles MR, Leigh MW, Ostrowski LE, Huang L, Carson JL, Hazucha MJ, et al. Exome sequencing identifies mutations in CCDC114 as a cause of primary ciliary dyskinesia. Am. J. Hum. Genet. 2013b;92:99–106. doi: 10.1016/j.ajhg.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarin GA, Haque IS, Nazareth S, Iori K, Patterson AS, Jacobson JL, et al. An empirical estimate of carrier frequencies for 400+ causal Mendelian variants: results from an ethnically diverse clinical sample of 23,453 individuals. Genet. Med. 2013;15:178–186. doi: 10.1038/gim.2012.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh MW, Zariwala MA. Knowles MR. Primary ciliary dyskinesia: improving the diagnostic approach. Curr. Opin. Pediatr. 2009;21:320–325. doi: 10.1097/MOP.0b013e328329cddb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh MW, Hazucha MJ, Chawla KK, Baker BR, Shapiro AJ, Brown DE, et al. Standardizing nasal nitric oxide measurement as a test for primary ciliary dyskinesia. Ann. Am. Thorac. Soc. 2013;10:574–581. doi: 10.1513/AnnalsATS.201305-110OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan KG, Feldman GL, Palomaki GE. Spector EB. Technical standards and guidelines for reproductive screening in the Ashkenazi Jewish population. Genet. Med. 2008;10:57–72. doi: 10.1097/GIM.0b013e31815f6eac. [DOI] [PubMed] [Google Scholar]
- Schwabe GC, Hoffmann K, Loges NT, Birker D, Rossier C, de Santi MM, et al. Primary ciliary dyskinesia associated with normal axoneme ultrastructure is caused by DNAH11 mutations. Hum. Mutat. 2008;29:289–298. doi: 10.1002/humu.20656. [DOI] [PubMed] [Google Scholar]
- Svobodova T, Djakow J, Zemkova D, Cipra A, Pohunek P. Lebl J. Impaired growth during childhood in patients with primary ciliary dyskinesia. Int. J. Endocrinol. 2013;2013:731423. doi: 10.1155/2013/731423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson EB. Probable inference, the law of succession, and statistical inference. J. Am. Stat. Assoc. 1927;22:209–212. [Google Scholar]
- Zariwala MA, Leigh MW, Ceppa F, Kennedy MP, Noone PG, Carson JL, et al. Mutations of DNAI1 in primary ciliary dyskinesia: evidence of founder effect in a common mutation. Am. J. Respir. Crit. Care Med. 2006;174:858–866. doi: 10.1164/rccm.200603-370OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zariwala MA, Knowles MR. Omran H. Genetic defects in ciliary structure and function. Annu. Rev. Physiol. 2007;69:423–450. doi: 10.1146/annurev.physiol.69.040705.141301. [DOI] [PubMed] [Google Scholar]