Abstract
Targeting of the Gag polyprotein to the plasma membrane (PM) for assembly is a critical event in the late phase of immunodeficiency virus type-1 (HIV-1) infection. Gag binding to the PM is mediated by interactions between the myristoylated matrix (MA) domain and PM lipids. Despite the extensive biochemical and in vitro studies of Gag and MA binding to membranes over the last two decades, the discovery of the role of phosphatidylinositol-4,5-bisphosphate [PI(4,5)P2] in Gag binding to the PM has sparked a string of studies aimed at elucidating the molecular mechanism of retroviral Gag–PM binding. Electrostatic interactions between a highly conserved basic region of MA and acidic phospholipids have long been thought to be the main driving force for Gag–membrane interactions. However, recent studies suggest that the mechanism is rather complex since other factors such as the hydrophobicity of the membrane interior represented by the acyl chains and cholesterol also play important roles. Here we summarize the current understanding of HIV-1 Gag–membrane interactions at the molecular and structural levels and briefly discuss the underlying forces governing interactions of other retroviral MA proteins with the PM.
Keywords: HIV-1; Gag; matrix; myristoyl; NMR; plasma membrane; PI(4,5)P2
Introduction
Prior to assembly on the plasma membrane (PM), the human immunodeficiency virus type-1 (HIV-1) Gag polyprotein adopts a compact “folded over” conformation and exists in the monomeric or low-order oligomeric states (Datta et al., 2007a, 2011; Kutluay and Bieniasz, 2010; Kutluay et al., 2014). Whereas it is established that the nucleocapsid (NC) domain of Gag specifically recognizes motifs in the viral RNA genome for packaging (Jouvenet et al., 2009; Kutluay et al., 2014), there is compelling evidence that the matrix (MA) domain also binds to cellular RNA to prevent premature Gag targeting to intracellular membranes (Figure 1; Chukkapalli et al., 2010, 2013; Chukkapalli and Ono, 2011; Hogue et al., 2012; Inlora et al., 2014; Kutluay et al., 2014; Olety and Ono, 2014). Upon transport of Gag to the PM, the interaction of MA with RNA is exchanged for an interaction of MA with PM components (Figure 1; Chukkapalli et al., 2010, 2013; Chukkapalli and Ono, 2011; Hogue et al., 2012; Inlora et al., 2014; Olety and Ono, 2014). This molecular switch induces an extended conformation of Gag, leading to formation of high-order Gag oligomers on the PM (Datta et al., 2007a,b, 2011; Munro et al., 2014). The key to understanding this essential switch is elucidating at the molecular level the interaction of MA with specific PM components. Our current understanding of Gag–PM interaction is incomplete because of the lack of molecular details on how various membrane components contribute to the overall binding and how they control this molecular switch.
FIGURE 1.
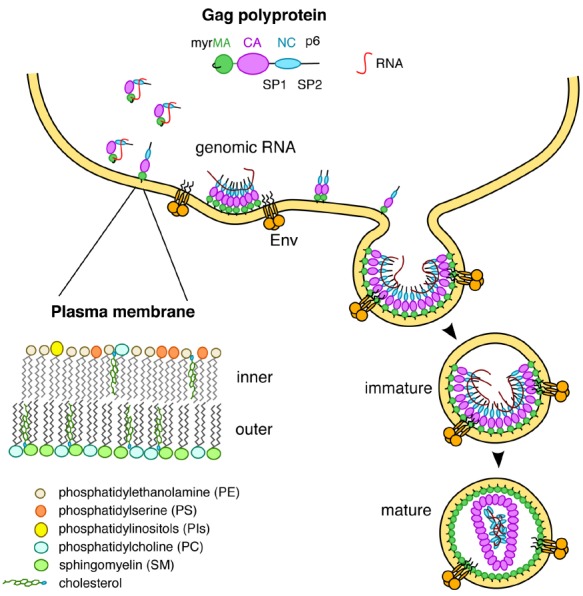
The MA domain of Gag is involved in multiple interactions including RNA and lipids on the inner leaflet of the PM.
Factors that Control HIV-1 Gag Assembly on the PM
For most retroviruses, assembly of the Gag proteins occurs on the PM of the infected cell (Ono et al., 2004; Jouvenet et al., 2006, 2008; Finzi et al., 2007; Welsch et al., 2007; Chukkapalli et al., 2008, 2010; Chu et al., 2010; Chukkapalli and Ono, 2011). The role of MA domain in Gag–PM binding is indispensable. Several factors can influence Gag–membrane binding including the myristoyl (myr) group, a conserved basic region in MA, protein multimerization, cellular RNA, and phosphatidylinositol-4,5-bisphosphate [PI(4,5)P2] (Ono and Freed, 1999; Ono et al., 2004; Jouvenet et al., 2006; Dalton et al., 2007; Chukkapalli et al., 2008, 2010, 2013; Alfadhli et al., 2011; Chukkapalli and Ono, 2011; Ghanam et al., 2012). The finding that Gag binds to membranes more efficiently than the isolated MA protein led to the hypothesis that the myr group is exposed in Gag and sequestered in the MA protein, which has become known as “the myr switch mechanism” (Zhou and Resh, 1996; Spearman et al., 1997; Hermida-Matsumoto and Resh, 1999; Ono and Freed, 1999; Paillart and Gottlinger, 1999). For over two decades, biochemical, in vivo, in vitro, and genetic data have provided invaluable insights on multiple factors that modulate Gag–membrane binding. However, only recently the molecular and structural determinants of this interaction have begun to emerge (Saad et al., 2006, 2007b, 2008; Shkriabai et al., 2006; Chukkapalli et al., 2008, 2010, 2013; Chukkapalli and Ono, 2011; Prchal et al., 2012; Vlach and Saad, 2013). Nuclear magnetic resonance (NMR) and analytical ultracentrifugation studies revealed that the myr group can adopt sequestered and exposed conformations in the MA protein, that the MA protein resides in monomer-trimer equilibrium, and that myr exposure is coupled with protein trimerization (Tang et al., 2004; Saad et al., 2007b). Exposure of the myr group is also modulated by other factors including the solution pH, inclusion of the CA domain, and binding of calmodulin (Tang et al., 2004; Fledderman et al., 2010; Ghanam et al., 2010). There is now convincing evidence that binding of RNA to MA prevents Gag from interacting with intracellular membranes (Chukkapalli et al., 2010, 2013; Chukkapalli and Ono, 2011; Inlora et al., 2014). As a consequence, RNA is considered as a negative regulator of Gag–membrane binding. Recent studies have shown that the MA domain binds almost exclusively to specific tRNAs in the cytosol (Kutluay et al., 2014). Incorporation of PI(4,5)P2 in membranes inhibits the interaction between MA and cellular RNA (Chukkapalli et al., 2010, 2013; Chukkapalli and Ono, 2011). Binding of Gag to membranes induces an extended conformation in the absence (Datta et al., 2011) or presence (Datta et al., 2007b) of inositol phosphates. Altogether, these studies indicate that particle assembly is regulated by coordinated interactions between the MA and NC domains of Gag with RNA and membrane lipids.
Structural Studies of HIV-1 MA Binding to PM Lipids
Proper targeting of HIV-1 Gag to the PM is dependent on specific interactions between the MA domain and PI(4,5)P2 (Ono et al., 2004; Chukkapalli et al., 2008, 2010; Chukkapalli and Ono, 2011; Inlora et al., 2014). The most abundant form of PI(4,5)P2 contains saturated 18-carbon 1′ and 20-carbon unsaturated 2′ fatty acid chains (Dudley and Spector, 1986), which promote micelle formation in aqueous solution (Janmey et al., 1987). Interactions of HIV-1 Gag and MA with PI(4,5)P2 have been detected by mass spectrometric protein footprinting (Shkriabai et al., 2006). Titration of native PI(4,5)P2 into MA samples led to severe broadening and loss of NMR signals. Therefore, soluble analogs of PI(4,5)P2 with truncated 1′- and 2′-acyl chains (C4 or C8) have been used (Saad et al., 2006, 2007a, 2008). NMR studies have shown that soluble analogs of PI(4,5)P2 bind directly to HIV-1 MA, inducing a conformational change that promotes myr exposure (Saad et al., 2006). The solution structure of the MA–PI(4,5)P2 complex revealed that the 2′-acyl chain is inserted in a hydrophobic cleft, whereas the inositol group is packed against a highly basic region of MA (Figure 2A). The 1′-acyl chain, however, is not involved in binding and is exposed to solvent. It was suggested that PI(4,5)P2 can function as both an allosteric trigger for myr exposure and as a direct membrane anchor (Saad et al., 2006). The involvement of the acyl chain of PI(4,5)P2 in MA and Gag binding has been confirmed by surface plasmon resonance methods (Anraku et al., 2010). Based on the NMR studies, a structural model for Gag bound to PM has been proposed. In this model, MA is anchored to the membrane by the myr group and 1′-acyl chain of PI(4,5)P2, which bracket a patch of conserved basic residues that can interact with the negatively charged surface of PM (Saad et al., 2006). A molecular model of MA bound to native PI(4,5)P2 shows that a longer 2′-acyl chain (18 carbons) can be accommodated into the hydrophobic cleft (Saad et al., 2006). Structural studies have yet to determine the precise mode of MA binding to native PI(4,5)P2 within the context of a membrane bilayer.
FIGURE 2.
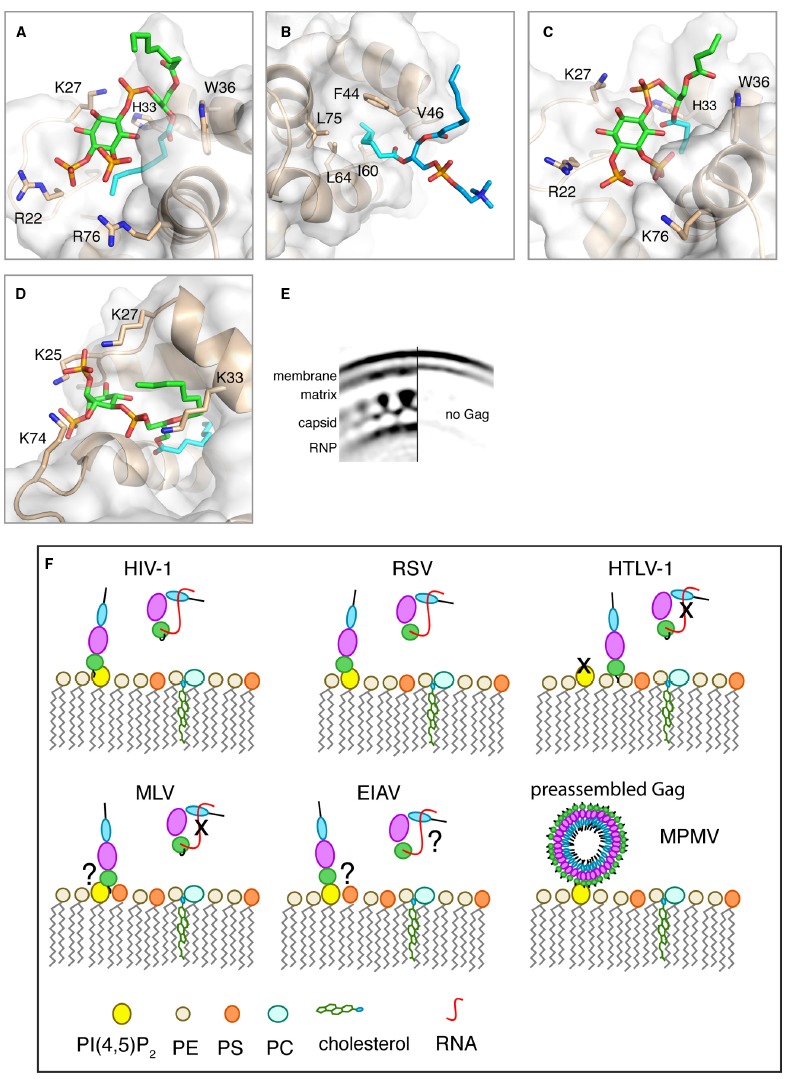
(A–D) Close-up views of structures of HIV-1, HIV-2, and M-PMV MA proteins bound to lipids. MA proteins are shown in ribbon and surface representations with residues implicated in binding shown as sticks. Phospholipids are shown as green [PI(4,5)P2] or blue (PC) sticks and their acyl chains involved in binding are shown in cyan. (A) HIV-1 myr(–)MA bound to di-C8-PI(4,5)P2 (PDB ID: 2H3V). (B) HIV-1 myr(–)MA bound to di-C8-PC (PDB ID: 2LYA). (C) HIV-2 MA bound di-C4-PI(4,5)P2 (PDB ID: 2K4I). (D) M-PMV MA bound to di-C8-PI(4,5)P2 structure provided by Prchal et al. (2012). (E) Cryoelectron microscopy reconstruction of HIV-1 immature particle in a section with (left) or without (right) Gag polyprotein present. Used with permission [Copyright (2009) National Academy of Sciences, USA (Briggs et al., 2009)]. (F) A schematic representation of potential mechanisms for Gag binding to the PM for different retroviruses. Gag binding to membranes is regulated by PI(4,5)P2 and RNA for some but not all retroviruses. Binding of Gag to membranes via the MA domain displaces RNA, which binds non-specifically to the basic region of MA. Data are not in agreement on the role of PI(4,5)P2 in MLV Gag binding to the PM. For EIAV, Gag binding to membranes appears to have no specific requirement for PI(4,5)P2 since other phosphoinositides may also play a role. For M-PMV, Gag assembly occurs in the cytoplasm prior to transport to the PM where MA specifically recognizes PI(4,5)P2.
In addition to PI(4,5)P2, other lipids have been implicated in HIV-1 Gag–membrane binding (Dalton et al., 2007; Ono, 2010a; Waheed and Freed, 2010; Chan et al., 2011; Chukkapalli and Ono, 2011; Chukkapalli et al., 2013). The affinity of Gag and MA to membranes is increased upon increasing the stoichiometry of phosphatidylserine (PS; Ehrlich et al., 1996; Zhou and Resh, 1996; Scarlata et al., 1998; Dalton et al., 2007; Chukkapalli et al., 2008, 2010, 2013; Alfadhli et al., 2009; Chan et al., 2011). NMR studies on HIV-1 MA binding to soluble analogs of PM lipids revealed that PS, phosphatidylethanolamine (PE) and phosphatidylcholine (PC) bind directly to MA via a distinct site that is adjacent to the PI(4,5)P2 binding site (Vlach and Saad, 2013). All three phospholipids interact with MA via sequestration of the 2′-acyl chain into a cavity formed by hydrophobic residues in helices α2 and α3, leaving the 1′-acyl exposed (Figure 2B). Surprisingly, the polar heads of PE, PS, or PC do not appear to play a major role in stabilizing the complex. Structural studies have been conducted with single lipids. It is likely that in a membrane environment the lipid polar heads will orient to make favorable electrostatic contacts with the basic domain of MA. Consistent with the hydrophobic nature of binding, the affinities of all three phospholipids to MA increased by two orders of magnitude upon extending the acyl chains from hexanoyl to octanoyl (Vlach and Saad, 2013). Intriguingly, the myr group is readily exposed when MA is bound to membrane mimetics such as micelles and bicelles in the absence of PI(4,5)P2. Exposure of the myr group does not appear to be triggered by an allosteric mechanism as in the case of PI(4,5)P2 binding, but rather by the mere presence of a lipid agglomerate. Based on these studies, we have proposed a trio engagement model by which HIV-1 Gag is anchored to the PM via the 1′-acyl chains of PI(4,5)P2 and PS/PE/PC lipids, and the myr group, which collectively bracket a basic patch projecting toward the polar leaflet of the membrane (Vlach and Saad, 2013). Exposure of the myr group by a membrane-like environment, independent of PI(4,5)P2 binding, was also observed in a reverse micellar system (Valentine et al., 2010). Spontaneous exposure of the myr group prior to association to membranes has also been observed by using coarse-grained simulation approaches (Charlier et al., 2014). The authors reported that insertion of the myr group into the bilayer is necessary for the orientation of MA to allow for the experimentally identified region of MA to interact with the PI(4,5)P2 head group. Flipping of the lipid acyl chain out of the membrane environment to the hydrophobic groove in MA was, however, not observed. Further studies are needed to answer yet unresolved questions: What is the actual trigger and mechanism of myr exposure in vivo? Are acyl chains of either PI(4,5)P2 or other phospholipids involved in MA binding to the PM? The Vogt lab has recently shown that Gag and MA strongly preferred lipids with both acyl chains unsaturated over those with one chain unsaturated (Dick et al., 2012). Furthermore, Gag can sense cholesterol in the membrane bilayer allowing for a more efficient Gag binding to membranes (Dick et al., 2012). However, it is not yet known whether cholesterol can bind directly to Gag. Recent studies have also suggested that interactions can occur between the isolated NC and p6 domains of Gag with membrane models (Solbak et al., 2013; Kempf et al., 2015). However, the biological significance of these interactions has yet to be established. Further studies are needed to determine the synergy and interplay between PM components and their role in modulating the conformational switch of HIV-1 Gag during assembly (Ono, 2010b).
Other Retroviral Gag and MA Interactions with Lipids and Membranes
Like HIV-1, the site of HIV-2 assembly in vivo is also dependent on PI(4,5)P2 (Saad et al., 2008). Structural studies show that the HIV-1 and HIV-2 MA structures are very similar and that PI(4,5)P2 binds to both proteins in an identical manner (Figures 2A,C; Saad et al., 2008). However, in contrast to HIV-1 MA the position of the myr group in HIV-2 MA is less sensitive to factors that modulate myr exposure in HIV-1 MA such as protein concentration and binding of di-C4-PI(4,5)P2 (Saad et al., 2008). Data are not in agreement on the role of PI(4,5)P2 in other retroviruses. Previous studies have shown that murine leukemia virus (MLV) Gag targeting to the PM is mediated by PI(4,5)P2 and PS, suggesting a synergy between these two lipids to modulate Gag–PM binding (Hamard-Peron et al., 2010). The Ono laboratory, however, has recently shown that MLV Gag–PM binding is not dependent on PI(4,5)P2 (Inlora et al., 2014). The Gag proteins of Rous sarcoma virus (RSV) and equine infectious anemia virus (EIAV) lack the myr group and it is thought that Gag–membrane interaction is driven mainly by electrostatic interactions (Erdie and Wills, 1990; Provitera et al., 2000; Dalton et al., 2005; Chen et al., 2008; Fernandes et al., 2011). Association of RSV MA and Gag with liposomes of defined composition is dependent on the presence of a biologically relevant concentration of negatively charged lipids like PS (Dalton et al., 2005). The Vogt laboratory has shown that RSV Gag has no specific requirement for PI(4,5)P2 for PM association in vivo or liposome interaction in vitro (Chan et al., 2011). Parent and colleagues, however, have shown that depletion of intracellular PI(4,5)P2 and phosphatidylinositol-(3,4,5)-trisphosphate [PI(3,4,5)P3] levels impaired RSV Gag PM localization (Nadaraia-Hoke et al., 2013). It was suggested that differences are partially attributed to the sensitivity of assays utilized in these studies (Dick and Vogt, 2014). Most recent in vitro and in vivo studies from the Ono laboratory have shown that Gag–membrane binding via RSV MA is both PI(4,5)P2 dependent and is susceptible to RNA-mediated inhibition (Inlora et al., 2014). No structural studies are currently available on how RSV MA interacts with PM lipids.
The Carter laboratory has reported that a soluble analog of PI(4,5)P2 interacts directly with EIAV MA (Chen et al., 2008). More recently, it was shown that EIAV Gag is not only present on the PM but also in compartments enriched in phosphatidylinositol 3,5-biphosphate [PI(3,5)P2] (Fernandes et al., 2011). In contrast to HIV-1, release of EIAV particles is not significantly diminished by co-expression with 5-phosphatase IV, which may suggest that PI(4,5)P2 does not play a critical role in EIAV Gag assembly on the PM. Additional NMR studies have shown that phosphatidylinositol 3-phosphate [PI(3)P] binds to EIAV MA tighter than that of PI(4,5)P2 (Fernandes et al., 2011). Altogether, it is likely that EIAV Gag membrane targeting proceeds via a mechanism that is different from that of HIV-1, HIV-2, MLV, or RSV.
The Gag precursor of deltaretrovirus human T-lymphotropic virus type 1 (HTLV-1), Pr53Gag, localizes at the cell surface and intracellular compartments in HeLa cells (Le Blanc et al., 1999, 2002; Heidecker et al., 2004). It has been shown that subcellular localization of HTLV-1 Gag and VLP release are minimally or not sensitive to depletion of PI(4,5)P2 from the PM, suggesting that the interaction of HTLV-1 MA with PI(4,5)P2 is not essential for HTLV-1 Gag–membrane binding and particle assembly (Inlora et al., 2011, 2014). Single amino acid substitutions that confer a large basic patch rendered HTLV-1 MA susceptible to the RNA-mediated block, a phenotype similar to that observed for HIV-1 and RSV. It remains unknown whether other PM lipids play important roles in HTLV-1 Gag assembly as further studies are needed to identify the precise molecular requirements for HTLV-1 Gag binding to membranes.
Assembly and particle production of Mason-Pfizer monkey virus (M-PMV) is perhaps the most distinct from all cases discussed above. M-PMV is thought to catalyze the membrane envelopment of a preassembled spherical capsid shell to release infectious virions (Sfakianos and Hunter, 2003). Assembly of M-PMV capsids occurs in a pericentriolar region of the cytoplasm prior to transport to the PM for budding. It is thought that both MA and CA domains of M-PMV Gag interact with the PM (Sfakianos and Hunter, 2003). Initial evidence for a potential role of PI(4,5)P2 in M-PMV Gag–PM binding has been reported by Hunter and colleagues (Stansell et al., 2007). Depletion of PI(4,5)P2 from the PM led to dramatic decrease of particle release from M-PMV infected cells. Direct interaction between M-PMV MA and a soluble analog of PI(4,5)P2 has been detected (Prchal et al., 2012). The solution structure of M-PMV MA protein and its binding mode to di-C8-PI(4,5)P2 have been determined by NMR and molecular docking methods (for more details, see Prchal et al., 2012, 2014). The hallmark of the binding mode is the deep penetration of one of the C8 chains into a hydrophobic pocket formed between helices α1, α2, and α4 (Figure 2D). The position of the myr group of M-PMV MA was not affected by di-C8-PI(4,5)P2 binding and remained sequestered in the protein core. Interestingly, no binding was detected between the unmyristoylated MA protein of M-PMV and soluble analogs of PI(4,5)P2 (Prchal et al., 2012). The authors attributed this observation to the structural differences between MA and unmyristoylated MA, in which the hydrophobic cavity is absent in the latter. Altogether, the mechanisms of retroviral Gag assembly appear to be complex and require more detailed investigation at the molecular level (Figure 2F).
Gaps in Our Understanding of Gag Binding to the PM and Future Directions
Structural studies of HIV-1 MA interactions with PI(4,5)P2, PS, PE, and PC have provided novel insights into the molecular mechanism of Gag assembly on the PM. These studies have utilized lipids with truncated acyl chains. The precise mode of binding of native lipids with longer chains to MA has yet to be elucidated. The lack of an atomic snapshot for MA when bound to membranes containing physiologically relevant lipid composition remains a major gap in our understanding of virus assembly. Investigation of the molecular rearrangements of Gag in the immature and mature HIV particles have heavily relied on cryoelectron microscopy (cEM) data. Although details of the hexameric CA lattice exist, cEM studies have not provided a clear picture of the membrane-associated MA domain of Gag in either of the immature or mature particles (Fuller et al., 1997; Briggs et al., 2006, 2009; Wright et al., 2007). cEM data show that the membrane bilayer is thicker in regions where the Gag lattice is present and that MA density appears to “penetrate” the membrane bilayer (Figure 2E; Briggs et al., 2009). Earlier cEM tomography studies have suggested that the membrane-bound MA domain lacks periodicity (Wright et al., 2007). We believe that future studies may benefit from hybrid structural and biophysical methods to construct a model of Gag and/or MA bound to membranes. These may include small-angle x-ray scattering, atomic force microscopy and high-resolution cEM. Understanding the molecular basis of Gag assembly will not only shed light on the assembly of HIV-1 particles but will likely provide insight into the control of assembly in other retroviruses that assemble at the PM. Such studies are of great importance as these interactions may serve as pharmacological targets to inhibit HIV assembly. Structure-based studies have identified small molecule inhibitors that bind to the PI(4,5)P2 site of HIV-1 MA and possess modest potency in reducing virus production (Zentner et al., 2013a,b; LaPlante et al., 2014). Although considered weak binders, these inhibitors may serve as a template for future studies aimed at the development of novel inhibitors of retroviral assembly.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
This work is supported by the National Institutes of Health (1R01AI087101).
Glossary
Abbreviations
- MA
matrix protein
- NMR
nuclear magnetic resonance
- PS
phosphatidylserine
- PI(4,5)P2
phosphatidylinositol-4,5-bisphosphate.
References
- Alfadhli A., Mcnett H., Tsagli S., Bächinger H. P., Peyton D. H., Barklis E. (2011). HIV-1 matrix protein binding to RNA. J. Mol. Biol. 410, 653–666. 10.1016/j.jmb.2011.04.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfadhli A., Still A., Barklis E. (2009). Analysis of human immunodeficiency virus type 1 matrix binding to membranes and nucleic acids. J. Virol. 83, 12196–12203. 10.1128/JVI.01197-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anraku K., Fukuda R., Takamune N., Misumi S., Okamoto Y., Otsuka M., et al. (2010). Highly sensitive analysis of the interaction between HIV-1 Gag and phosphoinositide derivatives based on surface plasmon resonance. Biochemistry 49, 5109–5116. 10.1021/bi9019274 [DOI] [PubMed] [Google Scholar]
- Briggs J. A., Johnson M. C., Simon M. N., Fuller S. D., Vogt V. M. (2006). Cryo-electron microscopy reveals conserved and divergent features of gag packing in immature particles of Rous sarcoma virus and human immunodeficiency virus. J. Mol. Biol. 355, 157–168. 10.1016/j.jmb.2005.10.025 [DOI] [PubMed] [Google Scholar]
- Briggs J. A., Riches J. D., Glass B., Bartonova V., Zanetti G., Krausslich H. G. (2009). Structure and assembly of immature HIV. Proc. Natl. Acad. Sci. U.S.A. 106, 11090–11095. 10.1073/pnas.0903535106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J., Dick R. A., Vogt V. M. (2011). Rous sarcoma virus gag has no specific requirement for phosphatidylinositol-(4,5)-bisphosphate for plasma membrane association in vivo or for liposome interaction in vitro. J. Virol. 85, 10851–10860. 10.1128/JVI.00760-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlier L., Louet M., Chaloin L., Fuchs P., Martinez J., Muriaux D., et al. (2014). Coarse-grained simulations of the HIV-1 matrix protein anchoring: revisiting its assembly on membrane domains. Biophys. J. 106, 577–585. 10.1016/j.bpj.2013.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Bachtiar I., Piszczek G., Bouamr F., Carter C., Tjandra N. (2008). Solution NMR characterization of oligomerization and dynamics of equine infectious anemia virus matrix protein and its interaction with PIP2. Biochemistry 47, 1928–1937. 10.1021/bi701984h [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H., Wang J. J., Spearman P. (2010). Human immunodeficiency virus type-1 gag and host vesicular trafficking pathways. Curr. Top. Microbiol. Immunol. 339, 67–84. 10.1007/978-3-642-02175-6_4 [DOI] [PubMed] [Google Scholar]
- Chukkapalli V., Hogue I. B., Boyko V., Hu W.-S., Ono A. (2008). Interaction between HIV-1 Gag matrix domain and phosphatidylinositol-(4,5)-bisphosphate is essential for efficient Gag-membrane binding. J. Virol. 82, 2405–2417. 10.1128/JVI.01614-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chukkapalli V., Inlora J., Todd G. C., Ono A. (2013). Evidence in support of RNA-mediated inhibition of phosphatidylserine-dependent HIV-1 Gag membrane binding in cells. J. Virol. 87, 7155–7159. 10.1128/JVI.00075-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chukkapalli V., Oh S. J., Ono A. (2010). Opposing mechanisms involving RNA and lipids regulate HIV-1 Gag membrane binding through the highly basic region of the matrix domain. Proc. Natl. Acad. Sci. U.S.A. 107, 1600–1605. 10.1073/pnas.0908661107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chukkapalli V., Ono A. (2011). Molecular determinants that regulate plasma membrane association of HIV-1 gag. J. Mol. Biol. 410, 512–524. 10.1016/j.jmb.2011.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton A. K., Ako-Adjei D., Murray P. S., Murray D., Vogt M. V. (2007). Electrostatic interactions drive membrane association of the human immunodeficiency virus type 1 gag MA domain. J. Virol. 81, 6434–6445. 10.1128/JVI.02757-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton A. K., Murray P. S., Murray D., Vogt V. M. (2005). Biochemical characterization of rous sarcoma virus MA protein interaction with membranes. J. Virol. 79, 6227–6238. 10.1128/JVI.79.10.6227-6238.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S. A., Heinrich F., Raghunandan S., Krueger S., Curtis J. E., Rein A., et al. (2011). HIV-1 Gag extension: conformational changes require simultaneous interaction with membrane and nucleic acid. J. Mol. Biol. 406, 205–215. 10.1016/j.jmb.2010.11.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S. A. K., Curtis J. E., Ratcliff W., Clark P. K., Crist R. M., Lebowitz J., et al. (2007a). Conformation of the HIV-1 gag protein in solution. J. Mol. Biol. 365, 812–824. 10.1016/j.jmb.2006.10.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S. A. K., Zhao Z., Clark P. K., Tarasov S., Alexandratos J. N., Campbell S. J., et al. (2007b). Interactions between HIV-1 gag molecules in solution: an inositol phosphate-mediated switch. J. Mol. Biol. 365, 799–811. 10.1016/j.jmb.2006.10.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick R. A., Goh S. L., Feigenson G. W., Vogt V. M. (2012). HIV-1 Gag protein can sense the cholesterol and acyl chain environment in model membranes. Proc. Natl. Acad. Sci. U.S.A. 109, 18761–18767. 10.1073/pnas.1209408109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick R. A., Vogt V. M. (2014). Membrane interaction of retroviral Gag proteins. Front. Microbiol. 5:187. 10.3389/fmicb.2014.00187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley D. T., Spector A. A. (1986). Inositol phospholipid arachidonic acid metabolism in GH3 pituitary cells. Biochem. J. 236, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich L. S., Fong S., Scarlata S., Zybarth G., Carter C. (1996). Partitioning of HIV-1 gag and gag-related proteins to membranes. Biochemistry 35, 3933–3943. 10.1021/bi952337x [DOI] [PubMed] [Google Scholar]
- Erdie C. R., Wills J. W. (1990). Myristoylation of Rous sarcoma virus Gag protein does not prevent replication in avian cells. J. Virol. 64, 5204–5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes F., Chen K., Ehrlich L. S., Jin J., Chen M. H., Medina G. N., et al. (2011). Phosphoinositides direct equine infectious anemia virus gag trafficking and release. Traffic 12, 438–451. 10.1111/j.1600-0854.2010.01153.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finzi A., Orthwein A., Mercier J., Cohen E. A. (2007). Productive human immunodeficiency virus type 1 assembly takes place at the plasma membrane. J. Virol. 81, 7476–7490. 10.1128/JVI.00308-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fledderman E. L., Fujii K., Ghanam R. H., Waki K., Prevelige P. E., Freed E. O., et al. (2010). Myristate exposure in the HIV-1 Matrix Protein is modulated by pH. Biochemistry 49, 9551–9562. 10.1021/bi101245j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller S. D., Wild T., Gowen B. E., Krausslich H.-G., Vogt V. M. (1997). Cryo-electron microscopy reveals ordered domains in the immature HIV-1 particle. Curr. Biol. 7, 729–738. 10.1016/S0960-9822(06)00331-9 [DOI] [PubMed] [Google Scholar]
- Ghanam R. H., Fernandez T. F., Fledderman E. L., Saad J. S. (2010). Binding of calmodulin to the HIV-1 matrix protein triggers myristate exposure. J. Biol. Chem. 285, 41911–41920. 10.1074/jbc.M110.179093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanam R. H., Samal A. B., Fernandez T. F., Saad J. S. (2012). Role of the HIV-1 matrix protein in Gag intracellular trafficking and targeting to the plasma membrane for virus assembly. Front. Microbiol. 3:55. 10.3389/fmicb.2012.00055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamard-Peron E., Juilliard F., Saad J. S., Roy C., Roingeard P., Summers M. F., et al. (2010). Targeting of MuLV Gag to the plasma membrane is mediated by PI(4,5)P2/PS and a polybasic region in the Matrix. J. Virol. 84, 503–515. 10.1128/JVI.01134-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidecker G., Lloyd P. A., Fox K., Nagashima K., Derse D. (2004). Late assembly motifs of human T-cell leukemia virus type 1 and their relative roles in particle release. J. Virol. 78, 6636–6648. 10.1128/JVI.78.12.6636-6648.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermida-Matsumoto L., Resh M. D. (1999). Human immunodeficiency virus type 1 protease triggers a myristoyl switch that modulates membrane binding fo Pr55gag and p17MA. J. Virol. 73, 1902–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogue I. B., Llewellyn G. N., Ono A. (2012). Dynamic association between HIV-1 gag and membrane domains. Mol. Biol. Int. 2012, 13. 10.1155/2012/979765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inlora J., Chukkapalli V., Derse D., Ono A. (2011). Gag localization and virus-like particle release mediated by the matrix domain of human T-lymphotropic virus type 1 Gag are less dependent on phosphatidylinositol-(4,5)-bisphosphate than those mediated by the matrix domain of HIV-1 Gag. J. Virol. 85, 3802–3810. 10.1128/JVI.02383-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inlora J., Collins D. R., Trubin M. E., Chung J. Y., Ono A. (2014). Membrane binding and subcellular localization of retroviral Gag proteins are differentially regulated by MA interactions with phosphatidylinositol-(4,5)-bisphosphate and RNA. mBio 5, e02202. 10.1128/mBio.02202-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janmey P. A., Iida K., Yin H. L., Stossel T. P. (1987). Polyphosphoinositide micelles and polyphosphoinositide-containing vesicles dissociate endogenous gelsolin-actin complexes and promote actin assembly from the fast-growing end of actin filaments blocked by gelsolin. J. Biol. Chem. 262, 12228–12236. [PubMed] [Google Scholar]
- Jouvenet N., Bieniasz P. D., Simon S. M. (2008). Imaging the biogenesis of individual HIV-1 virions in live cells. Nature 454, 236–240. 10.1038/nature06998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouvenet N., Neil S. J. D., Bess C., Johnson M. C., Virgen C. A., Simon S. M., et al. (2006). Plasma membrane is the site of productive HIV-1 particle assembly. PLoS Biol. 4:e435. 10.1371/journal.pbio.0040435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouvenet N., Simon S. M., Bieniasz P. D. (2009). Imaging the interaction of HIV-1 genomes and Gag during assembly of individual viral particles. Proc. Natl. Acad. Sci. U.S.A. 106, 19114–19119. 10.1073/pnas.0907364106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempf N., Postupalenko V., Bora S., Didier P., Arntz Y., De Rocquigny H., et al. (2015). The HIV-1 nucleocapsid protein recruits negatively charged lipids to ensure its optimal binding to lipid membranes. J. Virol. 89, 1756–1767. 10.1128/JVI.02931-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutluay S. B., Bieniasz P. D. (2010). Analysis of the initiating events in HIV-1 particle assembly and genome packaging. PLoS Pathog. 6:e1001200. 10.1371/journal.ppat.1001200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutluay S. B., Zang T., Blanco-Melo D., Powell C., Jannain D., Errando M., et al. (2014). Global changes in the RNA binding specificity of HIV-1 gag regulate virion genesis. Cell 159, 1096–1109. 10.1016/j.cell.2014.09.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaPlante S. R., Forgione P., Boucher C., Coulombe R., Gillard J., Hucke O., et al. (2014). Enantiomeric atropisomers inhibit HCV polymerase and/or HIV matrix: characterizing hindered bond rotations and target selectivity. J. Med. Chem. 57, 1944–1951. 10.1021/jm401202a [DOI] [PubMed] [Google Scholar]
- Le Blanc I., Blot V., Bouchaert I., Salamero J., Goud B., Rosenberg A. R., et al. (2002). Intracellular distribution of human T-cell leukemia virus type 1 Gag proteins is independent of interaction with intracellular membranes. J. Virol. 76, 905–911. 10.1128/JVI.76.2.905-911.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Blanc I., Rosenberg A. R., Dokhelar M. C. (1999). Multiple functions for the basic amino acids of the human T-cell leukemia virus type 1 matrix protein in viral transmission. J. Virol. 73, 1860–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro J. B., Nath A., Färber M., Datta S. A., Rein A., Rhoades E., et al. (2014). A conformational transition observed in single HIV-1 Gag molecules during in vitro assembly of virus like particles. J. Virol. 8, 3577–3585. 10.1128/JVI.03353-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadaraia-Hoke S., Bann D. V., Lochmann T. L., Gudleski-O’Regan N., Parent L. J. (2013). Alterations in the MA and NC domains modulate phosphoinositide-dependent plasma membrane localization of the Rous sarcoma virus Gag protein. J. Virol. 87, 3609–3615. 10.1128/JVI.03059-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olety B., Ono A. (2014). Roles played by acidic lipids in HIV-1 Gag membrane binding. Virus Res. 193, 108–115. 10.1016/j.virusres.2014.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A. (2010a). HIV-1 assembly at the plasma membrane. Vaccine 28S, B55–B59. 10.1016/j.vaccine.2009.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A. (2010b). Relationships between plasma membrane microdomains and HIV-1 assembly. Biol. Cell. 102, 335–350. 10.1042/BC20090165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A., Ablan S. D., Lockett S. J., Nagashima K., Freed E. O. (2004). Phosphatidylinositol (4,5) bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc. Natl. Acad. Sci. U.S.A. 101, 14889–14894. 10.1073/pnas.0405596101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A., Freed E. O. (1999). Binding of human immunodeficiency virus Type 1 gag to membrane: role of the matrix amino terminus. J. Virol. 73, 4136–4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paillart J.-C., Gottlinger H. G. (1999). Opposing effects of human immunodeficiency virus type 1 matrix mutations support a myristyl switch model of Gag membrane targeting. J. Virol. 73, 2604–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prchal J., Kroupa T., Ruml T., Hrabal R. (2014). Interaction of Mason-Pfizer monkey virus matrix protein with plasma membrane. Front. Microbiol. 4:423. 10.3389/fmicb.2013.00423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prchal J., Srb P., Hunter E., Ruml T., Hrabal R. (2012). The structure of myristoylated mason-pfizer monkey virus matrix protein and the role of phosphatidylinositol-(4,5)-bisphosphate in its membrane binding. J. Mol. Biol. 423, 427–438. 10.1016/j.jmb.2012.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provitera P., Bouamr F., Murray D., Carter C., Scarlata S. (2000). Binding of equine infectious anemia virus matrix protein to membrane bilayers involves multiple interactions. J. Mol. Biol. 296, 887–898. 10.1006/jmbi.1999.3482 [DOI] [PubMed] [Google Scholar]
- Saad J. S., Ablan S. D., Ghanam R. H., Kim A., Andrews K., Nagashima K., et al. (2008). Structure of the myristylated HIV-2 MA protein and the role of phosphatidylinositol-(4,5)-bisphosphate in membrane targeting. J. Mol. Biol. 382, 434–447. 10.1016/j.jmb.2008.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad J. S., Kim A., Ghanam R. H., Dalton A. K., Vogt M. V., Wu Z., et al. (2007a). Mutations that mimic phosphorylation of the HIV-1 matrix protein do not perturb the myristyl switch. Protein Sci. 16, 1793–1797. 10.1110/ps.072987607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad J. S., Loeliger E., Luncsford P., Liriano M., Tai J., Kim A., et al. (2007b). Point mutations in the HIV-1 matrix protein turn off the myristyl switch. J. Mol. Biol. 366, 574–585. 10.1016/j.jmb.2006.11.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad J. S., Miller J., Tai J., Kim A., Ghanam R. H., Summers M. F. (2006). Structural basis for targeting HIV-1 Gag to virus assembly sites on the plasma membrane. Proc. Natl. Acad. Sci. U.S.A. 103, 11364–11369. 10.1073/pnas.0602818103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarlata S., Ehrlich L. S., Carter C. A. (1998). Membrane-induced alterations in HIV-1 gag and matrix protein-protein interactions. J. Mol. Biol. 277, 161–169. 10.1006/jmbi.1997.1615 [DOI] [PubMed] [Google Scholar]
- Sfakianos J. N., Hunter E. (2003). M-PMV capsid transport is mediated by Enc/Gag intractions at the pericentriolar recycling endosome. Traffic 4, 671–680. 10.1034/j.1600-0854.2003.00126.x [DOI] [PubMed] [Google Scholar]
- Shkriabai N., Datta S. A., Zhao Z., Hess S., Rein A., Kvaratskhelia M. (2006). Interactions of HIV-1 Gag with assembly cofactors. Biochemistry 45, 4077–4083. 10.1021/bi052308e [DOI] [PubMed] [Google Scholar]
- Solbak S. M., Reksten T. R., Hahn F., Wray V., Henklein P., Henklein P., et al. (2013). HIV-1 p6 – a structured to flexible multifunctional membrane-interacting protein. Biochim. Biophys. Acta 1828, 816–823. 10.1016/j.bbamem.2012.11.010 [DOI] [PubMed] [Google Scholar]
- Spearman P., Horton R., Ratner L., Kuli-Zade I. (1997). Membrane binding of human immunodeficiency virus type 1 matrix protein in vivo supports a conformational myristyl switch mechanism. J. Virol. 71, 6582–6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stansell E., Apkarian R., Haubova S., Diehl W. E., Tytler E. M., Hunter E. (2007). Basic residues in the Mason-Pfizer monkey virus gag matrix domain regulate intracellular trafficking and capsid-membrane interactions. J. Virol. 81, 8977–8988. 10.1128/JVI.00657-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C., Loeliger E., Luncsford P., Kinde I., Beckett D., Summers M. F. (2004). Entropic switch regulates myristate exposure in the HIV-1 matrix protein. Proc. Natl. Acad. Sci. U.S.A. 101, 517–522. 10.1073/pnas.0305665101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine K. G., Peterson R., Saad J. S., Summers M. F., Xu X., Ames J. B., et al. (2010). Reverse micelle encapsulation of membrane anchored proteins for solution NMR studies. Structure 18, 9–16. 10.1016/j.str.2009.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlach J., Saad J. S. (2013). Trio engagement via plasma membrane phospholipids and the myristoyl moiety governs HIV-1 matrix binding to bilayers. Proc. Natl. Acad. Sci. U.S.A. 110, 3525–3530. 10.1073/pnas.1216655110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waheed A. A., Freed E. O. (2010). The role of lipids in retrovirus replication. Viruses 2, 1146–1180. 10.3390/v2051146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsch S., Keppler O. T., Habermann A., Allespach I., Krijnse-Locker J., Kräusslich H.-G. (2007). HIV-1 buds predominantly at the plasma membrane of primary human macrophages. PLoS Pathog. 3:e36. 10.1371/journal.ppat.0030036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright E. R., Schooler J. B., Ding H. J., Kieffer C., Fillmore C., Sundquist W. I., et al. (2007). Electron crytomography of immature HIV-1 virions reveals the structure of the CA and SP1 Gag shells. EMBO J. 26, 2218–2226. 10.1038/sj.emboj.7601664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentner I., Sierra L. J., Fraser A. K., Maciunas L., Mankowski M. K., Vinnik A., et al. (2013a). Identification of a small-molecule inhibitor of HIV-1 assembly that targets the phosphatidylinositol (4,5)-bisphosphate binding site of the HIV-1 matrix protein. ChemMedChem 8, 426–432. 10.1002/cmdc.201200577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentner I., Sierra L. J., Maciunas L., Vinnik A., Fedichev P., Mankowski M. K., et al. (2013b). Discovery of a small-molecule antiviral targeting the HIV-1 matrix protein. Bioorg. Med. Chem. Lett. 23, 1132–1135. 10.1016/j.bmcl.2012.11.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Resh M. D. (1996). Differential membrane binding of the human immunodeficiency virus type 1 matrix protein. J. Virol. 70, 8540–8548. [DOI] [PMC free article] [PubMed] [Google Scholar]


