Abstract
Avian photoreceptors are a diverse class of neurons, comprised of four single cones, the two members of the double cone, and rods. The signaling events and transcriptional regulators driving the differentiation of these diverse photoreceptors are currently unknown. In addition, many distinctive features of photoreceptor subtypes, including spectral tuning, oil droplet size and pigmentation, synaptic targets and spatial patterning, have been well characterized, but the molecular mechanisms underlying these attributes have not been explored. To identify genes specifically expressed in distinct chicken (Gallus gallus) photoreceptor subtypes, we developed fluorescent reporters that label photoreceptor subpopulations, isolated these subpopulations using fluorescence-activated cell sorting and subjected them to next-generation sequencing. By comparing the expression profiles of photoreceptors labeled with rhodopsin, red opsin, green opsin, and violet opsin reporters, we have identified hundreds of differentially expressed genes that may underlie the distinctive features of these photoreceptor subtypes. These genes are involved in a variety of processes, including phototransduction, transcriptional regulation, cell adhesion, maintenance of intra- and extra-cellular structure, and metabolism. Of particular note are a variety of differentially expressed transcription factors, which may drive and maintain photoreceptor diversity, and cell adhesion molecules that may mediate spatial patterning of photoreceptors and act to establish retinal circuitry. These analyses provide a framework for future studies that will dissect the role of these various factors in the differentiation of avian photoreceptor subtypes.
Keywords: Chicken, Photoreceptor, Rod, Cone, RNA-Seq, Transcriptome
Introduction
Diurnal organisms typically have a much higher proportion of cones relative to rods and a greater diversity of cone subtypes than nocturnal organisms (Walls, 1942). The primary feature that distinguishes cone photoreceptor subtypes is their spectral tuning, which arises from the expression of different opsin genes (Bruhn and Cepko, 1996; Okano et al., 1989; Yen and Fager, 1984). However, cone subtypes also have a variety of additional specialized adaptations that allow for improved color discrimination and functional vision in bright light (Walls, 1942). While these adaptations have been thoroughly characterized by electron microscopy, electrophysiology, biochemical assays, and physical models (Bowmaker and Knowles, 1977; Goldsmith et al., 1984; Hart and Vorobyev, 2005; Kram et al., 2010; Morris, 1970; Morris and Shorey, 1967), transcriptomic diversity between cone subtypes is largely unstudied. With recent advances in next-generation sequencing, we are now able to assay genome-wide differences in gene expression between relatively small populations of cells, a necessary step in understanding the molecular mechanisms that drive cone photoreceptor diversification.
The diurnal avian retina is an excellent model for studying functional diversity in photoreceptors because it contains a variety of different types of photoreceptors. These include four single cones, the principal and accessory members of the double cone, and rods, each of which has a number of specialized properties (Fig. 1A). For example, cone photoreceptors contain an optical organelle in the inner segment, the oil droplet, which acts as a long-pass filter to narrow the spectrum of light reaching the outer segment and enhance color discrimination (Goldsmith et al., 1984; Hart and Vorobyev, 2005; Lind and Kelber, 2009; Partridge, 1989). Oil droplets in each cone subtype are pigmented by a unique set of carotenoids (the exception being the absence of carotenoids in the clear violet cone oil droplets), resulting in spectral filtering specifically tuned for the opsin present in that subtype (Bowmaker and Knowles, 1977; Goldsmith et al., 1984; Hart and Vorobyev, 2005). Photoreceptors also differ in their interactions with neighboring cells. Each photoreceptor subtype tiles the retina in an independent, semi-regular mosaic (Jiao et al., 2014; Kram et al., 2010) and forms synapses with specific horizontal and bipolar cells at different levels of the outer plexiform layer (Mariani, 1987; Wahlin et al., 2008). The molecules mediating these various features of avian photoreceptor subtype specialization have not been identified.
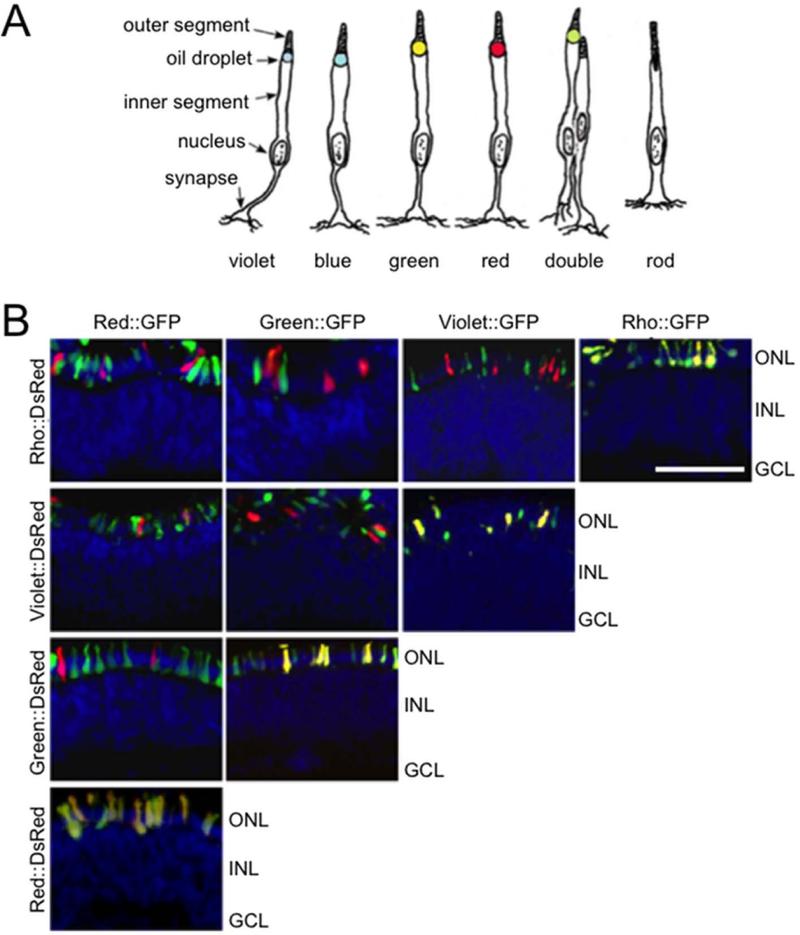
Figure 1.
Rhodopsin and cone opsin promoters drive expression in non-overlapping subsets of avian photoreceptors. A: The complement of avian photoreceptors includes the violet, blue, green and red single cones, the two components of the double cone and rods (this panel is adapted from Kram et al., 2010). In addition to expressing different opsins, these cells differ in a number of morphologic features including inner and outer segment structure, oil droplet size and pigmentation, axon length and synaptic targets. B: To isolate specific subpopulations of avian photoreceptors, fluorescent reporters were constructed by placing GFP and DsRed under the control of 1.5 – 3 kb upstream promoters of rhodopsin, red opsin, green opsin, and violet opsin. These reporters were electroporated pairwise into embryonic day 6 retinas, which were then grown in culture for 8 days prior to harvesting and processing the tissue for frozen section. Reporters driving GFP are listed along the top (B), while reporters driving DsRed are listed along the side. Each reporter drives expression restricted to the outer nuclear layer (ONL), with little expression seen in the inner nuclear layer (INL) or ganglion cell layer (GCL). Strong co-localization occurs when the same promoter drives both GFP and DsRed (B, diagonal). Only rare co-expression is detected when different promoters drive GFP and DsRed. Scale bar = 50 μm.
Furthermore, subtype-specific transcription factors likely drive photoreceptor differentiation and regulate the expression of the genes that mediate the specialized properties described above. The transcriptional regulation of photoreceptor differentiation has been relatively well studied in the rod-dominant mammalian retina, in which RORB, NRL and NR2E3 drive rod differentiation, and THRB, RXRG, SALL3, ONECUT1, and COUP-TFI and II are involved in regulating cone differentiation (de Melo et al., 2011; Emerson et al., 2013; Mears et al., 2001; Ng et al., 2001; Peng and Chen, 2005; Roberts et al., 2005; Roberts et al., 2006; Satoh et al., 2009; Srinivas et al., 2006; Yoshida et al., 2004). However, the role of these genes in the cone-dominant avian retina has not been thoroughly explored. In addition, it is likely that additional, as yet unstudied, transcription factors are necessary to generate the greater diversity of photoreceptors found in birds.
To discover genes that may drive the differentiation and diversification of avian photoreceptors, we conducted gene expression profiling of developing avian photoreceptor subtypes. We successfully identified hundreds of genes enriched in specific photoreceptor subtypes, including transcription factors, signal transduction molecules, cell adhesion molecules and lipid metabolizing enzymes. The spatiotemporal expression profiles of selected transcripts were determined using in situ hybridization, which confirmed expression in photoreceptors at various developmental stages. Identification of these differentially expressed genes yields insight into the molecular mechanisms driving avian photoreceptor diversification, setting the stage for further mechanistic analyses.
Methods
Animal Husbandry
All procedures were carried out in accordance with an animal protocol approved by the Animal Studies Committee of Washington University (No. 20110089). Pathogen-free White Leghorn eggs were obtained from Charles River (North Franklin, CT) and stored at 14°C upon arrival. Eggs were warmed to room temperature for 2 hours, then incubated in a humidified, rocking chamber held at 38°C for up to 20 days for embryonic time points, or were hatched and used for post-embryonic time points.
Construction of Fluorescent Reporters
The upstream promoter regions of rhodopsin, red opsin, green opsin and violet opsin were isolated by PCR of genomic DNA (gDNA). Rhodopsin and green opsin promoters were isolated from chicken (Gallus gallus) gDNA (nucleotides -1949 to -88 relative to the transcription start site [TSS]= +1, corresponding to chr12:19,499,638-19,501,514 in galGal4 and nucleotides 2975 to -8, corresponding to chr26:4,504,913-4,501,931 in galGal4 respectively). Since the red and violet opsin loci are not fully represented in the current chicken genome assembly (galGal4), we screened a chicken genomic bacterial artificial chromosome (BAC) library with red and violet opsin probes, without success. As an alternate approach, we isolated the red opsin promoter from Carolina anole lizard (Anolis carolinensis) gDNA (nucleotides -1797 to +87, corresponding to chr2:88663108-88664997 in anoCar2.0) and the violet opsin promoter from zebra finch (Taeniopygia guttata) gDNA (nucleotides -1946 to +45, corresponding to chr1A_random:206453-208444 in taeGut1). These regions were selected to include both the proximal promoter and phylogenetically conserved regions upstream of the TSS. Primers (Integrated DNA Technologies) include restriction enzyme sites used for sub-cloning and are listed in Supplemental Table 1. The PCR products were digested and ligated into the previously described “no basal DsRed” or “no basal GFP” reporter vectors, which are derived from pCAGGS (Hsiau et al., 2007). Restriction digestion and Sanger sequencing were used to confirm the sequences of the final constructs.
Electroporation of Fluorescent Reporters
Plasmid DNA precipitation, re-suspension, and electroporation were conducted essentially as previously described (Montana et al., 2013). Briefly, plasmid DNA was precipitated by adding 0.1 volumes 3 M sodium acetate, followed by 3 volumes 100% ethanol and washed in 70% ethanol before re-suspending to a final concentration of 0.25 μg/μl (rhodopsin and red opsin reporters) or 0.5 μg/μl (green and violet opsin reporters) in 1X PBS. A higher concentration of green and violet opsin reporters was used as they expressed more weakly than the rhodopsin and red opsin reporters.
Following 6 days of rocking incubation at 38°C, Hamburger and Hamilton (H&H) Stage 26-28 embryos (Hamburger and Hamilton, 1951) were harvested and the eyes removed. The sclera, choroid and RPE were dissected away from the retina, leaving the anterior segment attached. Retinas were then immersed in DNA solution and electroporated with an ECM830 Electro Square Porator (Harvard Apparatus) delivering 5 pulses at 30 V for 50 ms with a 950 ms interval. After electroporation, retinas were transferred to Whatman nucleopore filters floating on media (DMEM with 10% FBS, 2% chicken serum, 100 units/ml penicillin, 100 μg/ml streptomycin and 0.29 mg/ml lglutamine) and maintained in culture at 37°C, 5% CO2 for up to 15 days.
Each experiment consisted of electroporation of two reporters, one driving DsRed and the other driving GFP, and was conducted in triplicate. Experiments included: (1) rhodopsin vs. green opsin, embryonic day 6 (E6) + 9 days in vitro (DIV), (2) red opsin vs. green opsin, E6 + 9DIV, (3) red opsin vs. rhodopsin, E6 + 15DIV, (4) red opsin vs. green opsin, E6 + 15DIV, (5) red opsin vs. violet opsin, E6 + 15DIV. Expression driven by the violet opsin reporter was not robust until E6 + 12DIV, so it was not used at the earlier time point. Each replicate for the early time point comparing the rhodopsin and green opsin reporter was derived from eggs originating from a different flock, and sequenced individually. All other experiments were conducted using samples derived from the same flock that were processed in parallel (see below).
Retinal dissociation and FACS
For experiments at 9 DIV, 6 electroporated retinas were pooled, while 12 retinas were pooled at 15 DIV, as fluorescent cell recovery was reduced at this later time point because the tissue was more resistant to dissociation. This may have introduced some bias against more mature photoreceptors with longer processes, but should have impacted all cell populations similarly. Fluorescent regions of explanted retinas were dissected in calcium- and magnesium-free PBS and washed twice in the same buffer. Retinas were then dissociated in 1 mg/ml trypsin (Sigma) at 37°C for 10-12 minutes, with flicking every 2 minutes. Dissociation was stopped by addition of 1 mg/ml trypsin inhibitor (Sigma) and retinas were then treated with 0.2 mg/ml DNaseI (Sigma) for 5 minutes at 37°C. 3 volumes of media were then added and the tissue was triturated 5-10 times with a P1000 pipette (Rainin). Tissue was pelleted at 1,500 × g for 30 seconds, the supernatant removed, and cells re-suspended by flicking. 500 μl of sorting buffer (1% FBS, 0.1 mM EDTA in CMF) was added to the cells, which were passed twice through cell-strainer caps (12 × 75 mm) of 5 ml polystyrene round-bottom tubes (BD Biosciences) using a plastic transfer pipet. Cells were collected in RLT buffer supplied with the RNeasy mini kit (Qiagen) using a FACSAria II (BD Biosciences). Yield ranged from 12,000 to 200,000 cells per sample.
RNA isolation, cDNA amplification and sequencing
RNA was isolated from sorted cells using the RNeasy mini kit (Qiagen) as per the manufacturers protocol, with on-column DNase treatment, eluted in 30 μl elution buffer and stored immediately at -80°C. RNA quantity was determined using a Qubit 2.0 Fluorometer, with an estimated yield of 0.5 – 1.0 pg RNA/cell. RNA quality was assessed using an Agilent 2100 Bioanalyzer, and all samples had an RNA integrity number (RIN) ≥ 8. Five (5) μl of total RNA was used as input for the NuGen Ovation RNA-Seq kit, which was implemented as per the manufacturers protocol to generate complementary DNA (cDNA). Prior to cDNA purification, 1 μl was set aside for qPCR (see below). cDNA was then purified using the Qiagen MinElute PCR purification kit, eluted in 22 μl EB, and stored immediately at -20°C. 4 μg cDNA was submitted to the Genome Technology Access Center at Washington University for adapter ligation and sequencing on an Illumina HiSeq 2500. The three replicates comparing the rhodopsin and green opsin reporters at E6 + 9DIV were sequenced separately. The quality scores for two of these replicates were obtained using Casava v1.7 with Solexa 64 bit offset base quality scores, and the quality scores for the third replicate were obtained using Casava v1.8 with Sanger style 33 bit offset. All other experiments were sequenced in a single lane and employed Casava v1.8 to determine quality scores. Sequencing depth ranged from 21,000,000 to 82,000,000 reads per sample.
Sequencing data analysis
Reads were aligned to the chicken genome (galGal4) and splice junctions mapped using TopHat v1.4.0, RRID:OMICS_01257 (Trapnell et al., 2009), with the following options: anchor length of 5, splice mismatch of 1, intron length 10-500,000, 20 max transcriptome hits, segment-length of 21, and micro-exon search ‘on’. The first two replicates of the rhodopsin vs. green opsin early time point comparison were run with an additional option to use solexa1.3 quality scores.
Both Cuffdiff, RRID:OMICS_01969 (Trapnell et al., 2010) and edgeR, RRID:OMICS_01308 (Robinson et al., 2010) were used to identify differentially expressed genes. Cufflinks v1.3.0 (RRID:OMICS_01304) was run with a max intron length of 500,000 and compatible hits normalization, followed by Cuffmerge, and Cuffdiff with multi-read correction, upper quartile normalization, and fragment bias correction (Trapnell et al., 2010, RRID: OMICS_01969). Significance was scored at a cutoff false discovery rate (FDR) of 0.05. To generate input for edgeR v3.4.2, RRID:OMICS_01308 (Robinson et al., 2010), read counts were derived from the accepted_hits.bam file (generated by Bowtie, RRID:OMICS_00653 (Langmead et al., 2009)) using SAMtools, RRID:nlx_154607 (Li et al., 2009) and HTSeq, RRID:OMICS_01053 (Anders et al., 2014) and then compiled into a count table. Transcripts with 0 hits in four or more of the six samples (three replicates, two populations) were filtered out, and significance was scored at a cutoff FDR of 0.05. The differentially expressed genes presented in this manuscript were scored “significant” by both Cuffdiff and edgeR at 0.05 FDR, were > 2-fold enriched, and were expressed at ≥ 5 fragments per kilobase per million reads (FPKM) in the enriched population. Because red, blue, and violet opsin are not included in the current genome assembly, enrichment for these genes was determined by aligning reads to the NCBI reference sequence (NM_205440.1, NM_205517.1, and NM_205517.1 respectively) along with 21 other photoreceptor-specific genes including the other opsins (Supplemental Table 2) using Bowtie v0.12.7 (Langmead et al., 2009, RRID:OMICS_00653). The Bowtie output was then merged with the rest of the dataset and included in differential expression analysis using edgeR (Robinson et al., 2010, RRID:OMICS_01308). Raw sequencing data and differential expression data are available through NCBI’s Gene Expression Omnibus (Edgar et al., 2002), and can be accessed through GEO Series accession number GSE59850.
A heat map was used to display relative expression levels for the subset of differentially expressed genes with an FPKM value ≥ 20 and fold change ≥ 3. Gene expression was directly compared in all cell populations assayed at each time point (i.e. populations isolated with the Rho::DsRed, Red::DsRed, and Green::GFP at E6 + 9DIV were compared, as were the populations isolated with Rho::GFP, Red::Red, Green::GFP, and Violet::GFP at E6 + 15DIV). This was accomplished by using the equation shown below to calculate the log2 fold change of each transcript x in a specific population y relative to the mean expression of that transcript at the relevant time point. The average FPKM was used for populations measured more than once at a given time point (i.e. the green opsin-enriched population at the early time point and the red opsin-enriched population at the late time point).
Reverse transcription and quantitative PCR
Standard curves were generated using total RNA that was isolated from E16 chicken retinas using phenol/chloroform extraction, quantified on a NanoDrop 2000 UV-vis spectrophotometer (Thermo Scientific) and stored at -80°C. Reverse transcription of 5μg of total RNA was conducted for 50 minutes at 55°C in a total volume of 20 μl, using 200 units Superscript III, 0.5 mM dNTPs, 5 mM DTT, 40 units RNaseOUT, 1X FS buffer (Life Technologies Corp.), and 2.5 μM Oligo(dT)20 primer (Integrated DNA Technologies), followed by heat inactivation for 5 min at 85°C, and a 20 minute digestion with 2 units RNase H (Life Technologies Corp.) at 37°C. cDNA was purified using a PCR purification kit (Life Technologies Corp.) and eluted into 30 μl elution buffer. cDNA quality met the thresholds of 260/280 ≥ 2.0 and 260/230 ≥ 2.3, as determined by measurement on a NanoDrop 2000 UV-vis spectrophotometer (Thermo Scientific). Primers (Integrated DNA Technologies) were designed to target 80-120 bp regions spanning exon/exon junctions towards the 3’ end of the transcript to select for cDNA, were tested for specificity using BLAST (NCBI) and in silico PCR (UCSC genome bioinformatics), and are listed in Supplemental Table 1.
Quantitative PCR was conducted on a StepOne Plus Real Time PCR System (Life Technologies Corp.). A five point standard curve with 1:10 serial dilution and three technical replicates was measured in a total reaction volume of 20 μl, with 2 μl DNA, 0.5 μM primers (Integrated DNA Technologies) and 1X SYBR green PCR master mix (Life Technologies Corp.). Cycling conditions included a 10 minute denaturation at 95°C, 40 cycles of 95°C for 15 seconds, 60°C for 1 minute, followed by a melt curve measured at 0.3°C increments ranging from 60°C to 95°C. The melt curve was used to confirm specificity, and the slope of the calibration curve confirmed efficiencies of 90 – 110% in the range of Ct values relevant to the samples being measured. The most dilute point was not included in the calculation if the Ct value was > 35. Within a dilution point, outliers of > 1 cycle were also excluded. Finally, “no template control” Ct values were > 35, confirming that no significant amplification occurred in negative control wells.
qPCR was conducted on 0.5 μl 1:40 diluted cDNA per well, prepared as described for RNA-Seq, with the reaction and cycling conditions described above. Each plate included the reference gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH), which was expressed at comparable levels in all samples. Data analysis was conducted in Excel, and fold change was calculated using the equation below, where E is efficiency determined using the slope of the standard curve (E = 10[–1/slope]), GOI is the gene of interest, ref is GAPDH, CP is the crossing point or Ct value, and ΔCP reflects the difference in crossing point between the population with lower expression (control) and the population with higher expression (sample) (Pfaffl, 2001). This method was selected because it incorporates primer efficiencies and is therefore more precise than the comparative Ct method alone.
In Situ Hybridization and Imaging
Probe templates were isolated as 500-1000 bp fragments by PCR of E16 chicken cDNA (prepared as described above for qPCR standardization). PCR primers (Integrated DNA Technologies) included EcoRI sites for sub-cloning into the bluescript vector pBSK+ (Supplemental Table 1). The MAFA probe targets 612 nucleotides beginning with the transcription start site and was cloned with NcoI sites. Following digestion and ligation into pBSK+, Sanger sequencing was used to confirm insertion and orientation, and PCR with T7 and T3 primers was used to generate a template for DIG-labeled probe synthesis (Trimarchi et al., 2007).
Whole-mount in situ hybridization was conducted as previously described (Bruhn and Cepko, 1996). For frozen and paraffin section in situ hybridization, retinas were fixed in 4% paraformaldehyde (wt/vol) in 1X PBS overnight at 4°C, and either equilibrated overnight in 30% sucrose (wt/vol) in 1XPBS and embedded in Tissue-Tek OCT compound (Sakura, Torrance, CA), or dehydrated in an ethanol series and embedded in paraffin. Sections were taken at 12 μm, air-dried for 30 minutes at room temperature, and stored for no longer than 1 week at -20°C. In situ hybridization was then conducted as previously described (Trimarchi et al., 2007). The probes were used at 1 or 2 μl per 100 μl hybridization buffer, pH 7.5 (Trimarchi et al., 2007). Images were taken at 400x using an Olympus BX-51 compound microscope fitted with a DIC filter (Olympus, Tokyo, Japan), and processed using Adobe Photoshop CS5 to adjust brightness and contrast. For fluorescence imaging, 4',6-diamidino-2-phenylindole (DAPI) was briefly applied at a 1:1,000 concentration to counter-stain nuclei.
Results
Engineering opsin reporters to isolate photoreceptor subpopulations
To identify genes specific to photoreceptor subtypes in the chicken, we used fluorescence-activated cell sorting (FACS) of individual photoreceptor subpopulations followed by RNA-Seq. Opsins are the best-characterized markers of avian photoreceptor subtypes (Adler and Raymond, 2008). We therefore employed opsin promoters driving the expression of fluorescent reporters to selectively label individual photoreceptor subtypes. We obtained functional rhodopsin (RH1) and green opsin (RH2) promoters from chicken genomic DNA by PCR, but were unable to isolate promoters for red (LWS), blue (SWS2), and violet (SWS1) opsin from this species because the corresponding genomic regions are not represented in the current genome assembly and attempts to clone these regions from a bacterial artificial chromosome (BAC) library failed (data not shown). Instead, we obtained functional red and violet opsin promoters from Carolina anole lizard (Anolis carolinensis) and zebra finch (Taeniopygia guttata), respectively. We also obtained blue opsin promoter fragments from both zebra finch (Taeniopygia guttata) and platypus (Ornithorhynchus anatinus), but these promoters expressed weakly and inconsistently and thus were not used further (data not shown).
To verify that the opsin reporters drive expression in subsets of photoreceptors, pairs of fluorescent reporters driving GFP and DsRed were electroporated into embryonic day 6 (E6) retinas. This time point is at the peak of photoreceptor neurogenesis and immediately prior to cell cycle exit (Morris, 1973), a critical stage at which the plasmids can enter the nuclei during mitosis but will not be diluted out by numerous subsequent cell divisions. The electroporated retinas were then grown in explant culture for eight days, processed and visualized (Fig. 1B). In all cases, fluorescence was limited to the outer nuclear layer (ONL), indicating that the reporter-driven expression is restricted to photoreceptors. High levels of GFP and DsRed co-expression are present when the same promoter drives both reporters (Fig. 1B, diagonal). In contrast, when two different promoters drive GFP and DsRed, minimal co-expression is observed, indicating successful labeling of distinct photoreceptor subpopulations.
Identification of differentially expressed genes at two stages of photoreceptor development
We first isolated photoreceptors at a time point equivalent to E15, a developmental stage at which the induction of opsin expression, synaptogenesis, and outer segment elaboration occur (Bruhn and Cepko, 1996; Hanawa et al., 1976; McLaughlin, 1976; Meller and Tetzlaff, 1976; Wahlin et al., 2008). This was accomplished by co-electroporating six E6 retinas with two different reporters and harvesting dissociated cells by FACS after 9 days in culture. All comparisons consisted of three replicates and were conducted pairwise between cells isolated from the same tissue to control for variation in developmental stage, location and efficiency of electroporation, growth conditions and gender. Co-electroporation with Rho::DsRed and Green::GFP generated two distinct single-positive populations that were isolated by FACS using the gating shown in Fig. 2A. Similar results were obtained with the Red::DsRed and Green::GFP reporters (Fig. 2B). The yield from FACS ranged from 30,000 to 200,000 cells per sample. Fewer cells were obtained with the Green::GFP reporter relative to the Red::DsRed and Rho::DsRed reporters, consistent with the lower density of green cones relative to red cones and rods (Kram et al., 2010).
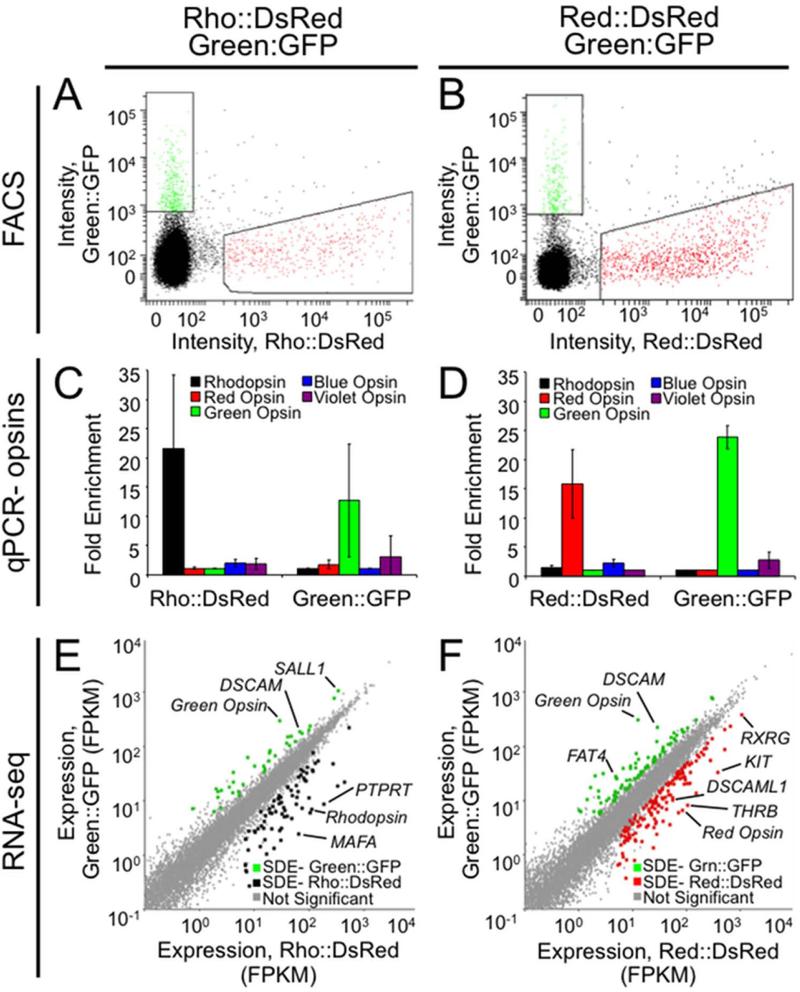
Figure 2.
Expression profiling at an intermediate developmental stage identifies several hundred differentially expressed genes. Retinas were co-electroporated with either Rho::DsRed and Green::GFP (A,C,E) or Red::DsRed and Green::GFP (B,D,F) at embryonic day 6 and cultured in vitro as explants for 9 days. A,B: Six retinas for each of three replicates were dissociated, and FACS was used to isolate fluorescent cells. Gating is shown for a representative replicate for each experiment. C,D: To verify enrichment of desired cell types, qPCR was conducted on amplified cDNA from sorted populations to determine the relative prevalence of opsin transcripts. Fold enrichment has been normalized to GAPDH, and error bars represent the standard deviation between the three biological replicates. E,F: Sequencing data was analyzed using TopHat to determine the expression level for each transcript (fragments per kilobase per million reads, FPKM). Each transcript is represented as a point plotted with expression levels (FPKM) in either Rho::DsRed (E) or Red::DsRed (F) on the x-axis and in Green::GFP on the y-axis. Cuffdiff was used to identify significantly differentially expressed (SDE) genes at 5% FDR. These positive hits were filtered to include only genes also called significant by edgeR, with ≥ 5 FPKM, and with ≥ 2 fold change between populations and are represented as colored dots for Rho::DsRed (E, black), Red::DsRed (F, red) and Green::GFP (E,F green).
Following RNA isolation and cDNA production and amplification, enrichment of specific photoreceptor populations was confirmed by using qPCR to determine the endogenous levels of the opsin transcripts. Strong enrichment for rhodopsin and green opsin transcript were observed for the Rho::DsRed- and Green::GFP-positive populations respectively (Fig. 2C), indicating successful enrichment for rods and green cones. Similarly, the Red::DsRed and Green::GFP reporters yielded strong enrichment for red and green opsin in the appropriate populations (Fig. 2D). Because red opsin is expressed in red single cones and in both members of the double cone (Araki et al., 1990), we expect enrichment of a mixed population of red single and double cones with Red::DsRed.
In order to identify genes that may underlie functions specific to photoreceptor subtypes, we conducted RNA-Seq and differential expression analysis on the enriched photoreceptor populations (Robinson et al., 2010; Trapnell et al., 2010). We identified 117 differentially expressed genes between the Rho::DsRed- and Green::GFP-positive populations, and 231 differentially expressed genes with Red::DsRed and Green::GFP, with a false discovery rate (FDR) of 5%, normalized expression level of ≥ 5 fragments per kilobase per million reads (FPKM) and ≥ 2-fold change between populations (Fig. 2E-F). A complete list of differentially expressed genes at E6 +9DIV is available in Supplemental Tables 3 and 4.
Next, we sought to identify genes that drive and maintain the diverse functions of developmentally advanced photoreceptors. We therefore conducted three separate comparisons of photoreceptor populations enriched at a later time point, E6 + 15 days in culture, which roughly corresponds to the time of hatching. The distribution of fluorescent cells observed with FACS of the Red::DsRed vs. Green::GFP populations are largely consistent with the earlier time point, and the gating was therefore kept the same (Fig. 2B, 3B). Two distinct populations were clearly visible for Red::DsRed and Rho::GFP (Fig. 3A), but there were apparent double positives with co-electroporation of Red::DsRed and Violet::GFP (Fig. 3C). Because these double positives are more frequent at higher levels of DsRed intensity, we suspect that this is artifact but cannot rule out either leakiness of the reporters or co-expression of two opsins within a single photoreceptor. We therefore employed more conservative gating to collect the Red::DsRed population from the Red::DsRed vs. Rho::GFP and Red::DsRed vs. Violet::GFP experiments (Fig. 3A,C).
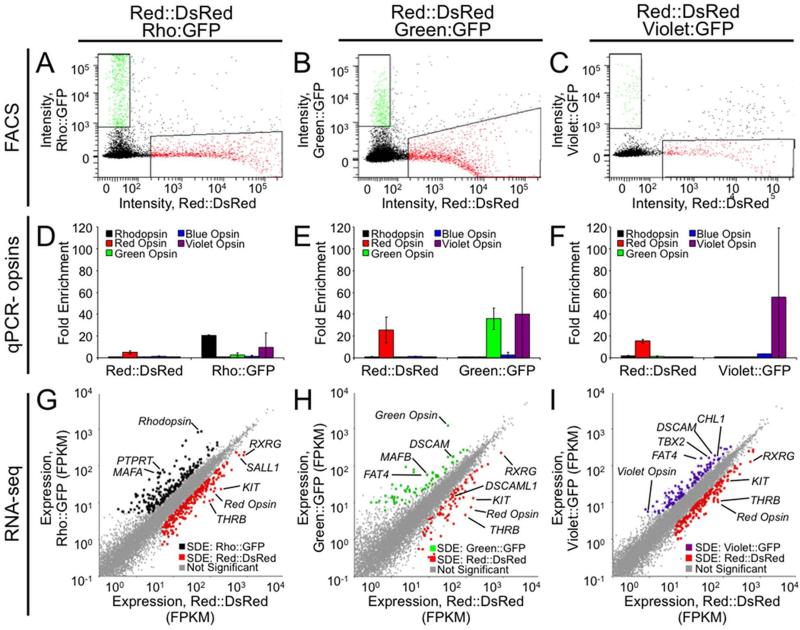
Figure 3.
Expression profiling at a late stage of photoreceptor development identifies several hundred differentially expressed genes. Retinas were co-electroporated with Red::DsRed and Rho::GFP (A,D,G), Red::DsRed and Green::GFP (B,E,H) or Red::DsRed and Violet::GFP (C,F,I) at embryonic day 6 and cultured in vitro as explants for 15 days. A-C: Twelve retinas for each of three replicates were dissociated, and fluorescent cells were isolated using FACS with the indicated gating. The downward curve of the data points at high DsRed intensities is an artifact that can occur when applying compensation controls, a necessary correction due to the overlapping excitation/emission spectra of DsRed and GFP. D-F: To verify enrichment of desired cell types, qPCR was used to determine the relative prevalence of opsin transcripts in the sorted populations. Fold enrichment has been normalized to GAPDH, and error bars represent the standard deviation between the three biological replicates. G-I: Sequencing data was analyzed using TopHat to determine the expression level for each transcript in FPKM. Each transcript is represented as a point plotted with expression levels in Red::DsRed on the x-axis (G-I) and in Rho::GFP (G), Green::GFP (H), or Violet::GFP (I) on the y-axis. Significantly differentially expressed (SDE) genes are represented as colored dots for Red::DsRed (G, H, I, red), Rho::GFP (G, black), Green::GFP (H, green) and Violet::GFP (I, violet).
Successful enrichment of photoreceptor populations using FACS was again confirmed by using qPCR to measure levels of endogenous opsin transcript. Red opsin and rhodopsin were enriched with Red::DsRed and Rho::GFP respectively (Fig. 3D), red and green opsin with Red::DsRed and Green::GFP respectively (Fig. 3E), and red and violet opsin with Red::DsRed and Violet::GFP respectively (Fig. 3F). These data indicate that our reporters were expressed in rods (Rho::GFP), red single and double cones (Red::DsRed), green cones (Green::GFP) and violet cones (Violet::GFP). However, we also observed 10-fold enrichment of violet opsin in the Rho::DsRed population, and 43-fold enrichment in the Green::DsRed population. These differences may be exaggerated as the levels of violet opsin are at the lower limit of detectability by qPCR and therefore more subject to variation. RNA-Seq indicated a less pronounced enrichment of violet opsin transcript (2-fold for Rho::GFP, 10-fold for Green::GFP, and 7-fold for Violet::GFP, Fig. 4C-E). The observed enrichment of violet opsin in the Rho::GFP population may therefore not be biologically meaningful. However, the strong enrichment of violet opsin in the Green::GFP population indicates that the Green::GFP population is enriched for both green and violet opsin transcripts.
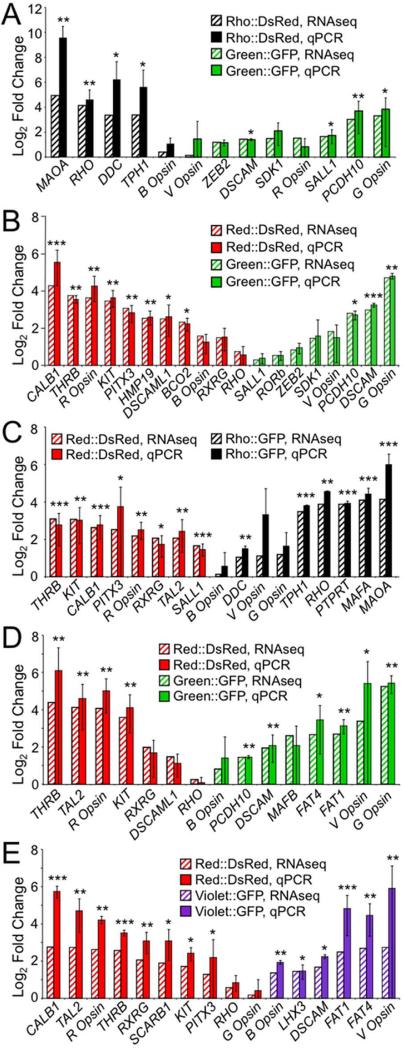
Figure 4.
Strong correlation between fold enrichment based on qPCR and RNA-Seq. For each experiment, differences in expression of ~ 15 genes across a range of fold changes and expression levels were further quantified by qPCR. Results are shown for experiments conducted at both the early time point, A: Rho::DsRed vs. Green::GFP and B: Red::DsRed vs. Green::GFP, and at the later time point, C: Red::DsRed vs. Rho::GFP, D: Red::DsRed vs. Green::GFP, and E: Red::DsRed vs. Violet::GFP. Fold change as calculated by RNA-Seq is shown as striped bars, while fold change as determined by qPCR is shown as solid bars (A-E). Fold enrichment for qPCR was normalized to GAPDH, and qPCR error bars represent 95% confidence intervals based on the standard error of the mean. A two-sided t-test was used to assess significant enrichment in the qPCR data, with p-values indicated as follows: p < 0.05 (*), p < 0.01 (**), and p < 0.001 (***).
Enrichment for violet opsin could have resulted either from leakiness of the reporter or transient co-expression of the two opsins within individual developing photoreceptors. We rarely saw co-expression of the violet opsin reporter with other reporters at E6 + 9DIV (Fig. 1B). To further address this issue, we co-electroporated Violet::GFP with each of the other reporters driving DsRed, harvested the explants at E6 + 15DIV, and took whole-mount images of three retinas per group at 200x magnification. We did not observe high levels of co-expression for any of the reporters aside from Violet::DsRed. Specifically, 2.5% (7/499) of Green::DsRed+ cells were also positive for Violet::GFP, as were 1.8% (28/1525) of Red::DsRed+ cells and 1.1% (13/1150) of Rho::DsRed+ cells. In contrast, 100% (398/398) of Violet::DsRed+ cells were also GFP+. Therefore, we conclude that reporter leakiness does not occur in a sufficiently high percentage of the cells to result in violet opsin enrichment in the Green::GFP population. Instead, the strong enrichment for violet opsin in the Green::GFP population is likely due to ectopic expression of violet opsin. Our attempts to prove expression of violet opsin in Green::GFP+ cells by staining electroporated explants with a violet opsin antibody were unsuccessful, as the antibody did not have activity against the retinal explants. However, opsin co-expression does occur in vitro when chicken photoreceptors are exposed to exogenous signaling molecules (Bradford et al., 2005). Therefore, it may be that differences between explant culture conditions and the signaling environment in vivo resulted in ectopic expression of violet opsin.
To identify genes enriched in specific photoreceptor populations at the later time point (E6 + 15 DIV), we conducted RNA-Seq and differential expression analysis. We identified 466 differentially expressed genes between Red::DsRed- and Rho::GFP-positive populations, 134 differentially expressed genes between Red::DsRed- and Green::GFP-positive populations, and 298 differentially expressed genes between Red::DsRed- and Violet::GFP-positive populations (5% FDR, ≥ 5 FPKM, ≥ 2-fold change). Differentially expressed genes include the relevant opsins, as well as transcription factors and cell adhesion molecules discussed in more detail below (Fig. 3G-I). A complete list of differentially expressed genes is provided in Supplemental Tables 5-7.
Validation of photoreceptor transcriptome profiling results
We next sought to validate the RNA-Seq results by performing qPCR for selected genes. We selected 31 transcripts across a range of expression levels and fold changes and assessed their abundance in the amplified cDNA samples. The fold changes detected by RNA-Seq and qPCR were tightly correlated for both datasets collected at the early time point, E6 + 9DIV (Fig. 4A-B). The largest discrepancies between RNASeq and qPCR occur for transcripts present at low levels, such as monoamine oxidase A (MAOA) (Fig. 4A) and calbindin 1 (CALB1) (Fig. 4B), for which RNA-Seq indicates more modest differences in expression between samples. Comparable fold changes were also found using samples from the later time point, E6 + 15DIV (Fig. 4C-E). Again, the greatest discrepancies between the two methods are observed with lowly expressed transcripts, including MAOA (Fig. 4C), Violet Opsin (Fig. 4C-E) and CALB1 (Fig. 4C,E). Overall, fold change as measured by RNA-Seq and qPCR were highly correlated (R2 = 0.74), confirming that RNA-Seq is a reliable means of assessing differences in gene expression in our system.
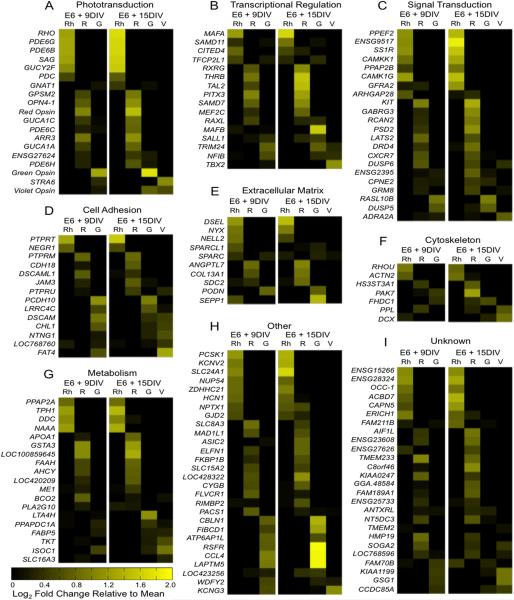
Identifying specific categories of differentially expressed genes can indicate cellular functions involved in diversification. Accordingly, differentially expressed genes were assigned to functional categories using gene ontology analysis (Huang da et al., 2009) and manual literature search. As expected, enrichment of genes involved in phototransduction was observed, including the opsins and rod- and cone-specific components of the phototransduction cascade (Fig. 5A). Interestingly, a large number of transcriptional regulators and signal transduction pathway components were differentially expressed (Fig. 5B-C). These genes may play a critical role in directing the specification of photoreceptors during differentiation, and may indicate particular signaling pathways involved in diversification. Other genes involved in cell adhesion and in forming the extracellular matrix and cytoskeleton may mediate synaptogenesis, spatial patterning, and outer segment elaboration (Fig. 5D-F). Of the large number of genes involved in metabolism, roughly half are specifically involved in lipid metabolism and could play critical roles in the formation and pigmentation of the oil droplets (Fig. 5G). Other categories were represented by only a few genes and include ion channels, transporters, immune response mediators, and protein-modifying enzymes (Fig 5H). Finally, some genes have no known function, but are nevertheless highly and differentially expressed (Fig. 5I).
Figure 5.
Differentially expressed genes across all datasets by functional category. Differentially expressed genes were filtered to include those with ≥ 20 average fragments per kilobase per million reads (FPKM) and ≥ 3 fold change and were combined into a single dataset. The average FPKM for each gene at the early and late time points were calculated. Then, the log2 fold change for each population (Rh = Rhodopsin, R = Red Opsin, G = Green Opsin, V = Violet Opsin) relative to the average was calculated and is shown above, with genes expressed at above-average levels shown in increasing yellow intensity (see color bar). Genes were sorted into functional categories using gene ontology (GO) annotation and manual literature search. Categories containing a large proportion of differentially expressed genes include A: phototransduction, B: transcriptional regulation, C: signal transduction, D: cell adhesion, E: extracellular matrix, F: cytoskeleton and G: metabolism. Some gene categories were represented by only a few genes and are grouped together under “other” (H), while the function of other genes is unknown (I).
Expression patterns of differentially expressed transcription factors, signal transducers, and cell adhesion molecules in the developing retina
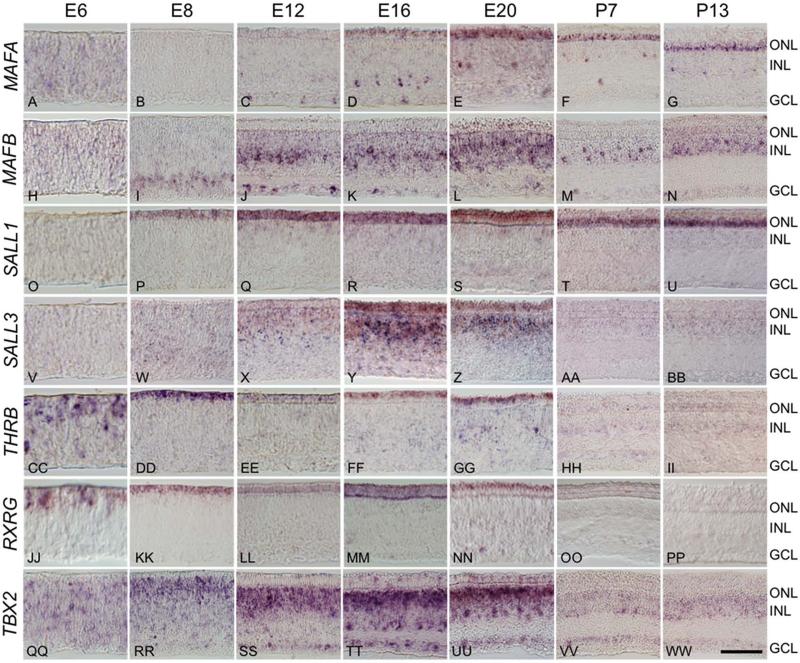
Transcription factors are of particular interest because they directly regulate the expression of large numbers of target genes and thereby coordinate differentiation. To verify expression of the most highly and differentially expressed transcription factors in developing photoreceptors, we conducted in situ hybridization over a developmental time course ranging from E6 to post-hatch day 13 (P13) (Fig. 6). The maf family member NRL plays a critical role in rod differentiation in mammals, but this gene is absent from all known avian genomes (Coolen et al., 2005; Mears et al., 2001; Peng and Chen, 2005; Yoshida et al., 2004). The absence of NRL has led to speculation that MAFA, a maf family member that is expressed in avian rods, acts as a key regulator of rod differentiation in the chicken, functionally substituting for NRL (Ochi et al., 2004). We first detected the MAFA transcript by in situ at E12 in post-mitotic, developing photoreceptors and in a small subset of cells in the inner nuclear layer (INL) and ganglion cell layer (GCL) (Fig. 6C). MAFA expression intensified at later time points, and persisted in the mature retina (Fig. 6D-G). We also observed that MAFA labeling is more widespread in the ventral than dorsal ONL, which parallels the distribution of rods in the avian retina (data not shown) (Bruhn and Cepko, 1996; Kram et al., 2010). The faint signal throughout the retina at E6 is likely non-specific background staining, but we cannot rule out low-level expression of MAFA at early developmental time points.
Figure 6.
Differentially expressed transcription factors have distinct spatiotemporal expression patterns. In situ hybridization across a developmental time course was used to assess the spatiotemporal expression patterns of a subset of differentially expressed transcription factors. Time points included embryonic days 6 (E6), E8, E12, E16, E20 and post-hatch days 7 (P7) and P13. A-G: Maf family member MAFA localizes to photoreceptors and small population of cells at the inner edge of the INL and GCL beginning at E12 and persisting through P13. H-N: Maf family member MAFB localizes to a subset of photoreceptors at E20, and to a subset of putative amacrine and ganglion cells beginning at E8 and persisting in the adult. O-U: Spalt family member SALL1 expression is largely restricted to the ONL, with faint staining at the outer border of the INL, beginning at E8 and persisting through P13, while SALL3 localizes strongly to the both the ONL and INL in a temporally restricted pattern from E12-E20 (V-BB). CC-PP: Thyroid hormone receptor β2 (THRB) and retinoid × receptor γ (RXRG) are both present as early as E6, persist through E20 and are restricted to the ONL. QQ-WW: T-box transcription factor TBX2 is found in a subset of photoreceptors at E12 through E20, and is widely expressed in the INL and GCL throughout development. ONL = outer nuclear layer, INL = inner nuclear layer, GCL = ganglion cell layer. Scale bar = 50 μm.
Another maf family member, MAFB, is enriched in the Green::GFP population at the late time point. While MAFB has not been previously reported to play a role in photoreceptor differentiation, the related family member NRL is critical for rod photoreceptor differentiation in mice, and MAFA may drive rod differentiation in birds, as discussed above (Mears et al., 2001; Peng and Chen, 2005; Yoshida et al., 2004). Like MAFA, MAFB transcripts localize to a subset of ganglion cells and amacrine cells, beginning at E8 and persisting into the adult (Fig. 6H-N). In addition, there is very faint, scattered signal in the ONL at E20. This suggests that if MAFB does play a role in green cone differentiation, it is limited to a short developmental time window.
Spalt family members play an important role in regulating photoreceptor diversification in both Drosophila and mammals (de Melo et al., 2011; Domingos et al., 2004a; Domingos et al., 2004b). Our dataset indicates that two spalt family members, SALL1 and SALL3, are strongly expressed in photoreceptors and are enriched in cones relative to rods, but are not restricted to a particular cone subtype. These genes have distinct spatiotemporal expression patterns. SALL1 expression is detected relatively early (by E8) is largely restricted to photoreceptors, and persists in the adult (Fig. 6OU). In contrast, SALL3 expression is first detected later (at E12) in both the outer and inner nuclear layers, but is undetected at post-embryonic time points (Fig. 6V-BB).
Nuclear receptors thyroid hormone receptor β2 (THRB) and retinoid × receptor γ (RXRG) are both enriched in Red::DsRed populations and direct the differentiation of long-wavelength cones in the mammalian and zebrafish retina (Ng et al., 2001; Roberts et al., 2005; Roberts et al., 2006; Suzuki et al., 2013). THRB and RXRG have similar expression patterns: they are both largely restricted to photoreceptors throughout development beginning as early as E6 and persisting through E20, but are not detected at post-embryonic time points (Fig. 6CC-PP).
T-box transcription factor TBX2 is enriched in the Violet::GFP population and is necessary for the differentiation of zebrafish UV cones, at the expense of rods (Alvarez-Delfin et al., 2009). We observed expression of TBX2 in the INL and GCL of the chicken retina throughout development, beginning at E6 and persisting through P14 (Fig. 6QQWW). However, TBX2 also localized to small population of photoreceptors at E12 through E20 (Fig. 6SS-UU).
Among many differentially expressed signal transduction molecules, the receptor tyrosine kinase KIT was the most enriched in the Red::DsRed population (Fig. 5C). In the mammalian retina, KIT is expressed in retinal progenitors, and later in a subset of amacrine and ganglion cells, but has not been reported in photoreceptors (Koso et al., 2007). We confirmed the expression of KIT in developing chicken photoreceptors, beginning at E8 and persisting through E16, and in a subset of inner retinal neurons at the outer border of the INL beginning at E12 and persisting in the adult (Fig. 7). This staining pattern suggests that KIT may play a direct role in avian photoreceptor differentiation, perhaps by acting upstream of transcription factors to direct the differentiation of red single and/or double cones. Interestingly, KIT ligand is mildly enriched in the Green::GFP population at both the early and late time point (by 2.2-fold and 2.8-fold, respectively). So, it is possible that green cones influence the differentiation of red single and/or double cones through the KIT pathway.
Figure 7.
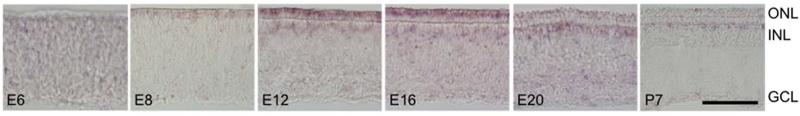
KIT expression is restricted to developing photoreceptors and developing and mature horizontal cells. The spatiotemporal expression pattern of KIT was assessed by frozen section in situ hybridization at embryonic days E6 (A), E8 (B), E12 (C), E16 (D), and E20 (E), and at post-hatch day 7 (F). Transcript is first detected at E8 at the outer edge of the outer neuroblast layer, and localizes to both the ONL and a subpopulation of INL cells at E12 and E16 (B-D). The signal in photoreceptors is diminished by E20 and absent by P7, while inner retinal staining remains distinct through P7 (E-F). ONL = outer nuclear layer, INL = inner nuclear layer, GCL = ganglion cell layer. Scale bar = 50 μm.
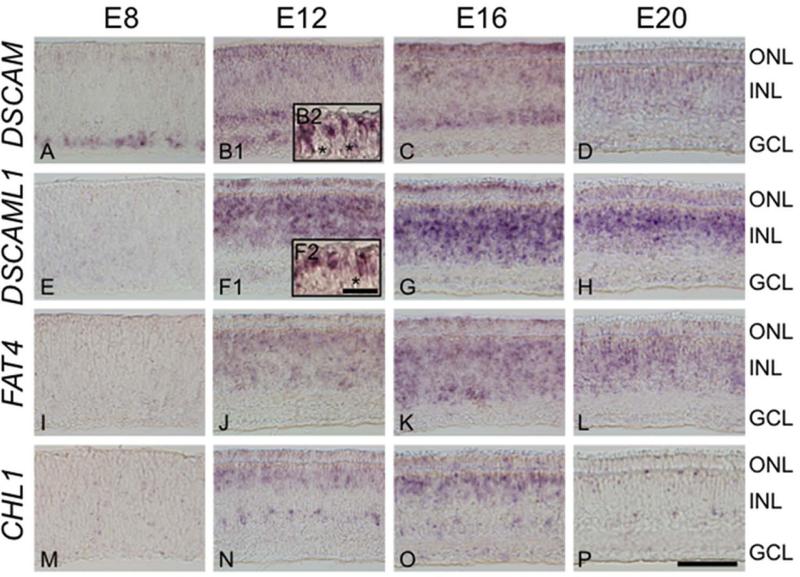
Synapse formation is critical for proper functioning of the visual system, and cell adhesion molecules are prime candidates for mediating the interaction between pre- and post-synaptic cells. The expression of strongly differentially expressed cell adhesion molecules in developing photoreceptors was assessed during the E8-E20 developmental window using in situ hybridization (Fig. 8). These time points coincide with chicken photoreceptor synaptogenesis, which begins at E13 and is complete by E20 (McLaughlin, 1976; Wahlin et al., 2008). Down syndrome cell adhesion molecules DSCAM, enriched in Green::GFP-, Rho::GFP- and Violet::GFP-positive populations, as well as DSCAML1, enriched in Red::DsRed-positive populations, were of particular interest because DSCAM homologs play a role in synaptic targeting and mosaic formation in the inner retina of mammals and birds and in the lamina and medulla of Drosophila (Fuerst et al., 2009; Fuerst et al., 2008; Hattori et al., 2008; Millard et al., 2010; Millard and Zipursky, 2008; Yamagata and Sanes, 2008). We detected DSCAM at E8 in photoreceptors and a subset of ganglion cells (Fig. 8A), and later in large subsets of cells at the inner and outer thirds of the inner nuclear layer (INL, Fig. 8B1). In contrast, DSCAML1 is first detected at E12 (Fig. 8F1) and strongly labels photoreceptors and the entire INL. INL expression of DSCAML1 becomes more restricted by E20, with a stronger signal in the outer half of the INL (Fig. 8H). Both DSCAM and DSCAML1 are detected in only a subset of photoreceptors at E12 (Fig. 8B2,F2).
Figure 8.
Differentially expressed cell adhesion molecules are restricted to photoreceptors and distinct populations of inner nuclear cells. The spatiotemporal expression pattern of differentially expressed cell adhesion molecules DSCAM (A-D), DSCAML1 (E-H), FAT4 (I-L) and CHL1 (M-P) were determined by in situ hybridization. Time points assessed were embryonic days 8 (E8), E12, E16 and E20, which include the critical period of synaptogenesis from E13-E20. Expression in the outer nuclear layer (ONL) is apparent for all adhesion molecules at E12 and E16. Strong expression is also detected within the inner nuclear layer (INL), with each gene localizing to distinct levels of the INL. Expression in the INL persists longer than in the ONL, and is present through E20. Localization of DSCAM and DSCAML1 in the ONL at E12 was assayed in paraffin sections at a higher magnification (B2,F2). Asterisks mark unlabeled photoreceptors. ONL= outer nuclear layer, INL = inner nuclear layer, GCL = ganglion cell layer. Scale bar = 50 μm for panels, 10 μm for insets.
Like DSCAMs, cadherin family members and L1CAM have been implicated in regulating synaptogenesis in the central nervous system (Gerrow and El-Husseini, 2006; Schmid and Maness, 2008). Accordingly, we further explored the expression patterns of the atypical cadherin FAT4 and of cell adhesion molecule with homology to L1CAM (CHL1), which were enriched in Green::GFP-positive and Violet::GFP-positive cells. FAT4 has an expression pattern that closely mimics DSCAML1, with induction at E12 and localization to photoreceptors and throughout the INL (Fig. 8I-L). Similarly, CHL1 is initially detected at E12 in photoreceptors and at the inner and outer borders of the INL (Fig. 8N). By E20, CHL1 is detected only in very rare populations at the extreme inner and outer edge of the INL (Fig. 8P).
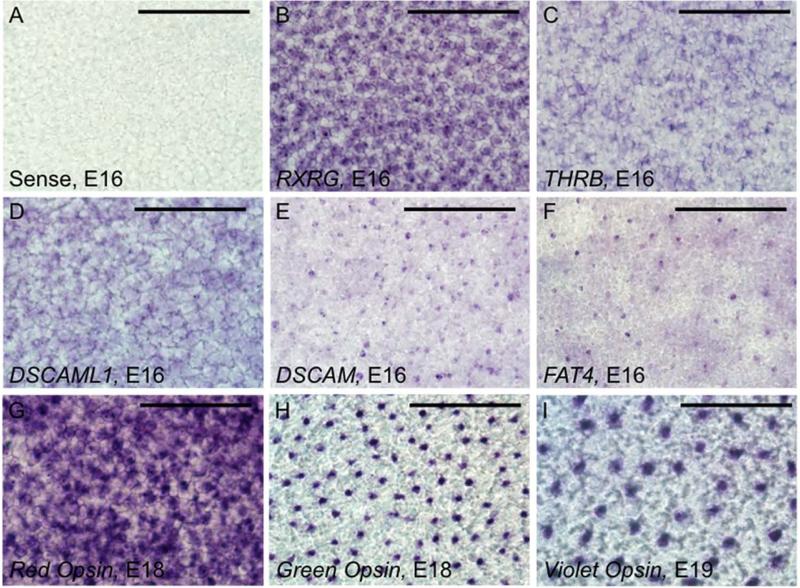
Finally, we sought to determine whether certain differentially expressed genes are in fact restricted to photoreceptor subpopulations. Subtype-specific antibodies are not available for all of the avian photoreceptors, so it was not feasible to use immunohistochemistry to unequivocally identify the photoreceptor subtypes expressing individual transcripts. Instead, we evaluated the spatial distribution of selected transcripts by using the in situ probes described above, and probes for red, green and violet opsin, to label whole-mounted retinas. Because individual photoreceptor subtypes are spatially arranged in mosaics (Bruhn and Cepko, 1996; Kram et al., 2010) we expect that genes restricted to photoreceptor subtypes will have a mosaic-like staining pattern. Furthermore, the photoreceptor subtypes are not present at equivalent densities: 40.7% are double cones, 21.1% green cones, 17.1% red cones, 12.6% blue cones, and 8.5% violet cones (Kram et al., 2010). We therefore expect that the density of labeling for differentially expressed genes should correspond to that of the appropriate cone opsin. To make the best assessment of population density, we chose time points at which each probe had robust staining, which was slightly later for the cone opsins than for the other probes (Fig. 9).
Figure 9.
Differentially expressed transcription factors and cell adhesion molecules localize to a subset of photoreceptors. Whole-mount in situ hybridization of embryonic day 16 retinas was used to visualize selected transcription factors (B,C) and cell adhesion molecules (D-F) at the level of the photoreceptors. A: No signal was observed with a sense control. B: Retinoid × receptor γ (RXRG), a transcription factor enriched with the red opsin reporter, was strongly detected in a large subset of photoreceptor cell bodies. C: Thyroid hormone receptor β2 (THRB) was also enriched with the red opsin reporter, and appears to localize to a smaller population of photoreceptor cell bodies than RXRG. D: Also enriched with the red opsin reporter, Down syndrome cell adhesion molecule like 1 (DSCAML1) shows a staining pattern similar to that of THRB and RXRG. E: Down syndrome cell adhesion molecule (DSCAM) was enriched with the green opsin reporter and localizes to a rare population of photoreceptors, with signal localized to the level of the inner segment, rather than the cell body. F: Atypical cadherin FAT4 was enriched with the violet opsin reporter and localizes to the inner segments of a very rare population of photoreceptors. G: Red opsin localizes to a large proportion of photoreceptors at E18. H: Green opsin localizes to a rare population of photoreceptors at E18. I: Violet opsin localizes to the sparsest population of photoreceptors at E19. The apparent larger size of cells labeled with violet opsin is due to differences in either developmental stage or subcellular localization, as magnification is equivalent for all panels. Scale bar = 50 μm.
We found that THRB, RXRG, DSCAML1, DSCAM and FAT4 transcripts are restricted to subpopulations of photoreceptors (Fig. 9B-F). The relative densities of DSCAM and green opsin (Fig. 9E,H) are comparable, as are those of FAT4 and violet opsin (Fig. 9F,I), supporting the hypothesis that these cell adhesion molecules are restricted to green and violet cones. Genes enriched with Red::DsRed could be expressed in red single cones, or double cones, or both. Because RXRG and DSCAML1 label a large population of cells comparable to the density of red opsin staining (Fig. 9B,D,G), they may be expressed in double and single cones, while THRB (which labels a smaller population of cells, Fig. 9C) may be expressed only in single or double cones. We could not obtain reliable signal in whole-mount preparations for the following probes: MAFA, SALL1, SALL3, or CHL1. However, a mosaic pattern was previously observed for MAFA using immunohistochemistry (Ochi et al., 2004).
Co-labeling experiments are needed to verify the localization of differentially expressed genes. Yet, despite repeated efforts, it was not possible to obtain convincing double fluorescent in situ hybridization (FISH) staining for individual cone opsin transcripts and other candidate cell type-specific transcripts. We attribute this failure to two causes. First, the sensitivity of the assay is such that reliable FISH signal could only be obtained for the highly expressed opsins. Second, the opsins could only be detected by FISH at late stages of embryogenesis, at time points when most of the other cell type-specific transcripts' levels were greatly reduced. Thus, both limitations in sensitivity and a limited overlap in the temporal windows of expression of the co-expressed transcripts precluded adequate evaluation by FISH.
Discussion
We conducted transcriptome profiling of developing avian photoreceptor subpopulations to identify genes that may regulate differentiation, mosaic formation, synaptogenesis, and oil droplet formation and pigmentation. To do this, we grew electroporated retinas in explant culture until they reached the desired developmental stage, a strategy that is more efficient than in vivo electroporation and was necessary to generate sufficient cell numbers for this study. We successfully identified hundreds of differentially expressed genes, and confirmed the localization of a number of differentially expressed transcription factors and cell adhesion molecules to subsets of endogenous developing photoreceptors using in situ hybridization. Furthermore, we found a number of intriguing parallels between differentially expressed genes identified in our study and known heterogeneity in other species, (further discussed below), that lend additional support to a role for these genes in avian photoreceptor diversification.
Transcriptional regulation of photoreceptor differentiation
NRL is a critical driver of rod differentiation in mammals, but a comprehensive study of maf family genes in vertebrates indicated that the NRL homolog is absent from all avian genomes (Coolen et al., 2005; Mears et al., 2001; Peng and Chen, 2005; Yoshida et al., 2004). Given the reported existence of NRL orthologs in fish and amphibians, it appears that NRL was specifically lost in the avian lineage (Coolen et al., 2005; McIlvain and Knox, 2007; Nelson et al., 2008). The maf family member MAFA was among the most highly enriched genes in cells expressing the rhodopsin reporter. In addition, the spatiotemporal expression pattern of MAFA is consistent with a role in driving rod differentiation and maintaining expression of rod-specific genes in the adult. These data suggest that a rewiring of the cis-regulatory network in avian rod photoreceptors has occurred in birds to allow MAFA to take on the role of NRL. Interestingly, MAFA is also expressed in a small subset of cells in the inner retina that are likely amacrine cells. The function of MAFA in these cells is unknown, but its presence suggests some divergence in function between MAFA and NRL, since NRL is restricted to rods in mammals.
Two of the most highly enriched transcription factors with the red opsin reporter are thyroid hormone receptor β2 (THRB) and retinoid × receptor γ (RXRG), which regulate long wavelength-sensitive (LWS) cone differentiation in mouse and zebrafish (Ng et al., 2001; Roberts et al., 2005; Roberts et al., 2006; Suzuki et al., 2013). While the expression of both RXRG and THRB in developing chicken photoreceptors has been previously demonstrated (Hoover et al., 1998; Trimarchi et al., 2008), the enrichment of these genes within a particular class of photoreceptors has not been described. This finding is significant in that (1) it demonstrates that the mechanisms driving the differentiation of long wavelength cones have been highly conserved and that (2) THRB and RXRG may serve as very early markers of avian photoreceptor specification. The latter point is important because the study of chicken photoreceptor cell fate has been limited by the inability to identify photoreceptor subtypes until a week after cell cycle exit, when opsin expression begins (Bruhn and Cepko, 1996). Therefore, the expression of RXRG and THRB during cell cycle exit at E6 (Fig. 6) provides a new entrée into studying the early aspects of avian photoreceptor specification and may allow for the dissection of cell fate decisions leading to the generation of red single and double cones.
There is no mammalian equivalent of the chicken green cone (which expresses RH2 and not OPN1MW), so chicken green cone differentiation may be regulated by transcription factors not involved in mammalian photoreceptor differentiation. The maf-family transcription factor MAFB was 5-fold enriched in green opsin-expressing photoreceptors at E6 + 15DIV, which suggests that MAFB may act to drive the expression of genes specific to green cones late in development. Of note, related family member MAFA is suspected to regulate the expression of rhodopsin, RH1, the vertebrate opsin with closest phylogeny to the chicken green opsin, RH2. The enrichment of two different maf family members in these closely related cell types suggests that the regulation of RH opsins by maf family transcription factors may be an evolutionarily ancient mechanism that predates the split between the RH1 and RH2 genes.
Spalt family members SALL1 and SALL3 were both enriched in cones relative to rods, but have distinct spatiotemporal expression patterns. SALL3 is expressed in the developing outer and inner nuclear layers, in a pattern similar to the mammalian SALL3, which regulates photoreceptor and horizontal cell development (de Melo et al., 2011). In contrast, SALL1 is largely restricted to photoreceptors and persists in the adult. These data suggest that the function of SALL3 may be conserved between birds and mammals, but that SALL1 may have diverged to take on a different role in regulating photoreceptor differentiation and function.
Orthologs of the chicken violet opsin, SWS1, include zebrafish UV opsin and mammalian blue opsin. Mutations in the zebrafish T-box transcription factor tbx2b cause UV cone precursors to differentiate as rods, resulting in the overproduction of the latter cell type (Alvarez-Delfin et al., 2009). In our study, TBX2 was strongly enriched in Violet::DsRed-positive cells, suggesting that TBX2 may drive the differentiation of the orthologous avian photoreceptor subtype, the violet cone. Given the potentially conserved role of TBX2 between these distant species, it is possible that TBX2 may play an important role in driving blue cone differentiation in the mammalian retina as well.
Synaptogenesis
Ribbon synapse formation is a critical aspect of photoreceptor differentiation that occurs between E13 and E20 in the outer plexiform layer (OPL) (McLaughlin, 1976; Wahlin et al., 2008). The avian OPL consists of three sublaminae, with rods and double cones synapsing in the outermost sublamina, red and green single cones in the middle sublamina, and blue and violet cones in the innermost sublamina (Mariani, 1987; Morris and Shorey, 1967; Wahlin et al., 2008). Of note, Whalin et al., found that dystrophin localizes specifically to the outer sublamina. In our dataset, dystrophin was enriched in both the rhodopsin and red opsin reporter-positive populations (Supplemental Tables 3, 6, and 7), suggesting that this extracellular matrix component is produced by rods and double cones and may contribute to the formation a distinct extracellular milieu in the outer sublamina of the OPL.
In addition to extracellular matrix components, differentially expressed adhesion molecules may be involved in both synaptic targeting and lateral spacing. DSCAM family members have been implicated in both processes in the inner retina of birds and mammals (Fuerst et al., 2009; Fuerst et al., 2008; Yamagata and Sanes, 2008), and in the lamina and medulla of Drosophila (Hattori et al., 2008; Millard et al., 2010; Millard and Zipursky, 2008). Furthermore, we found that DSCAM and DSCAML1 localize to a subset of cells in both the inner and outer retina during development, as do the atypical cadherin FAT4 and cell adhesion molecule with homology to L1CAM (CHL1). These data are consistent with a model in which these genes, along with other differentially expressed cell adhesion molecules, act in combination to regulate synaptogenesis and establish circuits throughout the thickness of the retina.
The receptor tyrosine phosphatase PTPRT and the alpha-liprin homolog PPFIA4 were both highly enriched in rhodopsin reporter-positive populations and may direct synaptic targeting in rods. Mutations in the Drosophila homologs of these genes disrupt targeting of R1-R6 and R7 photoreceptors within the lamina and medulla, respectively (Choe et al., 2006; Clandinin et al., 2001; Hofmeyer et al., 2006; Maurel-Zaffran et al., 2001). However, neither gene has been studied in the vertebrate retina to date. In addition to PTPRT, receptor tyrosine phosphatase isoforms PTPRF, PTPRG, PTPRM, PTPRR, and PTPRU were enriched in different cone populations (Supplemental Tables 3-7), indicating that this gene family may play a broad role in regulating photoreceptor synaptogenesis.
Paracrine factors can also mediate synaptogenesis. The expression of specific GABA receptor gamma subunits in cone photoreceptors has been reported in the salamander retina (Zhang et al., 2003), and Huang et al., speculated that gamma-aminobutyric acid (GABA) secreted by rabbit horizontal cells may act on cone photoreceptors to establish synaptic connections (Huang et al., 2000). Our observation of the enrichment of GABA A receptor γ 3 (GABRG3) in red opsin-expressing cone photoreceptors further supports a role for GABA-driven input in shaping synapse formation in specific photoreceptor subtypes, as it does elsewhere in the central nervous system (Akerman and Cline, 2007).
Oil droplet formation and pigmentation
The brilliantly colored oil droplets are a particularly striking feature of avian cone photoreceptors. Oil droplets in different photoreceptor subtypes differ both in size and in spectral absorbance (based on differential carotenoid accumulation) (Bowmaker and Knowles, 1977; Goldsmith et al., 1984). Although oil droplet formation does not occur in explant culture, possibly due to an absence of necessary carotenoid substrates, we anticipate that some genes important for oil droplet formation are still expressed. Cell death-inducing DFFA-like effector A (CIDEA) is essential for generating large lipid droplets in brown fat (Puri et al., 2008; Wu et al., 2014; Zhou et al., 2003) and is > 2-fold enriched in red- and green opsin expressing photoreceptors, both of which contain prominent oil droplets (Supplemental Tables 3 and 5). We have also identified two enzymes enriched in red opsin reporter-positive populations that may metabolize zeaxanthin to generate galloxanthin and ε-carotene, the carotenoids thought to be present in the double cone oil droplet (Goldsmith et al., 1984). Unpublished work in our lab suggests that beta-carotene oxygenase 2 (BCO2) can asymmetrically cleave zeaxanthin in the first step of galloxanthin biosynthesis (Matt Toomey, personal communication). In addition, glutathione S-transferase alpha 3 (GSTA3) isomerizes a steroid ring double bond during the synthesis of progesterone and testosterone (Johansson and Mannervik, 2001), analogous to the mechanism by which ε-carotene may be derived from zeaxanthin. Future studies will explore the potential role of these enzymes in carotenoid metabolism within avian cone subtypes.
Conclusions
We leveraged the power of next-generation sequencing to identify specific genes potentially involved in the transcriptional regulation of differentiation, synaptogenesis, mosaic formation, and oil droplet pigmentation in avian photoreceptor subtypes. We hope that these data will serve as a useful resource for the study of avian photoreceptor diversification, and expect that further inquiry into the function of particular differentially expressed genes will yield important insights into photoreceptor development.
Supplementary Material
Acknowledgements
The authors thank Timothy Laumann and Susan Shen for helpful discussion and comments on the manuscript and members of the Corbo lab for input and support during the course of the work. We also thank Matt Toomey and Kevin McGraw at Arizona State University for supplying us with zebra finch livers as a source of gDNA and Shane Campbell-Staton, Jonathan Losos and the Department of Herpetology in the Museum of Comparative Zoology at Harvard University for supplying us with Anolis carolinensis genomic DNA. We also thank Olga Malkova, Danielle Atibalentja, Erica Maria Lantelme and Dorjan Brinja and the Pathology and Immunology Flow Core Facility for sorting and help with flow cytometry analysis, and Dayna Oschwald, Toni Sinnwell, Paul Cliften, Eric Tycksen and the Genome Technology Access Center (GTAC) in the Department of Genetics at Washington University School of Medicine for conducting next-generation sequencing and help with genomic analysis. GTAC was partially supported by NCI Cancer Center Support Grant #P30 CA91842 to the Siteman Cancer Center and by ICTS/CTSA Grant# UL1 TR000448 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH), and NIH Roadmap for Medical Research. This project was supported by grants from the Human Frontier Science Program (RGP0017/2011 to JCC) and from the National Institute Of Neurological Disorders And Stroke (F31NS083170 to JME) and the National Eye Institute (5-T32EY013360-13 to JME and R01EY018826 to JCC) at the NIH. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or NCRR.
Role of authors
All authors had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Study concept and design: JME, KAL and JCC. Acquisition of data: JME, KAL, TH, JCC. Statistical analysis: JME. Analysis and interpretation of data: JME and JCC. Drafting of the manuscript: JME. Critical revision of the manuscript for important intellectual content: JME and JCC.
Research reported in this publication was supported by the Human Frontier Science Program (RGP0017/2011), the National Institute Of Neurological Disorders And Stroke (F31NS083170) and the National Eye Institute (R01EY018826 and 5-T32EY013360-13).
Footnotes
Conflict of Interest Statement
The authors have no financial conflicts of interest to report.
Literature Cited
- Adler R, Raymond PA. Have we achieved a unified model of photoreceptor cell fate specification in vertebrates? Brain research. 2008;1192:134–150. doi: 10.1016/j.brainres.2007.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akerman CJ, Cline HT. Refining the roles of GABAergic signaling during neural circuit formation. Trends in neurosciences. 2007;30(8):382–389. doi: 10.1016/j.tins.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Alvarez-Delfin K, Morris AC, Snelson CD, Gamse JT, Gupta T, Marlow FL, Mullins MC, Burgess HA, Granato M, Fadool JM. Tbx2b is required for ultraviolet photoreceptor cell specification during zebrafish retinal development. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(6):2023–2028. doi: 10.1073/pnas.0809439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. HTSeq — A Python framework to work with high-throughput sequencing data. BIoRxiv. 2014 doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki M, Fukada Y, Shichida Y, Yoshizawa T. Localization of iodopsin in the chick retina during in vivo and in vitro cone differentiation. Investigative ophthalmology & visual science. 1990;31(8):1466–1473. [PubMed] [Google Scholar]
- Bowmaker JK, Knowles A. Visual Pigments and Oil Droplets of Chicken Retina. Vision Res. 1977;17(7):755–764. doi: 10.1016/0042-6989(77)90117-1. [DOI] [PubMed] [Google Scholar]
- Bradford RL, Wang C, Zack DJ, Adler R. Roles of cell-intrinsic and microenvironmental factors in photoreceptor cell differentiation. Developmental biology. 2005;286(1):31–45. doi: 10.1016/j.ydbio.2005.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruhn SL, Cepko CL. Development of the pattern of photoreceptors in the chick retina. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1996;16(4):1430–1439. doi: 10.1523/JNEUROSCI.16-04-01430.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe KM, Prakash S, Bright A, Clandinin TR. Liprin-alpha is required for photoreceptor target selection in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(31):11601–11606. doi: 10.1073/pnas.0601185103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clandinin TR, Lee CH, Herman T, Lee RC, Yang AY, Ovasapyan S, Zipursky SL. Drosophila LAR regulates R1-R6 and R7 target specificity in the visual system. Neuron. 2001;32(2):237–248. doi: 10.1016/s0896-6273(01)00474-3. [DOI] [PubMed] [Google Scholar]
- Coolen M, Sii-Felice K, Bronchain O, Mazabraud A, Bourrat F, Retaux S, Felder-Schmittbuhl MP, Mazan S, Plouhinec JL. Phylogenomic analysis and expression patterns of large Maf genes in Xenopus tropicalis provide new insights into the functional evolution of the gene family in osteichthyans. Development genes and evolution. 2005;215(7):327–339. doi: 10.1007/s00427-005-0476-y. [DOI] [PubMed] [Google Scholar]
- de Melo J, Peng GH, Chen S, Blackshaw S. The Spalt family transcription factor Sall3 regulates the development of cone photoreceptors and retinal horizontal interneurons. Development. 2011;138(11):2325–2336. doi: 10.1242/dev.061846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingos PM, Brown S, Barrio R, Ratnakumar K, Frankfort BJ, Mardon G, Steller H, Mollereau B. Regulation of R7 and R8 differentiation by the spalt genes. Developmental biology. 2004a;273(1):121–133. doi: 10.1016/j.ydbio.2004.05.026. [DOI] [PubMed] [Google Scholar]
- Domingos PM, Mlodzik M, Mendes CS, Brown S, Steller H, Mollereau B. Spalt transcription factors are required for R3/R4 specification and establishment of planar cell polarity in the Drosophila eye. Development. 2004b;131(22):5695–5702. doi: 10.1242/dev.01443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic acids research. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson MM, Surzenko N, Goetz JJ, Trimarchi J, Cepko CL. Otx2 and Onecut1 promote the fates of cone photoreceptors and horizontal cells and repress rod photoreceptors. Developmental cell. 2013;26(1):59–72. doi: 10.1016/j.devcel.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst PG, Bruce F, Tian M, Wei W, Elstrott J, Feller MB, Erskine L, Singer JH, Burgess RW. DSCAM and DSCAML1 function in self-avoidance in multiple cell types in the developing mouse retina. Neuron. 2009;64(4):484–497. doi: 10.1016/j.neuron.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst PG, Koizumi A, Masland RH, Burgess RW. Neurite arborization and mosaic spacing in the mouse retina require DSCAM. Nature. 2008;451(7177):470–474. doi: 10.1038/nature06514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith TH, Collins JS, Licht S. The Cone Oil Droplets of Avian Retinas. Vision Res. 1984;24(11):1661–1671. doi: 10.1016/0042-6989(84)90324-9. [DOI] [PubMed] [Google Scholar]
- Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. Journal of morphology. 1951;88(1):49–92. [PubMed] [Google Scholar]
- Hanawa I, Takahashi K, Kawamoto N. A correlation of embryogenesis of visual cells and early receptor potential in the developing retina. Experimental eye research. 1976;23(6):587–594. doi: 10.1016/0014-4835(76)90216-5. [DOI] [PubMed] [Google Scholar]
- Hart NS, Vorobyev M. Modelling oil droplet absorption spectra and spectral sensitivities of bird cone photoreceptors. Journal of comparative physiology A, Neuroethology, sensory, neural, and behavioral physiology. 2005;191(4):381–392. doi: 10.1007/s00359-004-0595-3. [DOI] [PubMed] [Google Scholar]
- Hattori D, Millard SS, Wojtowicz WM, Zipursky SL. Dscam-mediated cell recognition regulates neural circuit formation. Annual review of cell and developmental biology. 2008;24:597–620. doi: 10.1146/annurev.cellbio.24.110707.175250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmeyer K, Maurel-Zaffran C, Sink H, Treisman JE. Liprin-alpha has LAR-independent functions in R7 photoreceptor axon targeting. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(31):11595–11600. doi: 10.1073/pnas.0604766103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover F, Seleiro EA, Kielland A, Brickell PM, Glover JC. Retinoid X receptor gamma gene transcripts are expressed by a subset of early generated retinal cells and eventually restricted to photoreceptors. The Journal of comparative neurology. 1998;391(2):204–213. [PubMed] [Google Scholar]
- Hsiau TH, Diaconu C, Myers CA, Lee J, Cepko CL, Corbo JC. The cis-regulatory logic of the mammalian photoreceptor transcriptional network. PloS one. 2007;2(7):e643. doi: 10.1371/journal.pone.0000643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Mitchell CK, Redburn-Johnson DA. GABA and GABA(A) receptor antagonists alter developing cone photoreceptor development in neonatal rabbit retina. Visual neuroscience. 2000;17(6):925–935. doi: 10.1017/s0952523800176126. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Lau T, Hatzikirou H, Meyer-Hermann M, Joseph CC, Torquato S. Avian photoreceptor patterns represent a disordered hyperuniform solution to a multiscale packing problem. Physical Review E. 2014;89(2):022721. doi: 10.1103/PhysRevE.89.022721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson AS, Mannervik B. Human glutathione transferase A3-3, a highly efficient catalyst of double-bond isomerization in the biosynthetic pathway of steroid hormones. The Journal of biological chemistry. 2001;276(35):33061–33065. doi: 10.1074/jbc.M104539200. [DOI] [PubMed] [Google Scholar]
- Koso H, Satoh S, Watanabe S. c-kit marks late retinal progenitor cells and regulates their differentiation in developing mouse retina. Developmental biology. 2007;301(1):141–154. doi: 10.1016/j.ydbio.2006.09.027. [DOI] [PubMed] [Google Scholar]
- Kram YA, Mantey S, Corbo JC. Avian cone photoreceptors tile the retina as five independent, self-organizing mosaics. PloS one. 2010;5(2):e8992. doi: 10.1371/journal.pone.0008992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing S The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind O, Kelber A. Avian colour vision: Effects of variation in receptor sensitivity and noise data on model predictions as compared to behavioural results. Vision Res. 2009;49(15):1939–1947. doi: 10.1016/j.visres.2009.05.003. [DOI] [PubMed] [Google Scholar]
- Mariani AP. Neuronal and synaptic organization of the outer plexiform layer of the pigeon retina. The American journal of anatomy. 1987;179(1):25–39. doi: 10.1002/aja.1001790105. [DOI] [PubMed] [Google Scholar]
- Maurel-Zaffran C, Suzuki T, Gahmon G, Treisman JE, Dickson BJ. Cell-autonomous and -nonautonomous functions of LAR in R7 photoreceptor axon targeting. Neuron. 2001;32(2):225–235. doi: 10.1016/s0896-6273(01)00471-8. [DOI] [PubMed] [Google Scholar]
- McIlvain VA, Knox BE. Nr2e3 and Nrl can reprogram retinal precursors to the rod fate in Xenopus retina. Developmental dynamics : an official publication of the American Association of Anatomists. 2007;236(7):1970–1979. doi: 10.1002/dvdy.21128. [DOI] [PubMed] [Google Scholar]
- McLaughlin BJ. A fine structural and E-PTA study of photoreceptor synaptogenesis in the chick retina. The Journal of comparative neurology. 1976;170(3):347–364. doi: 10.1002/cne.901700306. [DOI] [PubMed] [Google Scholar]
- Mears AJ, Kondo M, Swain PK, Takada Y, Bush RA, Saunders TL, Sieving PA, Swaroop A. Nrl is required for rod photoreceptor development. Nature genetics. 2001;29(4):447–452. doi: 10.1038/ng774. [DOI] [PubMed] [Google Scholar]
- Meller K, Tetzlaff W. Scanning electron microscopic studies on the development of the chick retina. Cell and tissue research. 1976;170(2):145–159. doi: 10.1007/BF00224296. [DOI] [PubMed] [Google Scholar]
- Millard SS, Lu Z, Zipursky SL, Meinertzhagen IA. Drosophila dscam proteins regulate postsynaptic specificity at multiple-contact synapses. Neuron. 2010;67(5):761–768. doi: 10.1016/j.neuron.2010.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard SS, Zipursky SL. Dscam-mediated repulsion controls tiling and self-avoidance. Current opinion in neurobiology. 2008;18(1):84–89. doi: 10.1016/j.conb.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montana CL, Myers CA, Corbo JC. Quantifying the activity of cis-regulatory elements in the mouse retina by explant electroporation. Methods in molecular biology. 2013;935:329–340. doi: 10.1007/978-1-62703-080-9_23. [DOI] [PubMed] [Google Scholar]
- Morris VB. Symmetry in a receptor mosaic demonstrated in the chick from the frequencies, spacing and arrangement of the types of retinal receptor. The Journal of comparative neurology. 1970;140(3):359–398. doi: 10.1002/cne.901400308. [DOI] [PubMed] [Google Scholar]
- Morris VB. Time differences in the formation of the receptor types in the developing chick retina. The Journal of comparative neurology. 1973;151(4):323–330. doi: 10.1002/cne.901510403. [DOI] [PubMed] [Google Scholar]
- Morris VB, Shorey CD. An electron microscope study of types of receptor in the chick retina. The Journal of comparative neurology. 1967;129(4):313–340. doi: 10.1002/cne.901290404. [DOI] [PubMed] [Google Scholar]
- Nelson SM, Frey RA, Wardwell SL, Stenkamp DL. The developmental sequence of gene expression within the rod photoreceptor lineage in embryonic zebrafish. Developmental dynamics : an official publication of the American Association of Anatomists. 2008;237(10):2903–2917. doi: 10.1002/dvdy.21721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng L, Hurley JB, Dierks B, Srinivas M, Salto C, Vennstrom B, Reh TA, Forrest D. A thyroid hormone receptor that is required for the development of green cone photoreceptors. Nature genetics. 2001;27(1):94–98. doi: 10.1038/83829. [DOI] [PubMed] [Google Scholar]
- Ochi H, Sakagami K, Ishii A, Morita N, Nishiuchi M, Ogino H, Yasuda K. Temporal expression of L-Maf and RaxL in developing chicken retina are arranged into mosaic pattern. Gene expression patterns : GEP. 2004;4(5):489–494. doi: 10.1016/j.modgep.2004.03.005. [DOI] [PubMed] [Google Scholar]
- Okano T, Fukada Y, Artamonov ID, Yoshizawa T. Purification of cone visual pigments from chicken retina. Biochemistry. 1989;28(22):8848–8856. doi: 10.1021/bi00448a025. [DOI] [PubMed] [Google Scholar]
- Partridge JC. The Visual Ecology of Avian Cone Oil Droplets. J Comp Physiol A. 1989;165(3):415–426. [Google Scholar]
- Peng GH, Chen S. Chromatin immunoprecipitation identifies photoreceptor transcription factor targets in mouse models of retinal degeneration: new findings and challenges. Visual neuroscience. 2005;22(5):575–586. doi: 10.1017/S0952523805225063. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic acids research. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri V, Ranjit S, Konda S, Nicoloro SM, Straubhaar J, Chawla A, Chouinard M, Lin C, Burkart A, Corvera S, Perugini RA, Czech MP. Cidea is associated with lipid droplets and insulin sensitivity in humans. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(22):7833–7838. doi: 10.1073/pnas.0802063105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts MR, Hendrickson A, McGuire CR, Reh TA. Retinoid X receptor (gamma) is necessary to establish the S-opsin gradient in cone photoreceptors of the developing mouse retina. Investigative ophthalmology & visual science. 2005;46(8):2897–2904. doi: 10.1167/iovs.05-0093. [DOI] [PubMed] [Google Scholar]
- Roberts MR, Srinivas M, Forrest D, Morreale de Escobar G, Reh TA. Making the gradient: thyroid hormone regulates cone opsin expression in the developing mouse retina. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(16):6218–6223. doi: 10.1073/pnas.0509981103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh S, Tang K, Iida A, Inoue M, Kodama T, Tsai SY, Tsai MJ, Furuta Y, Watanabe S. The spatial patterning of mouse cone opsin expression is regulated by bone morphogenetic protein signaling through downstream effector COUP-TF nuclear receptors. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2009;29(40):12401–12411. doi: 10.1523/JNEUROSCI.0951-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas M, Ng L, Liu H, Jia L, Forrest D. Activation of the blue opsin gene in cone photoreceptor development by retinoid-related orphan receptor beta. Molecular endocrinology. 2006;20(8):1728–1741. doi: 10.1210/me.2005-0505. [DOI] [PubMed] [Google Scholar]
- Suzuki SC, Bleckert A, Williams PR, Takechi M, Kawamura S, Wong RO. Cone photoreceptor types in zebrafish are generated by symmetric terminal divisions of dedicated precursors. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(37):15109–15114. doi: 10.1073/pnas.1303551110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature biotechnology. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimarchi JM, Harpavat S, Billings NA, Cepko CL. Thyroid hormone components are expressed in three sequential waves during development of the chick retina. BMC developmental biology. 2008;8:101. doi: 10.1186/1471-213X-8-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimarchi JM, Stadler MB, Roska B, Billings N, Sun B, Bartch B, Cepko CL. Molecular heterogeneity of developing retinal ganglion and amacrine cells revealed through single cell gene expression profiling. The Journal of comparative neurology. 2007;502(6):1047–1065. doi: 10.1002/cne.21368. [DOI] [PubMed] [Google Scholar]
- Wahlin KJ, Moreira EF, Huang H, Yu N, Adler R. Molecular dynamics of photoreceptor synapse formation in the developing chick retina. The Journal of comparative neurology. 2008;506(5):822–837. doi: 10.1002/cne.21582. [DOI] [PubMed] [Google Scholar]
- Walls GL. The Vertebrate Eye and Its Adaptive Radiation. The Cranbrook Institute of Science; Bloomfield Hills, Michigan: 1942. [Google Scholar]
- Wu L, Zhou L, Chen C, Gong J, Xu L, Ye J, Li D, Li P. Cidea controls lipid droplet fusion and lipid storage in brown and white adipose tissue. Science China Life sciences. 2014;57(1):107–116. doi: 10.1007/s11427-013-4585-y. [DOI] [PubMed] [Google Scholar]
- Yamagata M, Sanes JR. Dscam and Sidekick proteins direct lamina-specific synaptic connections in vertebrate retina. Nature. 2008;451(7177):465–469. doi: 10.1038/nature06469. [DOI] [PubMed] [Google Scholar]
- Yen L, Fager RS. Chromatographic resolution of the rod pigment from the four cone pigments of the chicken retina. Vision Res. 1984;24(11):1555–1562. doi: 10.1016/s0042-6989(84)80005-x. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Mears AJ, Friedman JS, Carter T, He S, Oh E, Jing Y, Farjo R, Fleury G, Barlow C, Hero AO, Swaroop A. Expression profiling of the developing and mature Nrl−/− mouse retina: identification of retinal disease candidates and transcriptional regulatory targets of Nrl. Human molecular genetics. 2004;13(14):1487–1503. doi: 10.1093/hmg/ddh160. [DOI] [PubMed] [Google Scholar]
- Zhang J, De Blas AL, Miralles CP, Yang CY. Localization of GABAA receptor subunits alpha 1, alpha 3, beta 1, beta 2/3, gamma 1, and gamma 2 in the salamander retina. The Journal of comparative neurology. 2003;459(4):440–453. doi: 10.1002/cne.10626. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Yon Toh S, Chen Z, Guo K, Ng CP, Ponniah S, Lin SC, Hong W, Li P. Cidea-deficient mice have lean phenotype and are resistant to obesity. Nature genetics. 2003;35(1):49–56. doi: 10.1038/ng1225. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.