Background: Conflicting results regarding the role of ARF6 in dendritic spine development have not been answered.
Results: ARF6-mediated Rac1 or RhoA activation via PLD pathway either positively or negatively regulates spine formation.
Conclusion: The key factor underlying conversion of the ARF6 effect during development is neuronal activity.
Significance: Activity dependence of ARF6-mediated spine formation may play a role in structural plasticity of mature neurons.
Keywords: ADP-ribosylation Factor (ARF), Dendritic Spine, Phospholipase D, Ras-related C3 Botulinum Toxin Substrate 1 (Rac1), Synaptic Plasticity, ARF6, Rac1, Neuronal Activity
Abstract
Recent studies have reported conflicting results regarding the role of ARF6 in dendritic spine development, but no clear answer for the controversy has been suggested. We found that ADP-ribosylation factor 6 (ARF6) either positively or negatively regulates dendritic spine formation depending on neuronal maturation and activity. ARF6 activation increased the spine formation in developing neurons, whereas it decreased spine density in mature neurons. Genome-wide microarray analysis revealed that ARF6 activation in each stage leads to opposite patterns of expression of a subset of genes that are involved in neuronal morphology. ARF6-mediated Rac1 activation via the phospholipase D pathway is the coincident factor in both stages, but the antagonistic RhoA pathway becomes involved in the mature stage. Furthermore, blocking neuronal activity in developing neurons using tetrodotoxin or enhancing the activity in mature neurons using picrotoxin or chemical long term potentiation reversed the effect of ARF6 on each stage. Thus, activity-dependent dynamic changes in ARF6-mediated spine structures may play a role in structural plasticity of mature neurons.
Introduction
Dendritic spines are actin-rich architectures that are important for receiving presynaptic inputs in most excitatory synapses in the central nervous system (1). These spines are flexible structures, and their formation and morphogenesis are dynamically regulated through actin cytoskeletal reorganization (2). The resulting changes are known to be cellular mechanisms that modify brain activity such as the formation of long term memory (3–5). Because of the presence of an extensive actin cytoskeleton and the central role of actin in inducing morphological changes of spines in response to a variety of neural events, proteins known to regulate actin cytoskeleton and the associated signal pathways have been the primary targets of studies focusing on spine formation and morphogenesis (6).
ADP-ribosylation factor 6 (ARF6)2 belongs to the ARF protein family, a Ras-related family of small GTPases that regulate actin cytoskeleton and membrane trafficking (7, 8). ARF6 has been known to play various roles in the regulation of cortical actin dynamics and in membrane exchanges between the plasma membrane and endocytic compartments (9). In the nervous system, ARF6 has been shown to regulate neurotransmitter release at the Xenopus neuromuscular junction via ARF GEF msec7-1 and facilitate AP-2 and clathrin recruitments to synaptic membranes (7). Furthermore, it was seen that ARF6 regulates early axonal and dendritic growth and branching (10, 11). Besides its involvement in early neuronal morphogenesis, ARF6 is known to play a role in later stages of neuronal development, and recent studies have reported that ARF6 affects dendritic spine formation and causes morphological changes (11–13). These studies, however, presented contradictory results regarding the role of ARF6 in spine development. Miyazaki et al. (12) found that a constitutively active mutant of ARF6 (ARF6-Q67L) displayed a severe spine reduction in mature hippocampal neurons (21 days in vitro (DIV 21)), whereas a dominant negative mutant of ARF6 (ARF6-T27N) resulted in a substantial increase in spine density. Choi et al. (13), however, showed that activation of ARF6 by overexpression of a fast cycling mutant of ARF6 (ARF6-T157A) increases the spine density in developing neurons (DIV 11), whereas a knockdown of ARF6 resulted in decreased spine formation. Although several explanations regarding this controversy have been suggested, it is still unclear whether ARF6 activation plays a positive or negative role in spine development.
Here we showed that an activation of ARF6 differentially regulates spine formation and maintenance depending on neuronal maturation and activity. In developing neurons, ARF6-T157A, a fast cycling mutant of ARF6, increased the number of dendritic spines via the ARF6/phospholipase D (PLD)/phosphatidic acid (PA) to PAK1-dependent (via guanine nucleotide dissociation inhibitor for Rho proteins (RhoGDI)/Rac1) and/or PAK1-independent pathway. Conversely, ARF6-T157A exhibited a completely opposite effect in mature neurons, reducing spine density by the balance between PLD-induced Rac1 and RhoA activation and bypassing the PAK/RhoGDI pathway. Consistent with these findings, genome-wide microarray analysis revealed that ARF6 activation in developing and mature neurons leads to opposite patterns of expression of a subset of genes that are involved in neuronal morphology. We further demonstrated that the key factor underlying the conversion of the ARF6 effect during development is neuronal activity because blocking neuronal activity in developing neurons using tetrodotoxin (TTX) or enhancing it in mature neurons using picrotoxin (PTX) or chemically induced long term potentiation (LTP) reversed the spine-promoting or spine-reducing effect of ARF6-T157A, respectively.
EXPERIMENTAL PROCEDURES
Ethics Statement
Animal experimental procedures were approved by the Institute of Animal Care and Use Committee of Seoul National University, Korea (approval identification number SNU-100930-5).
DNA Constructs and Reagents
Green fluorescent protein (GFP)-5-phosphatase domain of synaptojanin-1 (5Ptase) and GFP-RacDN (dominant negative mutant form; RacN17) were kindly provided by Dr. Pietro De Camilli (Yale University, New Haven, CT). GFP-PAK inhibitory domain (PID) was provided by Dr. Dong-Eun Park (Seoul National University, Seoul, South Korea), and GFP-RhoGDI1-S101A/S174A (mtRhoGDI) was provided by Dr. Eung-Gook Kim (Chungbuk National University, Cheongju, South Korea). C-terminally HA-tagged wild-type ARF6, ARF6-T27N (dominant negative mutant form), and ARF6-T157A (fast cycling mutant form) were provided by Dr. Eunjoon Kim (Korea Advanced Institute of Science and Technology, Daejeon, South Korea). ARF6-N48I and ARF6-T157A/N48I were generated by site-directed point mutations using a QuikChange® site-directed mutagenesis kit (Stratagene, La Jolla, CA). The fidelity of all constructs was verified by DNA sequencing. All other reagents were from Sigma.
Hippocampal Neuron Culture and Transfection
Primary rat hippocampal neurons were prepared as described (14). Briefly, hippocampi were dissected from embryonic day 18 Sprague-Dawley fetal rats, dissociated with papain, and triturated with a polished half-bore Pasteur pipette. The cells (2.5 × 105) in minimum Eagle's medium supplemented with 0.6% glucose, 1 mm pyruvate, 2 mm l-glutamine, 10% fetal bovine serum, and antibiotics were plated on poly-d-lysine-coated glass coverslips in a 60-mm Petri dish. Four hours after plating, the medium was replaced with Neurobasal (Invitrogen) supplemented with 2% B-27 and 0.5 mm l-glutamine. 4 μm 1-β-d-cytosine arabinofuranoside was added as needed. Neurons were transfected using the calcium phosphate method. Briefly, empty vector (pcDNA3.0-HA), ARF6-T27N, ARF6-T157A, ARF6-N48I, or ARF6-T157A/N48I with either GFP, GFP-5Ptase, GFP-RhoGDI, or GFP-RacDN was either co-transfected at DIV 11 and fixed at DIV 15 for developing stage or co-transfected at DIV 16 and fixed at DIV 20 for mature stage. Co-transfection was performed at a ratio of 1:3.
Pharmacological Treatments of Neurons at Either Developing Stage or Mature Stage for Activity-dependent Spine Changes
For global synaptic depression, 4 h after transfection at DIV 11 or 16, neurons were treated with 1 μm TTX for 4 days. Neurons were fixed in 4% paraformaldehyde, 4% sucrose, PBS for 15 min, mounted, and stored at 4 °C for further imaging analysis. For synaptic activity enhancement of mature neurons, 4 h after transfection at DIV 11 or 16, neurons were treated with 50 μm PTX for 4 days. Neurons were fixed in 4% paraformaldehyde, 4% sucrose, PBS for 15 min, mounted, and stored at 4 °C for further imaging analysis.
Chemical LTP Induction
Neurons were co-transfected with GFP with HA or ARF6-T157A at DIV 11 or 16 using he calcium phosphate method, and 9 h after transfection, these neurons were subjected to chemically induced long term potentiation (chem-LTP) as described (15). Briefly, transfected neurons were placed in Mg2+-free extracellular solution (110 mm NaCl, 2 mm CaCl2, 5 mm KCl, 10 mm HEPES, 30 mm glucose, 0.5 μm TTX, 1 μm strychnine, 20 μm bicuculline methiodide, pH 7.4) for 10 min at 37 °C in a CO2 incubator and then 200 μm glycine was applied for 3 min at 37 °C in a CO2 incubator to induce chem-LTP. After 20 min in extracellular solution, the neurons were transferred to a dish containing the original medium. The neurons were fixed at DIV 20, mounted, and stored at 4 °C for further imaging analysis.
Microscopy and Image Analysis
Fluorescence images were acquired on an Olympus IX-71 inverted microscope equipped with a UPlanApo 40×, 1.00 numerical aperture oil immersion objective (Olympus, Tokyo, Japan), a GFP-optimized filter set (Omega Optical, Brattleboro, VT), and an ORCA-R2 charge-coupled device camera (Hamamatsu Photonics, Hamamatsu, Japan) driven by MetaMorph Imaging software (Molecular Devices, Sunnyvale, CA). Cells were excited with 480 nm light (TouchBright X6, Live Cell Instrument, Seoul, South Korea). MetaMorph Imaging software was used for analysis. For three-dimensional structured illumination microscopy (3D-SIM), we used an N-SIM microscope system equipped with a Nikon CFI Plan Apo IR 60×, 1.27 numerical aperture water immersion objective (Nikon, Tokyo, Japan) and an Andor DU-897 X-5834 EM charge-coupled device camera (Andor, Belfast, UK). The channel was carefully aligned using an alignment parameter from control measurements with 0.1-μm-diameter fluorescent beads. Image stacks of typically 1.0-μm height with 33 images each and a z-distance of 0.03 μm were acquired and computationally reconstructed. Reconstructed images were generated in Blend Projection images with Imaris v7.5.2 (Bitplane, Zurich, Switzerland) including volume rendering, rotations, and display adjustment to eliminate background noise and brighten images. For analysis, well branched pyramidal or multipolar neurons were randomly selected, and the experiments were performed in a blinded manner. One to three secondary dendrites in each neuron were chosen. To determine the number of dendritic spines and filopodia, spines were defined as dendritic protrusions of <5 μm in length with a head and filopodia as dendritic protrusions of <10 μm in length without apparent heads. Spine head was defined as the tip structure that should be at least 2 times thicker than the spine neck. Spines can be morphologically classified into three types as follows: stubby, thin, and mushroom-shaped (16). To distinguish spine types, we defined spines as follows: the stubby type was defined as spines devoid of an apparent neck, the thin type was defined as spines having a thin neck and a small bulbous head, and the mushroom type was defined as spines having a neck and a large mushroom-shaped head. The density and types of spines from a single dendrite were grouped and averaged. Data are presented as means ± S.E. Statistical analysis was carried out with PASW Statistics 18 (formerly SPSS Statistics). For multiple conditions, we compared means by ANOVA followed by Tukey's HSD post hoc test.
Generation of Recombinant Sindbis Viruses and Infection
Sindbis viral constructs were kindly provided by Dr. Hyongkyu Kim (Chungbuk National University). Generation of recombinant Sindbis viruses expressing ARF6-T157A-His6 was performed as described previously (17). For genome-wide microarray analysis, rat hippocampal neuron cultures were infected at DIV 10 for developing stage and DIV 17 for mature stage and maintained for an additional 36 h. Infected neurons were washed twice with PBS and stored at −70 °C for further investigations. Samples were lysed, and RNA isolation was carried out using the RNA-spin total RNA extraction kit (iNtRON Biotechnology, Sungnam, Kyungki-Do, South Korea), and genome-wide microarray analysis was performed.
Microarray Experiments
We prepared total RNA independently from developing and mature neurons infected with Sindbis virus-expressing ARF6-T157A and empty (CTL) vectors using the RNeasy mini kit (Qiagen, Valencia, CA). We analyzed a total of eight samples composed of duplicate samples in the above four conditions. The RNA integrity of each sample was assessed using Bioanalyzer 2100 (Agilent technologies, Santa Clara, CA); the RNA integrity number was in the range between 9.3 and 10 for all the four samples. RNA was reverse transcribed, amplified, and then hybridized onto an Agilent SurePrint G3 Rat GE Microarray 8 × 60,000 (including 62,976 probes corresponding to 19,958 annotated genes) according to the Agilent standard protocols. The fluorescent signal on the array was measured using an Agilent SureScan microarray scanner. The probe intensities were converted to log2 intensities and then normalized using the quantile normalization method (18). The normalized data were deposited in the Gene Expression Omnibus (GEO) database under accession number GSE40937.
Statistical Analysis of Gene Expression Data
Using the normalized log2 intensities, the differentially expressed genes (DEGs) between two conditions were identified as described previously (19). Briefly, 1) Student's t test and log2 median ratio test were performed to compute T values and log2 median ratios for all the genes. 2) Empirical distributions of the null hypothesis were estimated by performing all possible combinations of random permutations of samples and then applying the Gaussian kernel density estimation method to T values and log2 median ratios resulting from the random permutations (20). 3) The adjusted p value of each gene for the individual tests was computed by the two-tailed test using the empirical null distribution. 4) The adjusted p values from the two tests were then combined using Stouffer's method (21). And finally, 5) the DEGs were identified as the present genes with a combined p value ≤0.1. To reduce false positives, we further selected only DEGs whose absolute log2 -fold changes were larger than the cutoff, the mean of 5th and 95th percentiles of the null distribution of log2 -fold changes (0.372). Finally, gene ontology biological processes enriched by a list of genes were identified as those with a p value <0.05 using DAVID software (22).
RNA Interference
Small hairpin RNA (shRNA) for RNA interference against rat PAK1 or rat ARF6 mRNA was designed based on the rat PAK1 (GenBankTM accession number NM_017198.1) or rat ARF6 (GenBank accession number NM_024152) cDNA sequence, respectively. PAK1 was targeted to the region of nucleotides 1222–1240. Complementary oligonucleotides were synthesized separately with the addition of a BamHI site at the 5′-end and an EcoRI site at the 3′-end. The forward targeting sequence of shRNA for PAK1 (shRNA-PAK1) was 5′-GGATTCTGTGCACAGATAA-3′. For ARF6 targeting, the region of nucleotides 305–323 was chosen. Complementary oligonucleotides were synthesized separately with the addition of a BamHI site at the 5′-end and an EcoRI site at the 3′-end. The forward targeting sequence of shRNA for ARF6 (shRNA-ARF6) was 5′-AGCTGCACCGCATTATCAA-3′. The annealed cDNA fragments were cloned into the BamHI-EcoRI sites of the vector pSIREN-DNR-DsRed-Express (BD Biosciences). The efficiency of shRNAs was tested on protein levels of HA-tagged rat PAK1 or rat ARF6 in HEK293T cells (data not shown). Both PAK1 and ARF6 targeting sequences have been already reported (13, 23). The fidelity of all constructs was verified by sequencing.
RESULTS AND DISCUSSION
The Number of Spines Increases Exponentially during DIV 11–15
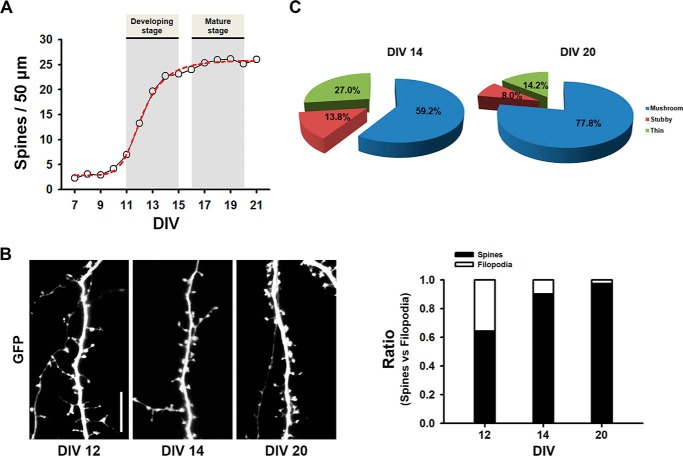
To evaluate dendritic spine formation during neuronal development, neurons were transfected with GFP at DIV 7, and spine numbers were counted during neuronal development. We defined dendritic spines as protrusions of 0.5–5 μm in length with a head. The number of spines increased during DIV 11–15 and reached a plateau at later stages (Fig. 1 A). We defined the developing stage of a neuron as when spine development shows exponential growth (DIV 11–15), whereas the mature stage was when it reaches a plateau (DIV 17–20). The ratio of spines versus filopodia also increased as neurons matured (Fig. 1B).
FIGURE 1.
Developmental changes of dendritic spine formation in rat primary cultured hippocampal neurons. A, cultured neurons were transfected with GFP at DIV 7 and then fixed at the indicated DIV. Spine density rises during DIV 11–15, and then the levels persist. Data were collected from three coverslips with each having two dendrites from three neurons at the indicated DIV. B, representative images for spines at the indicated DIV and a ratio graph of spines versus filopodia are depicted. C, pie graphs illustrating the changes in the portion of different morphological types of spine protrusions in the neurons expressing GFP depending on developmental stages.
Spines can be morphologically divided into three types as follows: thin, stubby, and mushroom-shaped. The thin type was defined as spines having a long thin neck and a small bulbous head, the stubby type was defined as those devoid of a neck, and the mushroom-shaped type was defined as those having a neck and a large head. The portion of different morphological types of spines changed with an increase in the proportion of mushroom-shaped spines and a concomitant reduction in the proportion of thin and stubby spines (Fig. 1C).
ARF6 Distinctly Modulates Dendritic Spine Formation at Each Stage
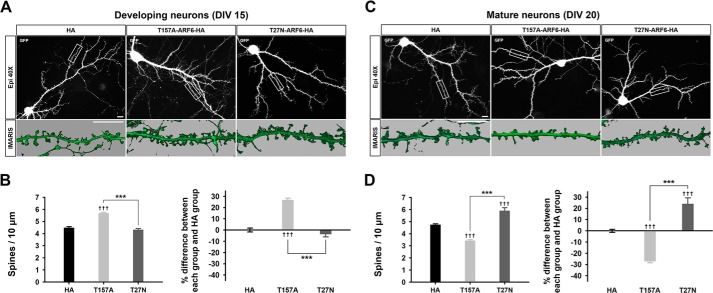
To determine the role of ARF6 in spine formation at the developing stage, we co-transfected neurons at DIV 11 with a fast cycling mutant of ARF6 (ARF6-T157A), a dominant negative mutant of ARF6 (ARF6-T27N), or an empty vector (HA) with GFP and fixed cells at DIV 15. Overexpression of ARF6-T157A significantly increased the number of spines, whereas overexpression of ARF6-T27N showed no change (Fig. 2, A and B, and Tables 1 and 2). These results are consistent with a previous report (13), confirming that ARF6 activation positively regulates spine formation in developing neurons.
FIGURE 2.
ARF6 differentially regulates dendritic spine formation in developing and mature neurons. A, representative images for developing neurons. Cultured hippocampal neurons were co-transfected at DIV 11 with T157A-ARF6-HA, T27N-ARF6-HA, or pcDNA3.0-HA (HA), and pEGFP-c1 and fixed at DIV 15. Inset images were acquired by three-dimensional structured illumination microscopy and were reconstructed by surface area rendering using Imaris software. Scale bars, 10 μm. B, quantification of the effect of ARF6 mutants on dendritic spine formation. The values in the spine number graph were recalculated to show percent differences between each group and HA. Detailed data are shown in Table 1. Data were collected for each group in three independent experiments. The values are means ± S.E. Daggers (†) indicate the significance between HA and each group, and asterisks (*) indicate the significance between each group. ††† and ***, p < 0.001 (ANOVA and Tukey's HSD post hoc test). C, representative images for mature neurons. Cultured hippocampal neurons were co-transfected at DIV 16 with T157A-ARF6-HA, T27N-ARF6-HA, or HA and GFP and fixed at DIV 20. Scale bars, 10 μm. D, quantification of the effect of ARF6 mutants on dendritic spine formation. The values in the spine number graph were recalculated to show the differences between each group and HA. Detailed data are shown in Table 2. Data were collected for each group in three independent experiments. The values are means ± S.E. ††† and ***, p < 0.001 (ANOVA and Tukey's HSD post hoc test). Error bars represent S.E.
TABLE 1.
Data for ARF6-T157A sets of developing neurons
| Sample name | No. of spines/10 μm (mean ± S.E.) | Percent differences (mean ± S.E.) | n (no. of coverslips) | No. of neurons | No. of dendrites |
|---|---|---|---|---|---|
| GFP + HA | 4.49 ± 0.08 | 0 ± 1.68 | 11 | 25 | 50 |
| GFP + T157A-HA | 5.69 ± 0.07 | 26.77 ± 1.59 | 13 | 28 | 56 |
| GFP + T27N-HA | 4.32 ± 0.10 | −3.81 ± 2.33 | 12 | 26 | 52 |
| GFP + N48I-HA | 4.39 ± 0.08 | −2.13 ± 1.86 | 5 | 14 | 28 |
| GFP + T157A/N48I-HA | 4.68 ± 0.04 | 4.20 ± 0.88 | 5 | 14 | 26 |
| GFP-PID + HA | 3.29 ± 0.10 | −26.62 ± 2.12 | 6 | 16 | 32 |
| GFP-PID + T157A-HA | 4.54 ± 0.11 | 1.04 ± 2.40 | 5 | 14 | 28 |
| GFP-mtRhoGDI + HA | 1.91 ± 0.05 | −57.42 ± 1.06 | 6 | 15 | 30 |
| GFP-mtRhoGDI + T157A-HA | 2.72 ± 0.06 | −39.44 ± 1.36 | 7 | 16 | 32 |
| GFP-RacDN + HA | 2.08 ± 0.21 | −53.56 ± 4.61 | 6 | 15 | 30 |
| GFP-RacDN + T157A-HA | 2.72 ± 0.15 | −39.32 ± 3.24 | 8 | 16 | 32 |
| GFP-5Ptase + HA | 3.58 ± 0.08 | −20.21 ± 1.85 | 4 | 14 | 28 |
| GFP-5Ptase + T157A-HA | 4.48 ± 0.11 | −0.21 ± 2.48 | 4 | 14 | 28 |
| TTX + GFP + HA | 5.16 ± 0.19 | 14.84 ± 4.31 | 10 | 18 | 36 |
| TTX + GFP + T157A-HA | 2.92 ± 0.10 | −34.86 ± 2.17 | 9 | 20 | 40 |
| PTX + GFP + HA | 5.22 ± 0.08 | 15.25 ± 2.77 | 5 | 15 | 30 |
| PTX + GFP + T157A-HA | 5.87 ± 0.18 | 28.05 ± 4.23 | 4 | 12 | 24 |
TABLE 2.
Data for ARF6-T157A sets of mature neurons
cLTP, chemically induced LTP.
| Sample name | No. of spines/10 μm (mean ± S.E.) | Percent differences (mean ± S.E.) | n (no. of coverslips) | No. of neurons | No. of dendrites |
|---|---|---|---|---|---|
| GFP + HA | 4.75 ± 0.06 | 0 ± 1.34 | 19 | 38 | 62 |
| GFP + T157A-HA | 3.44 ± 0.06 | −27.70 ± 1.33 | 16 | 32 | 64 |
| GFP + T27N-HA | 5.90 ± 0.25 | 24.01 ± 5.28 | 7 | 17 | 33 |
| GFP + N48I-HA | 4.65 ± 0.11 | −2.24 ± 2.25 | 5 | 14 | 26 |
| GFP + T157A/N48I-HA | 4.46 ± 0.19 | −6.12 ± 3.89 | 4 | 13 | 26 |
| GFP-PID + HA | 2.95 ± 0.11 | −38.02 ± 2.31 | 5 | 13 | 26 |
| GFP-PID + T157A-HA | 2.99 ± 0.09 | −37.10 ± 1.85 | 6 | 15 | 30 |
| GFP-mtRhoGDI + HA | 3.33 ± 0.06 | −29.91 ± 1.17 | 5 | 15 | 30 |
| GFP-mtRhoGDI + T157A-HA | 2.87 ± 0.05 | −39.65 ± 1.12 | 5 | 14 | 28 |
| GFP-RacDN + HA | 3.96 ± 0.11 | −16.61 ± 2.28 | 5 | 12 | 24 |
| GFP-RacDN + T157A-HA | 4.75 ± 0.15 | −0.08 ± 3.05 | 4 | 11 | 22 |
| GFP-5Ptase + HA | 3.42 ± 0.21 | −28.14 ± 4.34 | 6 | 13 | 26 |
| GFP-5Ptase + T157A-HA | 3.10 ± 0.31 | −34.85 ± 6.43 | 5 | 12 | 24 |
| TTX + GFP + HA | 4.15 ± 0.11 | −12.66 ± 2.41 | 5 | 13 | 26 |
| TTX + GFP + T157A-HA | 3.34 ± 0.06 | −29.70 ± 1.33 | 5 | 13 | 26 |
| PTX + GFP + HA | 5.08 ± 0.06 | 6.84 ± 1.21 | 5 | 14 | 28 |
| PTX + GFP + T157A-HA | 4.90 ± 0.13 | 2.97 ± 2.65 | 4 | 13 | 26 |
| cLTP + GFP + HA | 5.58 ± 0.17 | 17.44 ± 3.55 | 4 | 13 | 26 |
| cLTP + GFP + T157A-HA | 4.75 ± 0.09 | −0.04 ± 1.86 | 5 | 15 | 30 |
Next, we investigated the roles of active ARF6 at the mature stage of neurons. We co-transfected neurons with ARF6-T157A, ARF6-T27N, or HA with GFP at DIV 16, and the cells were fixed at DIV 20. In contrast to the results of developing neurons, ARF6-T157A significantly decreased the number of spines, whereas ARF6-T27N promoted spine formation (Fig. 2, C and D), which is consistent with a previous report (12), confirming that ARF6 activation negatively regulates spine formation in mature neurons.
Active ARF6 Promotes Dendritic Spine Formation in Developing Neurons
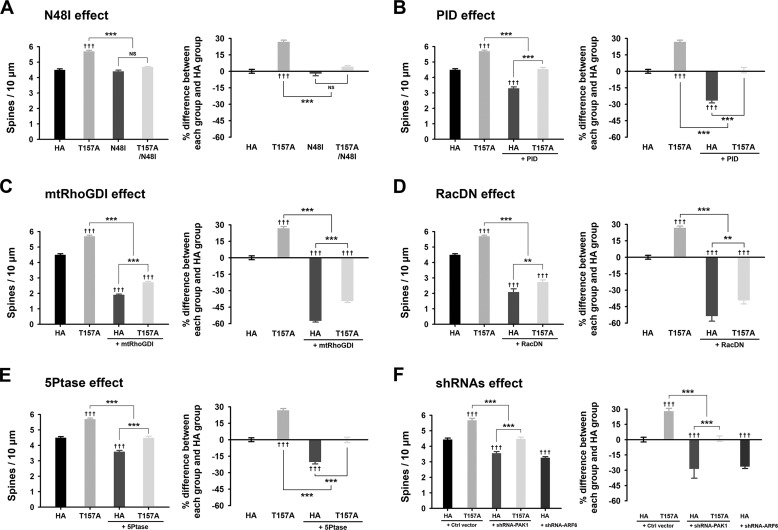
ARF6 is known to activate various downstream pathways including PLD, phosphatidylinositol 4-phosphate 5-kinases, and Rac1 (7). To test which pathway(s) is involved in the spine-promoting effect of ARF6 at the developing stage, we first co-transfected neurons with GFP and ARF6-N48I, which prevents activation of PLD, or ARF6-T157A/N48I at DIV 11, and the cells were fixed at DIV 15. We found that ARF6-T157A/N48I completely abrogated the spine-promoting effect of ARF6-T157A, whereas ARF6-N48I alone had no effect on spine formation (Fig. 3A). These results suggest that the spine-promoting effect of ARF6-T157A in developing neurons is mainly mediated through the PLD pathway.
FIGURE 3.
The effects of ARF6-T157A or ARF6 knockdown on dendritic spine formation in developing neurons. A–E, cultured hippocampal neurons were co-transfected at DIV 11 with T157A-ARF6-HA, T157A/N48I-ARF6-HA, N48I-ARF6-HA, or HA and GFP (A), T157A-ARF6-HA or HA and GFP or GFP-PID (B), T157A-ARF6-HA or HA and GFP or GFP-mtRhoGDI (C), T157A-ARF6-HA or HA and GFP or GFP-RacDN (D), or T157A-ARF6-HA or HA and GFP or GFP-5Ptase (E) and fixed at DIV 15. The effects of ARF6 mutants on dendritic spine formation were quantified to investigate the PLD/PA pathway. The values in the spine number graph were recalculated to show percent differences between each group and HA. Detailed data are shown in Table 1. Data were collected for each group in three independent experiments. ††† and ***, p < 0.001; **, p < 0.01; NS, not significant (ANOVA and Tukey's HSD post hoc test). F, cultured hippocampal neurons were co-transfected at DIV 11 with T157A-ARF6-HA or HA and pSIREN-empty (control (Ctrl) vector), shRNA-PAK1, or shRNA-ARF6 and fixed at DIV 15. The effect of ARF6-T157A with PAK1 depletion or ARF6 depletion on dendritic spine formation was quantified in developing neurons. The values in the spine number graph were recalculated to show the differences between each group and HA. Detailed data are shown in Table 3. Data were collected for each group in three independent experiments. The values are means ± S.E. ††† and ***, p < 0.001 (ANOVA and Tukey's HSD post hoc test). Error bars represent S.E.
The prominent downstream candidate for PLD/PA-mediated spine development is PAK1, a serine/threonine kinase that regulates the actin cytoskeleton (24). The role of PAK1 in spine regulation has been firmly established by a number of previous studies (25, 26). To test whether PAK1 is involved in ARF6/PLD-mediated spine regulation in developing neurons, we co-transfected neurons with ARF6-T157A and PID, an autoinhibitory domain of PAK1. PID alone reduced spine density, suggesting that in the resting state spine formation is regulated endogenously by PAK1 via an ARF6-independent pathway(s) (Fig. 3B). Co-expression of PID completely eliminated the effect of ARF6-T157A, indicating the involvement of the ARF6/PLD/PA/PAK1 pathway. The difference between PID alone and PID with ARF6-T157A also suggests that an ARF6-dependent but PAK1-independent pathway could be involved in ARF6-T157A-mediated spine formation (Fig. 3B; see also Fig. 7).
FIGURE 7.
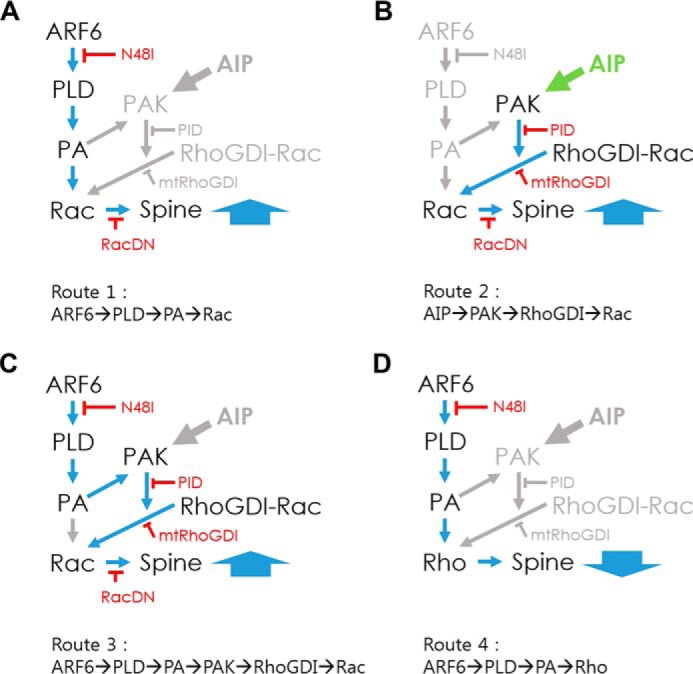
Hypothetical models of ARF6 signaling pathways for regulating spine formation. In developing neurons, three routes are in operation, and all converge on Rac1 to promote spine formation. A, Route 1: ARF6-dependent but PAK1-independent pathway (ARF6/PLD/PA/Rac) that explains the difference between PID alone and PID with ARF6-T157A in Fig. 3. B, Route 2: ARF6-independent but PAK1-dependent pathway (AIP/PAK/RhoGDI/Rac) that explains why PID alone reduced spine density in Fig. 3B. C, Route 3: ARF6- and PAK1-dependent pathway (ARF6/PLD/PA/PAK/RhoGDI/Rac). In mature neurons, another route is in operation. D, Route 4: ARF6-dependent and Rho-dependent pathway (ARF6/PLD/PA/Rho) antagonizes the effect of Rac1 and provides a strong negative impact on spine formation. The balance between ARF6-T157A-induced Rac1 and RhoA activation via the PLD pathway, bypassing PAK/RhoGDI pathway, plays a key role in the spine-reducing effect of ARF6-T157A in mature neurons. AIP, ARF6-independent pathway.
RhoGDI is known to be a downstream effector of PAK1 (27). RhoGDI phosphorylation by PAK1 dissociates Rac1 from RhoGDI, allowing subsequent Rac1 activation by Rac guanine nucleotide exchange factors (27). Because previous studies showed that the spine-promoting effect of ARF6 is partially blocked by a dominant negative Rac1 (13), we tested the involvement of the PAK1/RhoGDI pathway using ARF6-T157A. We co-transfected neurons with ARF6-T157A or HA and mtRhoGDI, which cannot be phosphorylated by PAK1. We found that mtRhoGDI completely abrogated the effect of ARF6-T157A (Fig. 3C). Consistently, we also found that RacDN largely blocked ARF6-T157A-induced spine formation (Fig. 3D).
PA can also activate phosphatidylinositol 4-phosphate 5-kinases, leading to an increase in the production of phosphatidylinositol 4,5-bisphosphate (PIP2) (28). We found that co-transfection of ARF6-T157A or HA and GFP-tagged 5Ptase, which reduces cellular PIP2 levels, reversed the effect of ARF6-T157A. In addition, 5Ptase alone reduced spine density (Fig. 3E). Because it is already known that PIP2 at the plasma membrane is required for proper GDP/GTP exchange on Rac1 where Rac1 regulates actin cytoskeleton (29), we hypothesized that the negating effect of 5Ptase on ARF6-T157A might be mediated by hampering Rac1 recruitment and activation at the plasma membrane.
These results suggest that the ARF6/PLD/PA to PAK1-dependent (via RhoGDI/Rac1) and/or PAK1-independent pathway is significant in the spine-promoting effect of ARF6 in developing neurons (see also Fig. 7). The above results were further corroborated by the results from knockdown of endogenous PAK1 (Fig. 3F and Table 3). Suppression of PAK1 expression by shRNA-PAK1 alone reduced spine density, and co-expression of shRNA-PAK1 and ARF6-T157A completely eliminated the effect of ARF6-T157A. Again, the difference between shRNA-PAK1 alone and shRNA-PAK1 with ARF6-T157A also suggests that an ARF6-dependent but PAK1-independent pathway could be involved in ARF6-T157A-mediated spine formation (Fig. 3F). The knockdown of endogenous ARF6 alone reduced spine density (Fig. 3F), which might be from side effects of knockdown considering the involvement of ARF6 in a wide variety of cellular processes.
TABLE 3.
Data for sets of shRNA-PAK1 and shRNA-ARF6 in both stages of neurons
Ctrl, control.
| Stage | Sample name | No. of spines/10 μm (mean ± S.E.) | Percent differences (mean ± S.E.) | n (no. of coverslips) | No. of neurons | No. of dendrites |
|---|---|---|---|---|---|---|
| Developing | Ctrl vector + HA | 4.43 ± 0.10 | 0 ± 2.33 | 8 | 20 | 40 |
| Ctrl vector + T157A-HA | 5.67 ± 0.13 | 27.91 ± 2.88 | 6 | 16 | 32 | |
| shRNA-PAK1 + HA | 3.55 ± 0.11 | −28.68 ± 9.17 | 8 | 20 | 40 | |
| shRNA-PAK1 + T157A-HA | 4.47 ± 0.12 | 1.00 ± 2.63 | 7 | 18 | 36 | |
| shRNA-ARF6 + HA | 3.25 ± 0.08 | −26.51 ± 1.74 | 5 | 14 | 28 | |
| Mature | Ctrl vector + HA | 4.80 ± 0.13 | 0 ± 2.69 | 9 | 24 | 48 |
| Ctrl vector + T157A-HA | 3.36 ± 0.11 | −29.95 ± 2.19 | 7 | 18 | 36 | |
| shRNA-PAK1 + HA | 4.24 ± 0.11 | −11.61 ± 2.19 | 5 | 14 | 28 | |
| shRNA-PAK1 + T157A-HA | 4.00 ± 0.14 | −16.67 ± 2.81 | 6 | 16 | 32 | |
| shRNA-ARF6 + HA | 3.75 ± 0.11 | −21.77 ± 2.26 | 8 | 20 | 40 |
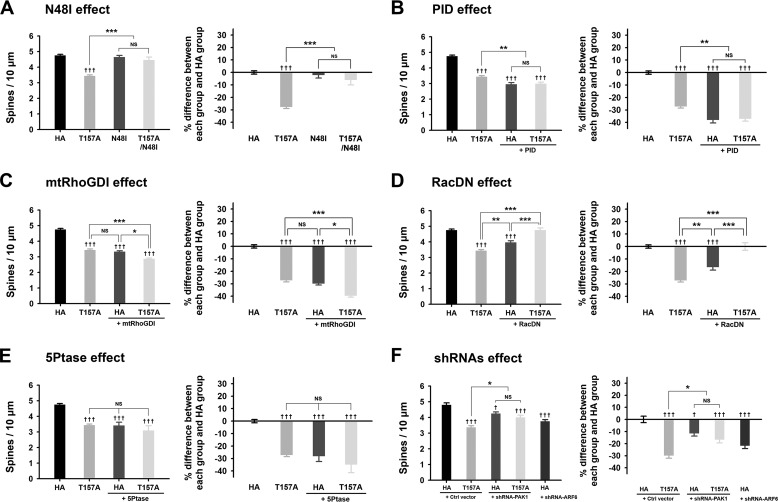
Active ARF6 Inhibits Dendritic Spine Formation in Mature Neurons
In mature neurons, the PLD pathway is also the major pathway for the spine-reducing effect of ARF6-T157A because ARF6-T157A/N48I largely abrogated the effect of ARF6-T157A, whereas ARF6-N48I alone had no effect on spine formation (Fig. 4 A). However, co-expression of PID did not eliminate the spine-reducing effect of ARF6-T157A (Fig. 4B). Also, introducing mtRhoGDI did not abolish the effect of ARF6-T157A. Interestingly, PID or mtRhoGDI itself reduced the spine density, which is consistent with the result from developing neurons (Figs. 3, B and C, and 4, B and C). These results suggest that in the resting state the ARF-independent PAK/RhoGDI pathway is important for spine maintenance in both developing neurons and mature neurons.
FIGURE 4.
The effects of ARF6-T157A or ARF6 knockdown on dendritic spine formation in mature neurons. A–D, Cultured hippocampal neurons were co-transfected at DIV 16 with T157A-ARF6-HA, T157A/N48I-ARF6-HA, N48I-ARF6-HA, or HA and GFP (A), T157A-ARF6-HA or HA and GFP or GFP-PID (B), T157A-ARF6-HA or HA and GFP or GFP-mtRhoGDI (C), T157A-ARF6-HA or HA and GFP or GFP-RacDN (D), or T157A-ARF6-HA or HA and GFP or GFP-5Ptase (E) and fixed at DIV 20. The effect of ARF6 mutants on dendritic spine formation was quantified to investigate the PLD/PA pathway. The values in the spine number graph were recalculated to show percent differences between each group and HA. Detailed data are shown in Table 2. Data were collected for each group in three independent experiments. The values are means ± S.E. Daggers (†) indicate the significance between HA and each group, and asterisks (*) indicate the significance between each group. ††† and ***, p < 0.001; **, p < 0.01; *, p < 0.05; NS, not significant (ANOVA and Tukey's HSD post hoc test). F, cultured hippocampal neurons were co-transfected at DIV 16 with T157A-ARF6-HA or HA and pSIREN-empty (control (Ctrl) vector), shRNA-PAK1, or shRNA-ARF6 and fixed at DIV 20. The effect of ARF6-T157A with PAK1 depletion or ARF6 depletion on dendritic spine formation was quantified in mature neurons. The values in the spine number graph were recalculated to show percent differences between each group and HA. Detailed data are shown in Table 3. Data were collected for each group in three independent experiments. †††, p < 0.001; † and *, p < 0.05; NS, not significant (ANOVA and Tukey's HSD post hoc test). Error bars represent S.E.
For mature neurons, we introduced another PLD downstream pathway, the RhoA pathway, in addition to the Rac1 pathway. Previously, we showed that unlike Rac1, which is expressed at high levels from the early stage and persists throughout development, RhoA expression levels are low at the early developmental stage but gradually increase, and after DIV 20, its expression levels are comparable with those of Rac1 (30). Besides, it is well known that RhoA and Rac1 exert antagonistic effects in spine development: RhoA plays a negative role, whereas Rac1 plays a positive role (31). Because the previous studies showed that PLD or PA is directly recruited to the plasma membrane and activates Rac1 or RhoA (32–34), we hypothesized that the balance between ARF6-T157A-induced Rac1 and RhoA activation via the PLD pathway, bypassing the PAK/RhoGDI pathway, plays a key role in the spine-reducing effect of ARF6-T157A in mature neurons (see also Fig. 7). Accordingly, RacDN alone reduced spine density and almost completely reversed the effect of ARF6-T157A in mature neurons (Fig. 4D). The phosphatidylinositol 4-phosphate 5-kinase/PIP2 pathway is not involved in the effect of ARF6 because co-transfection of ARF6-T157A with 5Ptase failed to recover spine density but rather induced a further decrease (Fig. 4E).
Suppression of PAK1 expression by shRNA-PAK1 alone reduced spine density, and co-expression of shRNA-PAK1 and ARF6-T157A did not abolish the effect of ARF6-T157A (Fig. 4F), which is consistent with the results from examination of the PID effect (Fig. 4B). The knockdown of endogenous ARF6 alone also reduced spine density in mature neurons (Fig. 4F).
Overexpression of Active ARF6 Alters Gene Transcription Profiles Differentially at Developing and Mature Stages
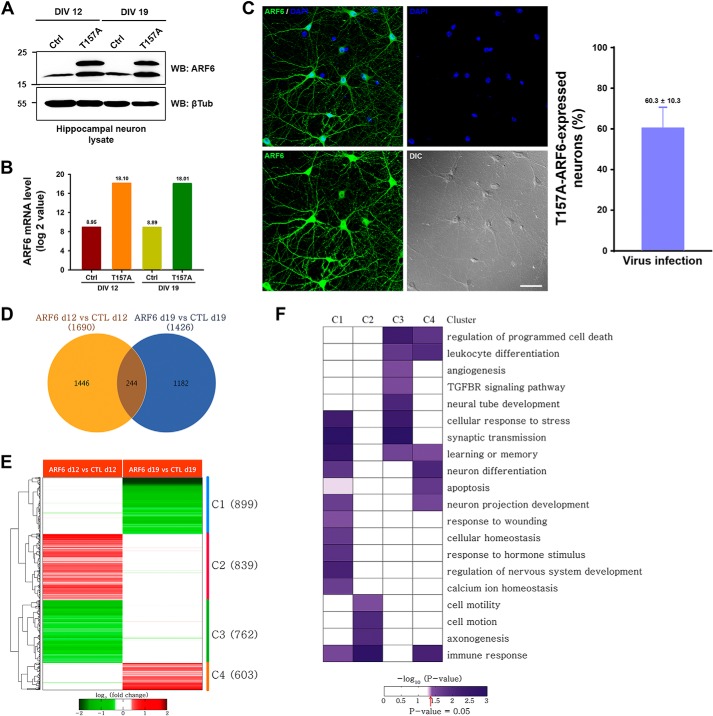
How does the same ARF activation mediate completely opposite effects on spine regulation depending on the level of neuronal maturation? To answer this question, we performed genome-wide gene expression profiling of developing (DIV 12) and mature neurons (DIV 19) infected with Sindbis virus expressing ARF6-T157A and CTL vectors to determine the downstream target genes affected by ARF6 in the two types of neurons. First, we generated Sindbis viruses and checked the expressions of ARF6 mRNA and protein levels. Neurons infected by Sindbis virus expressing ARF6-T157A exhibited strong expressions of both ARF6 mRNA and protein levels (Fig. 5, A and B), and the infection efficiency and cell viability of cells we had used were confirmed by immunostaining and the pyknotic viability test during a 36-h infection with Sindbis virus expressing ARF6-T157A-His6 (Fig. 5C and data not shown). We identified 1,690 and 1,426 DEGs from the following two comparisons (total of 2,872 DEGs): 1) ARF6-T157A versus CTL in developing neurons and 2) ARF6-T157A versus CTL in mature neurons (Fig. 5D). To systematically explore the association of the two sets of DEGs with the differential roles of ARF6 in spine formation, we categorized the 2,872 DEGs into eight possible clusters (C1–C8) based on up- and down-regulation patterns in the two comparisons. Among the eight clusters, we focused on four major clusters (C1–C4; Fig. 5E) containing greater than 100 genes. To understand cellular processes represented by the genes in these individual clusters, we performed enrichment analysis of gene ontology biological processes for the genes in each cluster (Fig. 5F). The results showed that DEGs are mainly involved in neuronal activity-related processes (synaptic transmission, learning or memory, and calcium ion homeostasis) and neuronal development and morphogenesis (neuron differentiation, regulation of nervous system development, axonogenesis, and cell motility). Intriguingly, neuronal morphology-related processes such as axonogenesis and cell motility involving actin cytoskeleton reorganization are regulated predominantly by the genes in C2, which are up-regulated by ARF6 in developing neurons but not in mature neurons. Conversely, the neuronal activity-related processes such as synaptic transmission and learning or memory are regulated predominantly by the genes in C3, which are down-regulated by ARF6 in developing neurons but not in mature neurons. These data indicate that ARF6 activation in developing and mature neurons leads to different expression patterns between developing and mature neurons for the aforementioned genes (C1 and C3) associated with actin cytoskeleton reorganization and neuronal activity-related processes.
FIGURE 5.
Differential gene expression patterns between developing and mature neurons. A, cultured hippocampal neurons were infected at DIV 10 or 17 with Sindbis empty virus (Ctrl) or T157A-ARF6-His6 virus (T157A). Thirty-six hours after infection, the cells were lysed and immunoblotted with anti-ARF6 antibody or anti-β-tubulin (βTub) antibody. WB, Western blot. B, the neuronal mRNAs were extracted from sister cultures, and the mRNA level of ARF6 expression was quantified. The values on top of bars indicate the mean of ARF6 mRNA level in each group. C, representative images of ARF6 expression after T157A infection. The efficiency of T157A infection in cultured hippocampal neurons was measured by counting neurons co-stained with anti-ARF6 antibody and DAPI or singly stained with DAPI. D, a Venn diagram showing relationships between DEGs from the following two comparisons: 1) ARF6-T157A versus CTL in developing neurons (ARF6 d12 vs CTL d12) and 2) ARF6-T157A versus CTL in mature neurons (ARF6 d19 vs CTL d19). E, a heat map showing up- and down-regulation of the genes in the four major clusters (C1–C4). Numbers in parentheses, numbers of the genes in the clusters. Red and green, up- and down-regulation of the genes, respectively. Color bar, gradient of log2 -fold changes. F, a heat map representing the degree of enrichment of the genes involved in each gene ontology biological process in the four major clusters. Color bar, gradient of proportion of the genes involved in each gene ontology biological process in a cluster. Error bars represent S.E. TGFBR, TGF-β receptor.
ARF6-mediated Spine Formation Is Regulated by Neuronal Activity
During the developing stage between DIV 11 and 15, neurons undergo drastic activity-dependent structural and functional rearrangements, which are later stabilized at the mature stage (35, 36). We reasoned that such neuronal activity could be the key factor for the conversion of the effect of ARF6 on spine development. We first tested this possibility in developing neurons. When we treated developing neurons with TTX, which blocks action potential generation in neurons and causes a global inhibition of neuronal activity (37), neurons having longer thin spinelike protrusions became a majority. We found that TTX treatment completely blocked the spine-promoting effect of ARF6-T157A, inducing longer thin spinelike protrusions (Fig. 6 A). To verify whether the effect of ARF6 on spine formation in mature neurons is dependent on neuronal activity, we treated mature neurons with PTX, a noncompetitive antagonist for the GABAA receptor, thus increasing neuronal activity (38). We discovered that PTX treatment completely blocked the spine-reducing effect of ARF6-T157A in mature neurons (Fig. 6D).
FIGURE 6.
ARF6-T157A regulates dendritic spine formation depending on neuronal activity. A, cultured hippocampal neurons were co-transfected at DIV 11 with T157A-ARF6-HA or HA and GFP, subsequently treated with or without 1 μm TTX, and then fixed at DIV 15. The effect of ARF6-T157A on dendritic spine formation with or without TTX treatment was quantified. B, cultured hippocampal neurons were co-transfected at DIV 11 with T157A-ARF6-HA or HA and GFP, subsequently treated with or without 50 μm PTX, and then fixed at DIV 15. The effect of ARF6-T157A on dendritic spine formation with or without PTX treatment was quantified. The values in the spine number graph were recalculated to show percent differences between each group and HA. Detailed data are shown in Table 1. Data were collected for each group in three independent experiments. The values are means ± S.E. ††† and ***, p < 0.001; †† and **, p < 0.01; *, p < 0.05; NS, not significant (ANOVA and Tukey's HSD post hoc test). C, cultured hippocampal neurons were co-transfected at DIV 16 with T157A-ARF6-HA or HA and GFP, subsequently treated with or without 1 μm TTX, and then fixed at DIV 20. The effect of ARF6-T157A on dendritic spine formation with or without TTX treatment was quantified. D, cultured hippocampal neurons were co-transfected at DIV 16 with T157A-ARF6-HA or HA and GFP, subsequently treated with or without 50 μm PTX, and then fixed at DIV 20. The effect of ARF6-T157A on dendritic spine formation with or without PTX treatment was quantified. E, cultured hippocampal neurons were co-transfected at DIV 16 with T157A-ARF6-HA or HA and GFP. Nine hours after transfection, transfected neurons were subjected to chem-LTP (cLTP) induction and fixed and analyzed at DIV 20. The effect of ARF6-T157A on dendritic spine formation in neurons with or without chem-LTP induction was quantified. The values in the spine number graph were recalculated to show percent differences between each group and HA. Detailed data are shown in Table 2. Data were collected for each group in three independent experiments. The values are means ± S.E. ††† and ***, p < 0.001; NS, not significant (ANOVA and Tukey's HSD post hoc test). Error bars represent S.E.
When we treated mature neurons with TTX or developing neurons with PTX, neither treatment affected the ARF6-T157A effect (Fig. 6, B and C). In addition, we used the protocol for chem-LTP, which has been shown previously to increase global neuronal activity in cultured neurons (39). After transfecting neurons with ARF6-T157A at DIV 16, chem-LTP was induced, and neurons were fixed and analyzed at DIV 20. Fig. 6E shows that chem-LTP successfully blocked the spine-reducing effect of ARF6-T157A in mature neurons.
ARF6 is the only member of the ARF family that can simultaneously regulate actin cytoskeleton changes and membrane exchange between the plasma membrane and endocytic compartments. ARF6 is present in the developing neurites and later on in dendrites, possibly suggesting a role for ARF6 in the processes of dendritic initiation, elongation, and branching (40, 41). Hernández-Deviez et al. (11) showed that overexpression of ARF6-T27N in young hippocampal neurons leads to a dramatic increase in the total number of dendrites, whereas ARF6-Q67L results in a reduction of dendritic tree complexity. Miyazaki et al. (12) reported that overexpression of wild-type or constitutively active Q67L mutant of ARF6 significantly decreases the number of spines in mature neurons, whereas overexpression of ARF6-T27N markedly increases the number of spines. In contrast, Choi et al. (13) showed that ARF6 activation promotes the conversion of dendritic filopodia to spines and increases spine density at an early stage, whereas depletion of endogenous ARF6 leads to a decrease in the number of spines and an increase in the number of filopodia. Raemaekers et al. (43) also confirmed the increase in spine number in hippocampal neurons overexpressing ARF6-T157A mutant. Various factors such as types of mutants, the spatiotemporal activation of GEFs or GTPase-activating proteins, and overexpression-induced artifacts may explain the conflicting results, but no clear answer has been suggested. We found that activation of ARF6 could either positively or negatively regulate dendritic spine formation depending on neuronal maturation and activity, thus providing a reconciling answer for the controversy on the role of ARF6.
We found that Rac1 activation via PLD is the coincident factor in developing or mature neurons, but in mature neurons, RhoA activation antagonizes the spine-promoting effect of Rac1. Levels of Rac1 in the mouse CNS are initially high in the embryo and decrease steadily after birth, suggesting that Rac1 activity in the CNS is required for spine formation and maturation in developing neurons (44). Previous studies also found that Rac1 appears to be important in two successive steps of spine morphogenesis: the conversion between longer filopodium-like protrusions and short, thin spines in developing stages and then later in spine head growth in mature stages either by inducing these processes or by inhibiting their reverse processes (45). On the contrary, the developmental expression pattern of RhoA appears to be the opposite in that its levels were low in the early stage of development and increase gradually during development (30). Because it is well known that RhoA and Rac1 have an antagonistic effect to each other (46), together with our current results, Rac1 activity seems to be a key determinant for spine formation and maturation in developing neurons, whereas in mature neurons, RhoA takes over the control, and the balance between RhoA and Rac1 activity regulates the formation and maintenance of mature spine structures.
In addition to Rac1, PIP2 is also involved in actin cytoskeleton rearrangement as well as membrane trafficking (47). PIP2 has been found in various intracellular compartments including the cytosolic face of the plasma membrane and is able to activate and/or recruit actin regulatory proteins such as myristoylated alanine-rich C kinase substrate and cofilin, suggesting that multiple functional roles of PIP2 could co-exist (48, 49). ARF6 can induce PIP2 production by modulating PLD/PA-dependent or -independent phosphatidylinositol 4-phosphate 5-kinase activation, leading to regulation of exo/endocytosis and changes in the actin cytoskeleton (7). Although we found that reducing PIP2 levels using 5Ptase reversed the effect of ARF6-T157A in developing neurons but failed to reverse the ARF6-T157A effect in mature neurons because 5Ptase alone mostly reduced spine density in both developing neurons and mature neurons, we assume that the effect of 5Ptase might not be related to ARF6, although we cannot rule out the possibility that ARF6 activation may exert effects on spine via PIP2.
Based on current and previously published results, here we propose signaling pathways mediated by ARF6 activation to regulate spine formation in developing and mature neurons (Fig. 7). In developing neurons, three routes are in operation, and all converge on Rac1 to promote spine formation: Route 1, ARF6-dependent but PAK1-independent pathway; Route 2, ARF6-independent but PAK1-dependent pathway; and Route 3, ARF6- and PAK1-dependent pathway. In mature neurons, another route is in operation: Route 4 in which an ARF6-dependent and Rho-dependent pathway provides a strong negative impact on spine formation. Using these routes, we are able to explain reasonably well the experimental data we obtained in Figs. 3 and 4. However, it is surely a too simplified abstraction considering the involvement of ARF6 in various other cellular signaling pathways and current microarray data in which thousands of genes were affected by ARF6 activation. A more detailed analysis of the levels of the signaling network is required in future work.
Neuronal activity such as LTP and long term depression induces changes in both motility and shape that are morphological correlates of synaptic plasticity (6). Here we demonstrate that TTX treatment of developing neurons or PTX treatment of mature neurons expressing ARF6-T157A completely abrogated the effect of ARF6-T157A on spine formation (Fig. 6, A and D). Therefore, our results suggest that neuronal activity is the key regulator for bidirectional regulation of ARF6-T157A in spine formation. Our results also suggest that Rac1 activity and neuronal activity are highly correlated in inducing spine formation and maintenance.
Growing evidence suggests that ARF6 activation is precisely regulated by ARF6 GEFs in a spatiotemporal manner, leading to neuronal development and synapse formation (7, 50). Cytohesin1 and -2, EFA6A–D, and brefeldin A-resistant ARF-GEF1-3 are well known positive regulators for ARF6 activation (51). Cytohesin2 (also called ARF nucleotide-binding site opener) inactivation causes an increase in dendritic branching, and EFA6A depletion decreases dendritic spine density in hippocampal neurons (11, 13). The brefeldin A-resistant ARF-GEFs are enriched in neuronal postsynaptic densities and have been linked to mental retardation and alterations in synaptic modulation (51). Recent studies further identified two downstream ARF6 effectors, telencephalin (also known as intercellular adhesion molecule-5) and vezatin (42, 43). Telencephalin mainly localizes to dendritic filopodia and negatively regulates spine development (43). ARF6-mediated endocytosis of telencephalin increases dendritic spine stability regulated at least in part by Rac1-mediated dephosphorylation and release of ezrin/radixin/moesin actin-binding proteins from telencephalin (43). Vezatin depletion significantly decreases total dendritic length and arborization, whereas its overexpression increases the length (42). Considering various ARF6 GEFs and GTPase-activating proteins that are activated to induce myriad downstream effects, many questions including the specificity and redundancy of the various ARF6 GEFs and GTPase-activating proteins as well as the exact method of ARF6 regulation of dendritic development require further detailed investigations.
All in all, we have provided evidence that ARF6 activation bidirectionally regulates dendritic spine formation depending on neuronal maturation and activity. Our results raise the possibility that activity-dependent dynamic changes in ARF6-mediated spine structures may play a role in structural plasticity of mature neurons.
Acknowledgments
We thank Hyangsoo Lee for critical reading of the manuscript. Confocal microscopy data for this study were acquired and analyzed in the Biomedical Imaging Center at Seoul National University College of Medicine.
This work was supported by Biomembrane Plasticity Research Center Grant 20100029395 (to S. C.) from the Ministry of Science, the Information and Communications Technology, and Future Planning (MSIP); Korea Health Technology Research and Development Project Grant A092058 (to S. C.) funded by the Ministry of Health and Welfare; and Research Fellow Grant NRF-2013R1A1A2059850 (to Y. K.) funded by the MSIP.
- ARF
- ADP-ribosylation factor
- GEF
- guanine nucleotide exchange factor
- DIV
- days in vitro
- PLD
- phospholipase D
- PA
- phosphatidic acid
- RhoGDI
- guanine nucleotide dissociation inhibitor for Rho proteins
- TTX
- tetrodotoxin
- PTX
- picrotoxin
- LTP
- long term potentiation
- 5Ptase
- 5-phosphatase domain of synaptojanin-1
- DN
- dominant negative
- PID
- PAK inhibitory domain
- PAK
- p21-activated kinase
- chem-LTP
- chemically induced long term potentiation
- ANOVA
- analysis of variance
- HSD
- honest significant difference
- CTL
- empty
- DEG
- differentially expressed gene
- mtRhoGDI
- RhoGDI1-S101A/S174A
- PIP2
- phosphatidylinositol 4,5-bisphosphate.
REFERENCES
- 1. Harris K. M., Kater S. B. (1994) Dendritic spines: cellular specializations imparting both stability and flexibility to synaptic function. Annu. Rev. Neurosci. 17, 341–371 [DOI] [PubMed] [Google Scholar]
- 2. Yuste R., Bonhoeffer T. (2004) Genesis of dendritic spines: insights from ultrastructural and imaging studies. Nat. Rev. Neurosci. 5, 24–34 [DOI] [PubMed] [Google Scholar]
- 3. Segal M., Andersen P. (2000) Dendritic spines shaped by synaptic activity. Curr. Opin. Neurobiol. 10, 582–586 [DOI] [PubMed] [Google Scholar]
- 4. Carlisle H. J., Kennedy M. B. (2005) Spine architecture and synaptic plasticity. Trends Neurosci. 28, 182–187 [DOI] [PubMed] [Google Scholar]
- 5. Tada T., Sheng M. (2006) Molecular mechanisms of dendritic spine morphogenesis. Curr. Opin. Neurobiol. 16, 95–101 [DOI] [PubMed] [Google Scholar]
- 6. Hayashi Y., Majewska A. K. (2005) Dendritic spine geometry: functional implication and regulation. Neuron 46, 529–532 [DOI] [PubMed] [Google Scholar]
- 7. Jaworski J. (2007) ARF6 in the nervous system. Eur. J. Cell Biol. 86, 513–524 [DOI] [PubMed] [Google Scholar]
- 8. D'Souza-Schorey C., Chavrier P. (2006) ARF proteins: roles in membrane traffic and beyond. Nat. Rev. Mol. Cell Biol. 7, 347–358 [DOI] [PubMed] [Google Scholar]
- 9. Chavrier P., Goud B. (1999) The role of ARF and Rab GTPases in membrane transport. Curr. Opin. Cell Biol. 11, 466–475 [DOI] [PubMed] [Google Scholar]
- 10. Hernández-Deviez D. J., Roth M. G., Casanova J. E., Wilson J. M. (2004) ARNO and ARF6 regulate axonal elongation and branching through downstream activation of phosphatidylinositol 4-phosphate 5-kinase α. Mol. Biol. Cell 15, 111–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hernández-Deviez D. J., Casanova J. E., Wilson J. M. (2002) Regulation of dendritic development by the ARF exchange factor ARNO. Nat. Neurosci. 5, 623–624 [DOI] [PubMed] [Google Scholar]
- 12. Miyazaki H., Yamazaki M., Watanabe H., Maehama T., Yokozeki T., Kanaho Y. (2005) The small GTPase ADP-ribosylation factor 6 negatively regulates dendritic spine formation. FEBS Lett. 579, 6834–6838 [DOI] [PubMed] [Google Scholar]
- 13. Choi S., Ko J., Lee J. R., Lee H. W., Kim K., Chung H. S., Kim H., Kim E. (2006) ARF6 and EFA6A regulate the development and maintenance of dendritic spines. J. Neurosci. 26, 4811–4819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chang S., De Camilli P. (2001) Glutamate regulates actin-based motility in axonal filopodia. Nat. Neurosci. 4, 787–793 [DOI] [PubMed] [Google Scholar]
- 15. Zakharenko S. S., Zablow L., Siegelbaum S. A. (2001) Visualization of changes in presynaptic function during long-term synaptic plasticity. Nat. Neurosci. 4, 711–717 [DOI] [PubMed] [Google Scholar]
- 16. Peters A., Kaiserman-Abramof I. R. (1970) The small pyramidal neuron of the rat cerebral cortex. The perikaryon, dendrites and spines. Am. J. Anat. 127, 321–355 [DOI] [PubMed] [Google Scholar]
- 17. Kim J., Dittgen T., Nimmerjahn A., Waters J., Pawlak V., Helmchen F., Schlesinger S., Seeburg P. H., Osten P. (2004) Sindbis vector SINrep(nsP2S726): a tool for rapid heterologous expression with attenuated cytotoxicity in neurons. J. Neurosci. Methods 133, 81–90 [DOI] [PubMed] [Google Scholar]
- 18. Bolstad B. M., Irizarry R. A., Astrand M., Speed T. P. (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19, 185–193 [DOI] [PubMed] [Google Scholar]
- 19. Lee H. J., Suk J. E., Patrick C., Bae E. J., Cho J. H., Rho S., Hwang D., Masliah E., Lee S. J. (2010) Direct transfer of α-synuclein from neuron to astroglia causes inflammatory responses in synucleinopathies. J. Biol. Chem. 285, 9262–9272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bowman A. W., Azzalini A. (1997) Applied Smoothing Techniques for Data Analysis: the Kernel Approach with S-Plus Illustrations, Oxford University Press, Oxford [Google Scholar]
- 21. Hwang D., Rust A. G., Ramsey S., Smith J. J., Leslie D. M., Weston A. D., de Atauri P., Aitchison J. D., Hood L., Siegel A. F., Bolouri H. (2005) A data integration methodology for systems biology. Proc. Natl. Acad. Sci. U.S.A. 102, 17296–17301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huang da W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 23. Yi C., Wilker E. W., Yaffe M. B., Stemmer-Rachamimov A., Kissil J. L. (2008) Validation of the p21-activated kinases as targets for inhibition in neurofibromatosis type 2. Cancer Res. 68, 7932–7937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hofmann C., Shepelev M., Chernoff J. (2004) The genetics of Pak. J. Cell Sci. 117, 4343–4354 [DOI] [PubMed] [Google Scholar]
- 25. Hayashi K., Ohshima T., Hashimoto M., Mikoshiba K. (2007) Pak1 regulates dendritic branching and spine formation. Dev. Neurobiol. 67, 655–669 [DOI] [PubMed] [Google Scholar]
- 26. Asrar S., Meng Y., Zhou Z., Todorovski Z., Huang W. W., Jia Z. (2009) Regulation of hippocampal long-term potentiation by p21-activated protein kinase 1 (PAK1). Neuropharmacology 56, 73–80 [DOI] [PubMed] [Google Scholar]
- 27. DerMardirossian C., Schnelzer A., Bokoch G. M. (2004) Phosphorylation of RhoGDI by Pak1 mediates dissociation of Rac GTPase. Mol. Cell 15, 117–127 [DOI] [PubMed] [Google Scholar]
- 28. Funakoshi Y., Hasegawa H., Kanaho Y. (2011) Regulation of PIP5K activity by Arf6 and its physiological significance. J. Cell. Physiol. 226, 888–895 [DOI] [PubMed] [Google Scholar]
- 29. Fleming I. N., Batty I. H., Prescott A. R., Gray A., Kular G. S., Stewart H., Downes C. P. (2004) Inositol phospholipids regulate the guanine-nucleotide-exchange factor Tiam1 by facilitating its binding to the plasma membrane and regulating GDP/GTP exchange on Rac1. Biochem. J. 382, 857–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim Y., Ha C. M., Chang S. (2013) SNX26, a GTPase-activating protein for Cdc42, interacts with PSD-95 protein and is involved in activity-dependent dendritic spine formation in mature neurons. J. Biol. Chem. 288, 29453–29466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tashiro A., Minden A., Yuste R. (2000) Regulation of dendritic spine morphology by the rho family of small GTPases: antagonistic roles of Rac and Rho. Cereb. Cortex 10, 927–938 [DOI] [PubMed] [Google Scholar]
- 32. Anders N., Jürgens G. (2008) Large ARF guanine nucleotide exchange factors in membrane trafficking. Cell. Mol. Life Sci. 65, 3433–3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chae Y. C., Kim J. H., Kim K. L., Kim H. W., Lee H. Y., Heo W. D., Meyer T., Suh P. G., Ryu S. H. (2008) Phospholipase D activity regulates integrin-mediated cell spreading and migration by inducing GTP-Rac translocation to the plasma membrane. Mol. Biol. Cell 19, 3111–3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Choi H. J., Chang B. J., Han J. S. (2012) Phospholipase D1 is an important regulator of bFGF-induced neurotrophin-3 expression and neurite outgrowth in H19–7 cells. Mol. Neurobiol. 45, 507–519 [DOI] [PubMed] [Google Scholar]
- 35. Zuo Y., Lin A., Chang P., Gan W. B. (2005) Development of long-term dendritic spine stability in diverse regions of cerebral cortex. Neuron 46, 181–189 [DOI] [PubMed] [Google Scholar]
- 36. Holtmaat A. J., Trachtenberg J. T., Wilbrecht L., Shepherd G. M., Zhang X., Knott G. W., Svoboda K. (2005) Transient and persistent dendritic spines in the neocortex in vivo. Neuron 45, 279–291 [DOI] [PubMed] [Google Scholar]
- 37. Burrone J., O'Byrne M., Murthy V. N. (2002) Multiple forms of synaptic plasticity triggered by selective suppression of activity in individual neurons. Nature 420, 414–418 [DOI] [PubMed] [Google Scholar]
- 38. Papa M., Segal M. (1996) Morphological plasticity in dendritic spines of cultured hippocampal neurons. Neuroscience 71, 1005–1011 [DOI] [PubMed] [Google Scholar]
- 39. Lu W., Man H., Ju W., Trimble W. S., MacDonald J. F., Wang Y. T. (2001) Activation of synaptic NMDA receptors induces membrane insertion of new AMPA receptors and LTP in cultured hippocampal neurons. Neuron 29, 243–254 [DOI] [PubMed] [Google Scholar]
- 40. Albertinazzi C., Za L., Paris S., de Curtis I. (2003) ADP-ribosylation factor 6 and a functional PIX/p95-APP1 complex are required for Rac1B-mediated neurite outgrowth. Mol. Biol. Cell 14, 1295–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gauthier-Campbell C., Bredt D. S., Murphy T. H., El-Din El-Husseini A.(2004) Regulation of dendritic branching and filopodia formation in hippocampal neurons by specific acylated protein motifs. Mol. Biol. Cell 15, 2205–2217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Danglot L., Freret T., Le Roux N., Narboux Nême N., Burgo A., Hyenne V., Roumier A., Contremoulins V., Dauphin F., Bizot J. C., Vodjdani G., Gaspar P., Boulouard M., Poncer J. C., Galli T., Simmler M. C. (2012) Vezatin is essential for dendritic spine morphogenesis and functional synaptic maturation. J. Neurosci. 32, 9007–9022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Raemaekers T., Peric A., Baatsen P., Sannerud R., Declerck I., Baert V., Michiels C., Annaert W. (2012) ARF6-mediated endosomal transport of telencephalin affects dendritic filopodia-to-spine maturation. EMBO J. 31, 3252–3269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bongmba O. Y., Martinez L. A., Elhardt M. E., Butler K., Tejada-Simon M. V. (2011) Modulation of dendritic spines and synaptic function by Rac1: a possible link to fragile X syndrome pathology. Brain Res. 1399, 79–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tashiro A., Yuste R. (2004) Regulation of dendritic spine motility and stability by Rac1 and Rho kinase: evidence for two forms of spine motility. Mol. Cell. Neurosci. 26, 429–440 [DOI] [PubMed] [Google Scholar]
- 46. Nakamura F. (2013) FilGAP and its close relatives: a mediator of Rho-Rac antagonism that regulates cell morphology and migration. Biochem. J. 453, 17–25 [DOI] [PubMed] [Google Scholar]
- 47. Takenawa T., Itoh T. (2001) Phosphoinositides, key molecules for regulation of actin cytoskeletal organization and membrane traffic from the plasma membrane. Biochim. Biophys. Acta 1533, 190–206 [DOI] [PubMed] [Google Scholar]
- 48. McLaughlin S., Wang J., Gambhir A., Murray D. (2002) PIP2 and proteins: interactions, organization, and information flow. Annu. Rev. Biophys. Biomol. Struct. 31, 151–175 [DOI] [PubMed] [Google Scholar]
- 49. Kusano K., Abe H., Obinata T. (1999) Detection of a sequence involved in actin-binding and phosphoinositide-binding in the N-terminal side of cofilin. Mol. Cell. Biochem. 190, 133–141 [PubMed] [Google Scholar]
- 50. Sakagami H. (2008) The EFA6 family: guanine nucleotide exchange factors for ADP ribosylation factor 6 at neuronal synapses. Tohoku J. Exp. Med. 214, 191–198 [DOI] [PubMed] [Google Scholar]
- 51. Hongu T., Kanaho Y. (2014) Activation machinery of the small GTPase Arf6. Adv. Biol. Regul. 54C, 59–66 [DOI] [PubMed] [Google Scholar]