Background: Yeast synthesizes coenzyme Q via a macromolecular protein complex.
Results: The Q biosynthetic complex includes Coq8 and the uncharacterized protein YLR290C; the ylr290cΔ mutant exhibits impaired Q synthesis.
Conclusion: YLR290C (Coq11) is a novel protein required for efficient yeast Q biosynthesis.
Significance: Discovery and characterization of yeast Coq biosynthetic proteins leads to an improved understanding of coenzyme Q biosynthesis and regulation.
Keywords: Mass Spectrometry (MS), Mitochondrial Metabolism, Protein Complex, Proteomics, Saccharomyces cerevisiae, Ubiquinone, Yeast, Q Biosynthetic Intermediates, Coenzyme Q, Immunoprecipitation
Abstract
Coenzyme Q (Q or ubiquinone) is a redox active lipid composed of a fully substituted benzoquinone ring and a polyisoprenoid tail and is required for mitochondrial electron transport. In the yeast Saccharomyces cerevisiae, Q is synthesized by the products of 11 known genes, COQ1–COQ9, YAH1, and ARH1. The function of some of the Coq proteins remains unknown, and several steps in the Q biosynthetic pathway are not fully characterized. Several of the Coq proteins are associated in a macromolecular complex on the matrix face of the inner mitochondrial membrane, and this complex is required for efficient Q synthesis. Here, we further characterize this complex via immunoblotting and proteomic analysis of tandem affinity-purified tagged Coq proteins. We show that Coq8, a putative kinase required for the stability of the Q biosynthetic complex, is associated with a Coq6-containing complex. Additionally Q6 and late stage Q biosynthetic intermediates were also found to co-purify with the complex. A mitochondrial protein of unknown function, encoded by the YLR290C open reading frame, is also identified as a constituent of the complex and is shown to be required for efficient de novo Q biosynthesis. Given its effect on Q synthesis and its association with the biosynthetic complex, we propose that the open reading frame YLR290C be designated COQ11.
Introduction
Coenzyme Q (ubiquinone or Q)2 is a small lipophilic molecule consisting of a hydrophobic polyisoprenoid chain and a fully substituted benzoquinone ring. The length of the polyisoprenoid group is species-specific; with 6, 8, 9, and 10 isoprenyl units in Saccharomyces cerevisiae, Escherichia coli, mice, and humans, respectively (1); and localizes Q to biological membranes. The primary function of Q is to serve as an electron carrier in respiratory electron transport, where it sequentially accepts up to two electrons from NADH at complex I or succinate at complex II and donates them to cytochrome c at complex III (2). S. cerevisiae lacks complex I and instead uses internal or external NADH:Q oxidoreductases on the inner mitochondrial membrane, which unlike complex I, do not facilitate proton transport across the inner membrane (3). Q also functions as an electron carrier in other aspects of metabolism including dihydroorotate dehydrogenase (pyrimidine synthesis), fatty acyl-CoA dehydrogenase (fatty acid β-oxidation), and other dehydrogenases that oxidize various substrates including choline, sarcosine, sulfide, and glycerol-3-phosphate (4–6). QH2 has also been demonstrated to function as a lipid-soluble antioxidant capable of mitigating lipid peroxidation and regenerating α-tocopherol (vitamin E) (7, 8). Primary deficiencies in Q biosynthesis have been identified in several human patients and manifest a variety of symptoms including encephalomyopathy, ataxia, cerebellar atrophy, myopathy, and steroid-resistant nephrotic syndrome (9).
In S. cerevisiae Q synthesis is dependent on the products of 11 known genes: COQ1–COQ9, YAH1, and ARH1 (Fig. 1). COQ1–COQ9 were identified as complementation groups of Q-deficient yeast mutants (10, 11), whereas YAH1 and ARH1 were identified through lipid analysis of Yah1- and Arh1-depleted strains under control of galactose-inducible promoters (12). Deletion of any of these COQ genes results in a loss of Q synthesis and consequently a failure to respire and grow on a nonfermentable carbon source (12, 13). Deletion of either YAH1 or ARH1 results in a loss in viability because the biosynthesis of iron-sulfur clusters and heme A requires these gene products (14–18). Although required for Q biosynthesis, the functions of Coq4, Coq8, and Coq9 are not fully understood, although Coq8 has been proposed to function as a protein kinase (19), and Coq9 is required for the replacement of the ring amino group with a hydroxyl group and for the efficient function of Coq6 and Coq7 (20). There are steps in the yeast Q biosynthetic pathway (Fig. 1), such as the decarboxylation reaction and subsequent ring hydroxylation, for which no enzyme has yet been characterized.
FIGURE 1.

Proposed coenzyme Q biosynthetic pathway in S. cerevisiae. Yeast utilize either 4HB or pABA to synthesize the first lipid intermediates of the pathway, HHB or HAB, respectively, through the actions of Coq1 and Coq2. These intermediates then undergo a series of ring modifications catalyzed by the enzymes denoted above each step to yield Q. Two steps in the pathway, denoted with question marks, remain uncharacterized. The step at which the amino group is converted to the hydroxyl group remains unclear and is denoted by dashed arrows. Two late stage intermediates, DMQ6 and IDMQ6, are readily detected in yeast. The hexaprenyl moiety is abbreviated as R.
Genetic and biochemical experiments have demonstrated the interdependence of several of the Coq proteins and have shown the existence of a high molecular mass Coq protein complex. Each of the coq3–coq9Δ mutants accumulate only the early intermediates 3-hexaprenyl-4-hydroxybenzoic acid (HHB) and 3-hexaprenyl-4-aminobenzoic acid (HAB), resulting from prenylation of 4-hydroxybenzoic acid (4HB) and para-aminobenzoic acid (pABA), respectively (20). Steady-state levels of Coq3, Coq4, Coq6, Coq7, and Coq9 are reduced in several coq1–coq9Δ mutants, although levels of Coq3 were stabilized in coq4–coq9Δ mutant samples prepared with phosphatase inhibitors (20–22). This has been observed in other mitochondrial complexes such as the cytochrome bc1 complex and ATP synthase in which the absence or mutation of one component protein results in reduced steady-state levels of the other subunits (23, 24). Gel filtration chromatography and blue native-PAGE have shown that several of the Coq proteins exist in high molecular mass complexes, and the O-methyltransferase activity of Coq3 can be detected in these complexes (21, 25–27). Co-precipitation experiments have demonstrated the physical association of several of the Coq proteins; biotinylated Coq3 was shown to co-precipitate Coq4 and HA-tagged Coq9 co-purified Coq4, Coq5, Coq6, and Coq7 (21, 25). Although Coq8 has not been shown to be associated in a complex with the other Coq proteins, its putative role as a kinase is important for the stability of several other Coq proteins because overexpression of COQ8 in various coqΔ mutants stabilizes several of the other Coq proteins, as well as multisubunit Coq polypeptide complexes, and leads to the accumulation of later stage Q biosynthetic intermediates diagnostic of the mutated step (20, 28, 29).
Overexpression of COQ8 in the coq1Δ or coq2Δ mutants, which do not produce any polyisoprenylated intermediates, fails to stabilize steady-state levels of sensitive Coq polypeptides (20). When diverse polyprenyl diphosphate synthases from prokaryotic species that do not synthesize Q are expressed in yeast coq1Δ mutants, Q biosynthesis and steady-state levels of sensitive Coq polypeptides are restored (30), suggesting that a polyisoprenylated compound is essential for complex stability. Analysis of gel filtration fractions has also shown the association of demethoxy-Q6 (DMQ6) with the complexes (25). Supplementation of exogenous Q6 was shown to stabilize the steady-state levels of Coq3 and Coq4 in the coq7Δ mutant (27), enhance DMQ6 production in the coq7Δ mutant (29), and stabilize steady-state levels of Coq9 in the coq3Δ, coq4Δ, coq6Δ, coq7Δ mutants and steady-state levels of Coq4 in the coq3Δ, coq6Δ, and coq7Δ mutants (28), suggesting that associated Q6 plays a role in the stability of the complex.
The functions of certain Coq polypeptides in Q biosynthesis remain unclear, and there are reactions in the Q biosynthetic pathway for which an enzyme has not been assigned. In this study, the composition of the Q biosynthetic complex is investigated through the use of C-terminal tandem affinity purification tags coupled to proteomic analysis. The presence of associated Q6 and Q6 intermediates with the complex is also assessed. Two new candidate proteins associated with the complex are characterized via analysis of their null mutants. One of these candidates, YLR290C, was further analyzed by tandem affinity purification and shown to be associated with the Q biosynthetic complex. This observation along with the decrease in de novo Q6 synthesis in the ylr290cΔ mutant has led us to propose that YLR290C be designated Coq11.
EXPERIMENTAL PROCEDURES
Yeast Strains and Growth Media
S. cerevisiae strains used in this study are described in Table 1. Medium was prepared as described (31) and included YPD (2% glucose, 1% yeast extract, 2% peptone), YPgal (2% galactose, 0.1% glucose, 1% yeast extract, 2% peptone), and YPG (3% glycerol, 1% yeast extract, 2% peptone). Synthetic dextrose/minimal medium was prepared as described (32) and consisted of all components (SD-complete) or all components minus histidine (SD−His). Drop out dextrose medium (DOD) was prepared as described (33), except that galactose was replaced with 2% dextrose. Plate medium was prepared with 2% bacto agar.
TABLE 1.
Genotype and source of yeast strains
| Strain | Genotype | Source or reference |
|---|---|---|
| W303-1A | Mat a ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 | R. Rothsteina |
| W303coq8Δ | Mat a ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 coq8::HIS3 | Ref. 81 |
| CNAP3 | Mat a ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 COQ3::COQ3-CNAP-HIS3 | This study |
| CNAP6 | Mat a ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 COQ6::COQ6-CNAP-HIS3 | This study |
| CNAP9 | Mat a ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 COQ9::COQ9-CNAP-HIS3 | This study |
| CA-1 | Mat a ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 YLR290C::YLR290C-CNAP-HIS3 | This study |
| BY4741 | MAT a his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Ref. 82 |
| BY4741coq3Δ | MAT a his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 coq3::kanMX4 | Ref. 83 |
| BY4741ilv6Δ | MAT a his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ilv6::kanMX4 | Ref. 84b |
| BY4741ylr290cΔ | MAT a his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ylr290c::kanMX4 | Ref. 84b |
a Dr. Rodney Rothstein, Department of Human Genetics, Columbia University.
b GE Healthcare Yeast Knockout Collection, available online.
Construction of Integrated CNAP-tagged Yeast Strains
The coding sequence for the CNAP (consecutive nondenaturing affinity purification) tag, HHHHHHHHHHGGAGGEDQVDPRLIDGK (His10 tag in bold, protein C (PC) epitope underlined) (34) and HIS3 ORF were amplified from pFACNAPHISMX (Dr. C. M. Koehler, Department of Chemistry and Biochemistry, UCLA) using primers with 5′-flanking regions corresponding to 50 bp upstream of the stop codon and 50 bp downstream of the stop codon of the gene of interest. Following PCR, the product was purified using the PureLink quick PCR purification kit (Life Technologies) and used to transform W303-1A yeast via the lithium acetate/PEG method (35) to allow integration of the CNAP and HIS3 coding sequence into the endogenous stop codon of the target gene. Integrants were selected on SD−His and screened by colony PCR to verify the presence of the CNAP-HIS3 insert at the correct locus; the wild-type ORF lacking the insert was not detected. Primer sequences are available upon request.
Mitochondrial Purification from Yeast
Yeast strains were grown in 5 ml of YPgal precultures and inoculated into 600-ml YPgal cultures for overnight growth in a shaking incubator (30 °C, 250 rpm). Cells were harvested at an A600 of 3.5–4.0, and mitochondria were purified as described (36), with the addition of Complete EDTA-free protease inhibitor mixture (Roche) and phosphatase inhibitor cocktails I and II (Calbiochem). Purified mitochondria were flash frozen in liquid nitrogen and stored at −80 °C. Protein concentration was measured with a BCA assay using bovine serum albumin as the standard (Thermo Scientific).
Tandem Affinity Purification of CNAP-tagged Proteins
Purified mitochondria (15 mg of protein) were pelleted by centrifugation at 12,000 × g for 10 min and solubilized at 2 mg/ml with solubilization buffer (20 mm HEPES-KOH, pH 7.4, 100 mm NaCl, 20 mm imidazole, 10% glycerol, 1 mm CaCl2, Complete EDTA-free protease inhibitor mixture (Roche), phosphatase inhibitor cocktails I and II (Calbiochem), 6 mg/ml digitonin (Biosynth)) for 1 h on ice with mixing every 10 min. The soluble supernatant fraction was then separated from the insoluble pellet by centrifugation at 100,000 × g for 10 min in a Beckman Airfuge. The soluble fraction was then mixed with 8 ml of lysis buffer (50 mm NaH2PO4, pH 8.0, 300 mm NaCl, 10 mm imidazole) and 800-μl bed volume Ni-NTA resin (Qiagen) pre-equilibrated with lysis buffer and incubated at 4 °C for 90 min with mixing by rotation. The Ni-NTA slurry was then applied to a flowthrough column and washed twice with 12 ml of Ni-NTA wash buffer (50 mm NaH2PO4, pH 8.0, 300 mm NaCl, 20 mm imidazole, 1 mg/ml digitonin), followed by elution of bound protein with 8 ml of Ni-NTA elution buffer (20 mm HEPES-KOH, pH 7.4, 100 mm NaCl, 300 mm imidazole, 10% glycerol, 1 mm CaCl2, 1 mg/ml digitonin). The Ni-NTA eluate was applied directly to 1 ml of anti-PC resin (Roche) pre-equilibrated with anti-PC equilibration buffer (20 mm HEPES-KOH, pH 7.4, 100 mm NaCl, 1 mm CaCl2) and incubated overnight at 4 °C with mixing by rotation. The anti-PC slurry was then applied to a flowthrough column and washed twice with 15 ml of anti-PC wash buffer (20 mm HEPES-KOH, pH 7.4, 100 mm NaCl, 1 mm CaCl2, 1 mg/ml digitonin). Bound protein was eluted twice with 1 ml of anti-PC elution buffer (20 mm HEPES-KOH, pH 7.4, 100 mm NaCl, 5 mm EDTA, 1 mg/ml digitonin). In the first elution (E1), the anti-PC resin was incubated at 4 °C for 15 min followed by incubation at room temperature for 15 min. In the second elution (E2), the anti-PC resin was incubated for 15 min at room temperature. Eluates were stored at −20 °C.
SDS-PAGE and Immunoblot Analysis
Protein samples incubated with SDS sample buffer (50 mm Tris-HCl, pH 6.8, 10% glycerol, 2% SDS, 0.1% bromphenol blue, 1.33% β-mercaptoethanol) were separated on 12% Tris-glycine SDS-polyacrylamide gels by electrophoresis (37) followed by transfer to Immobilon-P PVDF membranes (Millipore) at 100 V for 1.5 h. Membranes were then blocked overnight in 3% nonfat milk, phosphate-buffered saline (140.7 mm NaCl, 9.3 mm Na2HPO4, pH 7.4), 0.1% Tween 20. Membranes were then probed with primary antibodies (Table 2) in 2% nonfat milk, phosphate-buffered saline, 0.1% Tween 20 at the dilutions stated (Table 2). Goat anti-rabbit secondary antibody conjugated to horseradish peroxidase (Calbiochem) was used at 1:10,000 dilutions. Blots were visualized using Supersignal West Pico chemiluminescent substrate (Thermo).
TABLE 2.
Description and source of antibodies
| Antibody | Working dilution | Source |
|---|---|---|
| Atp2 | 1:2000 | C. M. Koehlera |
| Coq1 | 1:10,000 | Ref. 30 |
| Coq2 | 1:1000 | Ref. 21 |
| Coq3 | 1:1000 | Ref. 85 |
| Coq4 | 1:1000 | Ref. 86 |
| Coq5 | 1:5000 | Ref. 87 |
| Coq6 | 1:250 | Ref. 88 |
| Coq7 | 1:500 | Ref. 27 |
| Coq8 | (Affinity-purified; 1:30) | Ref. 21 |
| Coq9 | 1:1000 | Ref. 89 |
| Coq10 | (Affinity-purified; 1:15) | Ref. 90 |
| Mdh1 | 1:10,000 | L. McAlister-Hennb |
| Protein C | 0.7 μg/ml | Antibodies-online Inc. |
a Dr. C.M. Koehler, Department of Chemistry and Biochemistry, UCLA.
b Dr. L. McAlister-Henn, Department of Molecular Biophysics and Biochemistry, University of Texas Health Sciences Center, San Antonio, TX.
SYPRO Ruby Staining
Protein samples incubated with SDS sample buffer were separated on an 8–16% Criterion SDS-PAGE gel (Bio-Rad). Following electrophoresis, the gel was washed twice with water and fixed for 1 h with 100 ml of 10% methanol, 7% acetic acid. The gel was washed twice with water and then stained overnight at room temperature in the dark with 80 ml of a 1:1 mixture of fresh and used SYPRO Ruby (Life Technologies). Following staining, the gel was washed twice with water and destained for 4 h with 10% methanol, 7% acetic acid. Proteins were visualized with an FX Pro Plus Molecular Imager (Bio-Rad) with 532-nm excitation, and emission was measured with a 555-nm long pass filter.
In-gel Trypsin Digestion and Proteomic Analysis
Sample lanes from a SYPRO Ruby-stained gel were cut into 5-mm slices and subjected to in-gel trypsin digestion as described previously (38), with a few modifications. Gel slices were washed with 50 mm NH4HCO3, 50% acetonitrile followed by 100% acetonitrile, three alternating cycles total. Samples were then incubated with 10 mm dithiothreitol (MP Biomedicals) at 60 °C for 1 h to reduce disulfide bonds, followed by treatment with 50 mm iodoacetamide (Calbiochem) at 45 °C for 45 min in the dark to alkylate-free sulfhydryl groups. Gel slices were then washed once with 50 mm NH4HCO3, followed by three alternating washes with 100 mm NH4HCO3 and 100% acetonitrile. Gel slices were then dried by vacuum centrifugation (SpeedVac) at 60 °C for 10 min. Gel-entrapped proteins were then digested overnight at 37 °C using 20 ng/μl trypsin (Promega) in 50 mm NH4HCO3. Following digestion tryptic peptides were extracted from the gel slices with 50% acetonitrile, 0.1% TFA and dried by SpeedVac at 30 °C for 2 h.
LC-MS/MS analysis of the trypsin-digested peptides was performed as follows: Peptides were resuspended in 30 μl of 3% acetonitrile, 0.1% formic acid. The peptides were measured using a nanoACQUITY UPLC system coupled to a Xevo QTof (quadrupole time of flight) mass spectrometer (Waters). Peptides were eluted from the UPLC to the electrospray ionization mass spectrometer using a 5-μm 2G-VM Symmetry C18 180-μm × 20-mm trap column in-line with a 1.8-μm HSS T3, 75-μm × 150-mm C18 analytical column (Waters). The digested peptides were separated using a 0–60-min gradient, beginning at 3% acetonitrile, 0.1% formic acid to 40% acetonitrile, 0.1% formic acid, followed by a 40–95% gradient between 60 and 62 min at a flow rate of 0.3 μl min−1 (90-min total run time). [Glu1]-Fibrinopeptide B was used as a mass calibration standard (100 fmol/μl) and was infused via a separate electrospray ionization sprayer, and the standard peptide was measured every 45 s during data collection.
The mass spectrometer was operated in the MSE data-independent acquisition mode (39–41). ProteinLynx Global Server (version 3; Waters) was used for protein identification. The MS data were searched against the Uniprot S. cerevisiae (strain ATCC 204508/S288c) reference proteome data set (release date July 9, 2014, containing both Swiss-Prot (6718) and TrEMBL (9) entries). The database was further supplemented with sequences for keratin and trypsin. Low energy, elevated energy, and intensity thresholds were set to 250/100/1000 counts, respectively. The MS data files were searched following ProteinLynx Global Server workflow parameters: trypsin digestion with up to two allowed missed cleavages, peptide and fragment tolerances set automatically (∼11 ppm and 28 ppm, respectively), minimum number of peptide matches is one, minimum number of product ions per peptide is three, minimum number of product ions per protein is seven, and a maximum false positive rate of four percent. Carbamidomethylation of Cys residues was set as a fixed modification, and Met oxidation, Ser, Thr, and Tyr phosphorylation, and Cys propionamidation resulting from iodoacetamide treatment were set as variable modifications.
Metabolic Labeling of Q6 with 13C6-labeled Precursors
Yeast strains were grown overnight in 5 ml of SD-complete and diluted to an A600 of 0.05 in 50 ml of DOD-complete. Cultures were grown to an A600 of 0.5, corresponding to early log phase and then labeled with either 13C6-4HB or 13C6-pABA at a concentration of 5 μg/ml (250 μg total). Cells were grown an additional 3 h with label, harvested by centrifugation at 2000 × g for 10 min, and stored at −20 °C. Wet weights were determined for yeast cell pellets.
Analysis of Q6 and Q6 Intermediates
Lipid extraction of eluate samples and cells grown in DOD-complete was performed with methanol/petroleum ether and Q4 as an internal standard, and LC-MS/MS analysis of extracts was performed as described (33). Briefly, a 4000 QTRAP linear MS/MS spectrometer (Applied Biosystems, Foster City, CA) was used, and Analyst version 1.4.2 software (Applied Biosystems) was used for data acquisition and processing. A binary HPLC delivery system was used with a phenyl-hexyl column (Luna 5u, 100 × 4.60 mm, 5-μm; Phenomenex), with a mobile phase consisting of solvent A (methanol/2-propanol, 95:5, 2.5 mm ammonium formate) and solvent B (2-propanol, 2.5 mm ammonium formate). The percentage of solvent B was increased linearly from 0 to 5% over 6 min, and the flow rate was increased from 600 to 800 μl/min. The flow rate and mobile phase were changed back to initial conditions linearly over 7 min. All samples were analyzed in multiple reaction monitoring mode. Multiple reaction monitoring mode precursor to product ion transitions were detected as follows: 13C6-HAB m/z 552.4/156.0, 12C-HAB m/z 546.4/150.0, 13C6-HHB m/z 553.4/157.0, 12C-HHB m/z 547.4/151.0, IDMQ6 m/z 560.6/166.0, 13C6-DMQ6 m/z 567.6/173.0, 12C-DMQ6 m/z 561.6/167.0, 13C6-Q6 m/z 597.4/203.0, 13C6-Q6H2 m/z 616.4/203.0, 12C-Q6 m/z 591.4/197.0, and 12C-Q6H2 m/z 610.4/197.0.
Bioinformatic Analyses
Evolutionary relationships of amino acid sequences were inferred using the neighbor-joining method (42) with MEGA6 (43). The protein similarity network (44) was constructed as described previously (45) from all-versus-all blastp alignments using an E value cutoff of 1 × 10−21. Protein IDs analyzed are available in supplemental Data Set 1. Protein sequences were retrieved through BLAST of the Uniprot database (46). The multiple sequence alignment was built with MUSCLE (47) and visualized with EsPript (48).
RESULTS
Effects of CNAP Tag Integration on Respiration and Q Biosynthesis
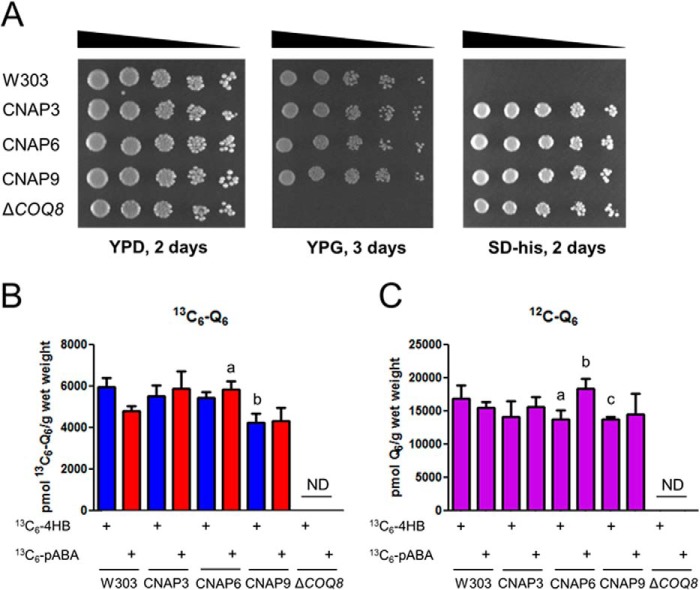
The CNAP tag was integrated into the 3′ end of the COQ3, COQ6, and COQ9 ORFs, generating the CNAP3, CNAP6, and CNAP9 strains, respectively. The ability of these strains to respire as measured by growth on a nonfermentable carbon source was assessed by plate dilution assay, using W3031B as the wild-type control and coq8Δ as the Q-less, respiratory defective control (Fig. 2A). All three tagged strains grew as well as wild type on the nonfermentable carbon source, YPG, whereas the Q-less mutant failed to grow. The three tagged strains and the Q-less mutant were generated by chromosomal integration of the HIS3 ORF and grew on SD−His, whereas the wild type did not. All five strains were able to grow on the permissible carbon source YPD.
FIGURE 2.
Expression of CNAP-tagged Coq3, Coq6, or Coq9 proteins preserves growth on a nonfermentable carbon source and de novo Q biosynthesis. A, designated yeast strains were grown overnight in 5 ml of YPD, diluted to an A600 of 0.2 with sterile PBS, and 2 μl of 5-fold serial dilutions were spotted onto each type of plate medium, corresponding to a final A600 of 0.2, 0.04, 0.008, 0.0016, and 0.00032. Plates were incubated at 30 °C, and growth is depicted after 2 days for both YPD and SD−His and 3 days for YPG. B and C, total 13C6-Q6 (13C6-Q6 + 13C6-Q6H2) (B) and total 12C-Q6 (12C-Q6 + 12C-Q6H2) (C) were measured in the designated yeast strains by HPLC-MS/MS. Yeast strains were grown overnight in 5 ml of SD-complete diluted to an A600 of 0.05 in 50 ml of DOD-complete, labeled with either 13C6-4HB or 13C6-pABA at an A600 of 0.5, and harvested after 3 h of labeling. Lipid measurements were normalized by the wet weight of extracted cells. Each bar represents the mean of four measurements from two biological samples with two injections each. Error bars represent standard deviations. Statistical significance was determined with the two-tailed Student's t test, and the lowercase letters above the bars are indicative of statistical significance. In B, the content of total 13C6-Q6 in CNAP3, CNAP6, CNAP9, and coq8Δ was compared with wild type with the corresponding 13C6-labeled precursor; 13C6-Q6 synthesized from 13C6-pABA in CNAP6 was 120% of wild type, whereas 13C6-Q6 synthesized from 13C6-4HB in CNAP9 was 70% of wild type (a, p = 0.0052; b, p = 0.0018). In C, the content of total 12C-Q6 in CNAP3, CNAP6, CNAP9, and coq8Δ was compared with wild type with the corresponding 13C6-labeled precursor; accumulated 12C-Q6 in CNAP6 was 80% of wild type under 13C6-4HB labeling, 120% in CNAP6 under 13C6-pABA labeling compared with wild type, and 80% of wild type in CNAP9 labeled with 13C6-4HB (a, p = 0.0423; b, p = 0.0179; c, p = 0.0224). ND, not detected.
De novo Q biosynthesis was measured by LC-MS/MS of cells grown in DOD-complete medium with the stable isotopically labeled ring precursors 13C6-4HB or 13C6-pABA (Fig. 2B). Levels of de novo 13C6-Q in CNAP3 synthesized from either precursor were not significantly different compared with wild type. 13C6-Q synthesized de novo from 13C6-4HB in CNAP6 and 13C6-Q synthesized de novo from 13C6-pABA in CNAP9 were not significantly different compared with wild type. 13C6-Q synthesized de novo from 13C6-pABA in CNAP6 was higher than wild type, and 13C6-Q synthesized de novo from 13C6-4HB in CNAP9 was lower than wild type. The coq8Δ strain did not produce detectable de novo 13C6-Q6. Total Q accumulation was also measured by analyzing unlabeled 12C-Q6 by LC-MS/MS (Fig. 2C). Levels of accumulated Q6 in CNAP3 grown with either precursor were not significantly different compared with wild type. The accumulated Q6 in CNAP9 grown with 13C6-pABA was not significantly different compared with wild type. Accumulated Q6 in CNAP6 was lower than wild type under 13C6-4HB labeling and higher under 13C6-pABA labeling when compared with wild type. Accumulated Q6 in CNAP9 labeled with 13C6-4HB was lower than wild type. The coq8Δ strain did not accumulate detectable Q6. The results indicate that CNAP-tagged Coq3, Coq6, or Coq9 polypeptides expressed from the integrated loci remain functional.
Co-precipitation of Coq Proteins with CNAP-tagged Coq Proteins
Solubilized mitochondria from W303, CNAP3, CNAP6, and CNAP9 were subjected to tandem affinity purification of the CNAP tag. Purified mitochondria (25 μg of protein) and fractions of the first anti-PC eluate (E1) were analyzed by immunoblot to identify interacting Coq proteins (Fig. 3). Membranes were probed with antibodies to Coq1–Coq10, the protein C epitope to identify the tagged protein, and Atp2 and Mdh1 to assess the purity of the eluates. CNAP3 co-precipitated Coq4, Coq5, Coq6, Coq7, and Coq9; CNAP6 co-precipitated Coq4, Coq5, Coq7, Coq8, and Coq9; and CNAP9 co-precipitated Coq4, Coq5, Coq6, and Coq7. The signals for Coq5 and Coq9 were much weaker with CNAP3 and CNAP6 compared with CNAP9. The predominant band in the Coq3 blot is a nonspecific band recognized by the antisera at the same molecular mass as Coq3 and is present in the coq3Δ, accounting for the apparent wild-type Coq3 band in the CNAP3 mitochondrial protein (M) sample; the primary purpose of the Coq3 blot is to visualize the shifted band in the CNAP3 E1 sample. All three tagged proteins are present in their respective blots at shifted molecular masses because of the tag (and are denoted by arrows in Fig. 3). The three tagged proteins are also present on the anti-PC blot at their appropriate molecular masses. No signal is visible in the eluate fractions for the nonspecific mitochondrial proteins Atp2 and Mdh1, which are 2–3 orders of magnitude more abundant than the Coq proteins, indicating the specificity of the co-precipitation.
FIGURE 3.
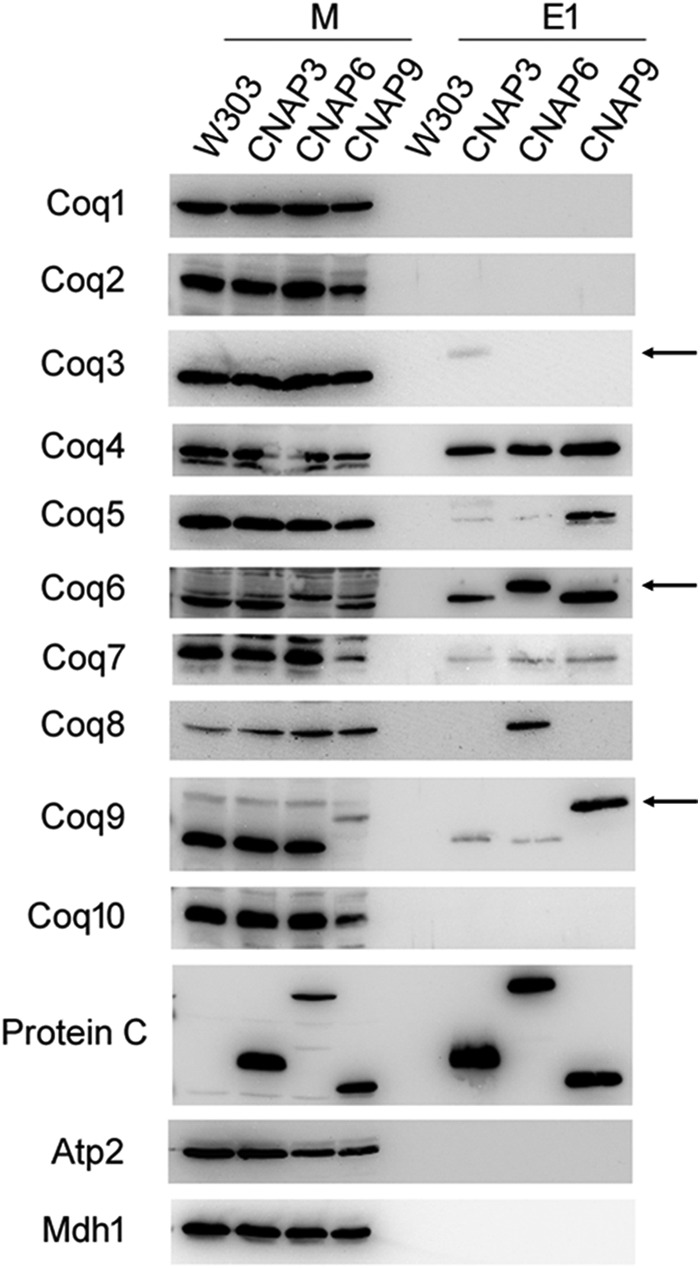
CNAP-tagged Coq proteins co-precipitate several other Coq proteins. W303, CNAP3, CNAP6, and CNAP9 purified mitochondria (15 mg of protein) were solubilized with digitonin and subjected to tandem affinity purification with Ni-NTA resin (Qiagen) followed by anti-PC-agarose (Roche). Samples were separated on 12% SDS-PAGE gels followed by transfer to PVDF membranes for immunoblotting with antisera to the designated yeast polypeptides. 25 μg of mitochondria protein were analyzed for each strain (M), and 2.5% of the first anti-PC elution (E1) volume was loaded per strain (25 μl). Arrows denote each tagged protein in their respective blots. The predominant band in the Coq3 blot detected in the mitochondrial samples represents a background protein and not Coq3, accounting for its presence in CNAP3 mitochondria.
Identification of Associated Lipids and Proteins by Mass Spectrometry
Aliquots of anti-PC eluates (29% E1 and 29% E2) were combined for each strain and subjected to lipid extraction (Fig. 4). Extracts were then analyzed by LC-MS/MS for Q6 and Q6 biosynthetic intermediates. The CNAP3, CNAP6, and CNAP9 eluates were all found to have significantly higher levels of associated Q6 compared with wild type (Fig. 4A). The late stage intermediate DMQ6 was also measured and found to be significantly higher in CNAP3, CNAP6, and CNAP9 compared with wild type (Fig. 4C). The late stage intermediate imino-demethoxy-Q6 (IDMQ6) was also found in measurable quantities in the CNAP3, CNAP6, and CNAP9 eluates, whereas no IDMQ6 was detected in the wild-type eluate (Fig. 4E). Representative HPLC traces of Q6, DMQ6, and IDMQ6 are shown (Fig. 4, B, D, and F). The early intermediates HHB and HAB were not detectable in any of the analyzed eluates (data not shown).
FIGURE 4.
Tandem affinity-purified Coq complexes co-purify with Q6 and late-stage Q6 intermediates. For each strain, aliquots of the anti-PC eluates (29% of E1 and 29% of E2) were combined and subjected to lipid extraction with methanol and petroleum ether. Lipid extracts were analyzed by HPLC-MS/MS for Q6 (A), DMQ6 (C), and IDMQ6 (E). Measured lipids were normalized by the extracted eluate volume. Bars represent the means of two measurements, and error bars represent the standard deviation. Statistical significance was determined with the two-tailed Student's t test, and the lowercase letters above the bars are indicative of statistical significance. In A, the content of Q6 in CNAP3, CNAP6, and CNAP9 was 11.7-, 8.6-, and 22.6-fold higher, respectively, compared with wild type (a, p = 0.0001; b, p = 0.0040; c, p = 0.0006). In C, the content of DMQ6 in CNAP3, CNAP6, and CNAP9 was 30-, 24-, and 97-fold higher, respectively, compared with wild type (a, p = 0.0110; b, p = 0.0113; c, p = 0.0010). In E, there was no detectable IDMQ6 in the wild type. ND, not detected. Representative overlaid traces of all four strains (W303, purple; CNAP3, green; CNAP6, red; CNAP9, blue) are shown for Q6 (B), DMQ6 (D), and IDMQ6 (F).
Aliquots of the anti-PC eluates (20% E1 and 20% E2) were combined for each strain, concentrated, and resuspended in 42 μl of SDS sample buffer. Protein was then separated on an 8–16% SDS-PAGE gel and stained with SYPRO Ruby to visualize total protein (Fig. 5). CNAP3, CNAP6, and CNAP9 each contain several unique protein bands compared with wild type, specifically at 25–37 and 50 kDa. Each sample lane was cut into 5-mm slices and subjected to in-gel trypsin digestion. Tryptic peptides were analyzed by LC-MS/MS, and the results from five trials (performed for W303, CNAP3, and CNAP9) or two trials (performed for CNAP6) are shown in Table 3. The results are ranked by sequence coverage of detected peptides and also indicated is the number of peptides recovered, which represents the total number of peptides detected for a given polypeptide across all trials. Several of the Coq proteins observed by immunoblotting were detected by mass spectrometry of the CNAP3, CNAP6, and CNAP9 eluates and are highlighted. Multiple incidents of recovery of Ilv6 in the CNAP6 and CNAP9 eluates and YLR290C in CNAP3 eluates seemed particularly noteworthy and were investigated further.
FIGURE 5.
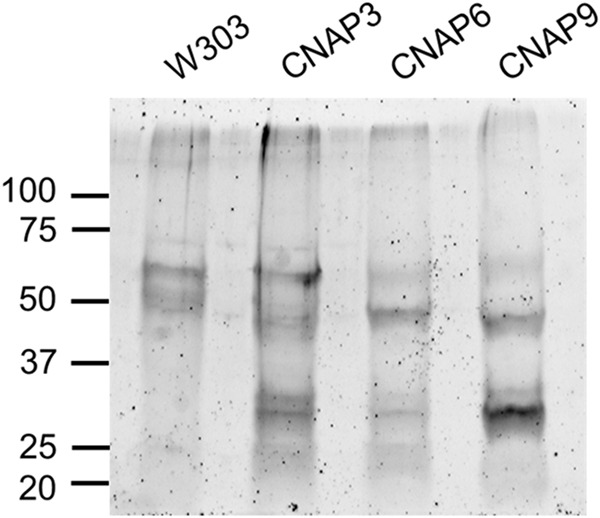
SYPRO Ruby staining for total protein reveals unique bands in CNAP tagged eluates. For each strain designated, aliquots of the anti-PC eluates (20% of E1 and 20% of anti-PC eluate two (E2)) were combined and dried with a SpeedVac at 60 °C for 2 h. Samples were resuspended in 42 μl of SDS sample buffer and separated with an 8–16% Criterion SDS-PAGE gel (Bio-Rad). Following separation proteins were fixed, and the gel was stained overnight with a 1:1 mixture of fresh and used SYPRO Ruby (Life Technologies). The gel was subsequently washed and visualized with an FX Pro Plus Molecular Imager (Bio-Rad) at 532-nm excitation, and emission was measured with a 555-nm long pass filter. The ladder denotes protein masses in kDa.
TABLE 3.
Top protein hits from proteomic analysis of tandem affinity purification eluates
Only proteins that were observed in duplicate injections of samples were considered top hits. Results are compiled from five replicates (W303, CNAP3, and CNAP9) or from two replicates (CNAP6).
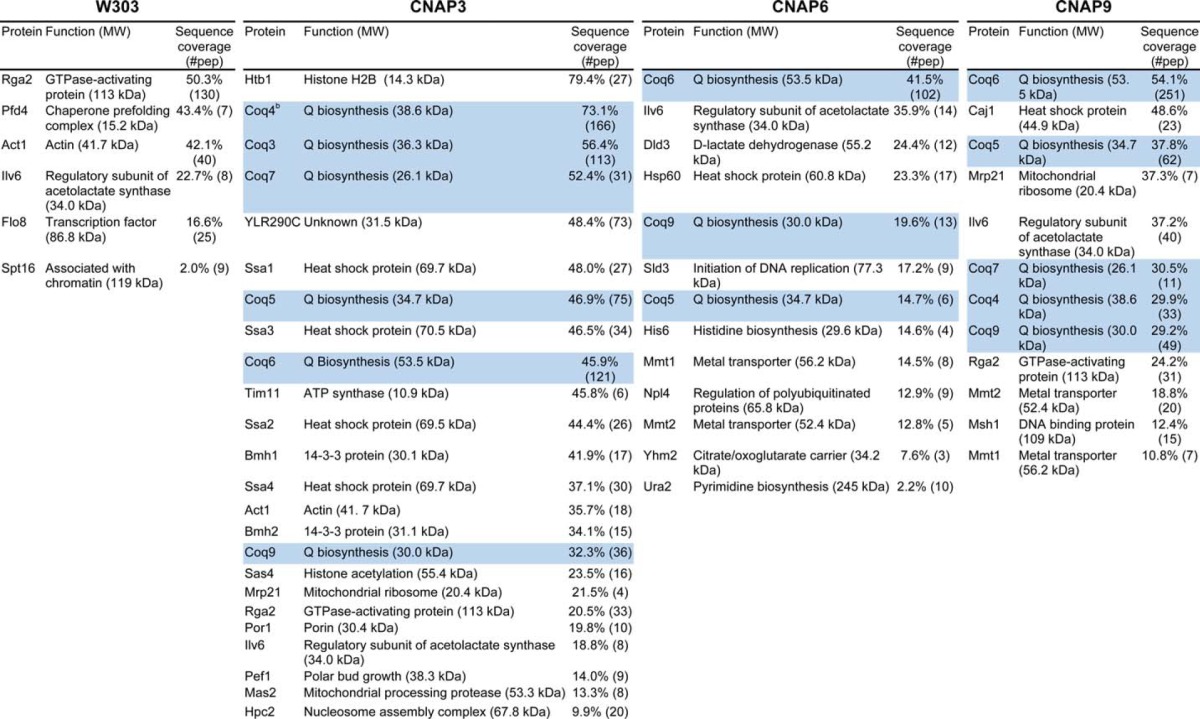
a Highlighted hits denote proteins confirmed by immunoblotting.
Analyses of the ilv6Δ and ylr290cΔ Mutants
Because of the high incidence of recovery of the Ilv6 and YLR290C polypeptides in the CNAP-tagged Coq protein eluates (Table 3), the ilv6Δ and ylr290cΔ mutants in the BY4741 background were purchased from the GE Healthcare yeast knock-out collection for phenotypic analysis. The ability of these null mutants to grow on a nonfermentable carbon source was assessed by plate dilution assay using BY4741 as the wild-type control and coq3Δ as the Q-less, mutant control (Fig. 6). Both the ilv6Δ and ylr290cΔ mutants grew as well as wild type on the nonfermentable carbon source YPG, whereas the coq3Δ mutant failed to grow. All strains grew on the permissible carbon source YPD.
FIGURE 6.
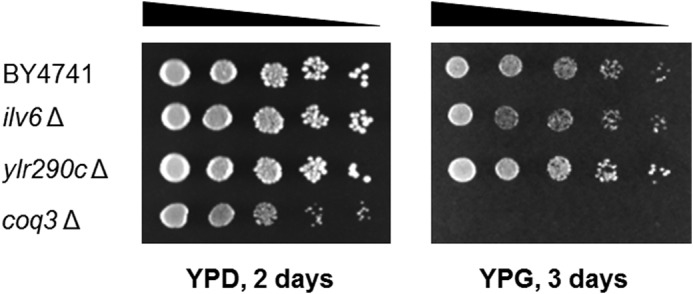
Yeast ilv6Δ and ylr290cΔ mutants retain the ability to grow on a nonfermentable carbon source. Designated yeast strains were grown overnight in 5 ml of YPD and diluted to an A600 of 0.2 with sterile PBS, and 2 μl of 5-fold serial dilutions were spotted onto each type of plate medium, corresponding to a final A600 of 0.2, 0.04, 0.008, 0.0016, and 0.00032. Plates were incubated at 30 °C, and growth is depicted after 2 days for YPD and 3 days for YPG.
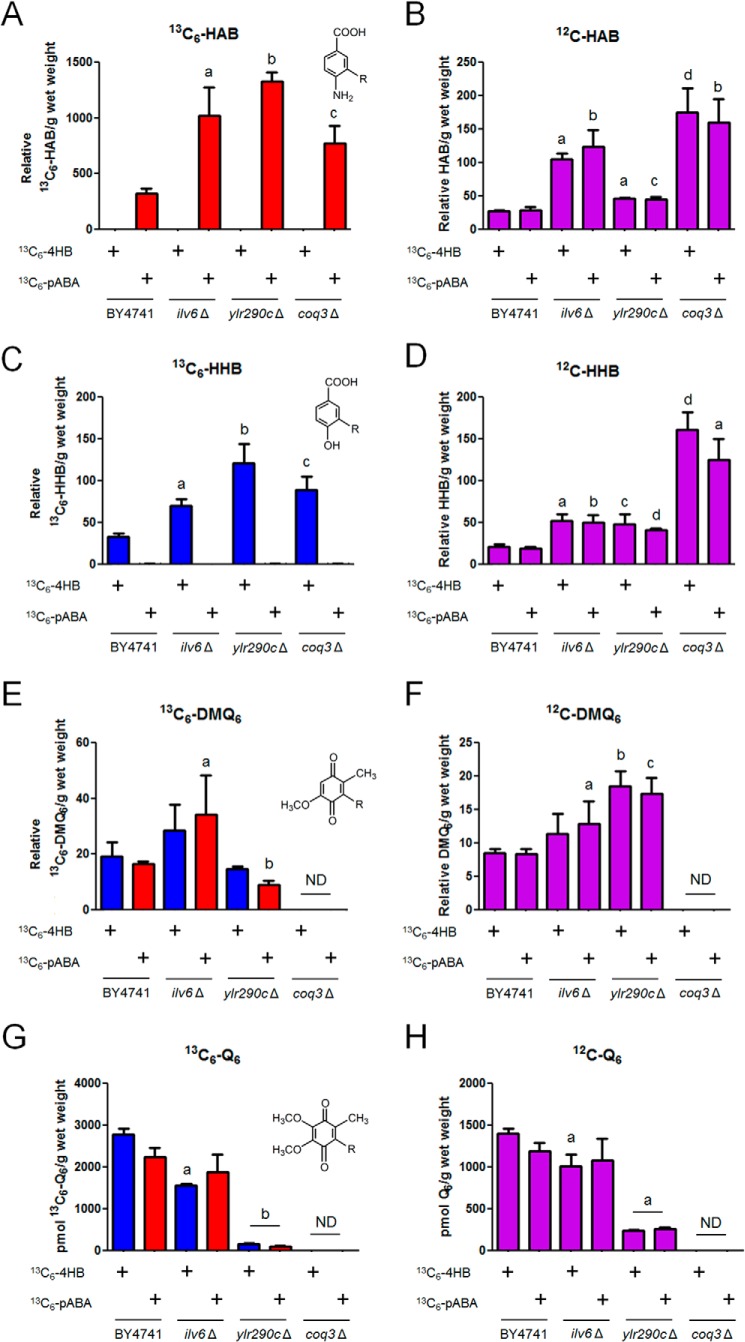
The levels of de novo 13C6-Q6 biosynthesis, as well as accumulation of Q6 and Q6 intermediates, were measured by LC-MS/MS of yeast cultures grown in DOD-complete medium labeled with the stable isotopically labeled precursors 13C6-4HB or 13C6-pABA (Fig. 7). The ilv6Δ mutant produced high levels of de novo 13C6-HAB and 13C6-HHB relative to wild type, similar to the coq3Δ mutant (Fig. 7, A and C). The ilv6Δ mutant also accumulated higher levels of 12C-HAB and 12C-HHB compared with wild type (Fig. 7, B and D). The ilv6Δ mutant had increased levels of the late stage intermediate 13C6-DMQ6 when labeled with 13C6-pABA (Fig. 7E), and accumulated higher levels of 12C-DMQ6 relative to wild type when labeled with 13C6-pABA (Fig. 7F). The late stage intermediate 13C6- or 12C-IDMQ6 was not detected in this analysis (data not shown). The ilv6Δ mutant produced decreased de novo 13C6-Q6 compared with wild type when labeled with 13C6-4HB (Fig. 7G) and accumulated decreased 12C-Q6 relative to wild type when labeled with 13C6-4HB (Fig. 7H). ILV6 encodes the regulatory subunit of acetolactate synthase; Ilv2 catalyzes this committed step of branched chain amino acid biosynthesis in yeast mitochondria (49). Ilv6 is not an essential protein and is not required for the catalytic activity of Ilv2 but regulates Ilv2 through feedback inhibition by valine, a final product of the branched chain amino acid biosynthetic pathway, an effect reversed by binding of ATP to Ilv6 (49, 50). There are functional homologs of Ilv6 in prokaryotes and plants but not other eukaryotes, making this regulatory scheme unique to prokaryotes, fungi, and plants (49, 51, 52).
FIGURE 7.
The yeast ylr290cΔ mutant, but not the ilv6Δ mutant, shows impaired de novo Q biosynthesis. 13C6-HAB (A), 12C-HAB (B), 13C6-HHB (C), 12C-HHB (D), 13C6-DMQ6 (E), 12C-DMQ6 (F), total 13C6-Q6 (13C6-Q6 + 13C6-Q6H2) (G), and total 12C-Q6 (12C-Q6 + 12C-Q6H2) (H) were measured in the designated yeast strains by HPLC-MS/MS. Yeast strains were grown overnight in 5 ml of SD-complete, diluted to an A600 of 0.05 in 50 ml of DOD-complete, labeled with either 13C6-4HB or 13C6-pABA at an A600 of 0.5, and harvested after 3 h of labeling. Lipid measurements were normalized by the wet weight of extracted cells. Each bar represents the mean of four measurements from two biological samples with two injections each. Error bars represent standard deviations. Statistical significance was determined with the two-tailed Student's t test, and the lowercase letters above the bars are indicative of statistical significance. In A, the relative content of 13C6-HAB in the three null mutants was compared with wild type with the corresponding 13C6-labeled precursor (a, p = 0.0017; b, p < 0.0001; c, p = 0.0014). In B, the relative content of 12C-HAB in the three null mutants was compared with wild type with the corresponding 13C6-labeled precursor (a, p < 0.0001; b, p = 0.0003; c, p = 0.0014; d, p = 0.0002). In C, the relative content of 13C6-HHB in the three null mutants was compared with wild type with the corresponding 13C6-labeled precursor (a, p = 0.0001; b, p = 0.0003; c, p = 0.0005). In D, the relative content of 12C-HHB in the three null mutants was compared with wild type with the corresponding 13C6-labeled precursor (a, p = 0.0002; b, p = 0.0004; c, p = 0.0047; d, p < 0.0001). In E, the relative content of 13C6-DMQ6 in the three null mutants was compared with wild type with the corresponding 13C6-labeled precursor (a, p = 0.0441; b, p < 0.0001). ND, not detected. In F, the relative content of 12C-DMQ6 in the three null mutants was compared with wild type with the corresponding 13C6-labeled precursor (a, p = 0.0453; b, p = 0.0002; c, p = 0.0003). ND, not detected. In G, the content of 13C6-Q6 in the three null mutants was compared with wild type with the corresponding 13C6-labeled precursor; the ilv6Δ mutant produced 60% 13C6-Q6 compared with wild type when labeled with 13C6-4HB, and the ylr290cΔ mutant produced 6.1 and 4.5% 13C6-Q6 compared with wild type when labeled with 13C6-4HB or 13C6-pABA, respectively (a, p = 0.0017; b, p < 0.0001). ND, not detected. In H, the content of 12C-Q6 in the three null mutants was compared with wild type with the corresponding 13C6-labeled precursor; the ylr290cΔ mutant accumulated 17.5 and 21.8% 12C-Q6 compared with wild type when labeled with 13C6-4HB or 13C6-pABA respectively (a, p < 0.0001). ND, not detected.
The ylr290cΔ mutant synthesized high levels of de novo 13C6-HAB and 13C6-HHB compared with wild type (Fig. 7, A and C) and also accumulated higher levels of 12C-HAB and 12C-HHB compared with wild type (Fig. 7, B and D). The late stage intermediate 13C6- or 12C-IDMQ6 was not detected in this analysis (data not shown); however, the ylr290cΔ mutant labeled with 13C6-pABA was found to have reduced levels of 13C6-DMQ6 relative to the wild type (Fig. 7E). In contrast, the ylr290cΔ mutant labeled with either precursor accumulated higher levels of 12C-DMQ6 relative to wild type (Fig. 7F). The ylr290cΔ mutant produced severely decreased de novo 13C6-Q6 when labeled with either 13C6-4HB or 13C6-pABA (Fig. 7G) and also accumulated significantly less 12C-Q6 when labeled with 13C6-4HB or 13C6-pABA compared with the wild type (Fig. 7H). These results indicate that the ylr290cΔ mutant accumulates higher levels of Q6 intermediates and has defective Q synthesis compared with wild type. Because of its dramatic effect on Q synthesis, we chose to focus our efforts on further characterization of YLR290C.
Analysis of CNAP-tagged YLR290C
The CNAP tag was integrated into the 3′ end of the YLR290C open reading frame in W303-1A to yield the strain designated CA-1. The levels of de novo and accumulated Q6 were measured in this strain to assess the impact of the tag on Q6 biosynthesis (Fig. 8). The CA-1 strain produced modestly reduced levels of de novo 13C6-Q6 with both 13C6-4HB and 13C6-pABA. Levels of accumulated 12C-Q6 were also decreased in CA-1 when labeled with either 13C6-4HB or 13C6-pABA.
FIGURE 8.
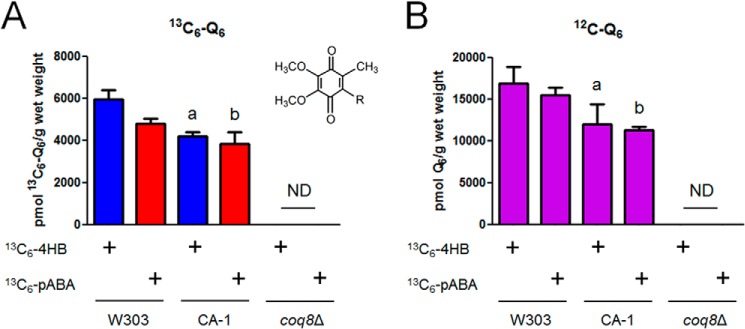
Expression of CNAP-tagged YLR290C preserves de novo Q biosynthesis. Levels of total 13C6-Q6 (13C6-Q6 + 13C6-Q6H2) (A) and total 12C-Q6 (12C-Q6 + 12C-Q6H2) (B) were measured in the designated yeast strains by HPLC-MS/MS. Yeast strains were grown overnight in 5 ml of SD-complete, diluted to an A600 of 0.05 in 50 ml of DOD-complete, labeled with either 13C6-4HB or 13C6-pABA at an A600 of 0.5, and harvested after 3 h of labeling. Lipid measurements were normalized by the wet weight of extracted cells. Each bar represents the mean of four measurements from two biological samples with two injections each. Error bars represent standard deviations. Statistical significance was determined with the two-tailed Student's t test, and the lowercase letters above the bars are indicative of statistical significance. In A, the content of 13C6-Q6 in CA-1 and coq8Δ was compared with wild type with the corresponding 13C6-labeled precursor; 13C6-Q6 synthesized by CA-1 was 70% of wild type and 80% of wild type with 13C6-4HB and 13C6-pABA, respectively (a, p = 0.0005; b, p = 0.0196). ND, not detected. In B, the content of 12C-Q6 in CA-1 and coq8Δ was compared with wild type with the corresponding 13C6-labeled precursor; accumulated 12C-Q6 in CA-1 was 70% of wild type when labeled with either 13C6-4HB or 13C6-pABA (a, p = 0.0202; b, p = 0.0001). ND, not detected.
Solubilized CA-1 mitochondria (15 mg of protein) were subjected to tandem affinity purification of the CNAP tag (Fig. 9). Purified mitochondria (25 μg of protein) and fractions of the first anti-PC eluate (E1) were analyzed by immunoblotting to identify interacting Coq proteins. Membranes were probed with antibodies to Coq1–Coq9, the protein C epitope to identify the tagged protein, and Atp2 and Mdh1 to assess the purity of the eluates. Coq4, Coq5, and Coq7 were observed to co-precipitate with YLR290C-CNAP in the CA-1 strain, whereas the nonspecific mitochondrial proteins Atp2 and Mdh1 were not present in the eluate. Although yielding the weakest signal, Coq7 was observed in the CA-1 eluate in independent blots. The anti-PC blot verifies the presence of the CNAP-tagged protein.
FIGURE 9.
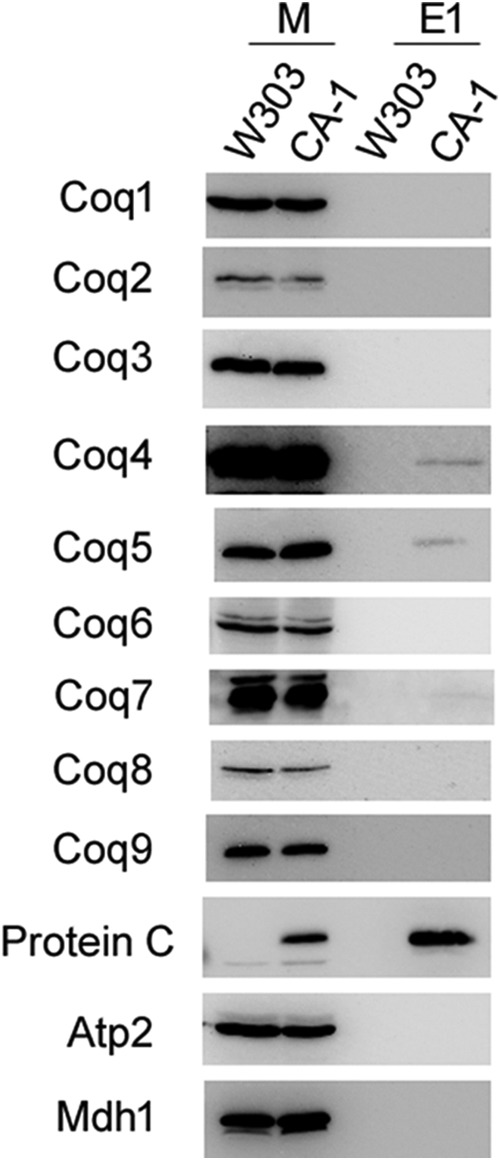
Coq4, Coq5, and Coq7 co-precipitate with YLR290C-CNAP. Purified mitochondria from W303 and CA-1 (15 mg of protein) were solubilized with digitonin and subjected to tandem affinity purification using Ni-NTA resin (Qiagen) followed by anti-PC-agarose (Roche). Samples were separated on 12% SDS-PAGE gels followed by transfer to PVDF membranes for immunoblotting. Mitochondria (25 μg of protein) (M) and 2.5% of the first anti-PC elution (E1) were analyzed for each of the two strains.
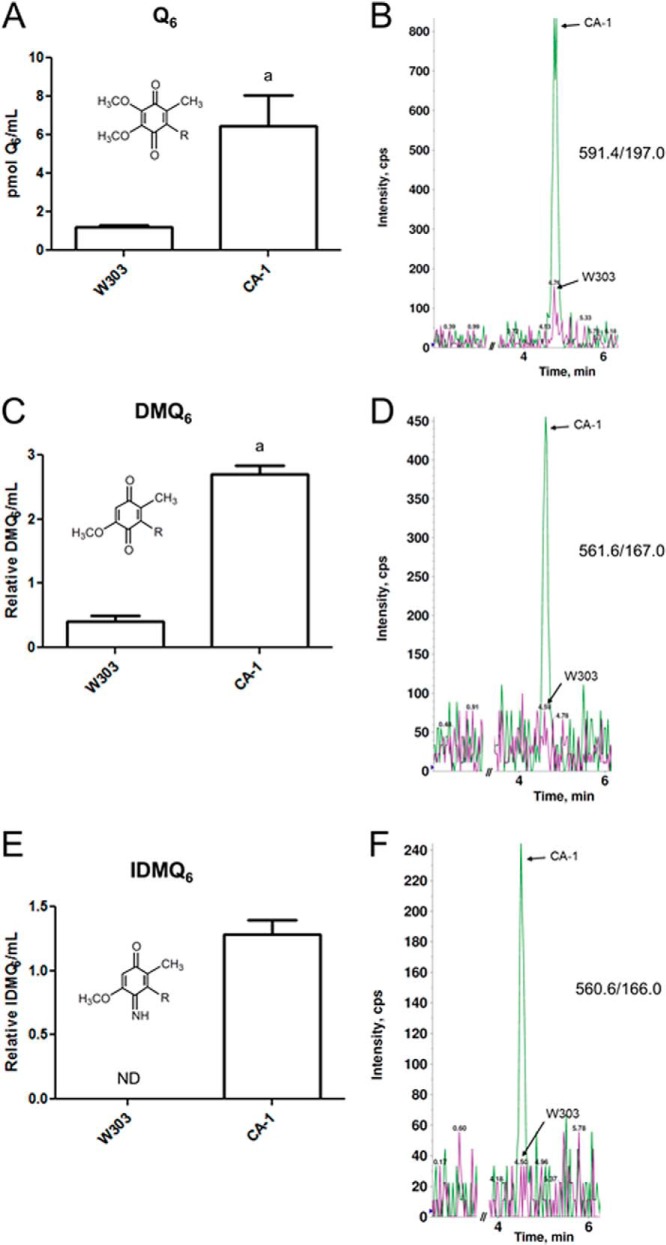
Aliquots of anti-PC eluates (29% E1 and 29% E2) were combined for the wild-type and CA-1 strains and subjected to lipid extraction (Fig. 10). Extracts were then analyzed by LC-MS/MS for Q6 and Q6 biosynthetic intermediates. The CA-1 eluate was found to have higher levels of associated Q6 compared with wild type (Fig. 10A). The late stage intermediate DMQ6 was also measured and found to be higher in CA-1 relative to wild type (Fig. 10C). The late stage intermediate IDMQ6 was also found in measurable quantities in the CA-1 eluate, whereas no IDMQ6 was detected in the wild-type eluate (Fig. 10E). Representative traces of Q6, DMQ6, and IDMQ6 are shown (Fig. 10, B, D, and F). The early intermediates HHB and HAB were not detectable in any of the analyzed eluates (data not shown).
FIGURE 10.
Tandem affinity-purified YLR290C-CNAP co-purifies with Q and late-stage Q intermediates. For each strain, aliquots of the anti-PC eluates (29% of E1 and 29% of E2) were combined and subjected to lipid extraction with methanol and petroleum ether. Lipid extracts were analyzed by HPLC-MS/MS for total Q6 (Q6 + Q6H2) (A), DMQ6 (C), and IDMQ6 (E). Measured lipids were normalized by the extracted eluate volume. The bars represent the mean of two measurements, and the error bars represent the standard deviation. Statistical significance was determined with the two-tailed Student's t test, and the lowercase letters above the bars are indicative of statistical significance. In A, the content of Q6 in CA-1 was 5.4-fold higher compared with wild type (a, p = 0.0418). In C, the content of DMQ6 in CA-1 was 6.7-fold higher compared with wild type (a, p = 0.0025). In E, there was no detectable IDMQ6 in the wild type. ND, not detected. Representative overlaid traces of both strains (W303, magenta; CA-1, green) are shown for Q6 (B), DMQ6 (D), and IDMQ6 (F).
In Silico Analysis of YLR290C and Related Proteins
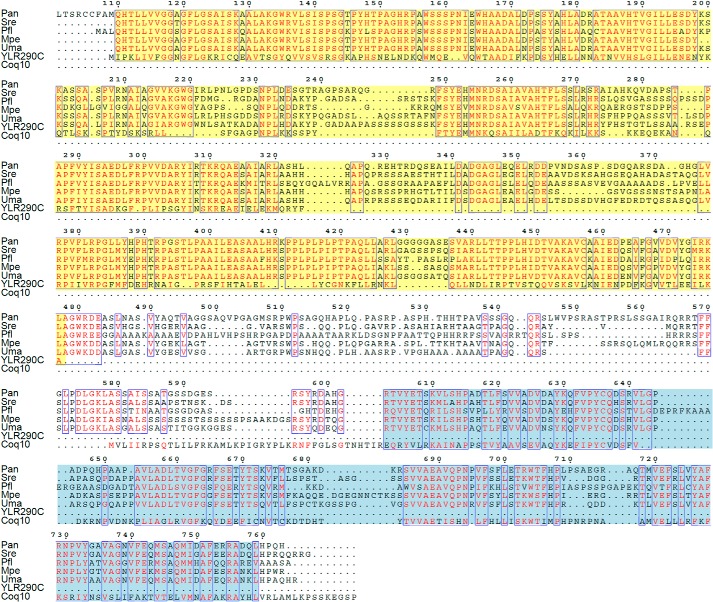
Based on RPS-BLAST to the NCBI Conserved Domain Database (53), YLR290C and closely related proteins (referred to as YLR290C-like) belong to subgroup 5 of the atypical short chain dehydrogenase/reductase (SDR) superfamily. A protein similarity network reveals that YLR290C-like proteins can be divided into three distinct clusters that are delineated largely by taxonomy with the NDUFA9 subfamily of SDR proteins representing the closest distinct similarity cluster (Fig. 11). YLR290C-like proteins were commonly found in fungal genomes and either photosynthetic organisms such as land plants and algae (green algae, stramenopiles, and Guillardia theta) or in the genomes of organisms that may have a photosynthetic ancestor such as the alveolates. The genome of the land plant Arabidopsis thaliana encodes three homologs of YLR290C; based on proteomics data, the ortholog of YLR290C, AT1G32220, is likely localized to the chloroplast and specifically to plastoglobules of the plastid, which are lipid bodies that contain a large amount of prenylquinones among other lipidic compounds (54). We were intrigued to find that YLR290C-like proteins in the fungi Ustilago maydis, Ustilago hordei, Pseudozyma flocculosa, Pseudozyma antarctica, Sporisorium reilianum, Pseudozyma hubeiensis, Pseudozyma aphidis, and Melanopsichium pennsylvanicum were fused to Coq10 (Figs. 11 and 12), suggesting a functional link between YLR290C-like proteins and Coq10.
FIGURE 11.
Sequence analysis of YLR290C and related proteins. In A, YLR290C resides in a protein cluster populated by homologous fungal sequences including proteins that are fused to Coq10. Plant-like homologs form a distinct cluster composed of proteins from photosynthetic organisms. Prokaryotic sequences form a third independent cluster. The closest identified SDR subfamily distinct from YLR290C-like proteins includes human NDUFA9, a subunit of complex I, and orthologs in animals, fungi, and plants. In B, sequences representing each cluster from the protein similarity network in A were used to build a neighbor-joining tree. The tree is drawn to scale, with branch lengths in the same units (amino acid substitutions per site) as those of the evolutionary distances used to infer the phylogenetic tree. A branch length scale bar is shown below. A number after an organism name signifies the presence of multiple homologs in the tree. Protein identifications can be found in the supplemental Data Set 1. A schematic of the YLR290C-Coq10 fusion found in U. maydis is shown in C.
FIGURE 12.
Amino acid alignment of selected fungal YLR290C fusion proteins with YLR290C and Coq10 from S. cerevisiae. The N-terminal YLR290C-like portion is highlighted in yellow, and the Coq10 portion near the C terminus is highlighted in blue. The poorly conserved N-terminal sequence found in P. antarctica (res 1–100) is not shown. The numbering at the top of the alignment corresponds to the P. antarctica sequence. Pan, P. antarctica; Sre, S. reilianum; Pfl, P. flocculosa; Mpe, M. pennsylvanicum; Uma, U. maydis.
DISCUSSION
This study characterized the composition of the Q biosynthetic complex (termed the CoQ-synthome) in yeast mitochondria. We used tandem affinity purification of C-terminally tagged Coq polypeptides. The CNAP tag was used to purify the complex from digitonin-solubilized mitochondria under gentle conditions to preserve the noncovalent associations between protein constituents of the complex to allow for identification by both immunoblotting and mass spectrometry analyses. Tandem affinity purification of tagged Coq3, Coq6, and Coq9 confirmed the association of Coq3, Coq4, Coq5, Coq6, Coq7, and Coq9 in one or more complexes (Fig. 3). These findings recapitulate the results from previous affinity purification experiments (21, 25), with the additional observation that Coq3 is now shown to be associated with the entire complex and not just Coq4. In addition Coq8 was observed to co-purify with a Coq6-containing complex, but not with tagged Coq3 or Coq9, suggesting a closer association with Coq6. Although Coq8 is required for the stability of the Coq polypeptides and the biosynthetic complex (20, 28), this is the first evidence showing direct association of Coq8 with the complex.
The eluates from tandem affinity purification were also subjected to proteomic analyses to identify potentially novel binding partners beyond the known Coq proteins, the results of which are shown in Table 3. Several of the Coq proteins were observed in the eluates for the CNAP-tagged Coq proteins, consistent with results observed by immunoblotting in Fig. 3. In addition to the Coq proteins, a few other proteins were detected at levels comparable with the Coq proteins: YLR290C in the CNAP3 eluate and Ilv6 in the CNAP9 eluate. Ilv6 was also detected in the wild-type control and the CNAP3 and CNAP6 eluates but was much more abundantly detected with the CNAP9 eluate. Although there were other candidate proteins found specifically in eluates of CNAP-tagged samples, Ilv6 is a known mitochondrial protein, and ILV6 was also found to have negative genetic interactions with YLR290C in a high throughput study (55). The potential function of these two proteins in Q biosynthesis was assessed by monitoring growth on a nonfermentable carbon source and measuring de novo Q synthesis using stable isotopically labeled ring precursors for the corresponding null mutants. The ilv6Δ and ylr290cΔ mutants did not display impaired growth on a nonfermentable carbon source (Fig. 6); however, both strains contained higher levels of Q intermediates compared with wild type. Moreover, the ylr290cΔ mutant was found to have severely decreased de novo Q synthesis and decreased total Q as measured by 13C6-Q6 and 12C-Q6, respectively (Fig. 7). The ability of the ylr290cΔ mutant to grow on a nonfermentable carbon source despite its decreased Q synthesis is not surprising, because other mutants with decreased Q synthesis have been characterized and retain the ability to grow on a nonfermentable carbon source. The coq10Δ mutant and particular yeast coq5 mutants expressing human COQ5 retain the ability to grow on a nonfermentable carbon source despite having significantly decreased Q synthesis (56, 57). However, the ylr290cΔ mutant appeared to synthesize wild-type levels of Q6 when grown in rich medium.3 Unlike several of the coqΔ mutants, steady-state levels of Coq4, Coq7, or Coq9 polypeptides were not decreased in the ylr290cΔ mutant in either rich or minimal medium.3
Because the ylr290cΔ mutant was partially defective in Q biosynthesis, the CNAP tag was integrated into the 3′ end of the YLR290C ORF to generate a C-terminal fusion protein, which was subjected to tandem affinity purification to identify co-purifying proteins. Immunoblotting showed that YLR290C co-purified with Coq4, Coq5, and Coq7, confirming the results of the proteomic analysis and demonstrating association of YLR290C with the CoQ-synthome (Fig. 9). The levels of co-purified proteins were less than observed for CNAP-tagged Coq3, Coq6, and Coq9, which is consistent with the observation that proteomic analysis of the CA-1 eluate did not identify additional binding partners (not shown). It may be that YLR290C is more transiently associated with the Q biosynthetic complex or is less stable in digitonin extracts of mitochondria, leading to a reduced yield of associated proteins. Alternatively, the C-terminal CNAP tag may disrupt the interactions of YLR290C with other constituents of the CoQ-synthome, accounting for these observations.
The function of YLR290C is presently unknown, but it was previously determined to be a mitochondrial protein in a high throughput study of Perocchi et al. (58). Although we were unable to find homologs in available animal genomes, YLR290C-like proteins are common to photosynthetic organisms, including those with both primary and secondary plastids (Fig. 11). The A. thaliana ortholog of YLR290C is localized to the plastid based on proteomics data (59), and several other reports have identified it in plastoglobules (54, 60–63), which are sites of prenylquinone synthesis and storage (64). Indeed, the finding of YLR290C homologs in plastoglobules is reminiscent of the Coq8/Abc1 atypical protein kinase family members (ABC1K), also present in plastoglobules (65).
Sequence analyses identified YLR290C as part of the SDR superfamily (Fig. 11), which includes a diverse family of oxidoreductases that catalyze reactions such as isomerization, decarboxylation, epimerization, imine reduction, and carbonyl-alcohol oxidoreduction (66). SDR proteins contain a conserved Rossmann fold, a structural motif found in proteins that bind nucleotide co-factors such as NAD(P), FAD, and FMN (67). The crystal structure of Pseudomonas aeruginosa UbiX, which catalyzes the decarboxylation step in Q biosynthesis, was shown to contain a Rossmann fold with a bound FMN (68). Similarly, the structure of E. coli Pad1, a paralog with 51% identity to E. coli UbiX, was shown to contain a typical Rossmann fold with a bound FMN (69). Although the function of the Rossmann fold in YLR290C remains unclear, it is tempting to speculate that it may also function to catalyze an FMN-dependent decarboxylation in yeast Q biosynthesis, potentially catalyzing the first step denoted by question marks in Fig. 1. The observation that the ylr290cΔ mutant still produces small amounts of Q6 is not fully consistent with this hypothesis; however, there may be redundant decarboxylases capable of bypassing the ylr290cΔ mutant such as Pad1 and Fdc1, two phenylacrylic acid decarboxylases with homology to the E. coli Q biosynthetic decarboxylases UbiD and UbiX (70, 71). Expression of yeast PAD1 in an E. coli ubiX mutant restored Q8 synthesis; however, yeast mutants lacking either PAD1, FDC1, or both produce wild-type levels of Q6 (71, 72), potentially because of their functions as redundant decarboxylases.
Yet another link connecting YLR290C to Q biosynthesis is the presence of YLR290C-Coq10 fusions encoded in five fungal genomes (Figs. 11 and 12). Such fusion proteins, also known as Rosetta Stone proteins, are often indicative of a functional link between two proteins including a physical interaction or residency in the same pathway (73). Coq10 was shown to be required for efficient de novo Q biosynthesis (56), and several studies indicate that yeast Coq10 and orthologs bind Q or DMQ via a hydrophobic START domain (56, 74, 75), leading to the hypothesis that Coq10 acts a Q chaperone, necessary for delivery of Q to the CoQ-synthome for efficient de novo Q synthesis and respiration (56). The fusion of YLR290C with Coq10 strongly suggests a functional interaction of YLR290C with this Q-binding protein. Interestingly, YLR290C was found to have a genetic correlation with both COQ2 and COQ10 in high throughput analyses (55). YLR290C was also identified along with COQ1, COQ2, COQ3, COQ4, COQ6, COQ7, and COQ10 as part of a set of mitochondrial genes required for doxorubicin resistance (76).
To further explore the association of associated lipids with the CoQ-synthome, eluates from tandem affinity purification were subjected to lipid extraction and LC-MS/MS to measure Q6 and Q6 intermediates. Q6 and the Q6 intermediates DMQ6 and IDMQ6 were all detected at levels significantly greater than the wild-type control in eluates recovered from the CNAP3, CNAP6, and CNAP9 strains (Fig. 4) and also in the CA-1 strain with the CNAP-tagged YLR290C-captured complex (Fig. 10). The early intermediates HHB and HAB were not detected in any of the eluates (data not shown). The observation that YLR290C co-purified with Q and late-stage Q intermediates is consistent with the immunoblot results showing the association of YLR290C with Coq4, Coq5, and Coq7 and supports the role of YLR290C as a component of the Q biosynthetic complex.
These results are consistent with the previous association of DMQ6 with the Q biosynthetic complex and with the ability of exogenous Q6 to stabilize several Coq proteins in certain coq mutants. For example, the late stage Q intermediate DMQ6 co-purifies with the Q biosynthetic complex (25), and exogenous Q6 has been shown to stabilize particular Coq proteins and lead to production of later stage biosynthetic intermediates in certain coq mutants (27–29), suggesting a function for Q or a Q intermediate in the stability of the CoQ-synthome. Coq4 was previously hypothesized to serve as a scaffolding protein for the complex, and the crystal structure of the Coq4 homolog Alr8543 from Nostoc sp. PCC7120 revealed a bound geranylgeranyl monophosphate, suggesting that Coq4 functions to stabilize the Q biosynthetic complex through its interactions with other Coq proteins and a polyisoprenoid lipid (26, 77). The crystal structure of human COQ9 was recently solved and shown to have a lipid-occupied cavity; mass spectrometry analyses of purified human COQ9 identified its association with phospholipids and Q (78). Moreover, human COQ9 was shown to bind human COQ7, indicating that a complex is required for Q biosynthesis in human cells (78). Finally, a recent study showed that a mutated form of ADCK3, a human Coq8 ortholog, possessed autophosphorylation activity (79), a finding corroborated by another recent study showing that a purified MBP-ADCK3 fusion exhibited ATPase activity in vitro (80). The authors suggested that ADCK3 may act in concert with Coq9, a hypothesis made particularly compelling by their observation that a fusion of COQ9 and ADCK3 is present in the protozoan Tetrahymena thermophila (79).
The findings presented here strongly indicate that YLR290C functions in Q biosynthesis, and we propose that this protein be designated Coq11. These results have been incorporated into an updated model of the Q biosynthetic complex (Fig. 13), which now depicts Coq8 in association with Coq6; Coq11 is shown in association with Coq4, Coq5, and Coq7. There were additional protein candidates from proteomic analysis of the tandem affinity purification eluates. Further characterization of these proteins may yield a more complete understanding of the proteins associated with the Q biosynthetic complex and the regulation of this pathway.
FIGURE 13.
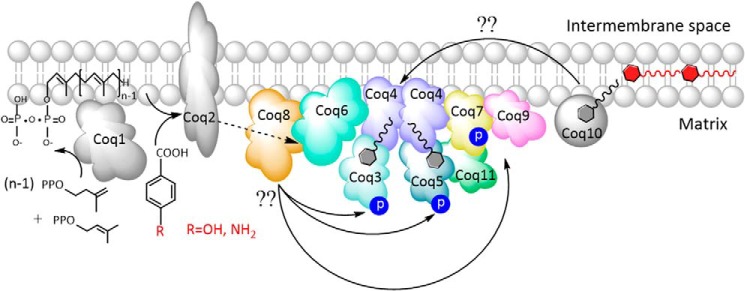
Model of the Q biosynthetic complex. The organization of the complex is based on co-precipitation experiments performed in previous work (21) and this study, as well as two-dimensional blue native-PAGE analysis (28). Coq1, Coq2, and Coq10 have not been shown to associate with the complex. Coq10 binds Q and particular Q intermediates and is postulated to function as a Q chaperone for efficient respiration and de novo Q biosynthesis (56). Coq8 is required for the phosphorylation of Coq3, Coq5, and Coq7 (19), and co-precipitation of CNAP-tagged Coq6 demonstrates their physical association. Co-precipitation of tagged YLR290C, here designated Coq11, showed its association with Coq4, Coq5, and Coq7. The number of copies of each Coq polypeptide depicted in the CoQ-synthome is hypothetical because the stoichiometry has not been determined. Lipid analysis of co-precipitation eluates demonstrated the association of Q6 and the late stage intermediates DMQ6 and IDMQ6 with the complex, illustrated as small molecules in association with Coq4.
Supplementary Material
Acknowledgments
We thank Dr. Carla Koehler (UCLA) for advice on generating the CNAP-tagged constructs used in this study. We thank Letian Xie and Kelly Quinn for help with analyses of steady-state levels of Coq polypeptides in the ylr290cΔ mutant yeast. We thank Dr. A. Tzagoloff (Columbia University) for the original yeast coq mutant strains. We acknowledge the UCLA Molecular Instrumentation Core proteomics facility for the use of QTRAP 4000 and the gel imager.
This work was supported, in whole or in part, by National Institutes of Health Grants S10RR024605 and R01GM103479 (to J. A. L.). This work was also supported by National Science Foundation Grant MCB-1330803 (to C. F. C.), Kirschstein National Research Service Award NIH GM100753 (to C. E. B.-H.), and Grant DE-FC02-02ER63421 from the Office of Science (Biological and Environmental Research), U.S. Department of Energy.

This article contains supplemental Data Set 1.
C. Allan, unpublished observations.
- Q
- coenzyme Q
- CNAP
- consecutive nondenaturing affinity purification
- DMQ6
- demethoxy-Q6
- HAB
- 3-hexaprenyl-4-aminobenzoic acid
- 4HB
- 4-hydroxybenzoic acid
- HHB
- 3-hexaprenyl-4-hydroxybenzoic acid
- IDMQ6
- imino-demethoxy-Q6
- MRM
- multiple reaction monitoring
- pABA
- para-aminobenzoic acid
- PC
- protein C
- DOD
- drop out dextrose medium
- Ni-NTA
- nickel-nitrilotriacetic acid
- SDR
- short chain dehydrogenase/reductase.
REFERENCES
- 1. Crane F. L. (1965) Distribution of ubiquinones. In Biochemistry of Quinones (Morton R. A., ed) pp. 183–206, Academic Press, London [Google Scholar]
- 2. Brandt U., Trumpower B. (1994) The protonmotive Q cycle in mitochondria and bacteria. Crit Rev Biochem. Mol. Biol. 29, 165–197 [DOI] [PubMed] [Google Scholar]
- 3. Grandier-Vazeille X., Bathany K., Chaignepain S., Camougrand N., Manon S., Schmitter J. M. (2001) Yeast mitochondrial dehydrogenases are associated in a supramolecular complex. Biochemistry 40, 9758–9769 [DOI] [PubMed] [Google Scholar]
- 4. Lenaz G., De Santis A. (1985) A survey of the function and specificity of ubiquione in the mitochondrial respiratory chain. In Coenzyme Q (Lenaz G., ed) pp. 165–199, John Wiley & Sons, Chichester, UK [Google Scholar]
- 5. Hildebrandt T. M., Grieshaber M. K. (2008) Three enzymatic activities catalyze the oxidation of sulfide to thiosulfate in mammalian and invertebrate mitochondria. FEBS J. 275, 3352–3361 [DOI] [PubMed] [Google Scholar]
- 6. Bentinger M., Tekle M., Dallner G. (2010) Coenzyme Q: biosynthesis and functions. Biochem. Biophys. Res. Commun. 396, 74–79 [DOI] [PubMed] [Google Scholar]
- 7. Frei B., Kim M. C., Ames B. N. (1990) Ubiquinol-10 is an effective lipid-soluble antioxidant at physiological concentrations. Proc. Natl. Acad. Sci. U.S.A. 87, 4879–4883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Turunen M., Olsson J., Dallner G. (2004) Metabolism and function of coenzyme Q. Biochim. Biophys. Acta 1660, 171–199 [DOI] [PubMed] [Google Scholar]
- 9. Laredj L. N., Licitra F., Puccio H. M. (2014) The molecular genetics of coenzyme Q biosynthesis in health and disease. Biochimie 100, 78–87 [DOI] [PubMed] [Google Scholar]
- 10. Tzagoloff A., Dieckmann C. L. (1990) PET genes of Saccharomyces cerevisiae. Microbiol. Rev. 54, 211–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Johnson A., Gin P., Marbois B. N., Hsieh E. J., Wu M., Barros M. H., Clarke C. F., Tzagoloff A. (2005) COQ9, a new gene required for the biosynthesis of coenzyme Q in Saccharomyces cerevisiae. J. Biol. Chem. 280, 31397–31404 [DOI] [PubMed] [Google Scholar]
- 12. Pierrel F., Hamelin O., Douki T., Kieffer-Jaquinod S., Mühlenhoff U., Ozeir M., Lill R., Fontecave M. (2010) Involvement of Mitochondrial Ferredoxin and para-Aminobenzoic acid in yeast coenzyme Q biosynthesis. Chem. Biol. 17, 449–459 [DOI] [PubMed] [Google Scholar]
- 13. Tran U. C., Clarke C. F. (2007) Endogenous synthesis of coenzyme Q in eukaryotes. Mitochondrion 7, S62–S71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Barros M. H., Nobrega F. G. (1999) YAH1 of Saccharomyces cerevisiae: a new essential gene that codes for a protein homologous to human adrenodoxin. Gene 233, 197–203 [DOI] [PubMed] [Google Scholar]
- 15. Manzella L., Barros M. H., Nobrega F. G. (1998) ARH1 of Saccharomyces cerevisiae: a new essential gene that codes for a protein homologous to the human adrenodoxin reductase. Yeast 14, 839–846 [DOI] [PubMed] [Google Scholar]
- 16. Lange H., Kaut A., Kispal G., Lill R. (2000) A mitochondrial ferredoxin is essential for biogenesis of cellular iron-sulfur proteins. Proc. Natl. Acad. Sci. U.S.A. 97, 1050–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li J., Saxena S., Pain D., Dancis A. (2001) Adrenodoxin reductase homolog (Arh1p) of yeast mitochondria required for iron homeostasis. J. Biol. Chem. 276, 1503–1509 [DOI] [PubMed] [Google Scholar]
- 18. Barros M. H., Nobrega F. G., Tzagoloff A. (2002) Mitochondrial ferredoxin is required for heme A synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 277, 9997–10002 [DOI] [PubMed] [Google Scholar]
- 19. Xie L. X., Hsieh E. J., Watanabe S., Allan C. M., Chen J. Y., Tran U. C., Clarke C. F. (2011) Expression of the human atypical kinase ADCK3 rescues coenzyme Q biosynthesis and phosphorylation of Coq polypeptides in yeast coq8 mutants. Biochim. Biophys. Acta 1811, 348–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Xie L. X., Ozeir M., Tang J. Y., Chen J. Y., Jaquinod S. K., Fontecave M., Clarke C. F., Pierrel F. (2012) Over-expression of the Coq8 kinase in Saccharomyces cerevisiae coq null mutants allows for accumulation of diagnostic intermediates of the Coenzyme Q6 biosynthetic pathway. J. Biol. Chem. 287, 23571–23581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hsieh E. J., Gin P., Gulmezian M., Tran U. C., Saiki R., Marbois B. N., Clarke C. F. (2007) Saccharomyces cerevisiae Coq9 polypeptide is a subunit of the mitochondrial coenzyme Q biosynthetic complex. Arch Biochem. Biophys 463, 19–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tauche A., Krause-Buchholz U., Rödel G. (2008) Ubiquinone biosynthesis in Saccharomyces cerevisiae: the molecular organization of O-methylase Coq3p depends on Abc1p/Coq8p. FEMS Yeast Res. 8, 1263–1275 [DOI] [PubMed] [Google Scholar]
- 23. Glerum D. M., Muroff I., Jin C., Tzagoloff A. (1997) COX15 codes for a mitochondrial protein essential for the assembly of yeast cytochrome oxidase. J. Biol. Chem. 272, 19088–19094 [DOI] [PubMed] [Google Scholar]
- 24. Tzagoloff A., Yue J., Jang J., Paul M. F. (1994) A new member of a family of ATPases is essential for assembly of mitochondrial respiratory chain and ATP synthetase complexes in Saccharomyces cerevisiae. J. Biol. Chem. 269, 26144–26151 [PubMed] [Google Scholar]
- 25. Marbois B., Gin P., Faull K. F., Poon W. W., Lee P. T., Strahan J., Shepherd J. N., Clarke C. F. (2005) Coq3 and Coq4 define a polypeptide complex in yeast mitochondria for the biosynthesis of coenzyme Q. J. Biol. Chem. 280, 20231–20238 [DOI] [PubMed] [Google Scholar]
- 26. Marbois B., Gin P., Gulmezian M., Clarke C. F. (2009) The yeast Coq4 polypeptide organizes a mitochondrial protein complex essential for coenzyme Q biosynthesis. Biochim. Biophys. Acta 1791, 69–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tran U. C., Marbois B., Gin P., Gulmezian M., Jonassen T., Clarke C. F. (2006) Complementation of Saccharomyces cerevisiae coq7 mutants by mitochondrial targeting of the Escherichia coli UbiF polypeptide: two functions of yeast Coq7 polypeptide in coenzyme Q biosynthesis. J. Biol. Chem. 281, 16401–16409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. He C. H., Xie L. X., Allan C. M., Tran U. C., Clarke C. F. (2014) Coenzyme Q supplementation or over-expression of the yeast Coq8 putative kinase stabilizes multi-subunit Coq polypeptide complexes in yeast coq null mutants. Biochim. Biophys Acta 1841, 630–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Padilla S., Tran U. C., Jiménez-Hidalgo M., López-Martín J. M., Martín-Montalvo A., Clarke C. F., Navas P., Santos-Ocaña C. (2009) Hydroxylation of demethoxy-Q6 constitutes a control point in yeast coenzyme Q6 biosynthesis. Cell Mol. Life Sci. 66, 173–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gin P., Clarke C. F. (2005) Genetic evidence for a multi-subunit complex in coenzyme Q biosynthesis in yeast and the role of the Coq1 hexaprenyl diphosphate synthase. J. Biol. Chem. 280, 2676–2681 [DOI] [PubMed] [Google Scholar]
- 31. Burke D., Dawson D., Stearns T. (2000) Methods in Yeast Genetics, pp. 177–186, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 32. Barkovich R. J., Shtanko A., Shepherd J. A., Lee P. T., Myles D. C., Tzagoloff A., Clarke C. F. (1997) Characterization of the COQ5 gene from Saccharomyces cerevisiae: evidence for a C-methyltransferase in ubiquinone biosynthesis. J. Biol. Chem. 272, 9182–9188 [DOI] [PubMed] [Google Scholar]
- 33. Marbois B., Xie L. X., Choi S., Hirano K., Hyman K., Clarke C. F. (2010) para-Aminobenzoic acid is a precursor in coenzyme Q(6) biosynthesis in Saccharomyces cerevisiae. J. Biol. Chem. 285, 27827–27838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Claypool S. M., Oktay Y., Boontheung P., Loo J. A., Koehler C. M. (2008) Cardiolipin defines the interactome of the major ADP/ATP carrier protein of the mitochondrial inner membrane. J. Cell Biol. 182, 937–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gietz R. D., Woods R. A. (2006) Yeast transformation by the LiAc/SS Carrier DNA/PEG method. Methods Mol. Biol. 313, 107–120 [DOI] [PubMed] [Google Scholar]
- 36. Glick B. S., Pon L. A. (1995) Isolation of highly purified mitochondria from Saccharomyces cerevisiae. Methods Enzymol. 260, 213–223 [DOI] [PubMed] [Google Scholar]
- 37. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 38. Shirasaki D. I., Greiner E. R., Al-Ramahi I., Gray M., Boontheung P., Geschwind D. H., Botas J., Coppola G., Horvath S., Loo J. A., Yang X. W. (2012) Network organization of the huntingtin proteomic interactome in mammalian brain. Neuron 75, 41–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Silva J. C., Denny R., Dorschel C., Gorenstein M. V., Li G. Z., Richardson K., Wall D., Geromanos S. J. (2006) Simultaneous qualitative and quantitative analysis of the Escherichia coli proteome: a sweet tale. Mol. Cell Proteomics 5, 589–607 [DOI] [PubMed] [Google Scholar]
- 40. Silva J. C., Gorenstein M. V., Li G. Z., Vissers J. P., Geromanos S. J. (2006) Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol. Cell Proteomics 5, 144–156 [DOI] [PubMed] [Google Scholar]
- 41. Geromanos S. J., Vissers J. P., Silva J. C., Dorschel C. A., Li G. Z., Gorenstein M. V., Bateman R. H., Langridge J. I. (2009) The detection, correlation, and comparison of peptide precursor and product ions from data independent LC-MS with data dependant LC-MS/MS. Proteomics 9, 1683–1695 [DOI] [PubMed] [Google Scholar]
- 42. Saitou N., Nei M. (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 [DOI] [PubMed] [Google Scholar]
- 43. Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Atkinson H. J., Morris J. H., Ferrin T. E., Babbitt P. C. (2009) Using sequence similarity networks for visualization of relationships across diverse protein superfamilies. PLoS One 4, e4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Blaby-Haas C. E., Merchant S. S. (2012) The ins and outs of algal metal transport. Biochim. Biophys. Acta 1823, 1531–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. UniProt C. (2014) Activities at the Universal Protein Resource (UniProt). Nucleic Acids Res. 42, D191–D198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Edgar R. C. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Robert X., Gouet P. (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cullin C., Baudin-Baillieu A., Guillemet E., Ozier-Kalogeropoulos O. (1996) Functional analysis of YCL09C: evidence for a role as the regulatory subunit of acetolactate synthase. Yeast 12, 1511–1518 [DOI] [PubMed] [Google Scholar]
- 50. Pang S. S., Duggleby R. G. (2001) Regulation of yeast acetohydroxyacid synthase by valine and ATP. Biochem. J. 357, 749–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hershey H. P., Schwartz L. J., Gale J. P., Abell L. M. (1999) Cloning and functional expression of the small subunit of acetolactate synthase from Nicotiana plumbaginifolia. Plant Mol. Biol. 40, 795–806 [DOI] [PubMed] [Google Scholar]
- 52. Lee Y. T., Duggleby R. G. (2001) Identification of the regulatory subunit of Arabidopsis thaliana acetohydroxyacid synthase and reconstitution with its catalytic subunit. Biochemistry 40, 6836–6844 [DOI] [PubMed] [Google Scholar]
- 53. Marchler-Bauer A., Lu S., Anderson J. B., Chitsaz F., Derbyshire M. K., DeWeese-Scott C., Fong J. H., Geer L. Y., Geer R. C., Gonzales N. R., Gwadz M., Hurwitz D. I., Jackson J. D., Ke Z., Lanczycki C. J., Lu F., Marchler G. H., Mullokandov M., Omelchenko M. V., Robertson C. L., Song J. S., Thanki N., Yamashita R. A., Zhang D., Zhang N., Zheng C., Bryant S. H. (2011) CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 39, D225–D229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ytterberg A. J., Peltier J. B., van Wijk K. J. (2006) Protein profiling of plastoglobules in chloroplasts and chromoplasts: a surprising site for differential accumulation of metabolic enzymes. Plant Physiol. 140, 984–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Costanzo M., Baryshnikova A., Bellay J., Kim Y., Spear E. D., Sevier C. S., Ding H. M., Koh J. L., Toufighi K., Mostafavi S., Prinz J., St. Onge R. P., VanderSluis B., Makhnevych T., Vizeacoumar F. J., Alizadeh S., Bahr S., Brost R. L., Chen Y., Cokol M., Deshpande R., Li Z., Lin Z. Y., Liang W., Marback M., Paw J., San Luis B. J., Shuteriqi E., Tong A. H., van Dyk N., Wallace I. M., Whitney J. A., Weirauch M. T., Zhong G., Zhu H., Houry W. A., Brudno M., Ragibizadeh S., Papp B., Pál C., Roth F. P., Giaever G., Nislow C., Troyanskaya O. G., Bussey H., Bader G. D., Gingras A. C., Morris Q. D., Kim P. M., Kaiser C. A., Myers C. L., Andrews B. J., Boone C. (2010) The genetic landscape of a cell. Science 327, 425–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Allan C. M., Hill S., Morvaridi S., Saiki R., Johnson J. S., Liau W. S., Hirano K., Kawashima T., Ji Z., Loo J. A., Shepherd J. N., Clarke C. F. (2013) A conserved START domain coenzyme Q-binding polypeptide is required for efficient Q biosynthesis, respiratory electron transport, and antioxidant function in Saccharomyces cerevisiae. Biochim Biophys Acta 1831, 776–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nguyen T. P., Casarin A., Desbats M. A., Doimo M., Trevisson E., Santos-Ocaña C., Navas P., Clarke C. F., Salviati L. (2014) Molecular characterization of the human COQ5 C-methyltransferase in coenzyme Q10 biosynthesis. Biochim. Biophys. Acta 1841, 1628–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Perocchi F., Jensen L. J., Gagneur J., Ahting U., von Mering C., Bork P., Prokisch H., Steinmetz L. M. (2006) Assessing systems properties of yeast mitochondria through an interaction map of the organelle. PLoS Genet. 2, e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sun Q., Zybailov B., Majeran W., Friso G., Olinares P. D., van Wijk K. J. (2009) PPDB, the Plant Proteomics Database at Cornell. Nucleic Acids Res. 37, D969–D974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Huang M., Friso G., Nishimura K., Qu X., Olinares P. D., Majeran W., Sun Q., van Wijk K. J. (2013) Construction of plastid reference proteomes for maize and Arabidopsis and evaluation of their orthologous relationships: the concept of orthoproteomics. J. Proteome Res. 12, 491–504 [DOI] [PubMed] [Google Scholar]
- 61. Vidi P. A., Kanwischer M., Baginsky S., Austin J. R., Csucs G., Dörmann P., Kessler F., Bréhélin C. (2006) Tocopherol cyclase (VTE1) localization and vitamin E accumulation in chloroplast plastoglobule lipoprotein particles. J. Biol. Chem. 281, 11225–11234 [DOI] [PubMed] [Google Scholar]
- 62. Lundquist P. K., Poliakov A., Bhuiyan N. H., Zybailov B., Sun Q., van Wijk K. J. (2012) The functional network of the Arabidopsis plastoglobule proteome based on quantitative proteomics and genome-wide coexpression analysis. Plant Physiol. 158, 1172–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zybailov B., Rutschow H., Friso G., Rudella A., Emanuelsson O., Sun Q., van Wijk K. J. (2008) Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS One 3, e1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Eugeni Piller L., Abraham M., Dörmann P., Kessler F., Besagni C. (2012) Plastid lipid droplets at the crossroads of prenylquinone metabolism. J. Exp. Bot. 63, 1609–1618 [DOI] [PubMed] [Google Scholar]
- 65. Lundquist P. K., Davis J. I., van Wijk K. J. (2012) ABC1K atypical kinases in plants: filling the organellar kinase void. Trends Plant Sci. 17, 546–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Marchler-Bauer A., Zheng C., Chitsaz F., Derbyshire M. K., Geer L. Y., Geer R. C., Gonzales N. R., Gwadz M., Hurwitz D. I., Lanczycki C. J., Lu F., Lu S., Marchler G. H., Song J. S., Thanki N., Yamashita R. A., Zhang D., Bryant S. H. (2013) CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res. 41, D348–D352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Rossmann M. G., Moras D., Olsen K. W. (1974) Chemical and biological evolution of a nucleotide-binding protein. Nature 250, 194–199 [DOI] [PubMed] [Google Scholar]
- 68. Kopec J., Schnell R., Schneider G. (2011) Structure of PA4019, a putative aromatic acid decarboxylase from Pseudomonas aeruginosa. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 67, 1184–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Rangarajan E. S., Li Y., Iannuzzi P., Tocilj A., Hung L. W., Matte A., Cygler M. (2004) Crystal structure of a dodecameric FMN-dependent UbiX-like decarboxylase (Pad1) from Escherichia coli O157: H7. Protein Sci. 13, 3006–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Mukai N., Masaki K., Fujii T., Kawamukai M., Iefuji H. (2010) PAD1 and FDC1 are essential for the decarboxylation of phenylacrylic acids in Saccharomyces cerevisiae. J. Biosci. Bioeng. 109, 564–569 [DOI] [PubMed] [Google Scholar]
- 71. Gulmezian M., Hyman K. R., Marbois B. N., Clarke C. F., Javor G. T. (2007) The role of UbiX in Escherichia coli coenzyme Q biosynthesis. Arch. Biochem. Biophys. 467, 144–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Gulmezian M. (2006) Characterization of Escherichia coli ubiX, Saccharomyces cerevisiae PAD1 and YDR539W, and a complex of polypeptides involved in coenzyme Q biosynthesis. Ph. D. thesis, UCLA, Los Angeles, CA [Google Scholar]
- 73. Marcotte E. M., Pellegrini M., Ng H. L., Rice D. W., Yeates T. O., Eisenberg D. (1999) Detecting protein function and protein-protein interactions from genome sequences. Science 285, 751–753 [DOI] [PubMed] [Google Scholar]
- 74. Cui T. Z., Kawamukai M. (2009) Coq10, a mitochondrial coenzyme Q binding protein, is required for proper respiration in Schizosaccharomyces pombe. FEBS J. 276, 748–759 [DOI] [PubMed] [Google Scholar]
- 75. Murai M., Matsunobu K., Kudo S., Ifuku K., Kawamukai M., Miyoshi H. (2014) Identification of the binding site of the quinone-head group in mitochondrial Coq10 by photoaffinity labeling. Biochemistry 53, 3995–4003 [DOI] [PubMed] [Google Scholar]
- 76. Tay Z., Eng R. J., Sajiki K., Lim K. K., Tang M. Y., Yanagida M., Chen E. S. (2013) Cellular robustness conferred by genetic crosstalk underlies resistance against chemotherapeutic drug doxorubicin in fission yeast. PLoS One 8, e55041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Rea S. L., Graham B. H., Nakamaru-Ogiso E., Kar A., Falk M. J. (2010) Bacteria, yeast, worms, and flies: exploiting simple model organisms to investigate human mitochondrial diseases. Dev. Disabil. Res. Rev. 16, 200–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Lohman D. C., Forouhar F., Beebe E. T., Stefely M. S., Minogue C. E., Ulbrich A., Stefely J. A., Sukumar S., Luna-Sánchez M., Jochem A., Lew S., Seetharaman J., Xiao R., Wang H., Westphall M. S., Wrobel R. L., Everett J. K., Mitchell J. C., López L. C., Coon J. J., Tong L., Pagliarini D. J. (2014) Mitochondrial COQ9 is a lipid-binding protein that associates with COQ7 to enable coenzyme Q biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 111, E4697–E4705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Stefely J. A., Reidenbach A. G., Ulbrich A., Oruganty K., Floyd B. J., Jochem A., Saunders J. M., Johnson I. E., Minogue C. E., Wrobel R. L., Barber G. E., Lee D., Li S., Kannan N., Coon J. J., Bingman C. A., Pagliarini D. J. (2015) Mitochondrial ADCK3 employs an atypical protein kinase-like fold to enable coenzyme Q biosynthesis. Mol. Cell 57, 83–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wheeler B., Jia Z. (2014) Preparation and characterization of human ADCK3, a putative atypical kinase. Protein Expr. Purif. 108C, 13–17 [DOI] [PubMed] [Google Scholar]
- 81. Do T. Q., Hsu A. Y., Jonassen T., Lee P. T., Clarke C. F. (2001) A defect in coenzyme Q biosynthesis is responsible for the respiratory deficiency in Saccharomyces cerevisiae abc1 mutants. J. Biol. Chem. 276, 18161–18168 [DOI] [PubMed] [Google Scholar]
- 82. Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., Hieter P., Boeke J. D. (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115–132 [DOI] [PubMed] [Google Scholar]
- 83. Winzeler E. A., Shoemaker D. D., Astromoff A., Liang H., Anderson K., Andre B., Bangham R., Benito R., Boeke J. D., Bussey H., Chu A. M., Connelly C., Davis K., Dietrich F., Dow S. W., El-Bakkoury M., Foury F., Friend S. H., Gentalen E., Giaever G., Hegemann J. H., Jones T., Laub M., Liao H., Liebundguth N., Lockhart D. J., Lucau-Danila A., Lussier M., M'Rabet N., Menard P., Mittmann M., Pai C., Rebischung C., Revuelta J. L., Riles L., Roberts C. J., Ross-MacDonald P., Scherens B., Snyder M., Sookhai-Mahadeo S., Storms R. K., Véronneau S., Voet M., Volckaert G., Ward T. R., Wysocki R., Yen G. S., Yu K., Zimmermann K., Philippsen P., Johnston M., Davis R. W. (1999) Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285, 901–906 [DOI] [PubMed] [Google Scholar]
- 84. Giaever G., Chu A. M., Ni L., Connelly C., Riles L., Véronneau S., Dow S., Lucau-Danila A., Anderson K., André B., Arkin A. P., Astromoff A., El-Bakkoury M., Bangham R., Benito R., Brachat S., Campanaro S., Curtiss M., Davis K., Deutschbauer A., Entian K. D., Flaherty P., Foury F., Garfinkel D. J., Gerstein M., Gotte D., Güldener U., Hegemann J. H., Hempel S., Herman Z., Jaramillo D. F., Kelly D. E., Kelly S. L., Kötter P., LaBonte D., Lamb D. C., Lan N., Liang H., Liao H., Liu L., Luo C., Lussier M., Mao R., Menard P., Ooi S. L., Revuelta J. L., Roberts C. J., Rose M., Ross-Macdonald P., Scherens B., Schimmack G., Shafer B., Shoemaker D. D., Sookhai-Mahadeo S., Storms R. K., Strathern J. N., Valle G., Voet M., Volckaert G., Wang C. Y., Ward T. R., Wilhelmy J., Winzeler E. A., Yang Y. H., Yen G., Youngman E., Yu K. X., Bussey H., Boeke J. D., Snyder M., Philippsen P., Davis R. W., Johnston M. (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature 418, 387–391 [DOI] [PubMed] [Google Scholar]
- 85. Poon W. W., Barkovich R. J., Hsu A. Y., Frankel A., Lee P. T., Shepherd J. N., Myles D. C., Clarke C. F. (1999) Yeast and rat Coq3 and Escherichia coli UbiG polypeptides catalyze both O-methyltransferase steps in coenzyme Q biosynthesis. J. Biol. Chem. 274, 21665–21672 [DOI] [PubMed] [Google Scholar]
- 86. Belogrudov G. I., Lee P. T., Jonassen T., Hsu A. Y., Gin P., Clarke C. F. (2001) Yeast COQ4 encodes a mitochondrial protein required for coenzyme Q synthesis. Arch. Biochem. Biophys. 392, 48–58 [DOI] [PubMed] [Google Scholar]
- 87. Baba S. W., Belogrudov G. I., Lee J. C., Lee P. T., Strahan J., Shepherd J. N., Clarke C. F. (2004) Yeast Coq5 C-methyltransferase is required for stability of other polypeptides involved in coenzyme Q biosynthesis. J. Biol. Chem. 279, 10052–10059 [DOI] [PubMed] [Google Scholar]
- 88. Gin P., Hsu A. Y., Rothman S. C., Jonassen T., Lee P. T., Tzagoloff A., Clarke C. F. (2003) The Saccharomyces cerevisiae COQ6 gene encodes a mitochondrial flavin-dependent monooxygenase required for coenzyme Q biosynthesis. J. Biol. Chem. 278, 25308–25316 [DOI] [PubMed] [Google Scholar]
- 89. Hsieh E. J., Dinoso J. B., Clarke C. F. (2004) A tRNA(TRP) gene mediates the suppression of cbs2–223 previously attributed to ABC1/COQ8. Biochem. Biophys. Res. Commun. 317, 648–653 [DOI] [PubMed] [Google Scholar]
- 90. Barros M. H., Johnson A., Gin P., Marbois B. N., Clarke C. F., Tzagoloff A. (2005) The Saccharomyces cerevisiae COQ10 gene encodes a START domain protein required for function of coenzyme Q in respiration. J. Biol. Chem. 280, 42627–42635 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.