Background: Dpb11 is required for the initiation of DNA replication. The replication fork helicase is composed of Cdc45, Mcm2-7, and GINS.
Results: Dpb11 recruits Cdc45 to Mcm2-7, and Dpb11 blocks GINS interaction with Mcm2-7. Dpb11 also binds to ssDNA, and this interaction releases Dpb11 from Mcm2-7.
Conclusion: Dpb11 helps control assembly of the replication fork helicase.
Significance: A mechanism for Dpb11 function is described.
Keywords: Cell Cycle, Cyclin-dependent Kinase (CDK), DNA, DNA Helicase, DNA Replication
Abstract
Dpb11 is required for the initiation of DNA replication in budding yeast. Dpb11 binds to S-phase cyclin-dependent kinase-phosphorylated Sld2 and Sld3 to form a ternary complex during S phase. The replication fork helicase in eukaryotes is composed of Cdc45, Mcm2-7, and GINS. We show here, using purified proteins from budding yeast, that Dpb11 alone binds to Mcm2-7 and that Dpb11 also competes with GINS for binding to Mcm2-7. Furthermore, Dpb11 binds directly to single-stranded DNA (ssDNA), and ssDNA inhibits the Dpb11 interaction with Mcm2-7. We also found that Dpb11 can recruit Cdc45 to Mcm2-7. We identified a mutant of the BRCT4 motif of Dpb11 that remains bound to Mcm2-7 in the presence of ssDNA (dpb11-m1,m2,m3,m5), and this mutant exhibits a DNA replication defect when expressed in budding yeast cells. Expression of this mutant results in increased interaction between Dpb11 and Mcm2-7 during S phase, impaired GINS interaction with Mcm2-7 during S phase, and decreased replication protein A (RPA) interaction with origin DNA during S phase. We propose a model in which Dpb11 first recruits Cdc45 to Mcm2-7. Dpb11, although bound to Cdc45·Mcm2-7, can block the interaction between GINS and Mcm2-7. Upon extrusion of ssDNA from the central channel of Mcm2-7, Dpb11 dissociates from Mcm2-7, and Dpb11 binds to ssDNA, thereby allowing GINS to bind to Cdc45·Mcm2-7. Finally, we propose that Dpb11 functions with Sld2 and Sld3 to help control the assembly of the replication fork helicase.
Introduction
Dpb11 is required for the initiation of DNA replication in budding yeast (1). The homologs of Dpb11 include Cut5 (Schizosaccharomyces pombe), Mus101 (Xenopus), and TopBP1 (human). In budding yeast, Sld2 (synthetic lethal with dpb11-1) and Sld3 bind to Dpb11 in a cyclin-dependent kinase (CDK)-dependent2 manner (2–4). Dpb11 (DNA polymerase B-associated protein) binds to CDK-phosphorylated Sld3 and Sld2, and the CDK phosphomimetic mutant of Sld2, Sld2T84D, binds to Dpb11 (2–4). The fusion of Sld3 with Dpb11, when combined with the CDK phosphomimetic mutant of Sld2 (Sld2T84D), bypasses the requirement for CDK in the budding yeast cell (4). These data suggest that formation of the Dpb11-Sld3-Sld2 ternary complex is essential for cell growth. Therefore, Dpb11 is currently viewed as a scaffolding protein for binding Sld2 and Sld3, but the molecular function of Dpb11 in replication initiation is not known.
Mcm2-7 forms a hexameric ring, and the subunit composition of this ring is known (Fig. 1B) (5, 6). Mcm2-7 loads onto double-stranded DNA origins during late M phase and G1 phase. The molecular details of this loading process have been elucidated (7, 8). The Mcm2-7 complex is loaded as a double hexamer, and the Mcm2-7 ring is cracked open at the Mcm2/Mcm5 interface during this process to encircle double-stranded DNA (9, 10). Mcm2-7 by itself is a very weak ATPase and helicase (5, 6, 11). In S phase, critical events function at the Mcm2-7 complex to activate the replication fork helicase.
FIGURE 1.
Dpb11 binds to Mcm2-7. A, 30 pmol of GST·Mcm2-7 (contains GST at the N terminus of Mcm3) or GST alone was incubated with glutathione-Sepharose and increasing concentrations of radiolabeled PKA-Dpb11 at 30 °C for 10 min in a GST pulldown assay as described under “Experimental Procedures.” The bound radioactive Dpb11 was analyzed by SDS/PAGE followed by phosphorimaging. These results were then quantified, averaged, and plotted as picomoles of Dpb11 bound versus the picomoles of Dpb11 input. B, arrangement of Mcm subunits forming the Mcm2-7 hexamer, along with the position of Cdc45 and GINS in the CMG complex (5, 15). C, 30 pmol of GST-Mcm2, GST-Mcm3, or GST-Mcm5 was incubated with increasing concentrations of radiolabeled Dpb11 in a GST pulldown assay. D, schematic of budding yeast Dpb11 protein indicating the four BRCT domains and the binding region of Sld3 and Sld2. E, different fragments of PKA-Dpb11 were incubated with 30 pmol of GST-Mcm3 in a GST pulldown assay to determine the region of Dpb11 involved in the Dpb11-Mcm3 interaction. FL, full-length. F, different fragments of PKA-Dpb11 were incubated with 30 pmol of GST-Mcm5 in a GST pulldown assay to determine the region of Dpb11 involved in the Dpb11-Mcm5 interaction. G, radiolabeled PKA-Dpb11 (253–430) was incubated with 30 pmol of GST-Mcm3, GST-Mcm5, or GST alone to determine whether amino acids 253–430 of Dpb11 bind GST-Mcm3 or GST-Mcm5. H, radiolabeled PKA-Dpb11 (430–615) was incubated with 30 pmol of GST-Mcm3, GST-Mcm5, or GST alone to determine whether amino acids 430–615 of Dpb11 bind GST-Mcm3 or GST-Mcm5.
During S phase, the Dbf4-dependent kinase phosphorylates components of the Mcm2-7 complex, and Cdc45 is recruited to Mcm2-7 (12–14). Cdc45 binds to the Mcm2 subunit of Mcm2-7 (Fig. 1B) (15). Also, during S phase, the Mcm2-7 ring transitions from surrounding double-stranded DNA to encircling single-stranded DNA (15, 16). Therefore, Mcm2-7 cracks open once again in S phase, also at the Mcm2-Mcm5 interface, to allow the extrusion of single-stranded DNA (17). This event is mediated by phosphorylation of Mcm2 by the Dbf4-dependent kinase (17).
During S phase, GINS associates with Cdc45·Mcm2-7, thereby completing the assembly of the Cdc45·Mcm2-7·GINS (CMG complex), the replication fork helicase in eukaryotes (18–21). GINS binds to the Mcm3 and Mcm5 subunits of Mcm2-7 (Fig. 1B) (15). The association of GINS with Cdc45·Mcm2-7 may require the activity of the S-phase·cyclin-dependent kinase (S-CDK) (22–24). Furthermore, the addition of Cdc45 and GINS to Mcm2-7 substantially stimulates the ATPase and helicase activities of Mcm2-7, suggesting that the assembly of the CMG complex is critical for activating the replication fork helicase (18). Finally, although the Mcm2-7 complex can crack open to encircle double-stranded DNA or single-stranded DNA, the CMG complex is a locked ring that encircles only single-stranded DNA (15).
It has been demonstrated previously that Sld3 helps recruit Cdc45 to Mcm2-7 (25–27). Furthermore, Sld2 or Sld3 can each bind directly to Mcm2-7, and Sld2 or Sld3 compete with GINS for binding to Mcm2-7 (26, 28). However, in the presence of single-stranded DNA, Sld2 or Sld3 dissociate from Mcm2-7, allowing GINS to bind Mcm2-7 (28, 29). Furthermore, GINS disrupts the interaction between Sld3 and Cdc45, suggesting an elegant mechanism to disengage Sld2 and Sld3 from Cdc45·Mcm2-7 (26).
In this work, we show, using purified proteins from budding yeast, that Dpb11 can bind directly to Mcm2-7, Cdc45, or single-stranded DNA. Furthermore, Dpb11 can recruit Cdc45 to Mcm2-7, and Dpb11 also competes with GINS for interaction with Mcm2-7. In the presence of single-stranded DNA (ssDNA), the interaction between Dpb11 and Mcm2-7 is disrupted, and this allows GINS to bind Mcm2-7. Single-stranded DNA also disrupts the interaction between Dpb11 and Cdc45. When mutant dpb11-m1,m2,m3,m5 (a BRCT4 motif mutant of Dpb11 that remains bound to Mcm2-7 in the presence of ssDNA) is expressed in yeast cells, the cells suffer severe growth and DNA replication defects. Expression of the dpb11-m1,m2,m3,m5 mutant results in decreased GINS·Mcm2-7 interaction during S phase and increased Dpb11 interaction with Mcm2-7 during S phase. These data suggest that Dpb11 functions to regulate assembly of the CMG complex. Expression of dpb11-m1,m2,m3,m5 also results in decreased RPA interaction with origin DNA during S phase. We propose a model in which Dpb11 first binds to Mcm2-7 to help recruit Cdc45 to Mcm2-7. Dpb11 also blocks the premature formation of the CMG complex by inhibiting GINS interaction with Mcm2-7. However, when single-stranded DNA is extruded from the central channel of Mcm2-7, Dpb11 dissociates from Mcm2-7 and Cdc45 to bind ssDNA, thereby allowing formation of the CMG complex.
EXPERIMENTAL PROCEDURES
Cloning and Purification of Proteins
The full-length DPB11 PCR product was cloned into the SpeI/XhoI sites of pET 41a vector and the NdeI/XhoI sites of pET 33b vector to contain an N-terminal GST tag or a PKA tag, respectively, as described previously (30). The cloning of DPB11 into pET 41a generates a two-tagged protein: a GST tag at the N terminus and a His-tag at the C terminus. Purification of GST-Dpb11 uses sequential nickel and glutathione resins. The details of purification of GST-Dpb11 and PKA-Dpb11 have been described previously (30). Fragments of Dpb11 and the various mutants of Dpb11 were each cloned into the NdeI/XhoI sites of the pET 33b vector and purified as described previously (30). GST alone was purified as described previously (30). The full-length MCM3 PCR product was cloned into the SpeI/XhoI site of the pET 41a vector, and the full-length MCM5 PCR product was cloned into the NcoI/XhoI site of the pET 41a vector to contain a GST tag at the N terminus of the protein. The full-length MCM3 PCR product was cloned into the pET 33b vector, and the full-length MCM5 PCR product was cloned into the NdeI/BglI sites of the pET 33b vector to contain a PKA tag at the N terminus of the protein. MCM2 was likewise cloned into the pET41a vector to obtain GST-tagged Mcm2. The purification of Mcm2-7 proteins has been described in detail elsewhere (5, 31). PKA-Cdc45, PKA-GINS, Sld2T84D, GST·Mcm2-7, and PKA·Mcm2-7 were purified as described previously (26, 30).
Kinase Labeling of Proteins
Kinase labeling was performed as described previously (30). Proteins with a PKA tag at the N terminus (Dpb11, Cdc45, Mcm2-7 (Mcm3), GINS (Psf1), Mcm3, Mcm5, and Sld2T84D) were labeled in a reaction volume of 100 μl that contained 3 μm of PKA-tagged protein with 0.017 mg/ml PKA and ATP in kinase reaction buffer (25 mm Tris-HCl (pH 7.5), 5 mm DTT, and 50 mm magnesium chloride). The reaction was incubated at 30 °C for 1 h and stopped with the addition of 2 μl of 0.5 m EDTA. The kinase was removed from the mixture by affinity purification before the pulldown assay was performed.
Radiolabeling DNA
ssDNA was end-labeled with T4 polynucleotide kinase (New England Biolabs), and radiolabeled ssDNA was purified over G-25 Sephadex columns for radiolabeled DNA purification (Roche). To make dsDNA substrates with radiolabeled ssDNA, 4 μl of 500 nm radiolabeled ssDNA was incubated with 4 μl of 500 nm complementary DNA in 4 μl of reaction buffer (20 mm Tris-HCl, 4% glycerol, 0.1 mm EDTA, 40 μg/ml BSA, 5 mm DTT, and 10 mm magnesium acetate) in a final volume of 12 μl. The reaction was incubated overnight at 37 °C. The reaction was then diluted with 20 mm Tris-HCl, 0.1 mm EDTA to a final volume of 40 nm radiolabeled DNA.
GST Pulldown Assays
The GST pulldown assays were performed as described previously (30). GST-tagged protein attached to prepared glutathione-Sepharose was incubated with varying concentrations of radiolabeled protein or DNA (as described in each figure) in GST binding buffer (40 mm Tris-HCl (pH 7.5), 100 mm NaCl, 0.1 mm EDTA, 10% glycerol, 0.1% Triton X-100, 1 mm DTT, 0.7 μg/ml pepstatin, 0.1 mm PMSF, and 0.1 mg/ml BSA) in a final reaction volume of 100 μl. The reactions were incubated at 30 °C for 10 min. Following incubation, the reactions were shifted to room temperature, and glutathione-Sepharose beads were allowed to settle. The supernatant was removed, and the beads were washed twice with GST binding buffer. After the last wash, beads were heated at 90 °C for 10 min in solution containing 2% SDS, 4% glycerol, 4 mm Tris-HCl, 2 mm DTT, and 0.01% bromphenol blue. The reactions were then analyzed by SDS/PAGE followed by phosphorimaging and quantification.
Biotin Pulldown Assays
Biotinylated DNA (4 pmol) conjugated to streptavidin-agarose magnetic beads was incubated with different concentrations of radiolabeled protein in a solution containing 20 mm Tris-HCl (pH 7.5), 100 μm EDTA, 10% glycerol, 40 μg/ml BSA, 10 mm magnesium acetate, and 200 μm DTT in a final reaction volume of 25 μl. The reactions were incubated at 30 °C for 10 min. After the incubation, the magnetic beads were collected at room temperature using a magnet (Dynal). The supernatant was removed, and the beads were washed twice with a solution containing 20 mm Tris-HCl (pH 7.5), 100 μm EDTA, 10% glycerol, and 40 μg/ml BSA. After the second wash, the beads were collected and heated at 90 °C for 10 min in a solution containing 2% SDS, 4% glycerol, 4 mm Tris-HCl, 2 mm DTT, and 0.01% bromphenol blue. The reactions were analyzed by SDS/PAGE. The gel was dried at 80 °C for 1 h and then exposed to a phosphorimaging screen for 30 min.
Yeast Strains and Plasmids
Degron strain dpb11-td (YJT70, MAT a ade2-1 ura3-1 his 3–11, 15 trp 1–1 leu2–3, 112 can 1–100 dpb11-td (DPB11 5′ upstream −100 to −1 is replaced with kanMX-tTA tetR-VP16-tetO2-Ub-DHFRts-HA-linker) UBR1::GAL-Ubiquitin-M-lacI fragment-Myc-UBR1 (HIS3) leu2–3,112::pCM244 (tetR′-SSN6, LEU2)) was a gift from John F. X. Diffley (London Research Institute, Cancer Research UK, London, UK (4)). The degron strain dpb11-td was transformed with a PRS416 vector containing an empty vector, DPB11 wild type, or dpb11-m1,m2,m3,m5 (a BRCT4 motif mutant that remains bound to Mcm2-7 in the presence of ssDNA) under the control of the native DPB11 promoter. Positive transformants were selected on CSM-Ura plates.
Serial Dilution Analysis
Serial dilution was performed as described previously (28). Yeast strains in overnight culture (CSM-Ura containing raffinose, 30 °C) were transferred into YPGal media containing 50 μg/ml doxycycline and incubated for 2 h at 37 °C. The 10-fold serial dilution was performed and spotted onto a plate containing CSM-Ura, which was incubated at 30 °C (permissive conditions) and a plate containing CSM-Ura + Gal + 50 μg of doxycycline, which was incubated at 37 °C (restrictive conditions) for 2 days.
FACS
FACS was performed as described previously (28). The strains were grown overnight in CSM-Ura media containing raffinose at 30 °C. For G1 arrest, 6 × 106 cells/ml were treated with α-factor (Zymo Research) for 3 h at 37 °C in YPGal media containing 50 μg/ml doxycycline. Following extensive washes and addition of 50 μg/ml Pronase (Calbiochem) to fresh YPGal + doxycycline, cells were further incubated at 37 °C. Cells were collected at the indicated time intervals and stained with propidium iodide. Cell cycle progression data were obtained using the BD FACSCanto Ruo special order system and analyzed using FACS Diva software.
Antibodies
Antibody directed against RPA was purchased (RPA-Pierce MA1-25889). Antibodies against Cdc45, GINS, Sld2, Dpb11, and Mcm2 were generated and purified as described previously (17, 28, 32, 33).
ChIP
For G1 arrest and release, 6 × 106 cells/ml were treated with α-factor (Zymo Research) for 3 h at 37 °C in YPGal media containing 50 μg/ml doxycycline. Following extensive washes and addition of 50 μg/ml Pronase (Calbiochem) to fresh YPGal + doxycycline, cells were further incubated at 37 °C for the indicated time. Chromatin immunoprecipitation was performed as described previously (32). We performed PCR with [32P-α]dCTP as a component of the PCR reaction to quantify the amplified product. Formaldehyde cross-linked cells were lysed with glass beads in a bead beater. DNA was fragmented by sonication (Branson 450). RPA antibody and magnetic protein A beads (Dynabeads protein A, Invitrogen, catalog no. 100.02D) were added to the cleared lysate to immunoprecipitate the DNA. Immunoprecipitates were washed extensively to remove nonspecific DNA. Eluted DNA was then subjected to PCR analysis using primers directed against ARS306 or a site midway between ARS306 and ARS305, as described previously (34). The radioactive band in the native gel, representing specific PCR-amplified DNA product, was quantified by phosphorimaging and normalized by a reference standard run in the same gel. The reference standard was a PCR reaction with a known quantity of template DNA replacing the immunoprecipitate.
Coimmunoprecipitation
For G1 arrest and release, 6 × 106 cells/ml were treated with α-factor (Zymo Research) for 3 h at 37 °C in YPGal media containing 50 μg/ml doxycycline. Following extensive washes and addition of 50 μg/ml Pronase (Calbiochem) to fresh YPGal + doxycycline, cells were further incubated at 37 °C for the indicated time. Coimmunoprecipitation was performed as described previously (33). Cells were collected and lysed at 4 °C with glass beads in IP buffer (100 mm HEPES-KOH (pH 7.9), 100 mm potassium acetate, 10 mm magnesium acetate, 2 mm sodium fluoride, 1 mm PMSF, 0.1 mm Na3VO4, 20 mm β-glycerophosphate, 1% Triton X-100, leupeptin, pepstatin, and 1× complete protease inhibitor mixture without EDTA (Roche)). Lysed material was treated with 200 units of Benzonase nuclease (Novagen, catalog no. 70746-3) at 4 °C for 1 h. Clarified extract was then mixed with 2 μl of specified antibody and rotated for 2 h at 4 °C. Following this, 7 μl of Dynabeads protein A (Invitrogen, catalog no. 100.01D) beads equilibrated with IP buffer was added to the extract and further rotated for 1 h at 4 °C. Beads were washed twice with 500 μl of IP buffer and finally resuspended in SDS sample buffer. Western blot analysis was performed, and blots were scanned using the LI-COR Odyssey infrared imager and analyzed in Image Studio 4.0 software.
RESULTS
Dpb11 Binds to Mcm2-7
Dpb11 is required for the initiation of DNA replication, and Dpb11 acts at a replication origin. Activation of the helicase is a critical step in the initiation process, and we investigated whether Dpb11 binds directly to the Mcm2-7 complex. We purified GST·Mcm2-7 by incubating GST-Mcm3 with native versions of the other five Mcm proteins and then assembled and purified the GST·Mcm2-7 complex by established methods (5, 31). We also purified Dpb11 with an N-terminal protein kinase A tag (amino acids LLRASV), followed by radiolabeling with protein kinase A and [γ-32P]ATP. The PKA tag is not physiologic. It is a tool to radiolabel the Dpb11 protein with 32P.
30 pmol of GST·Mcm2-7 or GST was incubated with varying amounts of radiolabeled PKA-Dpb11 (Fig. 1A). The reaction was incubated with glutathione-Sepharose, and the material bound to beads was analyzed by SDS/PAGE. We found that roughly one-sixth of the input Dpb11 was bound to GST·Mcm2-7, more than five times the signal for GST alone. These data suggest that Dpb11 binds directly to the Mcm2-7 complex. We postulated that the Dpb11 interaction may be important for assembly of the CMG complex because Dpb11 initiates DNA replication initiation and binds directly to Mcm2-7. Cdc45 binds to Mcm2 and GINS to Mcm3 and Mcm5 (Fig. 1B) (15). Because Mcm2, Mcm3, and Mcm5 are adjacent to each other on the Mcm2-7 complex (Fig. 1B) (5), we focused on this region of Mcm2-7 for further analysis.
We next investigated whether Mcm2, Mcm3, or Mcm5 alone bind directly to Dpb11. We cloned, overexpressed, and purified GST-Mcm2, GST-Mcm3, or GST-Mcm5. We then incubated each purified subunit individually with radiolabeled PKA-Dpb11. We quantified the Dpb11 bound by SDS/PAGE and found that GST-Mcm3 binds to the greatest fraction of input Dpb11, followed by Mcm5 (Fig. 1C). Binding to Mcm2 was equivalent to the GST background.
The BRCT4 Motif of Dpb11 Binds to Mcm3 or Mcm5
We wanted to determine which domain of Dpb11 binds to Mcm3 or Mcm5. Dpb11 has four BRCT motifs, BRCT1–4 (Fig. 1D). The C-terminal region is predicted to be unstructured, and it is not required for cell growth under normal conditions. We incubated GST-Mcm3 with full-length and three different fragments of Dpb11 (253–615, 1–253, and 616–764) (Fig. 1E). GST-Mcm3 pulled down similar levels of full-length Dpb11 and Dpb11·253–615, whereas GST-Mcm3 pulled down a substantially smaller fraction of Dpb11–1-253 or Dpb11·616–764. (Fig. 1E). These data suggest that Mcm3 binds to the region of Dpb11 encompassing amino acids 253–615. We next performed a similar analysis with GST-Mcm5 and fragments of Dpb11 (Fig. 1F). Similar to GST-Mcm3, GST-Mcm5 also pulled down similar levels of full-length Dpb11 and Dpb11·253–615. We then subdivided Dpb11·253–615 into two smaller fragments, 253–430 and 430–615. Both GST-Mcm3 and GST-Mcm5 pulled down Dpb11·253–430 equivalent to the GST background. However, binding of GST-Mcm3 and GST-Mcm5 to Dpb11·430–615 was significantly higher compared with the GST background, suggesting that the region 430–615 of Dpb11 binds to both Mcm3 and Mcm5 (Fig. 1, G and H). This region (amino acids 430–615) of Dpb11 encompasses BRCT domain 4 (Fig. 1D).
Dpb11 Binds to ssDNA
Because Dpb11 acts at a replication origin, we next investigated whether Dpb11 can bind directly to origin DNA. We incubated GST-Dpb11 with a region from ARS1, ARS1–3, which encompasses the B2 and B3 elements of the replication origin (30) and is 80 nucleotides in length. The sequence of ARS1–3 and its complement, ARS1–4, are shown in Fig. 2A. GST-Dpb11 pulled down a substantially greater fraction of single-stranded ARS1–3 compared with the double-stranded ARS1–3/ARS1–4 (Fig. 2B). We then performed the reverse reaction by incubating biotinylated DNA with 32P-labeled Dpb11 and it pulling down with streptavidin beads (Fig. 2C). Again, we found that single-stranded ARS1–3 pulled down a greater fraction of Dpb11 compared with the double-stranded ARS1–3/ARS1–4 (Fig. 2C). We then performed the same experiment with a sequence from ARS305, ARS305–1, encompassing the A and B1 elements of the replication origin (sequence shown in Fig. 2A) and found that single-stranded ARS305–1 pulls down a greater fraction of Dpb11 compared with the double-stranded ARS305–1/ARS305–2 (Fig. 2D). These data suggest that Dpb11 binds directly to ssDNA containing an origin sequence.
FIGURE 2.
Dpb11 binds to T-rich ssDNA of 40 nucleotides. A, oligonucleotide sequences used. B, purified GST-Dpb11 or GST were studied for interaction with radiolabeled single stranded (ss) or double stranded (ds) sequences from the ARS1 origin in budding yeast. 13 pmol of GST-Dpb11 was incubated with DNA and glutathione-Sepharose for 10 min at 30 °C. The glutathione-Sepharose beads were washed and analyzed as described under “Experimental Procedures.” The results from the experiment were quantified, averaged, and plotted as picomoles of DNA bound versus picomoles of input DNA. C and D, PKA-Dpb11 was radiolabeled and used in a biotin pulldown assay. Single-stranded or double-stranded DNA from yeast origins (ARS1 or ARS305) with a 3′ biotin tag was conjugated to magnetic streptavidin beads. 4 pmol of this biotinylated DNA was incubated with radiolabeled PKA-Dpb11 at 30 °C for 10 min. As a control, beads without DNA were incubated with PKA-Dpb11. The beads were washed and analyzed as described under “Experimental Procedures.” The results from the experiment were quantified, averaged, and plotted as picomoles of Dpb11 bound versus the picomoles of input Dpb11. E, GST Dpb11 or GST was studied for interaction with radiolabeled DNA (ssARS1–3 or modifications of ssARS1–3) as indicated in the graph. 13 pmol of GST Dpb11 or GST was incubated with glutathione-Sepharose and varying amounts of DNA at 30 °C for 10 min. The glutathione-Sepharose beads were washed and analyzed as described under “Experimental Procedures.” The results from the experiment were quantified, averaged, and plotted as picomoles of DNA bound versus picomoles of input DNA. F, PKA-Dpb11 was radiolabeled and used in a biotin pulldown assay. Single-stranded DNA of varying lengths (ss80dT, ss60dT, ss40dT, and ss20dT) with a 3′ biotin tag was conjugated to magnetic streptavidin beads. 4 pmol of this biotinylated DNA was incubated with radiolabeled PKA-Dpb11 at 30 °C for 10 min. As a control, beads without DNA were incubated with PKA-Dpb11. The beads were washed and analyzed as described under “Experimental Procedures.” The results from the experiment were quantified, averaged, and plotted as picomoles of Dpb11 bound versus picomoles of input Dpb11. G, similar to F, except matching nucleotides instead of matching picomoles of DNA. Therefore, 4 pmol of ss80dT, 5.3 pmol of ss60 dT, 8 pmol of ss40dT, or 16 pmol of ss20dT were used in the pulldowns.
To determine the sequence specificity for Dpb11-ssDNA interaction, we incubated GST-Dpb11 with radiolabeled ARS1–3 with the following nucleotide substitutions: T to A, C to A, A to C, or G to C. The sequences are shown in Fig. 2A. Substitution of ARS1–3 with T to A resulted in a substantial decrease in ssDNA binding (Fig. 2E). Substitution of C to A resulted in a modest decrease in ssDNA binding. In contrast, the A to C and G to C substitutions had only a slight effect on ssDNA binding. Dpb11 may be recognizing the thymine bases or simply the more flexible structure of a pyrimidine-rich sequence. In future studies we will investigate the nature of Dpb11-ssDNA sequence dependence more extensively.
Because we found that Dpb11 binds thymine-rich ssDNA, we constructed an artificial 3′-biotinylated 80dT sequence and incubated it with PKA-Dpb11. We then performed a biotin pulldown assay and found that 80dT bound to Dpb11 (Fig. 2F). We then incubated different lengths of ssdT with PKA-Dpb11 and found that, when we matched for oligomer concentration, the 80dT pulled down the greatest fraction of Dpb11 (Fig. 2F). We then repeated the experiment of Fig. 2F but matched for nucleotide concentration (Fig. 2G). When matched for nucleotide concentration, 40dT, 60dT, and 80dT each bound Dpb11, but 20dT binding was close to the background. These data suggest that Dpb11 binds to 21–40 nucleotides of ssdT.
Regions of Dpb11 That Bind to Single-stranded DNA
To determine the regions of Dpb11 that bind ssDNA, we first incubated biotinylated ssARS1–3 with full-length PKA-Dpb11 and two different fragments of Dpb11 (1–253 and 253–764) (Fig. 3A). Single-stranded ARS1–3 pulled down similar levels of full-length PKA-Dpb11 and PKA-Dpb11 (253–764) (Fig. 3A). We then subdivided Dpb11 (253–764) into three fragments (Dpb11 253–430 encompassing the BRCT3 region of Dpb11, Dpb11·430–615 encompassing the BRCT4 region of Dpb11, and Dpb11·616–764, which includes the unstructured C-terminal region of Dpb11), and incubated them with biotinylated ssARS1–3. PKA-Dpb11·253–430 bound to biotinylated ssARS1–3 with an affinity similar to that of mock biotin beads without any DNA sequence (Fig. 3B). However, PKA-Dpb11·430 to 615 and PKA-Dpb11·616–764 each bound to biotinylated ssARS1–3 with an affinity higher than the mock biotin beads without DNA (Fig. 3, C and D). These data suggests that the region encompassing BRCT4 and also the unstructured C-terminal region of Dpb11 bind to ssDNA.
FIGURE 3.

The BRCT 4 motif or the C-terminal region of Dpb11 binds to ssDNA. A–D, to determine the region of Dpb11 involved in the Dpb11-ssDNA interaction, single-stranded DNA from yeast origins (ARS1–3) with a 3′ biotin tag was conjugated to magnetic streptavidin beads. 4 pmol of this biotinylated DNA was incubated with the different fragments of radiolabeled PKA-Dpb11 (as indicated) at 30 °C for 10 min. As a control, beads without DNA were incubated with radiolabeled PKA-Dpb11 fragments. The beads were washed and analyzed as described under “Experimental Procedures.” The results from the experiment were quantified, averaged, and plotted as picomoles of Dpb11 bound versus picomoles of input Dpb11. FL, full-length.
Dpb11 Substantially Inhibits GINS Binding to Mcm2-7
Dpb11 binds to the Mcm3 and Mcm5 subunits of Mcm2-7 (Fig. 1), and the Mcm3 and Mcm5 subunits also bind to GINS (18). Therefore, we next investigated whether Dpb11 competes with GINS for binding to Mcm2-7. We incubated radiolabeled PKA-GINS with GST·Mcm2-7 and found a direct interaction between GINS and Mcm2-7 (Fig. 4A). We next added unlabeled Dpb11 to the reaction and found that the addition of Dpb11 substantially inhibited the interaction between GINS and Mcm2-7. These data suggest that Dpb11 competes with GINS for binding to Mcm2-7 (see Fig. 4A for illustration).
FIGURE 4.

Dpb11 competes with GINS for binding to Mcm2-7, and single-stranded DNA inhibits Dpb11 interaction with Mcm2-7, allowing GINS to bind Mcm2-7. A, 30 pmol of GST·Mcm2-7 (contains GST at the N terminus of Mcm3) was incubated with 30 pmol of radiolabeled PKA GINS and different amounts of Dpb11 in a GST pulldown assay. The beads were washed and analyzed as described under “Experimental Procedures.” The results from similar experiments were quantified, averaged, and plotted as picomoles of GINS bound versus picomoles of Dpb11 added. A schematic showing the competition between Dpb11 and GINS for Mcm2-7 binding is also shown. B, 30 pmol of GST Dpb11 was incubated with 30 pmol of radiolabeled PKA Mcm2-7 and varying amounts of ss40dT in a GST pulldown assay. The beads were washed and analyzed as described under “Experimental Procedures.” Results from similar experiments were quantified, averaged, and plotted as picomoles of Mcm2-7 bound versus the molar ratio of ss40dT/GST Dpb11. A model representing the inhibition of the Dpb11-Mcm2-7 interaction by single-stranded DNA is also shown. C, 30 pmol of GST·Mcm2-7 (contains GST at the N terminus of Mcm3) was incubated with 30 pmol of radiolabeled PKA GINS and different amounts of Dpb11 (0 and 10 pmol) at 30 °C for 10 min in a GST pulldown assay. Following this, 30 pmol of ss40dT was added to the reaction containing Mcm2-7, GINS, and Dpb11 and further incubated at 30 °C for 10 min. The beads were washed and analyzed as described under “Experimental Procedures.” The bound radioactive GINS was analyzed by SDS/PAGE followed by phosphorimaging. These results were then quantified, averaged, and plotted as picomoles of GINS bound versus picomoles of Dpb11 and picomoles of ss40dT added. A model representing the release of Dpb11 from Mcm2-7 upon addition of ssDNA and allowing GINS to bind Mcm2-7 is also shown.
Single-stranded DNA Competes with Dpb11 for Binding to Mcm2-7
Because the BRCT4 motif of Dpb11 binds to ssDNA and also to Mcm3 and Mcm5, we next investigated whether single-stranded DNA can displace Dpb11 from Mcm2-7 (Fig. 4B). We incubated GST-Dpb11 with radiolabeled PKA-Mcm2-7 and found a direct interaction between Dpb11 and Mcm2-7, as expected. Next, we added different concentrations of ss40dT and found that ss40dT disrupted the interaction between Dpb11 and Mcm2-7 (Fig. 4B). Similar results were found using the origin sequence ARS1–3 (data not shown). These data suggest that ssDNA specifically disrupts the interaction between Dpb11 and Mcm2-7 (see Fig. 4B for illustration).
Single-stranded DNA Releases Dpb11 from Mcm2-7, Allowing GINS to Bind Mcm2-7
Because Dpb11 inhibits GINS binding to Mcm2-7 and ssDNA disrupts the interaction between Dpb11 and Mcm2-7, we next investigated whether the addition of ssDNA to a reaction with Dpb11, GINS, and Mcm2-7 can rescue the binding of GINS to Mcm2-7. We incubated radiolabeled PKA-GINS with GST Mcm2-7 and found a direct interaction between GINS and Mcm2-7 (Fig. 4C). Next we added unlabeled Dpb11 to the reaction and found that the addition of Dpb11 inhibited the interaction between GINS and Mcm2-7 (Fig. 4C). To this, we further added ss40dT and found a substantial increase in the binding of GINS to Mcm2-7 (Fig. 4C). These data suggest that ssDNA releases Dpb11 from Mcm2-7, allowing GINS to bind Mcm2-7 (see Fig. 4C for illustration).
Dpb11 Binds Directly to Cdc45
Dpb11 binds to Mcm5, which is adjacent to Mcm2 in the Mcm2-7 ring (Fig. 1B). Furthermore, Cdc45 binds to Mcm2 (Fig. 1B). Therefore, Dpb11 is positioned to recruit Cdc45 to the Mcm2-7 ring. To investigate whether Dpb11 recruits Cdc45 to Mcm2-7, we first determined whether Dpb11 binds directly to Cdc45 (Fig. 5A). GST-Dpb11 was incubated with radiolabeled PKA-Cdc45, and a direct interaction was observed by a pulldown assay (Fig. 5A). To test whether Dpb11 can recruit Cdc45 to Mcm2-7, we next incubated GST-Cdc45 with radiolabeled PKA-Mcm2-7 and increasing concentrations of radiolabeled PKA-Dpb11 (Fig. 5B). We observed a direct, weak interaction between Cdc45 and Mcm2-7, as expected, but as Dpb11 was added in increasing concentrations, the amount of Cdc45 bound to Mcm2-7 increased substantially (Fig. 5B). Furthermore, because both Dpb11 and Mcm2-7 were radiolabeled in this reaction, we observed a simultaneous increase in binding of Dpb11 and Mcm2-7 to Cdc45 (Fig. 5B). These data suggest that Dpb11 can recruit Cdc45 to Mcm2-7 (see Fig. 5B for illustration).
FIGURE 5.
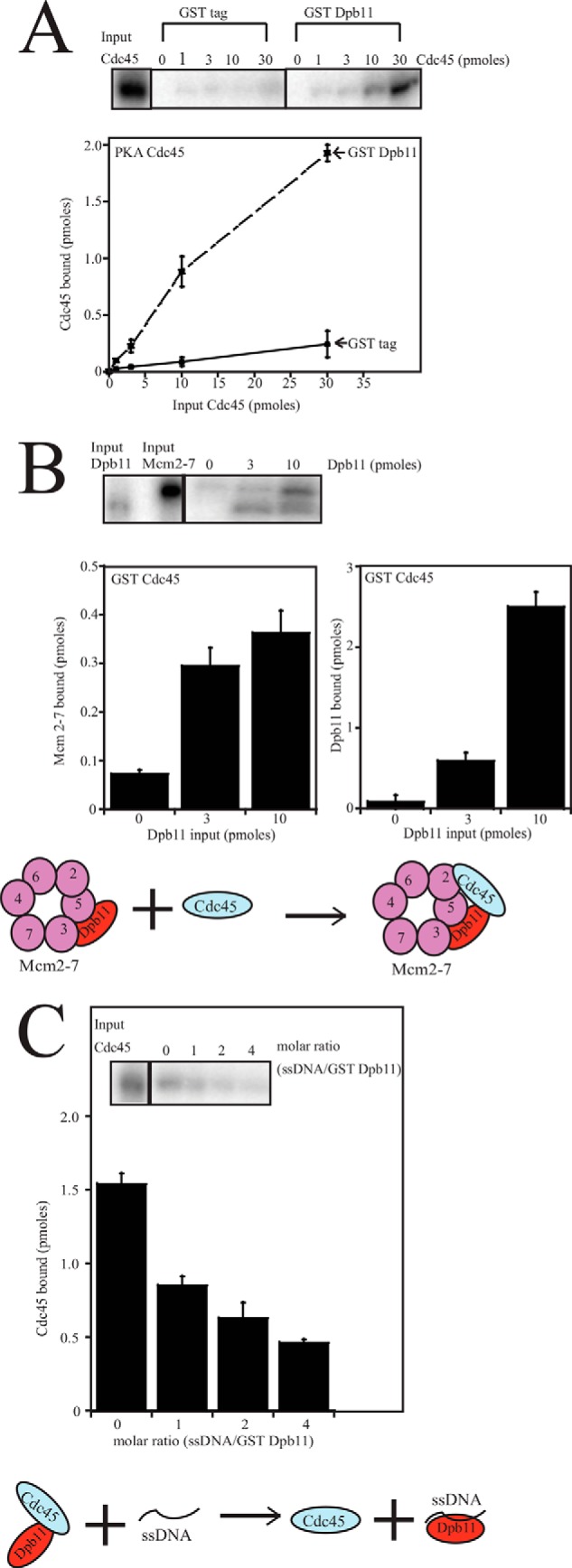
Dpb11 binds directly to Cdc45, Dpb11 recruits Cdc45 to the Mcm2-7 complex, and single-stranded DNA disrupts the interaction between Cdc45 and Mcm2-7. A, 30 pmol of GST tag or GST Dpb11 was incubated with glutathione-Sepharose and varying amounts of radiolabeled PKA-Cdc45 in a GST pulldown assay. The beads were washed and analyzed as described under “Experimental Procedures.” Results from similar experiments were quantified, averaged, and plotted as picomoles of Cdc45 bound versus picomoles of Cdc45 input. B, 10 pmol of GST-Cdc45 was incubated with 10 pmol of radiolabeled PKA·Mcm2-7 and varying amounts of radiolabeled PKA-Dpb11 in a GST pulldown assay. Results from similar experiments were quantified, averaged, and plotted as picomoles of Mcm2-7 bound versus picomoles of Dpb11 input and picomoles of Dpb11 bound versus picomoles of Dpb11 input. A model representing the recruitment of Cdc45 to Mcm2-7 with the help of Dpb11 is also shown. C, 30 pmol of GST Dpb11 was incubated with 30 pmol of radiolabeled PKA-Cdc45 and varying amounts of ss40dT in a GST pulldown assay. The beads were washed and analyzed as described under “Experimental Procedures.” Results from similar experiments were quantified, averaged, and plotted as picomoles of Cdc45 bound versus the molar ratio of ss40dT/GST-Dpb11. A model showing inhibition of the Dpb11-Cdc45 interaction by single-stranded DNA is also shown.
ssDNA Competes with Cdc45 for Dpb11 Interaction
Cdc45 and Dpb11 interaction must be transient because Cdc45 travels with the replication fork and Dpb11 does not (27). Therefore, some factor must dislodge Cdc45 from Dpb11. To determine whether single-stranded DNA disrupts Cdc45-Dpb11 interaction, we incubated GST-Dpb11 with radiolabeled PKA-Cdc45 and increasing concentrations of ss40dT. We found that ssDNA disrupts the interaction between Cdc45 and Dpb11 (Fig. 5C), suggesting a mechanism for dissociation in vivo (see Fig. 5C for illustration). Similar results were found using ssARS1–3 (data not shown).
Identification of the Dpb11 Mutant That Is Defective in ssDNA Binding
To study the effect of Dpb11-ssDNA binding in vivo, we identified mutations that disrupt Dpb11-ssDNA binding. Because we observed that the Dpb11·430–615 fragment bound to ssARS1–3, we performed a fungal alignment and found that there are eight conserved, positively charged residues in region 430–615 of Dpb11 (data not shown). Anticipating that a charge reversal mutation of a positively charged residue may inhibit interaction with the negatively charged ssDNA, we engineered six full-length Dpb11 mutants (m1, m2, m3, m4, m5, and m6) that contained one or more charge reversal mutations (Fig. 6A). We incubated biotinylated ssARS1–3 with the different mutants of Dpb11 and found that mutations in m1, m2, m3, and m5 partially decreased the interaction between Dpb11 and ssARS1–3 DNA (Fig. 6B). In contrast, incubation with dpb11-m4 or dpb11-m6 resulted in similar pulldown efficiency compared with wild-type Dpb11 (data not shown). To find a Dpb11 mutant that is defective in ssDNA binding, we combined mutations m1, m2, m3, and m5 to form a quadruple mutant. In addition, we also deleted the unstructured C-terminal region of Dpb11 (Dpb11·616–764) because we observed that Dpb11·616–764 also binds to ssDNA. This Dpb11 mutant (dpb11-m1,m2,m3,m5,ΔCTD, quadruple mutant plus C-terminal deletion) is defective in binding to ssDNA compared with the wild-type Dpb11, as observed in a biotin pulldown assay using biotinylated ssARS1–3 with radiolabeled Dpb11 (Fig. 6, C and D). However, the Dpb11 mutant (dpb11-m1,m2,m3,m5,ΔCTD, quadruple mutant plus C-terminal deletion) binds to Sld2T84D, Mcm2-7 and Cdc45 like the wild type full-length Dpb11 (Fig. 6, E–G).
FIGURE 6.
A Dpb11 mutant (Dpb11-m1,m2,m3,m5,ΔCTD) does not bind ssDNA but it binds Sld2T84D, Mcm2-7, and Cdc45 like wild type Dpb11. A, charge reversal mutations in the BRCT 4 region of Dpb11 (amino acids 430–615) that were studied for interaction with ssARS1–3. B, various concentrations of radiolabeled full-length PKA·Dpb11-wild-type, dpb11-m1, dpb11-m2, dpb11-m3, dpb11-m4 (not shown, binds like Dpb11·wild-type), dpb11-m5, or dpb11-m6 (not shown, binds like Dpb11·wild-type) were incubated with 4 pmol of ssARS1–3 in a biotin pulldown assay. The beads were washed and analyzed as described under “Experimental Procedures.” The results from the experiment were quantified, averaged, and plotted as picomoles of Dpb11 bound versus picomoles of input Dpb11. C, different concentrations of PKA-Dpb11 and PKA·dpb11-m1,m2,m3,m5,ΔCTD were incubated with 4 pmol of ssARS1–3 in a biotin pulldown assay. The beads were washed and analyzed as described under “Experimental Procedures.” D, results from experiments similar to C were quantified, averaged, and plotted as picomoles of Dpb11 bound versus picomoles of Dpb11 input. E, different concentrations of radiolabeled PKA-Sld2T84D were incubated with 30 pmol of GST-Dpb11 wild-type, GST·dpb11-m1,m2,m3,m5,ΔCTD, or GST tag alone in a GST pulldown assay as described under “Experimental Procedures.” The bound radioactive Sld2T84D was analyzed by SDS/PAGE followed by phosphorimaging. These results were then quantified, averaged, and plotted as picomoles of Sld2T84D bound versus picomoles of Sld2T84D input. F, different concentrations of radiolabeled PKA·Mcm2-7 was incubated with 10 pmol of GST-Dpb11 wild-type, GST·dpb11-m1,m2,m3,m5,ΔCTD, or GST tag alone in a GST pulldown assay as described under “Experimental Procedures.” The bound radioactive Mcm2-7 was analyzed by SDS/PAGE followed by phosphorimaging. These results were then quantified, averaged, and plotted as picomoles of Mcm2-7 bound versus picomoles of Mcm2-7 input. G, different concentrations of radiolabeled PKA-Cdc45 was incubated with 10 pmol of GST-Dpb11 wild-type, GST·dpb11-m1,m2,m3,m5,ΔCTD, or GST tag alone in a GST pulldown assay as described under “Experimental Procedures.” The bound radioactive Cdc45 was analyzed by SDS/PAGE followed by phosphorimaging. These results were then quantified, averaged, and plotted as picomoles of Cdc45 bound versus picomoles of Cdc45 input.
Expression of a BRCT4 Motif Mutant of Dpb11 That Remains Bound to Mcm2-7 in the Presence of ssDNA Exhibits Severely Inhibited Cell Growth and DNA Replication
Because the BRCT4 motif of Dpb11 binds to ssDNA and also to Mcm3 and Mcm5, we hypothesized that this BRCT 4 region of Dpb11 may be responsible for mediating competition between ssDNA and Dpb11 for Mcm2-7. To investigate whether the ssDNA binding residues of the BRCT4 motif are regulating Dpb11-Mcm2-7 interaction, we constructed a mutant with charge reversal mutations of the BRCT4 motif that are specifically defective in ssDNA binding (dpb11-m1,m2,m3,m5). This mutant is not completely defective in ssDNA binding because the C-terminal region of Dpb11 also binds to ssDNA (data not shown). However, this mutant (dpb11-m1,m2,m3,m5) binds to Mcm2-7, Mcm3, and Mcm5 like wild-type Dpb11 (Figs. 7, A–C). Furthermore, unlike wild-type Dpb11, mutant Dpb11 (dpb11-m1,m2,m3,m5) remains bound to Mcm2-7 in the presence of single-stranded DNA (Fig. 7D). In addition, mutant Dpb11 (dpb11-m1,m2,m3,m5) also binds to Sld2T84D and Cdc45 like the wild type full-length Dpb11 (Fig. 7, E and F), suggesting that the observed defect is specific.
FIGURE 7.
A BRCT4 motif mutant of Dpb11 (dpb11-m1,m2,m3,m5) remains bound to Mcm2-7 in the presence of single-stranded DNA. A, different concentrations of radiolabeled PKA·Mcm2-7 were incubated with 10 pmol of GST-Dpb11 wild-type, GST·dpb11-m1,m2,m3,m5, or GST tag alone in a GST pulldown assay as described under “Experimental Procedures.” The bound radioactive Mcm2-7 was analyzed by SDS/PAGE followed by phosphorimaging. These results were then quantified, averaged, and plotted as picomoles of Mcm2-7 bound versus picomoles of Mcm2-7 input. B, different concentrations of radiolabeled PKA-Mcm3 was incubated with 10 pmol of GST-Dpb11 wild-type, GST·dpb11-m1,m2,m3,m5, or GST tag alone in a GST pulldown assay as described under “Experimental Procedures.” The bound radioactive Mcm3 was analyzed by SDS/PAGE followed by phosphorimaging. These results were then quantified, averaged, and plotted as picomoles of Mcm3 bound versus picomoles of Mcm3 input. C, different concentrations of radiolabeled PKA-Mcm5 were incubated with 10 pmol of GST-Dpb11 wild-type, GST·dpb11-m1,m2,m3,m5, or GST tag alone in a GST pulldown assay as described under “Experimental Procedures.” The bound radioactive Mcm5 was analyzed by SDS/PAGE followed by phosphorimaging. These results were then quantified, averaged, and plotted as picomoles of Mcm5 bound versus picomoles of Mcm5 input. D, 30 pmol of GST·dpb11-m1,m2,m3,m5 was incubated with 30 pmol of radiolabeled PKA·Mcm2-7 and varying amounts ss40dT in a GST pulldown assay. The beads were washed and analyzed as described under “Experimental Procedures.” Results from similar experiments were quantified, averaged, and plotted as picomoles of Mcm2-7 bound versus the molar ratio of ss40dT/GST·dpb11-m1,m2,m3,m5. E, different concentrations of radiolabeled PKA-Sld2T84D were incubated with 30 pmol of GST-Dpb11 wild-type, GST·dpb11-m1,m2,m3,m5, or GST tag alone in a GST pulldown assay as described under “Experimental Procedures.” The bound radioactive Sld2T84D was analyzed by SDS/PAGE followed by phosphorimaging. These results were then quantified, averaged, and plotted as picomoles of Sld2T84D bound versus picomoles of Sld2T84D input. F, different concentrations of radiolabeled PKA-Cdc45 was incubated with 10 pmol of GST-Dpb11 wild-type, GST· dpb11-m1,m2,m3,m5, or GST tag alone in a GST pulldown assay as described under “Experimental Procedures.” The bound radioactive Cdc45 was analyzed by SDS/PAGE followed by phosphorimaging. These results were then quantified, averaged, and plotted as picomoles of Cdc45 bound versus picomoles of Cdc45 input.
To investigate in vivo the mutant of Dpb11 that remains bound to Mcm2-7 in the presence of ssDNA, we constructed plasmids with wild-type and mutant DPB11 (dpb11-m1,m2,m3,m5) under the control of its native promoter. These plasmids were then transformed into a temperature-sensitive Dpb11 degron yeast strain (dpb11-td, obtained from John Diffley (4)). We then studied these strains for cell growth by performing a 10-fold serial dilution and spotting onto CSM-Ura plates at 30 °C (permissive conditions) and CSM-Ura + Gal + doxycycline at 37 °C (restrictive conditions). At the permissive conditions, DPB11-WT, dpb11-m1,m2,m3,m5, and the vector-only control all grew to similar levels. However, at the restrictive conditions, there is a severe growth defect in the dpb11- m1,m2,m3,m5 and vector-only control compared with the DPB11-WT (Fig. 8A). These data suggests that the mutant Dpb11 protein cannot support yeast growth in the absence of wild-type DPB11. However, Dpb11 protein is expressed at equivalent levels in both the wild-type and mutant strains (Fig. 8B), suggesting that the mutant protein is properly transcribed and expressed.
FIGURE 8.
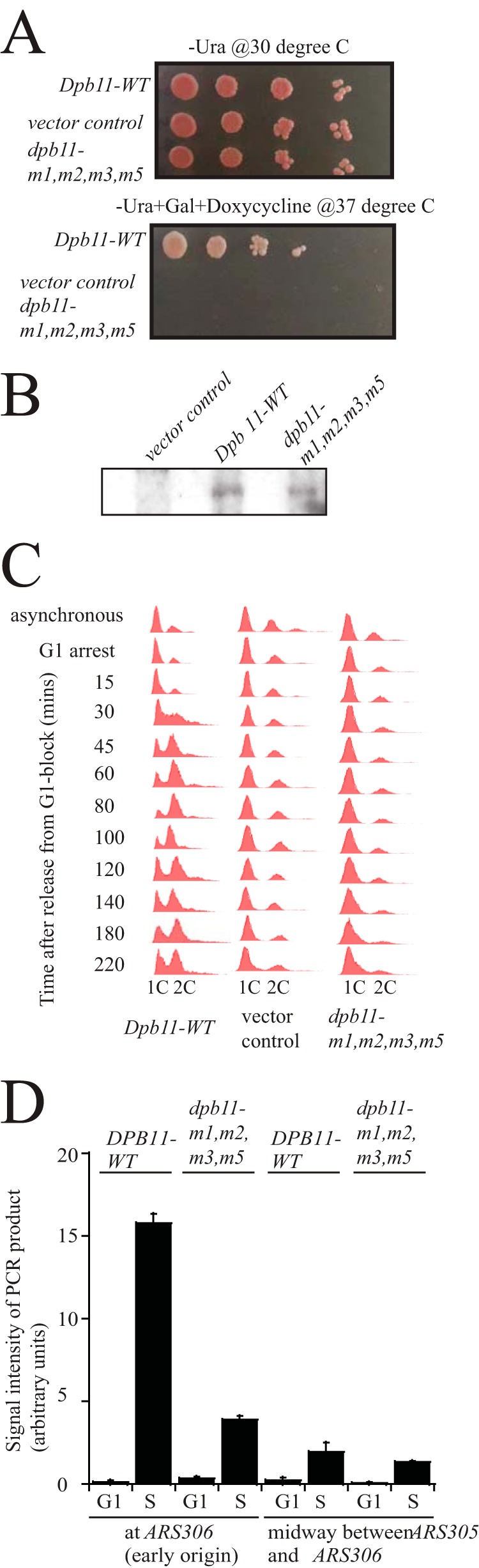
Cells expressing dpb11-m1,m2,m3,m5 exhibit a defect in cell growth and DNA replication. A, 10-fold serial dilutions of dpb11-td cells expressing DPB11-wild type, vector-only control, and dpb11-m1,m2,m3,m5 at permissive (CSM-Ura, 30 °C) or restrictive conditions (CSM-Ura + gal + doxycycline, 37 °C). B, Western blot analysis of DPB11-WT, dpb11-m1,m2,m3,m5, and vector-only control whole cell extracts showing equivalent levels of Dpb11 expression under restrictive conditions. C, FACS analysis was performed as described under “Experimental Procedures” on dpb11-td cells expressing DPB11-wild type, vector-only control, and dpb11-m1,m2,m3,m5. Cells were synchronized in G1 phase with α-factor and then released into medium lacking α-factor for the indicated time points. D, dpb11-td cells expressing DPB11-WT or dpb11-m1,m2,m3,m5 under restrictive conditions were arrested with α-factor and then released into medium lacking α-factor for 15 min. Chromatin immunoprecipitation was performed as described under “Experimental Procedures.” PCR primers were used that target the early yeast origin ARS306 or a region positioned midway between ARS305 and ARS306. [32P-α]dCTP was included in the PCR reaction for quantification. Radioactive PCR bands were quantified and plotted.
We further studied cell cycle progression into S phase in a FACS analysis experiment. We arrested cells in G1 with α-factor at the restrictive temperature (37 °C) for 3 h and then released them into growth media (YPGal + 50 μg/ml doxycycline) lacking α-factor at the restrictive temperature for 15–220 min. Cells expressing wild-type DPB11 accumulated two copies of DNA within 45 min after α-factor release. However, cells with the vector-only control and dpb11-m1,m2,m3,m5 exhibited a severe defect in progression into S phase (Fig. 8C), suggesting that dpb11-m1,m2,m3,m5 is defective in DNA replication.
Cells Expressing dpb11-m1,m2,m3,m5 Exhibit Decreased RPA Interaction with Origin DNA during S Phase
We next examined whether dpb11-m1,m2,m3,m5 is impaired in the accumulation of origin ssDNA coated by RPA, the yeast single-stranded binding protein. For this experiment, we used chromatin immunoprecipitation with antibodies directed against RPA (Fig. 8D). We then subjected the immunoprecipitate to quantitative PCR using oligonucleotides directed against the early origin ARS306 or a non-origin region located midway between the ARS305 and ARS306 origins (non-origin sequence). We arrested cells at the restrictive temperature in G1 with α-factor and then released them into medium lacking α-factor for 15 min to collect cells in S phase. No hydroxyurea was added to the medium. For wild-type cells, there is a substantial increase in origin PCR signal for S phase cells compared with G1 phase cells (Fig. 8D). There is also a slight increase in non-origin PCR signal for S phase cells compared with G1 phase cells. The increase in signal at the origin sequence may predominantly reflect the extrusion of ssDNA from the central channel of Mcm2-7 during S phase, and it may also include some signal for the initiation of DNA unwinding. For cells expressing dpb11-m1,m2,m3,m5, there is a substantially diminished signal for the origin sequence during S phase. There is also a slightly decreased signal for the non-origin sequence, consistent with the mutant slow progression through S phase. The decrease in RPA-ChIP signal at the origin sequence may reflect the importance of these BRCT4 residues (Lys-463, Lys-469, Arg-538, and Lys-549) for stabilizing the extruded strand of origin DNA. Alternatively, the decrease in RPA-ChIP signal may reflect that the mutant is defective in release of Dpb11 from Mcm2-7. A third possibility is that the decrease in signal for the mutant at an origin may reflect a decrease in unwinding during initiation.
Cells Expressing dpb11-m1,m2,m3,m5 Exhibit Decreased GINS-Mcm2-7 Interaction
We next performed co-IP analysis to study the assembly of proteins required for replication initiation in strains expressing the wild type-DPB11 and dpb11-m1,m2,m3,m5 at the restrictive conditions. Using the approach to isolate loaded Mcm2-7 complexes on chromosomal DNA as described previously (34), we studied the interaction between loaded Mcm2-7 and Dpb11 and also between Sld2 and Dpb11 (Fig. 9A). We arrested cells in G1 with α-factor at the restrictive temperature (37 °C) for 3 h and then released them into growth media (YPGal + 50 μg/ml doxycycline) lacking α-factor at the restrictive temperature for 0, 15, 30, or 45 min. We did not use any cross-linking reagent in these experiments, and no hydroxyurea was added to the medium. We made whole-cell extracts, probed them with antibodies against Mcm2 and Sld2, and found similar levels of Mcm2 and Sld2 proteins in wild-type compared with mutant cells (Fig. 9A). However, for cell extracts precipitated with antibodies against Dpb11 and then probed with Mcm2, we observed an increased Dpb11-Mcm2 interaction in dpb11-m1,m2,m3,m5 cells at the 15- and 30-min time points compared with DPB11-wild-type cells (Fig. 9A). Mcm2 precipitates as part of the loaded Mcm2-7 complex in these experiments, and the Dpb11-Mcm2 co-IP signal, therefore, reflects the interaction between Dpb11 and loaded Mcm2-7. These data suggest that the Dpb11-ssDNA interaction is required for the release of Dpb11 from loaded Mcm2-7 as cells progress into S phase. We next probed these Dpb11 immunoprecipitates with Sld2 antibody and found that there is a very weak signal at 0 min for both wild-type and mutant cells. However, at 15, 30, and 45 min, we observed a similar Dpb11-Sld2 interaction for wild-type and mutant cells. This is consistent with the CDK-dependent binding of Sld2 with Dpb11 and suggests that dpb11-m1,m2,m3,m5 binds normally with Sld2, similar to DPB11-WT (Fig. 9A).
FIGURE 9.
Cells expressing dpb11-m1,m2,m3,m5 exhibit no GINS·Mcm2-7 interaction. dpb11-td cells expressing DPB11-WT or dpb11-m1,m2,m3,m5 under restrictive conditions were arrested with α-factor and then released into medium lacking α-factor for the indicated times. A and B, left panels, whole cell extracts were analyzed by Western blot for expression of the indicated proteins. A, right panel, cell extracts were immunoprecipitated with antibodies directed against Dpb11, followed by Western analysis for Mcm2 or Sld2. B, right panel, cell extracts were prepared to isolate loaded Mcm2-7 complex as described previously (34). Antibodies directed against Mcm2 were used for immunoprecipitation, followed by Western blot analysis of Mcm2, Cdc45, or GINS.
We next probed for the interaction between loaded Mcm2-7 and Cdc45 or GINS using a similar approach (Fig. 9B). The whole cell extracts showed similar levels of Mcm2, Cdc45, or GINS proteins in cells expressing DPB11-WT or dpb11-m1,m2,m3,m5 forms of Dpb11 protein (Fig. 9B). These cell extracts were then precipitated with antibodies against Mcm2 and probed with Cdc45 or GINS antibodies. The interaction between loaded Mcm2 and Cdc45 was slightly affected in DPB11-WT and dpb11-m1,m2,m3,m5 cells (Fig. 9B), suggesting that these residues play some role in Cdc45·Mcm2-7 interaction. However, there is a substantially diminished interaction between loaded Mcm2 and GINS in dpb11-m1,m2,m3,m5 cells compared with wild-type cells, where GINS is associated with loaded Mcm2 15 and 30 min after G1 release (Fig. 9B). This suggests that release of Dpb11 from Mcm2-7 is important for GINS to bind loaded Mcm2-7 in S phase.
DISCUSSION
Summary of Results
We found, using purified proteins, that Dpb11 binds directly to Mcm2-7, Cdc45, or single-stranded DNA. Dpb11 can recruit Cdc45 to the Mcm2-7 complex, whereas Dpb11 inhibits the interaction between GINS and Mcm2-7. The recruitment of Cdc45 to Mcm2-7 may be conserved in higher organisms because it has been shown that Xmus101 may recruit Cdc45 to replication origins in Xenopus extract assays (35). Interestingly, ssDNA disrupts the interaction between Dpb11 and Mcm2-7, and ssDNA also disrupts the interaction between Dpb11 and Cdc45. Furthermore, the inhibition of GINS-Mcm2-7 interaction caused by Dpb11 can be overcome by the addition of ssDNA. Amino acids 430–615 of Dpb11 bind to Mcm3/Mcm5 and also ssDNA, suggesting a mechanism for competitive interaction between ssDNA and Mcm3/Mcm5 for binding to Dpb11. The region of amino acids 430–615 of Dpb11 is poorly conserved from yeast to humans, making it difficult to determine whether the residues involved in ssDNA binding are conserved from yeast to human. Dpb11 prefers single-stranded DNA binding to blunt duplex binding.
We also found that, when a mutant dpb11-m1,m2,m3,m5 (a BRCT4 motif mutant of Dpb11 that remains bound to Mcm2-7 in the presence of ssDNA) is expressed in yeast cells, the cells suffer severe growth and DNA replication defects in the absence of wild-type DPB11. Expression of dpb11-m1,m2,m3,m5 results in decreased RPA interaction with origin DNA compared with DPB11-WT. In addition, the dpb11-m1,m2,m3,m5 mutant exhibits a prolonged interaction between Dpb11 and Mcm2-7 after release from G1 phase. GINS also fails to associate with Mcm2-7 during S phase in the mutant cells. These data suggest that release of Dpb11 from Mcm2-7 is important for GINS-Mcm2-7 interaction in S phase.
Sld2 and Dpb11 Block the Premature Interaction between GINS and Mcm2-7 in G1 Phase
The Mcm2-7 ring cracks open in late M and G1 phase to encircle double-stranded DNA (data not shown) (10). However, the helicase is not active at this point because it is not bound to GINS in G1 phase. Sld3 binding to Mcm3/Mcm5 blocks GINS interaction with Mcm2-7 in vitro (26), Sld2 binding to Mcm3/Mcm5 blocks GINS interaction with Mcm2-7 in vitro and in vivo (28, 33), and Dpb11 interaction with Mcm3/Mcm5 blocks the interaction between GINS and Mcm2-7 in vitro (Fig. 4A) and in vivo (Fig. 9B). These data suggest that Sld3, Sld2, and Dpb11 can block the interaction between GINS and Mcm2-7 in G1 phase (Fig. 10A). Blocking premature CMG assembly may be important for two reasons. First, if GINS binds to Cdc45 prematurely, then the helicase may be prematurely activated. It is critical that helicase activation is precisely coordinated with polymerase DNA synthesis to prevent the accumulation of excess ssDNA and genome instability. Second, the CMG complex forms a sealed ring, and it cannot open to allow the extrusion of ssDNA (15, 36). Therefore, if the CMG complex were to form prior to ssDNA extrusion, the sealed CMG complex may block ssDNA extrusion from the central channel of Mcm2-7. Previous published work has demonstrated a preloading complex composed of Sld2, Dpb11, GINS, and Pol ϵ that is important for the recruitment of GINS to replication origins (37). Furthermore, Dpb11 has been shown to bind directly to GINS through an interacting region between BRCT motifs 2 and 3 (38). We are pointing out here that, although the preloading complex may be important for recruiting GINS to origins, the direct interaction between GINS and Mcm2-7 is blocked by Dpb11, Sld2, and, possibly, Sld3 (29, 33).
FIGURE 10.
Dpb11, Sld3, and Sld2 regulate the assembly of the CMG (Cdc45·Mcm2-7·GINS) helicase complex in budding yeast. A, Mcm2-7 loads as a double hexamer to encircle double-stranded DNA during late M and G1 phase. Dpb11, Sld3, or Sld2 bind to Mcm2-7, and binding of Dpb11, Sld3, or Sld2 to Mcm2-7 blocks the premature interaction between GINS and Mcm2-7 during G1 phase. Furthermore, Dpb11 or Sld3 may recruit Cdc45 to Mcm2-7, and this process may depend upon the Dbf4-dependent kinase. B, DDK phosphorylates Mcm2, Mcm4, and Mcm6. DDK phosphorylation of Mcm2 opens the Mcm2-Mcm5 gate, allowing single-stranded DNA to be extruded from the central channel of Mcm2-7 in S phase. In S phase, the S-phase cyclin-dependent kinase phosphorylates Sld2 and Sld3, and these phosphorylated proteins each bind to Dpb11 to form a ternary complex of Dpb11-Sld3-Sld2. This Dpb11-Sld3-Sld2 ternary complex binds to single-stranded DNA. The Dpb11-Sld3-Sld2 ternary complex will disengage from Mcm2-7 upon binding ssDNA. C, the disengagement of Dpb11-Sld3-Sld2 from Mcm2-7 will allow GINS to bind Cdc45·Mcm2-7. GINS binds to the Cdc45·Mcm2-7 complex, forming the closed Cdc45·Mcm2-7·GINS helicase complex that encircles single-stranded DNA.
Cdc45 Is Recruited to Mcm2-7 by Sld3 and, possibly, by Dpb11 and Sld7
Previous data suggest that Sld3 is responsible for the recruitment of Cdc45 to Mcm2-7 (Fig. 10A) (25). We show here that Dpb11 can also function to recruit Cdc45 to Mcm2-7 in vitro, suggesting an additional mechanism for Cdc45 recruitment to Mcm2-7 in vivo. Sld7 may also play a role in the recruitment of Cdc45 to Mcm2-7 (data not shown) (39). It has also been shown that DDK phosphorylates Mcm4, relieving an inhibitory function for the N terminus of Mcm4 (40). DDK phosphorylation of Mcm4 may also be important for the attachment of Cdc45 to loaded Mcm2-7 (13, 14). Furthermore, DDK phosphorylation of Mcm6 may also be important for cell growth (14, 41). Future work may help resolve how DDK stimulates Cdc45 recruitment to Mcm2-7.
The Sld3-Sld2-Dpb11 Ternary Complex in S Phase Binds to ssDNA, Allowing GINS to Bind Mcm2-7
In S phase, S-CDK phosphorylates Sld2 and Sld3 (3, 4). These phosphorylation events stimulate the interactions between Dpb11 and Sld2 and Sld3 to form the Dpb11-Sld3-Sld2 ternary complex (3, 4). The formation of the Sld3-Sld2-Dpb11 complex has been shown to be essential, and, in fact, the formation of this complex can bypass the requirement for CDK in the cell (3, 4).During S phase, the T-rich strand of origin DNA is extruded from the central channel of Mcm2-7 (42). Recent work has demonstrated that the Dbf4-dependent kinase phosphorylates Mcm2, opening the Mcm2-Mcm5 gate of the Mcm2-7 complex to allow for the extrusion of T-rich ssDNA from the central channel of Mcm2-7 (17). The Mcm2-Mcm5 gate, which may function to open and close the Mcm2-7 ring, appears to be conserved from yeast to Drosophila (10, 15, 43).
Dpb11, Sld3, or Sld2 each bind individually to ssDNA in vitro (29, 30). Furthermore, ssDNA disrupts the interaction between Dpb11 and Mcm3/Mcm5 and allows GINS to bind Mcm3/Mcm5 in vitro and in vivo (Figs. 4 and 9). Moreover, ssDNA also disrupts the interaction between Sld2 and Mcm3/Mcm5 in vitro and in vivo, allowing GINS to bind Mcm2-7 (28, 33). Finally, it has been shown, using purified proteins, that ssDNA disrupts the interaction between Sld3 and Mcm3/Mcm5, allowing GINS to bind Mcm2-7 (29). These data suggest that when ssDNA is extruded from the central channel of Mcm2-7 during S phase, the Dpb11-Sld3-Sld2 complex will bind to ssDNA instead of Mcm3/Mcm5 (Fig. 10B). The Dpb11-Sld3-Sld2 complex has three different binding sites for ssDNA that may all act together to form a very tight interaction with ssDNA. The disengagement of Dpb11, Sld3, and Sld2 from Mcm2-7 and the binding of the Dpb11-Sld3-Sld2 ternary complex to ssDNA may allow for the subsequent binding of GINS to Cdc45·Mcm2-7. The binding of GINS to Cdc45·Mcm2-7 completes the assembly of the CMG complex, appropriately positioned to encircle ssDNA (Fig. 10C). The ability of ssDNA to displace Dpb11, Sld3, and Sld2 from Mcm2-7 therefore fulfills an elegant regulatory mechanism to ensure that the CMG complex can only assemble when a single strand is extruded from Mcm2-7.
Acknowledgments
We thank Dr. Susan Taylor for purified PKA, Dr. Mike O'Donnell for expression constructs for GINS, and Ruth Didier for help with FACS analysis and data collection. We also thank Dr. Yoichi Kato for the use of the sonicator and Dr. Akash Gunjan and Yanchang Wang for helpful comments.
This work was supported by National Science Foundation Grant 1265431 (to D. L. K.).
- CDK
- cyclin-dependent kinase
- S-CDK
- S-phase cyclin-dependent kinase
- ssDNA
- single-stranded DNA
- IP
- immunoprecipitation
- DDK
- Dbf4-dependent kinase
- BRCT
- breast cancer susceptibility protein C-terminal domain
- RPA
- replication protein A
- CSM-Ura
- complete synthetic media minus uracil
- YPGal
- yeast-peptone-galactose.
REFERENCES
- 1. Araki H., Leem S. H., Phongdara A., Sugino A. (1995) Dpb11, which interacts with DNA polymerase II(ϵ) in Saccharomyces cerevisiae, has a dual role in S-phase progression and at a cell cycle checkpoint. Proc. Natl. Acad. Sci. U.S.A. 92, 11791–11795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tak Y. S., Tanaka Y., Endo S., Kamimura Y., Araki H. (2006) A CDK-catalysed regulatory phosphorylation for formation of the DNA replication complex Sld2-Dpb11. EMBO J. 25, 1987–1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tanaka S., Umemori T., Hirai K., Muramatsu S., Kamimura Y., Araki H. (2007) CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature 445, 328–332 [DOI] [PubMed] [Google Scholar]
- 4. Zegerman P., Diffley J. F. (2007) Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature 445, 281–285 [DOI] [PubMed] [Google Scholar]
- 5. Davey M. J., Indiani C., O'Donnell M. (2003) Reconstitution of the Mcm2-7p heterohexamer, subunit arrangement, and ATP site architecture. J. Biol. Chem. 278, 4491–4499 [DOI] [PubMed] [Google Scholar]
- 6. Bochman M. L., Bell S. P., Schwacha A. (2008) Subunit organization of Mcm2-7 and the unequal role of active sites in ATP hydrolysis and viability. Mol. Cell Biol. 28, 5865–5873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Remus D., Beuron F., Tolun G., Griffith J. D., Morris E. P., Diffley J. F. (2009) Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell 139, 719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Evrin C., Clarke P., Zech J., Lurz R., Sun J., Uhle S., Li H., Stillman B., Speck C. (2009) A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc. Natl. Acad. Sci. U.S.A. 106, 20240–20245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sun J., Fernandez-Cid A., Riera A., Tognetti S., Yuan Z., Stillman B., Speck C., Li H. (2014) Structural and mechanistic insights into Mcm2-7 double-hexamer assembly and function. Genes Dev. 28, 2291–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Samel S. A., Fernández-Cid A., Sun J., Riera A., Tognetti S., Herrera M. C., Li H., Speck C. (2014) A unique DNA entry gate serves for regulated loading of the eukaryotic replicative helicase MCM2-7 onto DNA. Genes Dev. 28, 1653–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bochman M. L., Schwacha A. (2008) The Mcm2-7 complex has in vitro helicase activity. Mol. Cell 31, 287–293 [DOI] [PubMed] [Google Scholar]
- 12. Labib K. (2010) How do Cdc7 and cyclin-dependent kinases trigger the initiation of chromosome replication in eukaryotic cells? Genes. Dev. 24, 1208–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sheu Y.-J., Stillman B. (2006) Cdc7-Dbf4 phosphorylates MCM proteins via a docking site-mediated mechanism to promote S phase progression. Mol. Cell 24, 101–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Masai H., Taniyama C., Ogino K., Matsui E., Kakusho N., Matsumoto S., Kim J. M., Ishii A., Tanaka T., Kobayashi T., Tamai K., Ohtani K., Arai K. (2006) Phosphorylation of MCM4 by Cdc7 kinase facilitates its interaction with Cdc45 on the chromatin. J. Biol. Chem. 281, 39249–39261 [DOI] [PubMed] [Google Scholar]
- 15. Costa A., Ilves I., Tamberg N., Petojevic T., Nogales E., Botchan M. R., Berger J. M. (2011) The structural basis for MCM2-7 helicase activation by GINS and Cdc45. Nat. Struct. Mol. Biol. 18, 471–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fu Y. V., Yardimci H., Long D. T., Ho T. V., Guainazzi A., Bermudez V. P., Hurwitz J., van Oijen A., Schärer O. D., Walter J. C. (2011) Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell 146, 931–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bruck I., Kaplan D. L. (2015) The Dbf4-Cdc7 kinase promotes Mcm2-7 ring opening to allow for single-stranded DNA extrusion and helicase assembly. J. Biol. Chem. 290, 1210–1221 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18. Ilves I., Petojevic T., Pesavento J. J., Botchan M. R. (2010) Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol. Cell 37, 247–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Moyer S. E., Lewis P. W., Botchan M. R. (2006) Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc. Natl. Acad. Sci. U.S.A. 103, 10236–10241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gambus A., Jones R. C., Sanchez-Diaz A., Kanemaki M., van Deursen F., Edmondson R. D., Labib K. (2006) GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat. Cell Biol. 8, 358–366 [DOI] [PubMed] [Google Scholar]
- 21. Pacek M., Tutter A. V., Kubota Y., Takisawa H., Walter J. C. (2006) Localization of MCM2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol. Cell 21, 581–587 [DOI] [PubMed] [Google Scholar]
- 22. Heller R. C., Kang S., Lam W. M., Chen S., Chan C. S., Bell S. P. (2011) Eukaryotic origin-dependent DNA replication in vitro reveals sequential action of DDK and S-CDK kinases. Cell 146, 80–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. On K., Beuron F., Frith D., Snijders A., Morris E., Diffley J. (2014) Prereplicative complexes assembled in vitro support origin-dependent and independent DNA replication. EMBO J. 18, 605–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gros J., Devbhandari S., Remus D. (2014) Origin plasticity during budding yeast DNA replication in vitro. EMBO J. 33, 621–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kamimura Y., Tak Y. S., Sugino A., Araki H. (2001) Sld3, which interacts with Cdc45 (Sld4), functions for chromosomal DNA replication in Saccharomyces cerevisiae. EMBO J. 20, 2097–2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bruck I., Kaplan D. (2011) GINS and Sld3 compete with one another for Mcm2-7 and Cdc45 binding. J. Biol. Chem. 286, 14157–14167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kanemaki M., Labib K. (2006) Distinct roles for Sld3 and GINS during establishment and progression of eukaryotic DNA replication forks. EMBO J. 25, 1753–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bruck I., Kanter D. M., Kaplan D. L. (2011) Enabling association of the GINS tetramer with the Mcm2-7 complex by phosphorylated Sld2 protein and single-stranded origin DNA. J. Biol. Chem. 286, 36414–36426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bruck I., Kaplan D. (2011) Origin single-stranded DNA releases Sld3 Protein from the Mcm2-7 complex, allowing the GINS tetramer to bind the Mcm2-7 complex. J. Biol. Chem. 286, 18602–18613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kanter D. M., Kaplan D. L. (2011) Sld2 binds to origin single-stranded DNA and stimulates DNA annealing. Nucleic Acids Res. 39, 2580–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kaplan D. L., Davey M. J., O'Donnell M. (2003) Mcm4,6,7 uses a “pump in ring” mechanism to unwind DNA by steric exclusion and actively translocate along a duplex. J. Biol. Chem. 278, 49171–49182 [DOI] [PubMed] [Google Scholar]
- 32. Bruck I., Kaplan D. (2013) Cdc45 protein-single-stranded DNA interaction is important for stalling the helicase during replication stress. J. Biol. Chem. 288, 7550–7563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bruck I., Kaplan D. (2014) The replication initiation protein sld2 regulates helicase assembly. J. Biol. Chem. 289, 1948–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. van Deursen F., Sengupta S., De Piccoli G., Sanchez-Diaz A., Labib K. (2012) Mcm10 associates with the loaded DNA helicase at replication origins and defines a novel step in its activation. EMBO J. 31, 2195–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Van Hatten R. A., Tutter A. V., Holway A. H., Khederian A. M., Walter J. C., Michael W. M. (2002) The Xenopus Xmus101 protein is required for the recruitment of Cdc45 to origins of DNA replication. J. Cell Biol. 159, 541–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Costa A., Renault L., Swuec P., Petojevic T., Pesavento J. J., Ilves I., MacLellan-Gibson K., Fleck R. A., Botchan M. R., Berger J. M. (2014) DNA binding polarity, dimerization, and ATPase ring remodeling in the CMG helicase of the eukaryotic replisome. eLife 3, e03273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Muramatsu S., Hirai K., Tak Y. S., Kamimura Y., Araki H. (2010) CDK-dependent complex formation between replication proteins Dpb11, Sld2, Pol ϵ, and GINS in budding yeast. Genes Dev. 24, 602–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tanaka S., Komeda Y., Umemori T., Kubota Y., Takisawa H., Araki H. (2013) Efficient initiation of DNA replication in eukaryotes requires Dpb11/TopBP1-GINS interaction. Mol. Cell Biol. 33, 2614–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tanaka S., Nakato R., Katou Y., Shirahige K., Araki H. (2011) Origin association of Sld3, Sld7, and Cdc45 proteins is a key step for determination of origin-firing timing. Curr. Biol. 21, 2055–2063 [DOI] [PubMed] [Google Scholar]
- 40. Sheu Y. J., Stillman B. (2010) The Dbf4-Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4. Nature 463, 113–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Randell J. C., Fan A., Chan C., Francis L. I., Heller R. C., Galani K., Bell S. P. (2010) Mec1 is one of multiple kinases that prime the Mcm2-7 helicase for phosphorylation by Cdc7. Mol. Cell 40, 353–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Geraghty D. S., Ding M., Heintz N. H., Pederson D. S. (2000) Premature structural changes at replication origins in a yeast minichromosome maintenance (MCM) mutant. J. Biol. Chem. 275, 18011–18021 [DOI] [PubMed] [Google Scholar]
- 43. Bochman M. L., Schwacha A. (2010) The Saccharomyces cerevisiae Mcm6/2 and Mcm5/3 ATPase active sites contribute to the function of the putative Mcm2-7 “gate”. Nucleic Acids Res. 38, 6078–6088 [DOI] [PMC free article] [PubMed] [Google Scholar]








