Background: Transit peptides direct the import of the majority of chloroplast-destined proteins into chloroplasts.
Results: Multiple unrelated Hsp70 recognition elements are able to substitute for the translocation determinant domain of transit peptides.
Conclusion: The majority of transit peptides utilize Hsp70-interacting property to initiate import.
Significance: This finding expands the mechanistic view of the chloroplast protein import process.
Keywords: 70-Kilodalton Heat Shock Protein (Hsp70), Arabidopsis thaliana, Cell Biology, Chloroplast, Molecular Motor, Protein Translocation, Transit Peptide
Abstract
Previously, we identified the N-terminal domain of transit peptides (TPs) as a major determinant for the translocation step in plastid protein import. Analysis of Arabidopsis TP dataset revealed that this domain has two overlapping characteristics, highly uncharged and Hsp70-interacting. To investigate these two properties, we replaced the N-terminal domains of the TP of the small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase and its reverse peptide with a series of unrelated peptides whose affinities to the chloroplast stromal Hsp70 have been determined. Bioinformatic analysis indicated that eight out of nine peptides in this series are not similar to the TP N terminus. Using in vivo and in vitro protein import assays, the majority of the precursors containing Hsp70-binding elements were targeted to plastids, whereas none of the chimeric precursors lacking an N-terminal Hsp70-binding element were targeted to the plastids. Moreover, a pulse-chase assay showed that two chimeric precursors with the most uncharged peptides failed to translocate into the stroma. The ability of multiple unrelated Hsp70-binding elements to support protein import verified that the majority of TPs utilize an N-terminal Hsp70-binding domain during translocation and expand the mechanistic view of the import process. This work also indicates that synthetic biology may be utilized to create de novo TPs that exceed the targeting activity of naturally occurring sequences.
Introduction
In plants, between 2000 and 5000 nucleus-encoded proteins are predicted to target to plastids (1). More than 92% of plastid-localized proteins contain an N-ter extension, TP,3 that has been added to the mature domain via evolution creating a precursor form (2, 3). TPs govern post-translational targeting of precursors into the plastid stroma via sequential interaction with the TOC and TIC (the translocons at outer and inner chloroplast envelopes), respectively (4, 5). Upon entry into the stroma, TPs are removed via proteolytic processing by the stromal processing peptidase. Despite identification >30 years ago (6), little is known about how TPs accomplish this function.
Although there is a plethora of programs to predict TPs (7–9), these tools do not provide insight into how TPs can function in a common import pathway. Moreover, recent bioinformatic analysis has not been fruitful in identifying the consensus motifs within TPs (10). However, when TPs are clustered into subgroups, a few short conserved peptide motifs were identified within each subgroup, but their roles remain largely uncharacterized (10). Despite the large number of TPs and short amino acid (aa) length, structural approaches have not provided much insight. This is due in part to TPs being highly unstructured in aqueous solutions (11, 12) hindering direct structure-function analysis. Nevertheless, three loosely defined regions in TPs have been identified as follows: (i) N-ter of ∼10 uncharged aa ending with Pro/Gly and preferably Ala as the second aa, (ii) central domain lacking acidic aa but rich in hydroxylated aa, and (iii) C-ter rich in Arg and possibly forming an amphiphilic β-strand (5, 13).
The importance of the highly uncharged TP N-ter (13) in plastid protein import has been shown both in vitro (14–17) and in vivo (10, 18–20). These studies have proposed that the TP N-ter of the small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase (SSU) and ferredoxin (FD) directs precursor binding during the import process by interacting with the envelope lipids or the Toc159 receptor. However, we generated synthetic TPs lacking the uncharged N-ter but still able to bind to plastids, similar to the wild-type TPs. These synthetic TPs of SSU (SStp) and FD (FDtp) contain the reversed aa sequences from C- to N-ter. Together with a series of their N-ter mutants, these constructs have confirmed the TP N-ter as an essential determinant for protein translocation into plastid stroma (21).
In addition to the uncharged properties, the TP N-ter has been shown to harbor a strong Hsp70-binding site (H70BS) (22–24). Based on the predictions, all of our import-competent mutants contain a strong H70BS at their N-ter, which the import-deficient mutants lack (21). Because the stromal Hsp70 was shown to act as a plastid translocation motor (25–29), we suspected that the N-ter H70BS in our import-competent constructs is required for the stromal Hsp70 interaction to trap and/or pull the precursor into plastids (21) similar to the proposed unfolding and pulling model of chloroplast protein import (30).
To further address these two N-ter properties, we have constructed new TPs based on the forward (native) and reverse constructs of SStp that are called SSF and SSR, respectively. The N-ter of these constructs was extended to include peptide sequences derived from a comprehensive study of 36 peptides where their affinities for multiple Hsp70s have been determined (31). Nine peptides were selected that are strongly interacting with all chaperones to noninteracting with any of them. We further determined the relative affinity of these peptides to the chloroplast Hsp70 CSS1. The effect of the N-ter alteration in preprotein import was assessed by in vivo and in vitro import assays. In addition, the recognition of physicochemical properties was further elucidated by reversing and scrambling the N-ter 10 aa. Collectively, these results demonstrate that the addition of a short, non-native, Hsp70-interacting domain that even includes charged residues at the N-ter of a TP can support preprotein translocation into plastids in vivo and in vitro. This work indicated that the majority of TPs utilize an Hsp70-binding N-ter during translocation and opens the way for synthetic biology to create de novo TPs that exceed the targeting activity of natural TPs.
EXPERIMENTAL PROCEDURES
Constructions of TP-YFP Fusion Protein Expression Plasmids
The N-ter mutants were generated based on two previously generated constructs, pBS-SSR10-MtoA-SSF-YFP and pBS-SSF10-MtoA-SSR-YFP (21), for plant expression. The DNA sequences of nine peptides, pp38, pp9, PepG, V10, A6R, HbS, np09, DRC8, and HA, were added on the PCR primers to replace SSR10 and SSF10. The reversed and scrambled N-ter mutants were produced similarly to replace the N-ter of SSF and FDF in pBS-SSF-YFP and pBS-FDF-YFP (21), respectively.
To ease the generation of Escherichia coli expression vectors, an NheI site was added to a pET-30a-based pET-SSF-YFP (21), producing pET-NheI-YFP. The expression cassette in pBS was subcloned into pET-NheI-YFP to generate pET constructs. The negative control, pET-m20-YFPHis6, containing the first 20 aa (m20) of the mature domain of SSU (mSSU) fused to YFP and His6 tag.
All of the primers are listed in supplemental Table S1. All constructs have been verified by sequencing.
In Vivo Plastid Import Assays
Arabidopsis thaliana Col-0 and onion (Allium cepa) cultivar Vidalia were used if not stated otherwise. Transient expression in onion epidermis peels and Arabidopsis seedlings using particle bombardment was done as reported previously (32). Epifluorescence imaging and the relative intensity ratio calculations were performed as described previously (21). The number of cells used is stated in the figure legend.
In Vitro Chloroplast Import Assays
The TP-YFP fusion proteins and m20-YFPHis6 control were produced from pET constructs using TnT T7 Coupled Wheat Germ Extract System (Promega, Madison, WI) with 0.8 μCi/μl [35S]methionine (MP Biomedical). Two batches of translation were used. The translation products were separated on SDS-polyacrylamide gel followed by digital autoradiography using Storage Phosphor Screen (Molecular Dynamics) or Molecular Imager FX (Bio-Rad) and quantified using Quantity One software (Bio-Rad). The relative concentration of each protein was determined from the radioactivity and the numbers of Met. The concentrations were equalized by adding 50% TnT Wheat Germ Extract (Promega). An equal volume of equalized proteins was used in the import assay.
The chloroplasts were isolated from 12- to 14-day-old seedlings of dwarf pea (Pisum sativum) cultivar Green Arrow as described previously (33). The import assays were performed using a modified method from Dabney-Smith et al. (34). Briefly, the assays were performed in a 500-μl reaction containing 0.25 mg of chlorophyll/ml chloroplasts, labeled protein, 2 mm l-Met, 10 mm DTT, 2 mm Mg-ATP, 0.5% BSA, 300 mm urea, 330 mm sorbitol, 50 mm HEPES-KOH, pH 8.0. The reactions were incubated at room temperature. A 150-μl sample was taken at 5, 10, and 15 min after the reaction was started and mixed with 600 μl of ice-cold buffer (330 mm sorbitol, 50 mm HEPES-KOH, pH 8.0). The chloroplasts were re-isolated and separated on a 15% SDS-polyacrylamide gel and quantified by digital autoradiography. Two sets of independent assays were performed.
The amount of imported proteins was normalized where the amount of mature domain of the SSF construct at 15 min was assigned as 100% for the SSF mutant series and the m20-YFPHis6 control. The amount of mature domain of FDF at 15 min was assigned as 100% for the FDF mutant series. The relative import rate of each time point was calculated from the relative amount of the mature proteins at that time point divided by the import time. The fastest rate among the three time points was assigned as the maximum import rate.
Pulse-Chase Assays
To avoid multiple products presented in the in vitro translation of TP-YFP constructs, we chose to use the construct containing TP followed by mSSU with C-ter 3×HA and His6 tags. The precursor of SSU from pET-11 vector (35) was subcloned into pET-30a. The C-ter tag was added by PCR. Primer sequences are listed in supplemental Table S1. The mSSU-3×HA-His6 region was subcloned into the pET constructs containing TP-YFP fusion producing TP-mSSU-3×HA-His6 constructs. The 35S-labeled precursors were expressed in E. coli BL21(DE3) and purified as described previously (33).
The chloroplasts were isolated from 12-day-old seedlings of dwarf pea cultivar Wando as described previously (33). The plants were kept in the dark for at least 14 h to deplete ATP, and all procedures were performed under dim light to prevent ATP synthesis. The precursors were incubated with isolated chloroplasts in the presence of 0.1 mm ATP in the buffer containing 10 mm DTT, 4 mm MgCl2, 1% BSA, 300 mm urea, 330 mm sorbitol, and 50 mm HEPES-KOH, pH 8.0, at room temperature for 20 min. In this condition (pulse), the precursors are bound to the TOC/TIC translocons as the import intermediate (21, 36). The intact chloroplasts were re-isolated using 40% Percoll (GE Healthcare). To initiate the translocation (chase), the re-isolated chloroplasts were suspended into the same buffer with the presence of 2 mm ATP and incubated at room temperature. Samples were taken at 0, 2, 4, 8, and 20 min and rapidly diluted with 3 volumes of ice-cold buffer (330 mm sorbitol, 50 mm HEPES-KOH, pH 8.0) to stop the import. The chloroplasts were re-isolated, and a fraction was used to determine total protein concentration using BCA assay (Thermo Fisher Scientific). Equal amounts of proteins were separated on a 10–20% SDS-polyacrylamide gel and quantified by autoradiography. Two sets of independent assays were performed.
Competitive mCSS1 Binding Assays
To express the mature domain of CSS1 precursor protein (mCSS1; residues 66–706) in E. coli, the mCSS1 cDNA was amplified together with the N-ter His6 tag and cloned into pET-30a generating pET-His-mCSS1. The rare Leu codon CTC in residues 167, 270, 373 and 652 was mutated to a CTG codon using QuikChange Lightning multisite-directed mutagenesis kit (Agilent Technologies) generating pET-His-mCSS1-mut. All primers are listed in supplemental Table S1. The His-mCSS1 was expressed using E. coli Rosetta 2 (DE3) (EMD Millipore), purified using PrepEase-high specificity kit (Affymetrix), and dialyzed into Buffer M (20 mm HEPES-KOH, pH 7.5, 20 mm NaCl, 2.5 mm magnesium acetate).
The substrate was produced from bovine α-lactalbumin (Sigma). The reduced carboxymethylated α-lactalbumin (RCMLA) was made based on the published method (37). To produce fluorescein-labeled RCMLA (F-RCMLA), the protein was dialyzed into 50 mm borate buffer, pH 8.50. NHS-fluorescein (Thermo Scientific) was added at 25-fold molar excess and incubated at room temperature for 1 h in the dark. F-RCMLA was then separated using a PD-10 column (GE Healthcare) into 20 mm potassium phosphate buffer, pH 7.40, and finally dialyzed into Buffer M.
The SSF peptide was expressed and purified as reported previously (21). The N-ter peptide competitors were synthesized and modified with N-ter acetylation and the C-ter amidation by New England Peptide, Inc. All peptides are 15 aa in length containing the Hsp70-binding domain (Fig. 2A) and part of SSR sequence similar to the first 15 aa of the N-ter mutant constructs based on SSR (Fig. 2D). These synthetic peptides were solubilized in DMSO before diluting with Buffer M containing 8 m urea to the final concentration of DMSO at 5%.
FIGURE 2.
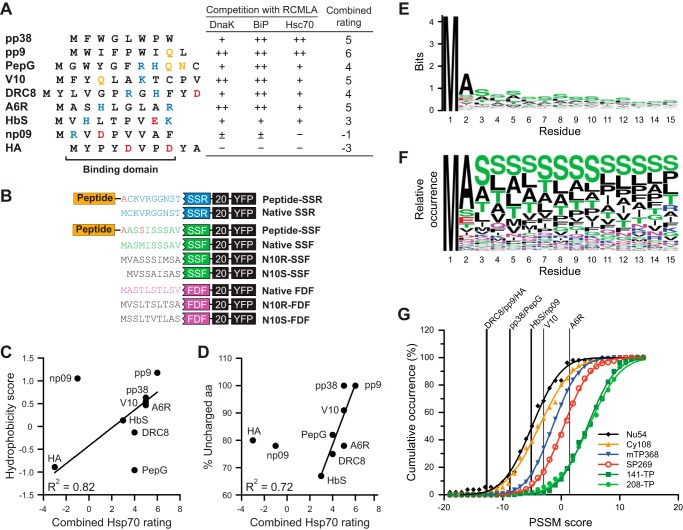
N-ter peptides, the mutant constructs, and aa distribution of the TP N-ter. A, sequences of the Hsp70-interacting and noninteracting peptides are shown. Polar, negative, and positive aa are colored yellow, red, and blue, respectively. Table shows the published ratings of peptide competitiveness in binding competition with RCMLA to three Hsp70s (31). The combined rating affinities were calculated from the summation of the ratings where + and − were assigned values of 1 and −1, respectively. B, representation of the N-ter mutant constructs. For the fusion peptide constructs, the internal Met was mutated to Ala or Ser (colored red). N10R and N10S denote the reversed and scrambled mutants. The second Met in N10S-SSF was deleted. The native sequences of SSR, SSF, and FDF are colored blue, green, and magenta, respectively. C, hydrophobicity of the peptides in A plotted against the combined rating. The correlation line was determined without np09 and PepG values. D, %UA of the peptides in A plotted against the combined rating. The correlation line was determined without HA and np09 values. E, logo plot of the N-ter 15 aa of the 208-TP set. The total height of each aa position corresponds to the conservation in that position. F, logo plot showing the relative occurrence of each aa at each position in the N-ter 15 aa of the 208-TP set. G, PSSM scores of the Hsp70-interacting and noninteracting peptides. The cumulative occurrence of the scores calculated from the N-ter domain of the sequences in the TargetP training set (Nu54, Cy108, mTP368, SP269, and 141-TP) is shown together with the scores from the 208-TP set (the training set for this PSSM). The lines were fitted based on a cumulative normal distribution.
The competitive binding assays were performed in a 40-μl reaction containing varying concentrations of SSF or 40 μm of the N-ter peptide, 10 μm His-mCSS1, 1 μm F-RCMLA, 2 mm ADP, 250 mm urea, 1.25% DMSO in Buffer M. The reaction was incubated at 37 °C for 30 min before separation using native PAGE. The gel with 6% acrylamide was used in 0.5× TBE buffer (44.5 mm Tris borate, pH 8.30, 1 mm EDTA). To increase detection sensitivity, the proteins in the gel were transferred onto a nitrocellulose membrane. The fluorescent signal was detected with Bioimaging Systems (UVP) equipped with 302 nm UV light source and SYBR Green emission filter. The image was captured using LabWorks software (UVP), and the quantitation of band intensity was performed using Quantity One software (Bio-Rad).
MALDI-TOF MS
The saturated sinapinic acid matrix in 33% acetonitrile and 0.1% trifluoroacetic acid was mixed with equal volume of protein sample. The mixture was spotted onto the stainless steel target plate and dried. MALDI-TOF MS was performed on a Bruker Daltonics Microflex mass spectrometer with positive ion mode.
TP and Other Protein Datasets
The 912- and 208-TP datasets were acquired from the studies by Chotewutmontri et al. (21) and Lee et al. (10), respectively. The plant protein training sets of TargetP, including 141-TP (called cTP141), mTP368, Nu54, Cy108, and SP269, were part of the work by Emanuelsson et al. (38). The numbers in the TargetP dataset names indicate the total number of sequences in that set.
Comparison of TP Datasets
To compare the TPs in different datasets, a standalone BLAST program version 2.2.25+ was utilized (39). The TP datasets were converted into BLAST databases. The TP sequences from each set were searched against each database to identify the same protein.
Percentage of Uncharged aa (%UA) Calculations—
The calculation was performed according to Chotewutmontri et al. (21). The %UA (not Asp, Glu, His, Lys, or Arg) was calculated within a subsequence in a window of length (L). The calculation was repeated from L of 5 to 17 aa. The %UA data within the first 30 aa was fitted to the inverted Boltzmann sigmoidal model using GraphPad Prism 5.0 (GraphPad Software).
Hydrophobic Accessible Surface Area (HASA) Calculations
The HASA of the hydrophobic aa (Ala, Ile, Leu, Met, Phe, Trp, Tyr, and Val) within an 8-aa subsequence was calculated by summation of the corresponding area of each aa based on the published total side-chain accessible surface area values (40). The 8-aa window was moved along the length of the sequences to show the HASA at different areas of the sequences. The Perl script for this analysis is available upon request.
H70BS Predictions
We utilized the previously created Perl scripts (21). The scripts are based on the random peptide phage display-derived algorithm (RPPD) (22, 41).
Clustering of Sequences Based on H70BS Patterns
For the 208-TP dataset (10), the N-ter 80-aa sequences were utilized. The RPPD algorithm was used to predict H70BSs for each TP. The MATLAB program (MathWorks) was used. The clustergram function from the bioinformatics toolbox was applied to cluster data based on the hierarchical clustering and generated a dendrogram along with a heat map of the clustering. To cluster the TPs according to their Hsp70-binding patterns, Pearson correlation was used in distance matrix calculation. Unweighted pair group method with arithmetic mean was used in dendrogram construction. The clustering results are listed in supplemental Table S2.
For the analysis in Fig. 1B, 20 random sequence datasets were generated based on the aa frequency of the entire 208-TP set. Each set contains 208 of 80-aa sequences. The H70BS was predicted based on the RPPD algorithm. The mean RPPD score as a function of aa position was calculated for an entire dset. The MATLAB (MathWorks) was used to cluster the mean RPPD score patterns of all the datasets similar to the above paragraph.
FIGURE 1.
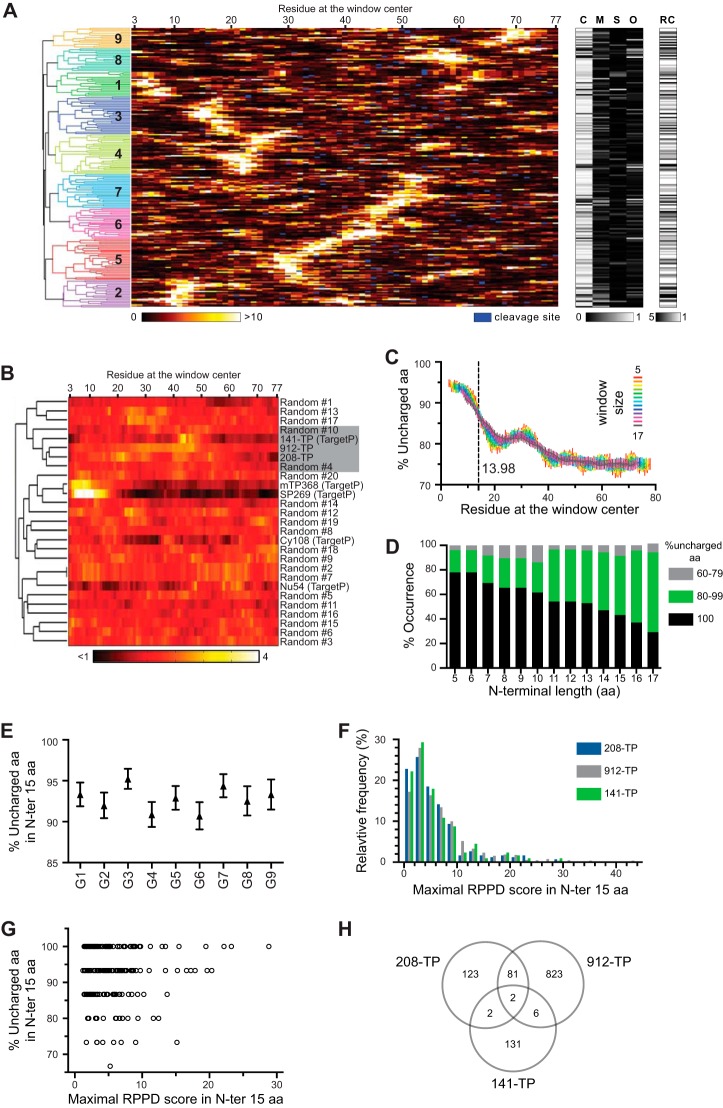
Bioinformatic analysis of the TP datasets. A, clustering of the 208-TP set. Each line represents a TP from the 208-TP set. Left panel shows a dendrogram of the hierarchical clustering of the TPs based on their predicted H70BS patterns. Nine clusters were formed as indicated by numbers. The orange-yellow heat map panel represents the predicted Hsp70-binding score based on RPPD where the higher score has higher affinity. The black-and-white heat maps show TargetP prediction results. The predicted cleavage sites were marked in blue in the RPPD heat map. C, M, S, and O are the TargetP probability scores for chloroplasts, mitochondria, secretory pathway, and other localizations, respectively. RC is the TargetP reliability class where the lower value means higher confidence of prediction. B, comparison of the average RPPD distribution of TP datasets and the random sequence datasets. The aa frequency of the 208-TP set was used to generate 20 random sequence sets. Two other TP datasets were included, 912-TP and 141-TP. Four non-TP datasets from TargetP training sets were also added as controls as follows: the datasets of mitochondrial (mTP368), nuclear (Nu54), cytosol (Cy108), and secretory pathway (SP269) localized proteins. Each line represents an average RPPD score as the function of aa position of an entire dataset. Hierarchical clustering was applied to the mean RPPD score patterns. The dendrogram of the clustering is shown. All three TP datasets (208-TP, 912-TP, and 141-TP) are found to cluster together. This TP cluster only contains two random sets (#4 and #10) despite the fact that the 20 random sets were derived from the 208-TP set. C, %UA within the window of size 5–17 aa along the length of TP was calculated from the 208-TP set. n = 208. Means ± S.E. are shown. D, fractions of TPs in each of the 3%UA groups observed in the TP N-ter of the 208-TP set based on the length of the N-ter region from 5 to 17 aa. E, %UA values calculated from the N-ter 15 aa were plotted based on the cluster group as in A. 16 ≤ n ≤ 31. Means ± S.E. are shown. F, distribution of the maximal RPPD score in the N-ter 15 aa of the 208-TP set. Frequency and cumulative distributions are shown with bars for left axis and blue line for right axis, respectively. G, %UA value versus the maximal RPPD score of each TP was plotted. Both values were calculated from in the N-ter 15 aa of the 208-TP set. H, comparison of three TP datasets: the 208-TP, the 912-TP, and the 141-TP datasets. Numbers of TPs are shown.
Hydrophobicity Calculations
The hydrophobicity of the peptides was estimated using an on-line program ProtScale on the ExPASy server (42). The hydrophobic scale of aa by Kyte and Doolittle (43) was applied.
Sequence Logo
To generate the logo, the sequence datasets were submitted to the on-line program WebLogo (44).
Evaluation of the N-ter Sequences Using Position-specific Scoring Matrix (PSSM)
To evaluate whether the N-ter peptide sequences were similar to the TP N-ter sequences, the PSSM method was used. This method is widely used in identifying the motifs or patterns within the sequences (45). Generally, the aa sequences containing the motif of interest were aligned. This alignment of the motif with the length of M aa was then used to generate a PSSM with a dimension of 20 aa × M positions. Each row represents an aa (i), and each column represents an aa position (j) of the motif. The matrix element contains a score (si,j). To identify the motif within a sequence, a sliding window of size M is scanned through the sequence. At each location, the score of the subsequence is calculated from the summation of the score si,j of each aa in the subsequence (46). A type of scoring used the relative frequency (Fi,j) of the aa i at position j and the background frequency (pi) of the aa i (46). The matrix element score (si,j) was calculated as the log ratio of these frequencies (fi,j/pi). The subsequence score (SS) can then be calculated from the summation of the si,j scores associated with the aa in the subsequences (46).
Because the peptides in the series are 8–12 aa long, a TP PSSM corresponding to the TP N-ter 10 aa was created. The N-ter sequences of TPs from the 208-TP set (10) were used without aligning the sequences. Because the first position is always Met (Fig. 2G), this position is ignored. Although position 2 had a distinct aa distribution, positions 3–12 had approximately the same distributions (Fig. 2E). For the first position of the PSSM matrix (j = 1), the calculated relative frequency of aa at the sequence position 2 (fi,l) showed that the 208-TP lacked Cys, His, Trp, and Tyr at this position. We instead calculated these frequencies from a larger set, the 912-TP set (21). However, we found no Trp at this position. We fixed this missing value by using the value of 0.01 as the number of Trp counts at this position. To avoid the same problem, the aa from the sequence position 3–12 were combined to calculate an averaged frequency (fi,a(3–12)), which was used as the frequencies of the PSSM matrix at positions 2–9. The aa frequencies of UniProt Release 2012_10 were used as the background frequencies (pi). We used log base 2 of the ratio of Fi,j over psi to calculate the matrix element si,j scores. The PSSM score of a sequence was then calculated from the summation of the si,j scores from nine positions corresponding to sequence positions 2–10. The frequencies and the si,j scores of this TP PSSM are reported in Table 1. A Perl script was written to perform this calculation and is available upon request.
TABLE 1.
The components of the TP PSSM for analysis of the N-ter sequences
These parameters were derived from the N-ter sequences of TP datasets. fi,j indicates relative frequency of the aa i at position j; pi indicates background frequency of the aa i derived from UniProt database; si,j indicates the matrix element score calculated from the log ratio of fi,j/pi.
| Amino acids | fi, 1 | fi, 2 to fi, 9 | pi | si, 1 | si, 2 to fi, 9 |
|---|---|---|---|---|---|
| Ala | 0.4693 | 0.1197 | 0.0863 | 2.4426 | 0.4716 |
| Cys | 0.0055 | 0.0192 | 0.0125 | −1.1912 | 0.6193 |
| Asp | 0.0241 | 0.0053 | 0.0532 | −1.1405 | −3.3300 |
| Glu | 0.0811 | 0.0082 | 0.0620 | 0.3883 | −2.9232 |
| Phe | 0.0143 | 0.0442 | 0.0403 | −1.4980 | 0.1357 |
| Gly | 0.0515 | 0.0288 | 0.0709 | −0.4604 | −1.2975 |
| His | 0.0033 | 0.0106 | 0.0221 | −2.7503 | −1.0653 |
| Ile | 0.0230 | 0.0500 | 0.0601 | −1.3839 | −0.2652 |
| Lys | 0.0154 | 0.0183 | 0.0531 | −1.7899 | −1.5387 |
| Leu | 0.0296 | 0.1240 | 0.0992 | −1.7452 | 0.3217 |
| Met | 0.0175 | 0.0279 | 0.0246 | −0.4899 | 0.1786 |
| Asn | 0.0362 | 0.0274 | 0.0412 | −0.1860 | −0.5869 |
| Pro | 0.0164 | 0.0510 | 0.0469 | −1.5108 | 0.1207 |
| Gln | 0.0219 | 0.0308 | 0.0396 | −0.8511 | −0.3625 |
| Arg | 0.0121 | 0.0178 | 0.0544 | −2.1727 | −1.6122 |
| Ser | 0.1075 | 0.2409 | 0.0666 | 0.6901 | 1.8546 |
| Thr | 0.0296 | 0.1014 | 0.0558 | −0.9140 | 0.8627 |
| Val | 0.0373 | 0.0635 | 0.0678 | −0.8629 | −0.0954 |
| Trp | 0.0000 | 0.0029 | 0.0129 | −10.2024 | −2.1631 |
| Tyr | 0.0044 | 0.0082 | 0.0305 | −2.8000 | −1.9020 |
Subcellular Localization Predictions
We predicted the subcellular localization of the proteins using Predotar (8), PredSL (9), TargetP (38, 47), WolF Psort (48) and YLoc (49).
Homology Modeling of Chloroplast Hsp70
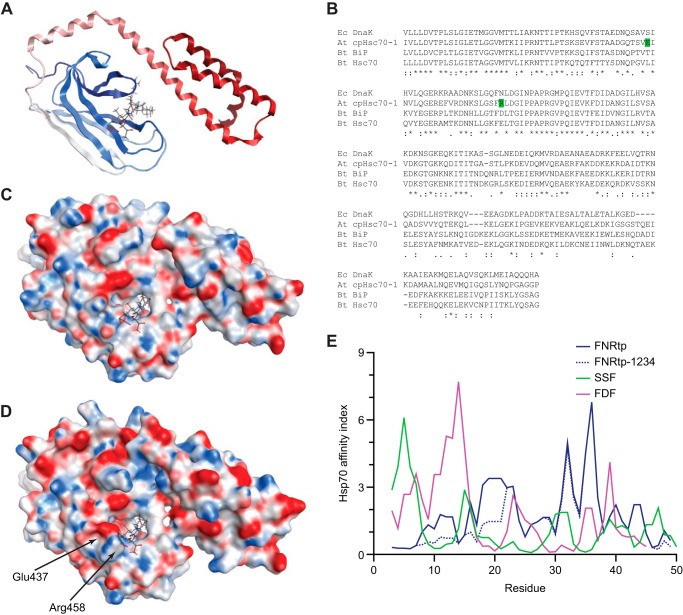
The crystal structure of substrate-binding domain (SBD) of E. coli DnaK with its peptide substrate (PDB 1DKZ) (50) was used as the template. First, the sequence of an Arabidopsis chloroplast Hsp70 (AtcpHsc70-1) was aligned to the other Hsp70 sequences using T-Coffee (51). The alignment of the SBD as shown in Fig. 10B was used to build the homology model of AtcpHsc70-1 on the Swiss-Model server (52). The electrostatic surfaces of the structures were produced from Molecular Operating Environment program (MOE; Chemical Computing Group) using CHARMM force field.
FIGURE 10.
Peptide binding surfaces of Hsp70s and H70BS prediction of FNRtp. A, crystal structure (Protein Data Bank code 1DKZ) of SBD of E. coli DnaK (shown as ribbon) together with the peptide substrate (shown as stick model). B, alignment of the SBD of Hsp70s. The aa of AtcpHsc70-1 possibly involved in the alteration of substrate binding are highlighted green. Ec, E. coli; At, Arabidopsis; Bt, Bos taurus. C and D, electrostatic surfaces of E. coli DnaK structure and the homology model of AtcpHsc70-1, respectively. The altered binding surfaces are indicated with arrows. E, RPPD was used to predict the H70BS. Although the strong N-ter H70BSs are present in SSF and FDF, the site is absent from FNRtp and its mutant FNRtp-1234.
Accession Numbers
Sequence data from this study can be found in the GenBankTM/EMBL data libraries under the following accession numbers: pea SSU (CAA25390), tobacco SSU (NP_054507); Silene latifolia FD (CAA26281), pea CSS1 (Q02028), pea FNR (P10933), E. coli DnaK (P0A6Y8), bovine BiP (NP_001068616), bovine Hsc70 (P19120), and AtcpHsc70-1 (NP_194159).
RESULTS
Bioinformatic Analysis of TP Datasets
Previously, Ivey et al. (22) suggested that ∼70% of TPs in the CHLPEP dataset (53) harbored a strong H70BS at their N-ter. However, because only 14 TPs in this set are from Arabidopsis and it is redundant (53 instances of SStp), we chose to revisit the H70BS analysis. Using the 208-TP dataset of experimentally verified chloroplast proteins from Arabidopsis (10), we analyzed the occurrence and placement of H70BSs with the RPPD algorithm (22, 41) that utilizes 6-aa window analysis. The analysis with the Rudiger et al. (54) algorithm using a 13-aa window was not performed because it is less accessible to the terminal regions and shows inconsistency with our previous results (21). Fig. 1A shows this analysis and TargetP prediction. The orange-yellow heat map in Fig. 1 represents levels of Hsp70 affinity predicted by RPPD with higher score (yellow/white) corresponding to higher affinity. The TPs were clustered into nine groups based on their H70BS patterns within the first 80 aa (TP and part of mature protein sequence) using Hierarchical clustering. These groups show pronounced differences in the position of their highest affinity, which varied from position 5 to 70 (5, 10, 17, 23, 32, 46, 52, 58, and 70 for groups 1–9, respectively). This fairly even distribution raised a concern that the H70BS was randomly distributed. To resolve this, we generated 20 random sequence datasets, each contain 208 sequences derived from the aa frequency of the 208-TP set. The RPPD score of each sequence were calculated. We then asked how similar are the mean RPPD scores of an entire set as a function of aa positions between the random sets, the 208-TP set, and the other six control sets. The control sets include a much larger set of 912 reliably predicted TP from Arabidopsis (21) and the training sets of TargetP (38) for chloroplast (141-TP), mitochondrial (mTP368), nuclear (Nu54), cytosol (Cy108), and secretory pathway (SP269) localized proteins. The number in the dataset name indicates the total sequences in the set. We applied hierarchical clustering to the mean RPPD score patterns of theses sets. All three TP datasets (208-TP, 912-TP, and 141-TP) were found to cluster together (Fig. 1B). This TP cluster only contains two random sets despite the fact that the 20 random sets were derived from the 208-TP set. Thus, the observation that the RPPD patterns among three separate TP sets are more similar than those of the random sets indicates that this distribution is not random and seems to be conserved among TPs. In addition, the TP groups in Fig. 1A were forms with distinct links between multiple H70BS. For example, group 1 has the highest H70BS at position 5 and weaker sites at position 30 and 60, whereas group 2 has the highest site at position 10 and weaker site at position 40. Fig. 1A also indicates that 32 and 47% of these TPs contain the strongest H70BS within the first 20 and 30 aa, respectively. The detailed results of Fig. 1A are shown in supplemental Table S2.
We further analyzed this dataset using TargetP (38). About 90% were predicted to localize to chloroplasts (C-M-S-O heat map in Fig. 1A) with 68% of these TPs classified into TargetP reliability classes 1 and 2 (white for class 1 in RC heat map of Fig. 1A). The 208-TP set has a predicted TP length of 52.2 ± 17.2 aa (mean ± S.D.).
Previously, the uncharged N-ter of TPs was identified by von Heijne et al. (13), and we have verified them using a larger set of 912 predicted TPs from Arabidopsis (21). The same analysis was applied to the 208-TP set. To show %UA along the length of TPs, we used a sliding window of analysis (from 5 to 17 aa) and observed that the %UA at the N-ter had a maximum value of 94% and dropped to 81% between aa 20 and 30 (Fig. 1C). The C-ter after aa 40 contained ∼75%UA. When fit to a sigmoidal curve, the transition mid-point between 94 to 81%UA occurred at aa 14, regardless of the window size.
To investigate distribution of the %UA of the TP N-ter, we segregated the N-ter into three groups based on their %UA (60–79%, 80–99, and 100%). The fraction of TPs observed in each group is shown as a function of N-ter length in Fig. 1D. The fraction of 100% uncharged N-ter reduced from ∼78 to 30% when the analyzed N-ter length increased from 5 to 17 aa. Inversely, the fraction of 80–99% uncharged N-ter increased from 18 to 65%. Yet, the fraction of 60–79% uncharged N-ter remained very low and fluctuated between 3 and 13%. Thus, >90% of the TPs in the 208-TP set contained a predominantly uncharged N-ter (>80%UA) regardless of analysis length. Moreover, >40% of the TPs have totally uncharged N-ter within their first 15 aa.
When separated into nine clusters as shown in Fig. 1A, each TP cluster had about the same average %UA in the N-ter 15 aa (Fig. 1E). We also plotted the frequency distribution of the maximal RPPD score within the N-ter 15 aa of the 208-TP set (Fig. 1F). Based on the finding that Hsp70-interacting peptides have an RPPD score above 2 (22), we found that 77% of TPs in the 208-TP set had an RPPD score above 2 within the first 15 aa, indicating that these TPs contain an N-ter H70BS. The same analysis was applied to the 912-TP set from Arabidopsis (21) and the 141-TP set containing TPs from various plant species (38), and we found that 82 and 78% of the TPs had an RPPD score above 2, respectively (Fig. 1F). Although Fig. 1A shows that 32–47% of TPs harboring the strongest H70BS at the N-ter, these analyses indicate that the majority of the N-ter of TPs (77–82%) is able to interact with Hsp70.
We then asked whether there is a correlation between %UA and predicted Hsp70 affinity of the TP N-ter. Using the 208-TP set, the maximal RPPD scores from the TP N-ter 15 aa were plotted against the %UA of the N-ter 15 aa (Fig. 1G). The RPPD scores of the TPs with different %UA were distributed throughout the range of 2 to 30, suggesting that there is no direct correlation between the %UA and Hsp70 affinity of the TP N-ter.
Different TP datasets have been used in various works, yet dataset comparison has not been done. We therefore compared three TP datasets, the 208-TP set (10), the 912-TP set (21), and the 141-TP training set of TargetP (38). We found that the majority of TPs in each set did not overlap with the others (Fig. 1H). This was expected for the 141-TP set where TPs were derived from all plants in Swiss-Prot, but it was unexpected for the 208-TP and the 912-TP sets because both were composed solely of Arabidopsis proteins. The discrepancy between the 208-TP and the 912-TP sets may result from the 912-TP set containing only TPs that are classified by TargetP with the top two reliability classes. Similar analysis of the 208-TP set revealed that only 125 TPs belong to classes 1 and 2. The large differences observed between the TargetP training set and the other two datasets clearly indicate that this set is not a good representation of higher plant TPs, and it provides motivation for a more systematic investigation of TP properties in general.
Construction of TP N-ter Mutants
We developed a quantitative in vivo plastid protein import assay as described in Chotewutmontri et al. (21). This assay utilizes the transient expression of different TP-YFP fusion proteins in onion epidermal cells to determine the plastid targeting efficiency of the TPs and is expressed as a ratio of plastid/cytosolic YFP signals. To evaluate the variability of this assay, we expressed SStp followed by m20 from tobacco fused to YFP and measured the ratios from three widely available onion cultivars. We observed that the ratios were reproducible within a given cultivar but differ significantly between cultivars (supplemental Fig. S1). For this work, we used only the Vidalia cultivar because it was used in our previous study (21).
Using this in vivo assay, we investigated the roles of uncharged and Hsp70-binding properties of the TP N-ter by utilizing two previously generated constructs. The first construct is based on a native TP, SSF, fused to m20 followed by YFP (SSF-20-YFP) and the second related construct that contains a dysfunctional synthetic TP coding for the reversed sequence of SSF called SSR-20-YFP. Although SSF-20-YFP localized to the plastids, SSR-20-YFP had a very low targeting efficiency (21). These two constructs served as the positive and negative starting vectors that provided import-competent and import-deficient constructs, respectively.
The N-ter of these two constructs were further modified by adding additional peptide sequences in front of SSF and SSR. To estimate the function of the N-ter peptides, we interpreted the results from SSF-20-YFP and SSR-20-YFP as the upper and lower limit of the real values. Using only the data from the dysfunctional SSR case may not be accurate because it is not a real TP and the fact that the fusion product can have unforeseen characteristics. These new peptide sequences were derived from a detailed study where their affinities for three different Hsp70s were experimentally determined (31). We selected nine peptides with a range of Hsp70 affinities. The sequence and the experimental affinity of these peptides are shown in Fig. 2A. These peptides originated from very different sources, yet none are from plants. We have truncated the original peptide to only the Hsp70-binding domains (31) creating peptides ranging from 8 to 12 aa in length. All truncated peptides contain polar and/or charged aa except for pp38 peptide. The affinities of full-length peptide to E. coli DnaK, bovine e luminal BiP, and bovine cytosolic Hsc70 had been determined as rating affinities of the competitiveness against RCMLA in binding to Hsp70s (31). We calculated the summation of the rating affinities to generate a combined Hsp70 rating affinity for each peptide (Fig. 2A). The TP N-ter mutants were generated by extending their original N-ter to include the truncated peptide sequences together with the substitution of internal Met to Ala or Ser (Fig. 2B).
To assess whether the TP N-ter recognition is based on local physicochemical properties, another series of mutants was generated. The N-ter 10 aa of both SSF in SSF-20-YFP and FDtp named FDF in FDF-20-YFP (21) were replaced with their reversed or scrambled sequences (Fig. 2B). These mutants should retain the same physicochemical properties as the original constructs.
Bioinformatic Analysis of the Hsp70-interacting and Noninteracting Peptides
To further characterize these nine peptides, we determined hydrophobicity and %UA of the peptides. The hydrophobicity scale of aa by Kyte and Doolittle (43) was used to calculate the hydrophobicity score. When the combined rating affinities were plotted against the hydrophobicity scores (Fig. 2C), seven of the nine peptides showed some correlation (R2 of 0.82; p value of 0.0046). However, PepG and np09 showed no correlation, and PepG had a strong Hsp70 affinity yet is hydrophilic. Inversely, np09 was very hydrophobic but has a weak affinity. The combined rating affinities versus %UA plot (Fig. 2D) also showed a correlation with R2 of 0.72 (p value of 0.015).
Although these peptides were not plant-derived, it was possible that they were simply functioning as a TP due to random sequence similarity, so we tested whether the utilized N-ter peptides were similar to TP N-ter. The PSSM method, which is heavily used in identification of motif or pattern in the sequences (45), was utilized, even though TP N-ter are poorly conserved with high levels of Ser and Thr (Fig. 2, E and F). The PSSM scores were calculated based on the aa distributions at specific positions in the TP sequences in comparison with the background frequencies (the aa frequencies of UniProt database). A total PSSM score greater than 0 indicates a higher chance of being a TP N-ter than a random sequence. About 90% of tested TP N-ter had the PSSM scores greater than 0 (Fig. 2G). The PSSM score distributions of nonchloroplast proteins (mTP368, Nu54, Cy108, and SP269 datasets mentioned earlier) were plotted as references. We found that DRC8, pp9, HA, pp38, PepG, HbS, np09, and V10 had the scores of −12.8, −12.7, −12.6, −8.8, −8.7, −5.2, −5.1, and −3.0, respectively, suggesting that they are not similar to TP N-ter (Fig. 2G). Only A6R had a positive score of 1.4, which makes it a possible TP N-ter. However, about 20% of known TPs, (both TargetP and 208-TP datasets) had scores less than 1.4 (Fig. 2G), which is consistent with this predictive tool having a specificity of ∼87% (55).
To further characterize these new TPs, we used five subcellular localization programs to predict localization of the mutants. The most frequent predictions indicated that all mutants are targeted to the chloroplasts (Table 2).
TABLE 2.
Subcellular localization predictions of the N-ter mutants and their experimental localizations
| Constructs | Predicted localizationsa |
In vivo localizationse |
In vitro chloroplast import in peah | |||||
|---|---|---|---|---|---|---|---|---|
| YLocb | WoLF PSORTc | TargetPb,d | Predotarb,d | PredSLb,d | Onionf | Arabidopsisg | ||
| SSF-20-YFP | Chloroplast (99.96%) | Chloroplast (12/13) | Chloroplast (0.676) | Chloroplast (0.96) | Chloroplast (1.00) | Chloroplast | Chloroplast | Imported |
| pp38-SSF | Chloroplast (99.99%) | Chloroplast (13.5/21) | Chloroplast (0.770) | Chloroplast (0.75) | Chloroplast (0.86) | Chloroplast & Mitochondria | Cytosol | Not imported |
| pp9-SSF | Chloroplast (99.95%) | Chloroplast (14/14) | Chloroplast (0.817) | Chloroplast (0.64) | Other | Chloroplast & Mitochondria | Chloroplast | Not imported |
| PepG-SSF | Chloroplast (99.97%) | Chloroplast (11/13) | Mitochondria (0.738) | Chloroplast (0.69) | Chloroplast (0.99) | Chloroplast | Chloroplast | Not imported |
| V10-SSF | Chloroplast (99.98%) | Chloroplast (12/13) | Chloroplast (0.831) | Mitochondria (0.50) | Chloroplast (0.99) | Chloroplast & Mitochondria | Chloroplast & Mitochondria | Imported |
| DRC8-SSF | Chloroplast (99.98%) | Chloroplast (11/13) | Chloroplast (0.563) | Chloroplast (0.35) | Chloroplast (1.00) | Chloroplast | Chloroplast | Imported |
| A6R-SSF | Chloroplast (99.95%) | Chloroplast (13/13) | Chloroplast (0.803) | Chloroplast (0.92) | Chloroplast (1.00) | Chloroplast | Chloroplast & Mitochondria | Imported |
| HbS-SSF | Chloroplast (99.95%) | Chloroplast (13/13) | Chloroplast (0.851) | Chloroplast (0.72) | Chloroplast (0.99) | Chloroplast | Chloroplast | Not imported |
| np09-SSF | Chloroplast (99.97%) | Chloroplast (14/14) | Chloroplast (0.811) | Chloroplast (0.57) | Chloroplast (1.00) | Not applicable | Not applicable | Not imported |
| HA-SSF | Chloroplast (99.98%) | Nucleus (9/13) | Chloroplast (0.651) | Chloroplast (0.39) | Chloroplast (0.99) | Cytosol | Cytosol | Not imported |
| SSR-20-YFP | Chloroplast (99.34%) | Peroxisome (7/14) | Chloroplast (0.668) | Other (0.93) | Chloroplast (0.97) | Cytosol | Cytosol | Imported |
| pp38-SSR | Chloroplast (99.97%) | Chloroplast (8/13) | Chloroplast (0.491) | Chloroplast (0.60) | Mitochondria (1.00) | Cytosol | Cytosol | Not imported |
| pp9-SSR | Chloroplast (99.86%) | Chloroplast (8/13) | Chloroplast (0.553) | Mitochondria (0.33) | Mitochondria (0.93) | Cytosol | Cytosol | Not imported |
| PepG-SSR | Chloroplast (99.81%) | Chloroplast (10/13) | Mitochondria (0.403) | Other (0.83) | Mitochondria (0.97) | Chloroplast & Mitochondria | Chloroplast | Not imported |
| V10-SSR | Chloroplast (99.86%) | Chloroplast (11/13) | Other (0.453) | Other (0.80) | Chloroplast (1.00) | Chloroplast | Chloroplast | Imported |
| DRC8-SSR | Chloroplast (99.86%) | Chloroplast (8/14) | Other (0.570) | Mitochondria (0.21) | Other | Chloroplast | Chloroplast | Imported |
| A6R-SSR | Chloroplast (99.50%) | Chloroplast (8/14) | Chloroplast (0.619) | Mitochondria (0.36) | Chloroplast (1.00) | Chloroplast | Chloroplast | Imported |
| HbS-SSR | Chloroplast (99.81%) | Cytosol (7/13) | Other (0.697) | Other (0.79) | Chloroplast (0.99) | Chloroplast | Chloroplast | Not imported |
| np09-SSR | Chloroplast (99.83%) | Chloroplast (11/14) | Other (0.551) | Other (0.96) | Chloroplast (0.99) | Cytosol | Cytosol | Imported |
| HA-SSR | Chloroplast (99.13%) | Peroxisome (5/13) | Other (0.812) | Other (0.82) | Chloroplast (0.73) | Cytosol | Cytosol | Not imported |
a The predicted localizations conflicting with experimental results are shown as italicized text.
b The probability scores are shown in parentheses.
c The numbers of the most localization sites to the numbers of total predictions are shown in parentheses.
d The other localizations indicate that the proteins are not chloroplastic, mitochondrial, or secretory pathway proteins.
e Data are based on the results of transient expression of YFP fusion proteins in this study. The localizations in onion and Arabidopsis were determined 12 h after transformation.
f The chloroplast localization was assigned when the constructs had the quantitative ratio measurement values shown in Fig. 4I above 3.
g The cytosol localization was assigned if the majority of the cells expressing the constructs had strong YFP signal in the nucleus as a result of YFP diffusion from the cytosol.
h The imported proteins were measured within 15 min of import. The precursors were assigned as not imported proteins if the import rates were lower than 20% of SSF-20-YFP import rate.
Affinity of the Peptides to the Chloroplast Hsp70 CSS1
To determine the affinity of the peptides, the mature domain of chloroplast CSS1 precursor protein from pea (56) was expressed in E. coli and purified with N-ter His tag (His-mCSS1, Fig. 3A). Native PAGE indicated that the majority of His-mCSS1 was in monomer form, although dimer and aggregated forms were also detected (Fig. 3B). We chose to work with the well characterized model Hsp70 substrate, RCMLA. We produced RCMLA from bovine α-lactalbumin and further labeled it with fluorescein, generating F-RCMLA. The MALDI spectra indicated all eight Cys residues were carboxymethylated, and every RCMLA was labeled with 2–5 fluoresceins (Fig. 3C).
FIGURE 3.
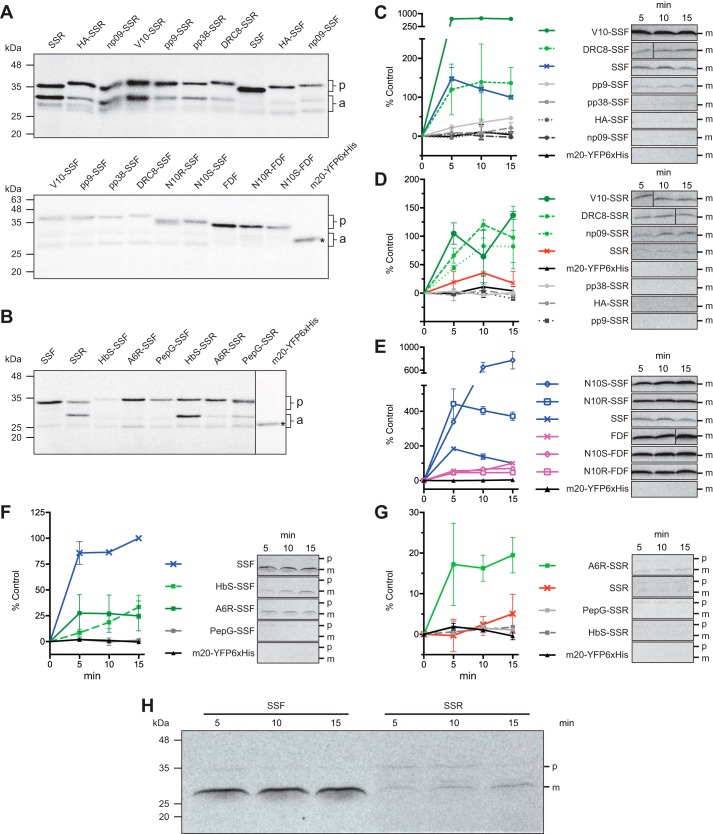
Relative affinities of the N-ter peptides to the chloroplast Hsp70. A, SDS-PAGE of purified chloroplast Hsp70, His-mCSS1. B, native PAGE of His-mCSS1 with BSA as control. C, MALDI-TOF spectra of bovine α-lactalbumin, RCMLA, and F-RCMLA. D and E, competitive binding assay of SSF against F-RCMLA binding to His-mCSS1. The assay concentration of SSF is indicated. The control lane (C) lacked His-mCSS1. D, representative of a nitrocellulose membrane of the native PAGE of the binding reactions at different concentrations of SSF. Top and bottom panels show the fluorescent signals of F-RCMLA (Flu) and the Coomassie Brilliant Blue (CBB)-stained His-mCSS1. R, F-RMCLA; A, aggregated His-mCSS1; D, His-mCSS1 dimer; M, His-mCSS1 monomer. E, quantification of the F-RCMLA-His-mCSS1 (R+M) complex from the fluorescent signals. n = 3. Means ± S.E. are shown. F and G, competitive binding assay of the N-ter peptides against F-RCMLA binding to His-mCSS1. The assays contain 40 μm of the N-ter peptide. F, representative of a nitrocellulose membrane of the native PAGE of the binding reactions. Top and bottom panels show the fluorescent signals of F-RCMLA (Flu) and the Ponceau S-stained His-mCSS1 (PSS). R, F-RMCLA; D, His-mCSS1 dimer; M, His-mCSS1 monomer. G, quantification of the F-RCMLA-His-mCSS1 (R+M) complex from the fluorescent signals. The negative control lane (Neg.) lacked the N-ter peptide. The fluorescent signal of R+M in the negative control lane was defined as 100% bound RCMLA. n = 6. Means ± S.E. are shown.
Using this modified RCMLA, we performed competitive binding assay using SSF against F-RCMLA binding to His-mCSS1. Native PAGE was used to separate the CSS1-bound and free forms of F-RCMLA. The proteins were electroblotted onto nitrocellulose membrane to increase sensitivity of fluorescent detection. Fig. 3, D and E, shows that the level of F-RCMLA bound to the monomeric His-mCSS1 (R+M) decreases when the higher concentration of SSF was added. At 40 μm SSF, the bound F-RCMLA was reduced to about 50% (Fig. 3E).
We then performed the competitive binding assay using short synthetic peptides corresponding to the first 15 aa of the N-ter mutant constructs based on SSR (Fig. 2, A and D). We did not use the peptide based on SSF constructs because the N-ter of SSF contains a strong H70BS and would complicate the results (22). A single concentration at 40 μm of the peptide was used. The level of F-RCMLA bound to the monomeric His-mCSS1 (R+M) was quantified and expressed as the percentage of reduction of the bound F-RCMLA from no competitor control (Fig. 3, F and G). These relative affinities of the peptides are in good agreement with the previously determined affinities to other Hsp70s (31). Both peptides derived from noninteracting peptides HA and np09 showed very weak affinities although other peptides derived from Hsp70-interacting peptides showed greater affinities to His-mCSS1. This result corroborated the prior results and confirmed their activity with the recombinant form of the major stromal Hsp70, His-mCSS1.
In Vivo Import Assays
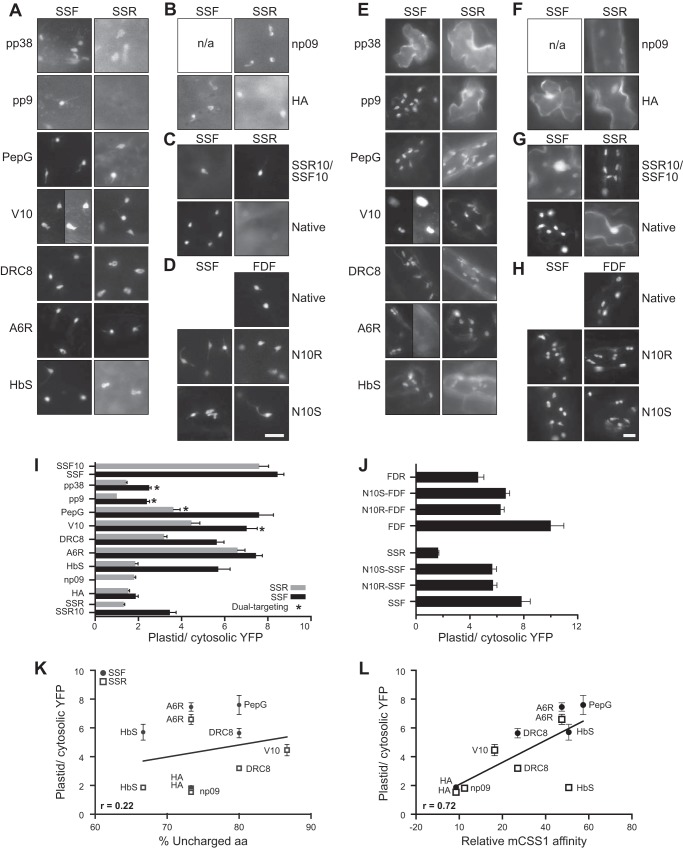
The N-ter peptide extensions were then tested for their ability to direct the import of YFP protein in vivo in both onion epidermal cells and Arabidopsis seedlings. All constructs are based on the same expression cassette differing by only about 10 aa from others, and we assumed that the expression should be very similar. The mature forms of the precursors are exactly the same and should have the same stability in the chloroplasts. Although a range in expression was observed in different cells as intrinsic variations of biolistic transformation, only a limited range suitable for fluorescent imaging detection was used. To account for variation, the images from at least 30 cells per construct were taken. The ratio analysis of the data also partially controls for varying expression levels. Fig. 4, A–D, shows representative images of the onion cells transiently expressing YFP 12 h after transformation. Although most of the constructs were able to target to plastids at different levels, pp9-SSR could not be detected in plastids, and np09-SSF was not expressed. Four constructs, pp38-SSF, pp9-SSF, PepG-SSR, and V10-SSF, had dual localization to plastids and mitochondria. Fig. 5 shows a representative of the dual-localized construct. V10-SSF was localized to the compartments with the same characteristics as those of plastid (around 5-μm compartments that sometimes form small tubular structures called stromules) and mitochondrial (small with dramatic morphology ranging from round spots to tubular shape) markers (32). To extend the targeting results from the non-green plastid found in the monocot (onion), the constructs were also transiently expressed in dicot chloroplasts using Arabidopsis seedlings. Fig. 4, E–H, shows the localization patterns of these constructs 2 days after transformation. More than 30 cells expressing each construct were observed. The construct was assigned as cytosol-localized if the majority of the cells showed a strong YFP signal in the nucleus resulting from diffusion of YFP in the cytosol. The localization observed in Arabidopsis is fairly similar to that observed in onion cells in terms of plastid targeting (Table 2). We considered the dual-targeted constructs as having the ability to target to plastids. However, the frequency of dual localization was significantly reduced and only observed from the V10-SSF and A6R-SSF constructs. This difference may reflect the more robust targeting to green plastids with the SSF TP than what is observed in the non-green, nonphotosynthetic leucoplasts found in onion cells.
FIGURE 4.
Targeting of the N-ter mutants in onion and Arabidopsis cells and the relationship with the properties of the N-ter. Representative images of transiently expressed N-ter mutant TP-YFP fusion proteins in onion (A–D) and Arabidopsis (E–H) cells. Left and right labels indicate the N-ter peptides used to generate the mutants. Top labels indicate the original constructs. N10R and N10S denote the reversed and scrambled mutants. The bars in D and H show the scales for A–D and E–H, respectively. Bars, 10 μm; n/a, not applicable. A and E, mutants containing Hsp70-inteacting peptides. For clarity of dual localization, two images taken from the same cell with different exposure time of V10-SSF are shown in A and E, and the two images of the same cells of A6R-SSF are shown in E. B and F, mutants containing noninteracting peptides. C and G, control constructs from previously published results (21). D and H, reversed (N10R) and scrambled (N10S) mutants. I and J, ratios of plastid/cytosolic YFP were measured from onion assays similar to those shown in A–D. Means ± S.E. are shown. 20 ≤ n ≤ 30. In addition, FDF, FDR, SSF10, SSR10, FDF10, and FDR10 from a previous study (21) are included. These constructs with suffix 10 contain the N-ter 10 aa of the opposite TP at their N-ter. I, ratios from the N-ter peptide constructs. J, ratios from the reversed (N10R) and scrambled (N10S) N-ter constructs. K, plastid targeting efficiency ratios were plotted against the %UA calculated from the N-ter 15 aa of the precursors. The correlation line was determined without the dual-localized mutants, pp9 and pp38 mutants. L, plastid targeting efficiency ratios were plotted against the relative mCSS1 affinity. The correlation line was determined without the dual-localized mutants, pp9 and pp38 mutants.
FIGURE 5.
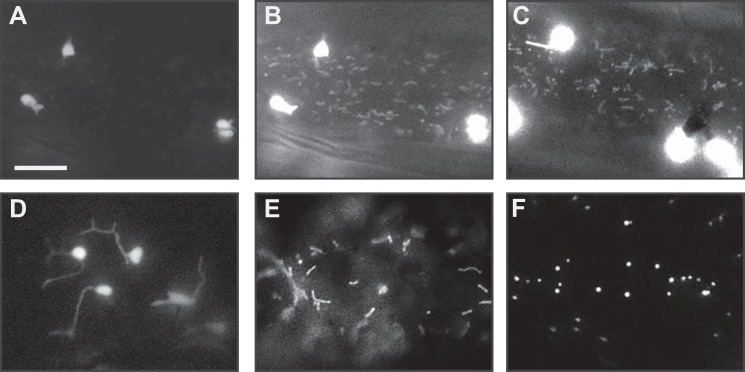
Localization of the dual-targeted construct. The signals were observed in onion epidermal cells transiently expressing the constructs. The images were captured 12 h after transformation. The representative images of the cells expressing dual-targeted constructs are shown (A–C) in comparison with the cells expressing the organelle markers (D–F). The signals in the dual-targeted constructs showed similar characteristics with the plastid and mitochondrial markers. A, cell expressing V10-SSF construct (the same cell as shown in Fig. 4A). B, same cell as in A except with longer exposure time. C, another cell expressing V10-SSF construct. D, cell expressing plastid marker construct (tobacco SStp-m20 fused to cyan fluorescent protein). E, cell expressing mitochondrial marker construct (the first 29 aa of Saccharomyces cerevisiae cytochrome c oxidase IV fused to cyan fluorescent protein). F, cell expressing peroxisome marker construct (cyan fluorescent protein fused with the peroxisomal targeting signal 1). All of the organelle markers were based on Nelson et al. (32). Bar in A indicates 10 μm for A–F.
To quantify the efficiencies of plastid targeting, images of onion cells similar to those of Fig. 4, A–D, were used. Although both onion and Arabidopsis showed similar qualitative results, we quantitated the targeting activity in onion because the distribution and uniform shape of the plastids permitted a robust quantitative analysis. We calculated the ratio of plastid/cytosolic YFP targeting for 2–3 plastids in each of ≥20 different cells. These values were averaged, and the targeting ratio is reported in Fig. 4, I and J. These ratios indicate that the mutants of both a weak Hsp70-binding peptide (np09) and a nonbinding peptide (HA) were unable to support the targeting of either SSF or SSR constructs and had the lowest targeting efficiencies at ∼1.7 (Fig. 4I). The addition of the peptides with strong Hsp70-binding affinities (PepG, V10, DRC8 and A6R) yielded the much higher ratios of ∼5.5 for both SSF and SSR. Consistent with this trend, the addition of the moderate Hsp70-binding peptide, HbS, was found to have intermediate ratios at ∼4.0 (the average of both SSF and SSR). Only two peptides behaved differently from their predicted and measured Hsp70 affinities; the two strong Hsp70-binding peptides, pp9 and pp38, displayed low targeting ratios at ∼2.0. We also found that all constructs based on SSF have higher efficiency than those with SSR. We used the ratio cutoff of three to assign the targeting of the constructs in onion cells (Table 2).
The reversed (N10R) and scrambled (N10S) mutants that we described in Fig. 2B show reduced targeting ratios compared with the original constructs (Fig. 4J). However, these ratios are still significantly higher than those of their reversed TP constructs (FDR and SSR) based on Tukey's test (α = 0.05), except N10R-FDF versus FDR with a p value of 0.0576. This result suggests that the recognition of the TP N-ter domain is based on physicochemical properties and is independent of sequence.
To further determine the relationship between two properties of the TP N-ter (%UA and mCSS1 affinity) and the targeting efficiency, we plotted the plastid/cytosolic YFP ratios of the mutants with unambiguous data. The ratios from the dual-targeting constructs were omitted because the defined cytosolic region in our analysis includes some mitochondria, and the measured cytosolic YFP signal includes the signal from mitochondrial YFP in these constructs. The plastid/cytosolic YFP signal ratio of these constructs is always lower than the true value. The ratios of pp9 and pp38 were also omitted because they lack a native property of TPs, which will be discussed below. When the plastid/cytosolic YFP ratios were plotted against %UA (Fig. 4K), %UA showed weak correlation with Pearson correlation coefficient (r) of 0.22 and a p value of 0.51. The targeting ratio versus mCSS1 affinity (Fig. 4L) showed a stronger correlation with r of 0.72 and p value of 0.01.
In Vitro Import Assays
To determine the import rate of the N-ter mutants, in vitro import assays using isolated pea chloroplasts were employed. The precursors were labeled with [35S]Met via in vitro translation. We have performed the assays in two batches. The translation products from each batch are shown in Fig. 6, A and B. Unfortunately, in vitro translations of these constructs produced both full-length proteins and some smaller species possibly due to the translation using internal ATG start sites. The translation from the ATG of the last residues of SSF, SSR, FDF, and FDR corresponds to the same 30-kDa proteins, and the translation from the ATG of the first residue of YFP corresponds to the 27-kDa proteins. To account for this variation, the translation products were analyzed by SDS-PAGE and autoradiography to allow the quantitation of the full-length proteins. We then tested each batch of precursors for import using equal input of full-length precursors in the assays. Fig. 6, C–H, shows the import time course of the precursors where the accumulation of the imported and processed mature domain was quantitated from the autoradiographs. The precursors and the mature proteins were well separated and easily identified. However, all of the translation products yield a similar sized preprotein whose apparent molecular weight is accurate based on the pre-stained protein standards. The mature band is also the correct molecular weight and also has a uniform size for the different constructs (Fig. 6H). The import of some precursors showed an early plateau after 5 min, although others did not plateau after 15 min of import. The maximum import rates determined from these time points were plotted in Fig. 7A. Surprisingly, V10 mutants import at a much higher rate than SSF-20-YFP in vitro but not as high in vivo. To compare the import rates with the targeting efficiencies, the in vivo targeting ratios of the precursors were plotted against the in vitro import rates (Fig. 7B). This plot indicates that the precursors with higher import rates have higher targeting efficiencies. The different amount of precursors used between two batches also resulted in different import rates between two batches. The variation in import in vitro versus in vivo for certain constructs such as V10 will require additional investigation.
FIGURE 6.
Autoradiographs and import time course of the N-ter mutant precursors. Two batches of translations were performed and separated by SDS-PAGE (A and B). For each translation batch, equal molar concentrations of precursor proteins were used in the import assays. The precursors from translation batches 1 and 2 were used in C–E and F and G, respectively. Two sets of assays were performed. Representative autoradiographs of the import-processed mature domains are shown on the right (C–G). The amounts of mature domains were measured at 5, 10, and 15 min after import started. The values from the SSF and SSR series were normalized where the amount of mature domain of SSF construct at 15 min was assigned as 100%. The amount of mature domain of FDF at 15 min was assigned as 100% for FDF series. A and B show the autoradiographs of 1-μl translation products from batches 1 and 2, respectively. Top labels, constructs; p, precursor size; a, additional products; asterisk, mature domain size. C, constructs based on SSF-20-YFP from the translation batch 1. D, constructs based on SSR-20-YFP from the translation batch 1. E, reversed and scrambled constructs from the translation batch 1. F, constructs based on SSF-20-YFP from the translation batch 2. G, constructs based on SSR-20-YFP from the translation batch 2. H, representative autoradiograph of the SDS-polyacrylamide gels before the images were cropped and represented in C–G. Each gel contained a lane with pre-stained standard marker. The region around 35 kDa is marked as precursor size (p), and the region around 28 kDa is marked as mature protein size (m).
FIGURE 7.
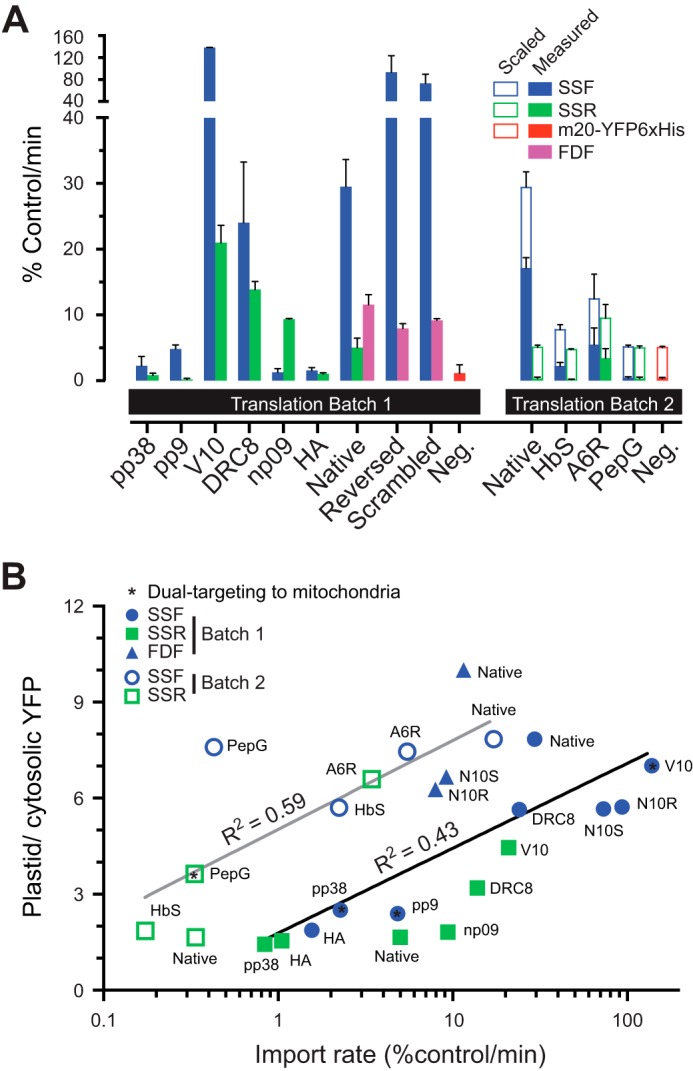
Maximum in vitro import rates of the N-ter mutant precursors. The in vitro import rates were derived from the fastest rates within 15 min of import. Two separate sets are shown based on the batch of in vitro translation of the precursors. The raw values from m20-YFPHis6, SSF, and SSR series were normalized to the raw value of mature domains from a 15-min import of SSF-20-YFP (100%). The FDF series were normalized to the raw value of mature domains from a 15-min import of FDF-20-YFP. The precursor without TP, m20-YFPHis6, was used as a negative control. n = 2. Means ± S.E. are shown. A, maximum import rates. The rates of batch 2 (filled bars) were scaled to match the batch 1 rates (opened bars) using the native SSF and SSR rates as controls. B, maximum import rates plotted against the plastid targeting efficiency ratios.
Analysis of pp9 and pp8 Constructs
We further investigated the constructs based on the two strong Hsp70-binding peptides, pp9 and pp38, which were unexpectedly import-deficient. The sequences of pp9 and pp38 contain multiple aromatic aa, including 2 and 3 Trp, respectively (Fig. 2A). Trp is the rarest aa in the TP N-ter (Table 1). Trp is not only hydrophobic but also has very large side chains. We developed a sliding window analysis that plots the total HASA of the peptide chain (Fig. 8A). It is clear that SSF and TPs as a group (the 208-TP set) have a relatively constant yet low HASA (<500 Å2/8 aa). However, when we measured pp9 and pp38 constructs, they have a much higher HASA at their N-ter compared with others (supplemental Fig. S2). The fact that this area is >2 times the TP average suggests that this property may interfere with the translocation.
FIGURE 8.
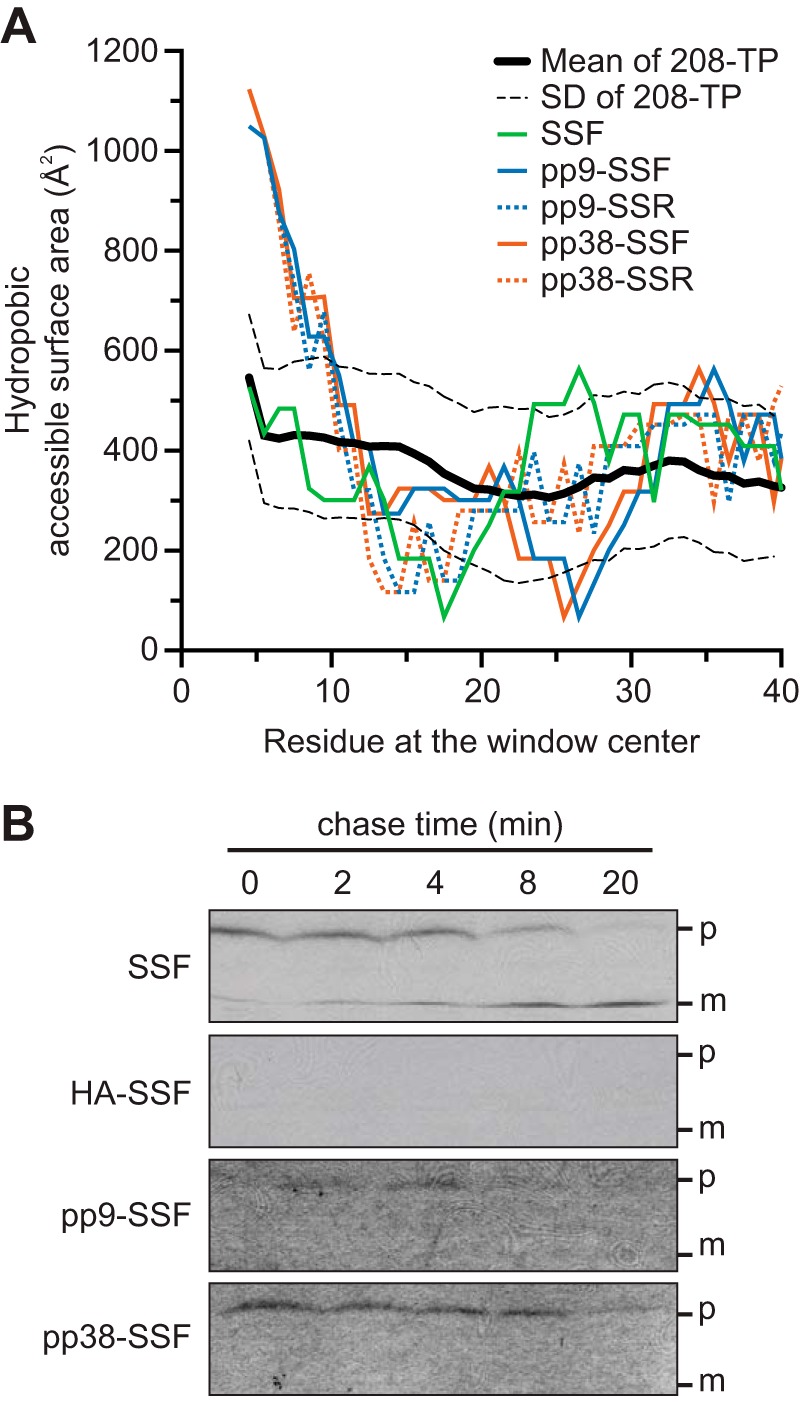
Hydrophobic accessible surface area of the mutant N-ter and the pulse-chase assays. A, HASA within an 8-aa window are plotted along the length of the aa sequences. The average HASA calculated from the 208-TP dataset is shown as a reference (black line) with S.D. as dashed lines. B, pulse-chase assays of the precursors of SSF, HA-SSF, pp9-SSF, and pp38-SSF fusion to mSSU followed by 3×HA and His6 tags. p, full-length precursors; m, imported and processed mature domains.
We investigated the failure of import of pp9 and pp38 constructs using pulse-chase assays (Fig. 8B). The 35S-labeled peptide-SSF fusion to mSSU followed by 3×HA and His6 tags was used as the precursor. The precursors were incubated with chloroplasts in the presence of 0.1 mm ATP, which allows the precursors to pulse as the import intermediates inside the TOC/TIC translocons and not translocated into the stroma (36). The chloroplasts with bound precursors were re-isolate, and the translocation was initiated with the presence of 2 mm ATP. The import was terminated at different times, and the chloroplasts were re-isolated. Equal amounts of total proteins were loaded on the gel in Fig. 8B. As expected for the wild-type SSF, the amount of precursor was reduced, although the amount of imported protein was increased over time. The import-deficient and non-Hsp70-interacting HA precursor failed to form the bound intermediate. Although the precursor forms of both pp9 and pp38 were reduced over time, the imported forms were not detected confirming the import deficiency of these constructs. This result also suggested that the pp9 and pp38 precursors were able to form the import intermediates but unable to translocate into the stroma, and the intermediates were released from the envelopes over time.
DISCUSSION
Our previous work demonstrated that the N-ter 10 aa of two well characterized TPs (SStp and FDtp) determined the translocation efficiency of the preprotein both in vivo and in vitro (21). When these 10 aa were added to the N-ter of an inactive precursor, both of these sequences restored the translocation of the preprotein in vitro and in vivo, suggesting that the critical element enabling translocation was found within these 10 aa. Bioinformatic analysis of these short N-ter indicated that both had a predicted strong H70BS, whereas the N-ter of inactive preproteins lacked this property (21). In this study, we directly confirm that it is the presence of N-ter H70BS that determines the ability of a precursor to be translocated by testing a set of nine unrelated peptides that possess a range of affinities for the chloroplast Hsp70 CSS1.
Locations of H70BSs Vary in TPs
Previous works had predicted that a large percentage of TPs contained an H70BS at the N-ter region based on the redundant TP datasets (22, 23). When this analysis was repeated using a nonredundant and experimentally confirmed set of chloroplast TPs, we observed a much greater distribution in the placement of H70BSs (Fig. 1A). Using a clustering algorithm, we can group these TPs into nine clusters where the position of the strongest H70BS is located in different regions. How this H70BS placement functions in relation to the other identified motifs (15, 18) and contributes to a general TP design (21, 57, 58) will be the subject for further study. It is now clear that only three clusters of the TPs have a strongest H70BS within their N-ter 20 aa and together represent 32% of the TPs. This is considerably less than 70% reported by our laboratory (22). However, this result may suggest that many of the well studied proteins that are highly abundant and associated with major photosynthetic processes, such as SSU and FD, may have evolved a TP organization that is very efficient to enable rapid translocation and that a common feature of these proteins is the N-ter placement of a strong H70BS. The inclusion of these proteins largely bias the previous TP sets because they are both abundant and very well studied (7, 23, 53). Despite 32% of the TPs having the strongest H70BS at the N-ter (Fig. 1A), our analysis also indicates that the majority of the TP N-ter (77–82%) is able to interact with Hsp70 (Fig. 1F). However, it is possible that these remaining TPs contain a N-ter-binding site for another molecular chaperone such as Hsp90 or Hsp100.
Role of N-ter Hydrophobicity Versus Hsp70 Recognition
Our earlier paper verified the observation that TP N-ter tends to be uncharged (13, 21). Because it has been shown that Hsp70 proteins recognize hydrophobic peptides (31, 41, 54), it was also important to show that translocation was dependent on an actual Hsp70 interaction and not just a consequence of their hydrophobicity. It is now clear that the hydrophobic core of a peptide/protein fits into the substrate cavity of Hsp70s, although the charged flanking regions interact with the surrounding area (59). Because most TPs contain an uncharged N-ter, it creates a challenge to separate the Hsp70-interacting function from the uncharged property (31, 41, 54). Using the database of 208 experimentally verified TPs, we observe a transition point between the highly to moderately uncharged regions at aa 14 (Fig. 1C), which is almost the same as aa 15 reported in the 912-TP set (21). Interestingly, comparison of the proteins that make up these two sets, as well as the TargetP training set, indicates that there is very little overlap in composition (<8% are common to any two sets). Even when restricted to only Arabidopsis protein datasets (208-TP and 912-TP), only 7.4% are common to both (Fig. 1H). However, in all three sets, the highly uncharged N-ter is a common feature. Considering only this highly uncharged 15 aa region, we found that 43% of the 208 TPs contain purely uncharged aa (Fig. 1C). When the TPs from clusters 1 to 3 (Fig. 1A) that contain the strongest H70BS within the first 20 aa were analyzed, 36% of them are completely uncharged in the N-ter 15 aa. These results indicate that more than a third of TPs have purely uncharged N-ter regions regardless of their N-ter Hsp70 affinities.
Non-native N-ter Are Not Similar to TP N-ter
The peptide set used here is composed of nine peptides that are 8–12 aa in length. For each peptide, two mutants were generated by fusing the peptide at the N-ter of SSF- and SSR-20-YFP fusion constructs (Fig. 2B). All peptides except A6R were predicted to be compositionally different from the TP N-ter (Fig. 2G). When we analyzed the complete set of 18 fusion proteins for their abilities to be predicted as TPs by five different subcellular localization programs (Table 2), 14 were predicted by ≥3 programs, 11 by ≥4 programs, and only 5 by all 5 programs. These results and the PSSM analysis suggest that these non-native peptides have functional properties not recognized by the leading prediction tools that are largely based on aa composition.
N-ter Peptides Restore Import in Vivo
We have selected nine tested peptides that were previously tested for binding to three functional and evolutionarily distinct Hsp70s as follows: E. coli DnaK, bovine cytosolic Hsc70, and bovine endoplasmic reticulum-localized BiP (31). We extended this assay for affinity to the major plastid Hsp70 CSS1 (Fig. 3, F and G); these peptides span a range of interaction strengths. The peptides, pp9 and pp38, are very nonpolar and contain no charged aa, and they strongly and moderately interact with CSS1, respectively (Fig. 2, A, C, and D, and 3G). The PepG, V10, DRC8, A6R, and HbS contain charged aa, range from 67–91%UA, and interact with CSS1. Peptides, np09 and HA, contain acidic aa and do not interact with CSS1. Thus, our peptide set contains an experimentally verified set of peptides with strong, moderate, and no interaction with CSS1.
We have previously designed and described a positive (SSF-20-YFP) and a negative SSR-20-YFP) construct that targets YFP to plastids (21). These constructs allowed the testing of targeting and translocation in multiple plants as follows: Arabidopsis, onion, and tobacco. We used these constructs as parental constructs to test the effect of the nine N-ter peptides described above and shown in Fig. 2A. The effects on YFP targeting were observed using in vivo plastid protein import assays in both onion (Fig. 4, A and B) and Arabidopsis (Fig. 4, E and F) cells. The results from both plants are similar in terms of plastid targeting (Table 2). We quantified the in vivo targeting efficiencies of these 18 constructs from onion cells. In each pair, the mutant based on SSF had higher efficiency than the SSR mutant (Fig. 4I). This small but reproducible difference indicated that the targeting efficiency of the constructs is not only affected by the N-ter peptide sequence but also from secondary contributions derived from the sequence of SSF or SSR. Nevertheless, the better ability to translocate preproteins containing two tandem H70BSs (N-ter peptide and N-ter of SSF) may be the result of cooperativity at one or more step during translocation.
By comparing in vivo import efficiency with %UA and CSS1 affinity values, we found a positive correlation in both %UA and CSS1 affinity (Fig. 4, K and L). These correlations were determined without the values from the dual-targeting constructs and the pp9 and pp38 constructs. In general, TPs with a higher %UA or CSS1 affinity N-ter had higher in vivo import efficiency. However, the correlation between CSS1 affinity and the import efficiency at r of 0.72 (p value of 0.01) is much greater than the correlation of %UA at r of 0.22 (p value of 0.51).
Fig. 9 summarizes the import assays for both in vivo (left axis) and in vitro (right axis) targeting results of each peptide when placed in front of either the SSF and SSR constructs. The N-ter peptides are shown according to their experimental CSS1 affinities (x axis). If we used a cutoff of 10 for the affinity, it is clear that the mutants with noninteracting N-ter cannot support the import, although five out of seven mutant sets with H70BS N-ter can. The pp9 and pp38 peptides failed to support import both in vivo and in vitro despite having strong Hsp70 affinities. The lack of protein import of the pp9 and pp38 constructs requires some additional analysis because both contain N-ter with 100%UA and strong Hsp70 affinity (Figs. 2, C and D, 3G, and 4I). This indicates that other properties of their N-ter either fail to comply with some other criteria for the import or possibly have some elements that block translocation. Our results (Fig. 4I) and previous results (21) showed that only precursors containing H70BS N-ter were competent in plastid targeting indicating that these TPs met the requirements for every step. Because the TP N-ter domain is required for the translocation step (21), we hypothesize that the failure of pp9 and pp38 peptides occurs during binding and/or intermediate steps.
FIGURE 9.
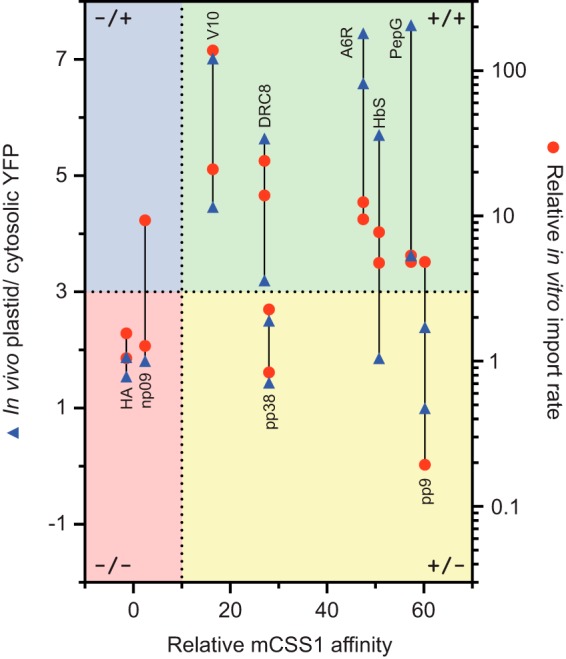
Summary of in vivo and in vitro results based on the relative mCSS1 affinity of the N-ter peptides. The in vivo ratio of 3, the relative in vitro import rate of 3, and the relative mCSS1 affinity of 10 were used to divide these data into four groups as follows: negative affinity-negative import (−/−), negative affinity-positive import (−/+), positive affinity-negative import (+/−), and positive affinity-positive import (+/+), which are highlighted in red, blue, yellow, and green areas, respectively. A line is drawn to connect the data points from the same N-ter sequence. Note that the scaled values from in vitro import of batch 2 were used here.
The sequences of pp9 and pp38 contain multiple aromatic aa (Fig. 2A). Aromatic aa are well known to interact directly with lipids in the membrane (60, 61). Trp and Phe are not only hydrophobic but also have very large side chains. Although TPs on average have a relatively constant HASA at <500 Å2/8 aa, pp9 and pp38 constructs have >2 times HASA (Fig. 8A and supplemental Fig. S2). Pulse-chase assays showed that these constructs were able to form the early import intermediates but failed to be translocated (Fig. 8B). It may be that these aa interact very strongly with the lipid bilayer in such a way that the TP cannot productively engage one or more components of the translocon. Further experimentation and mutagenesis may uncover how these peptides interact with artificial membranes mimicking the chloroplast outer envelope. It has already been shown that the N-ter of SStp can interact with lipids and undergo a conformational change (12, 14) that may be involved in the initial reversible binding step.
Plastid-localized Hsp70s May Have Different Affinity for Peptides
This work and previous studies (22–24) have shown that most TPs contain a strongly predicted H70BS. Analysis of early datasets such as the TargetP training set have indicated that these sequences are largely located at the N-ter; however, our current work as well as the work by Rial et al. (23) indicate that these sequences may be distributed throughout the TP sequence with only ∼35% being found within the N-ter third of the TP. One limitation of this type of analysis is that the algorithms used were developed from bacterial DnaK. It is possible that the structural properties of the SBD of the plastid Hsp70 may be quite different from DnaK. To investigate this, we built a model of the SBD (aa 460–682) of a plastid Hsp70 from Arabidopsis, cpHsc70-1, using the crystal structure of the E. coli DnaK SBD (aa 389–607) bound to a 7-aa peptide (Fig. 10A). This domain has 52% identity and 72% similarity to the SBD of E. coli DnaK. AtcpHsc70-1 is even less similar to the mammalian chaperones with only 50 and 49% identity to bovine Hsc70 and BiP, respectively. The alignment is shown in Fig. 10B.
Analysis of the homology model indicates that AtcpHsc70-1 has considerable differences in aa that line the peptide-binding channel (Fig. 10, C and D). Based on the bound peptide in DnaK structure, it is clear that in the AtcpHsc70-1 model several aa near the C-ter of the peptide are quite a bit larger and more charged (Glu-508 and Arg-529 compared with Ser-437 and Asn-458 of DnaK, respectively). It is possible that these structural changes could result in a significant difference in the affinity and specificity for how AtcpHsc70-1 binds peptides. Therefore, it is possible that the algorithms used to predict the H70BSs of TPs may not be fully accurate in predicting where the plastid Hsp70 binds TPs. Unfortunately, without any additional data, these existing prediction tools are the best we have to predict Hsp70 binding.
Multiple Chaperones May Drive Translocation
Although the mechanism of protein translocation into plastids is still poorly understood (4, 62–65), most current models illustrate a vectorial transfer of the TP into the organelle with the N-ter entering the stroma first and the C-ter and the mature domain remaining behind. It has also been suggested that the TP enters and is translocated via the TOC and TIC translocons as an extended linear peptide devoid of secondary structure (66–68). These models and our observation of an N-ter placement of a strong H70BS indicate that when the TP emerges from the translocon pore, the interaction with the stromal Hsp70 traps the precursor and initiates the translocation. This is consistent with the role of Hsp70s functioning as a molecular motor that couples ATP hydrolysis to the vectorial movement of preprotein translocation across the plastid membranes. This work is supported by experimental work that has shown reduced protein import upon disruption of plastid Hsp70 in several plants, including moss (28), pea, and Arabidopsis (29). It was clearly shown that knock-out mutants of either of the two stromal Hsp70s lead to a defect in precursor translocation across the membrane (29). However, protein import still partially functioned in these plants suggesting other ATP-dependent protein translocation mechanisms.
Although our current analysis did not preclude the involvement of cytosolic Hsp70s, our previous report indicated that the interaction of the TP N-ter with a component within the isolated chloroplasts is essential for the translocation process (21). Nevertheless, the cis-located or cytosolic Hsp70 may involve in this process in vivo.
In prokaryotes, the ATPases of the Hsp100/Clp family enable the ATP-dependent degradation of the ClpAP, ClpXP, and other proteases (69). The plastid stroma contains both ClpP and ClpC that function as two-component ATP-dependent protease (70, 71). It has been shown that some Hsp100 members, ClpB/Hsp104, are capable of disaggregating protein without proteolytic degradation, suggesting that ClpB functions independently of ClpP or another proteolytic subunit (72). Several studies indicate that ClpC (now called Hsp93) is involved in the import of preproteins into chloroplasts (73, 74). In Arabidopsis, there are two plastid forms of ClpC, AtHsp93V and AtHsp93III. Knock-out mutants of hsp93III have a normal phenotype, yet hsp93V mutant plants are defective in protein import (75, 76). It is believed that Hsp93 interacts with incoming preproteins in the Tic110-Tic40-Hsp93 complex (77).
Unfortunately, the details of peptide recognition by the Clp/Hsp100 chaperones are poorly understood. Earlier structural work with E. coli ClpA indicates it has three functional domains as follows: an N-ter domain and two ATPase domains (78). The N-ter is believed to function as the SBD. Both ClpA and ClpB have a prominent hydrophobic surface that lies between two helices (79). Interestingly, it was recently shown that it is the N-ter domain of chloroplast Hsp93 that is important for its function in the import (80). These data in combination with the structural data of ClpA suggest that the Hsp93 N-ter is responsible for binding both to substrates and possibly to TIC. Future work will be needed to elucidate whether Hsp93 has a similar hydrophobic patch that may bind to incoming TPs.
Concluding Remarks
Our results indicate that about 77% of TPs contain an N-ter placed H70BS, although about a third of the TPs contain the strongest HS70BS within their N-ter 20 aa. Although most models put Hsp93 as the motor that drives protein translocation, recent publications have suggested that a stromal Hsp70 is also involved in chloroplast protein import (28, 29, 57, 63, 81, 82). Thus import may be mediated by at least two motor proteins, Hsp93 and Hsp70. Interestingly, when Hsp93V is knocked out and Hsp93III is knocked down, plastids can still import proteins at 40–60% of the wild type, suggesting that Hsp70 or some other ATPase function as the motor (83). Stromal Hsp70 interaction may initiate the translocation process based on N-ter H70BS in TPs, although the TPs lacking an N-ter H70BS may utilize other stromal chaperones such as Hsp93. For example, the TP of ferredoxin-NADPH reductase (FNRtp) intrinsically lacks a strong N-ter H70BS based on our analysis in Fig. 10E. Both FNRtp and its mutant (FNRtp-1234) having all of the H70BSs mutated were still able to support import (84). FNRtp was recently found to interact with Hsp93 only when it is located at the N-ter of preprotein (85). This suggests that FNRtp may be an example of a TP that utilizes a N-ter Hsp93-binding site to initiate translocation.
It is clear that two chaperones either alone or together are responsible for the stromal ATP requirement during import (25). Although all known translocons involve one or more peripherally associated NTPase to drive translocation (86), the Hsp100 family has only been shown to participate in protein translocation in other systems like the endoplasmic reticulum and mitochondria where the Hsp70/BiP system is lost or compromised (87). This suggests that the plastid may be unique because it utilizes both chaperones as motors even in wild-type plants (81). Considering the possible dual involvement of Hsp93 and Hsp70, one can envision two modes of function as follows: (a) they bind TPs sequentially and independently or (b) they bind TPs simultaneously and synergistically (81). Recent work has shown some Hsp100 are intimately associated with DnaK, and it is now clear that disaggregation function of ClpB involved interaction with DnaK (88, 89). This observation would support synergistic model B. Many works have shown that the same chloroplast import intermediates were associated with both Hsp93 and Hsp70 (28, 29) and recently also with Hsp90 (90). It is possible that different TPs may utilize different modes of Hsp70/Hsp93/Hsp90 interaction that depend on the TP sequence and organization. These variations may result in different levels of translocation force and may be why the plastid translocation process is a more robust unfoldase than what is observed in mitochondria (91) or the AAA+ ATPases alone (92). It will be very interesting to develop single molecule force measurements or other methods that may reveal how different TPs are recognized by this multichaperone system. Our work has clearly shown that one class of TPs seems to require an Hsp70 recognition domain at the N-ter and indicates that other TPs may be initially recognized by the Hsp93 or Hsp90 chaperones by binding to currently unknown elements probably also placed at the N-ter.
Supplementary Material
Acknowledgments
The plasmid of CSS1 cDNA, pSP65, was kindly provided by Dr. Kenneth Keegstra. We thank Andreas Nebenführ for providing organelle marker constructs. We thank Richard Simmerman, Khoa Nguyen, Kristen Holbrook, and J. Ridge Carter for critically reading the manuscript.
This work was supported in part by National Science Foundation Program in Cell Biology Grants MCB-0628670 and MCB-0344601 (to B. D. B.), TN-SCORE, a multidisciplinary research program sponsored by NSF-EPSCOR Grant EPS-1004083 (to B. D. B.), and support from the Gibson Family Foundation (to B. D. B.).

This article contains supplemental Figs. S1 and S2 and Tables S1 and S2.
- TP
- transit peptide
- TOC and TIC
- translocons at outer and inner chloroplast envelopes
- aa
- amino acid
- SSU
- small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase
- FD
- ferredoxin
- SStp and FDtp
- TPs of SSU and FD
- H70BS
- Hsp70-binding site
- RPPD
- random peptide phage display-derived algorithm
- mSSU
- the mature domain of SSU
- m20
- first 20-aa of mSSU
- mCSS1
- the mature domain of CSS1
- RCMLA
- reduced carboxymethylated α-lactalbumin
- F-RCMLA
- fluorescein-labeled RCMLA
- SBD
- substrate-binding domain
- PSSM
- position-specific scoring matrix
- %UA
- percentage of uncharged aa
- HASA
- hydrophobic accessible surface area
- N-ter
- N-terminal or N terminus
- C-ter
- C-terminal or C terminus
- SSF
- forward (native SStp)
- FDF
- forward (native) FDtp
- FDR
- reversed FDtp
- SSR
- reversed SStp.
REFERENCES
- 1. Richly E., Leister D. (2004) An improved prediction of chloroplast proteins reveals diversities and commonalities in the chloroplast proteomes of Arabidopsis and rice. Gene 329, 11–16 [DOI] [PubMed] [Google Scholar]
- 2. Zybailov B., Rutschow H., Friso G., Rudella A., Emanuelsson O., Sun Q., van Wijk K. J. (2008) Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS One 3, e1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kleffmann T., Russenberger D., von Zychlinski A., Christopher W., Sjölander K., Gruissem W., Baginsky S. (2004) The Arabidopsis thaliana chloroplast proteome reveals pathway abundance and novel protein functions. Curr. Biol. 14, 354–362 [DOI] [PubMed] [Google Scholar]
- 4. Bruce B. D. (2000) Chloroplast transit peptides: structure, function and evolution. Trends Cell Biol. 10, 440–447 [DOI] [PubMed] [Google Scholar]
- 5. Bruce B. D. (2001) The paradox of plastid transit peptides: conservation of function despite divergence in primary structure. Biochim. Biophys. Acta 1541, 2–21 [DOI] [PubMed] [Google Scholar]
- 6. Dobberstein B., Blobel G., Chua N. H. (1977) In vitro synthesis and processing of a putative precursor for the small subunit of ribulose-1,5-bisphosphate carboxylase of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U.S.A. 74, 1082–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Emanuelsson O., Brunak S., von Heijne G., Nielsen H. (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2, 953–971 [DOI] [PubMed] [Google Scholar]
- 8. Small I., Peeters N., Legeai F., Lurin C. (2004) Predotar: A tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 4, 1581–1590 [DOI] [PubMed] [Google Scholar]
- 9. Petsalaki E. I., Bagos P. G., Litou Z. I., Hamodrakas S. J. (2006) PredSL: a tool for the N-terminal sequence-based prediction of protein subcellular localization. Genomics Proteomics Bioinformatics 4, 48–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lee D. W., Kim J. K., Lee S., Choi S., Kim S., Hwang I. (2008) Arabidopsis nuclear-encoded plastid transit peptides contain multiple sequence subgroups with distinctive chloroplast-targeting sequence motifs. Plant Cell 20, 1603–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. von Heijne G., Nishikawa K. (1991) Chloroplast transit peptides. The perfect random coil? FEBS Lett. 278, 1–3 [DOI] [PubMed] [Google Scholar]
- 12. Bruce B. D. (1998) The role of lipids in plastid protein transport. Plant Mol. Biol. 38, 223–246 [PubMed] [Google Scholar]
- 13. von Heijne G., Steppuhn J., Herrmann R. G. (1989) Domain structure of mitochondrial and chloroplast targeting peptides. Eur. J. Biochem. 180, 535–545 [DOI] [PubMed] [Google Scholar]
- 14. Pinnaduwage P., Bruce B. D. (1996) In vitro interaction between a chloroplast transit peptide and chloroplast outer envelope lipids is sequence-specific and lipid class-dependent. J. Biol. Chem. 271, 32907–32915 [DOI] [PubMed] [Google Scholar]
- 15. Pilon M., Wienk H., Sips W., de Swaaf M., Talboom I., van 't Hof R., de Korte-Kool G., Demel R., Weisbeek P., de Kruijff B. (1995) Functional domains of the ferredoxin transit sequence involved in chloroplast import. J. Biol. Chem. 270, 3882–3893 [DOI] [PubMed] [Google Scholar]
- 16. Rensink W. A., Pilon M., Weisbeek P. (1998) Domains of a transit sequence required for in vivo import in Arabidopsis chloroplasts. Plant Physiol. 118, 691–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. van den Wijngaard P. W., Vredenberg W. J. (1997) A 50-picosiemens anion channel of the chloroplast envelope is involved in chloroplast protein import. J. Biol. Chem. 272, 29430–29433 [DOI] [PubMed] [Google Scholar]
- 18. Lee D. W., Lee S., Lee G. J., Lee K. H., Kim S., Cheong G. W., Hwang I. (2006) Functional characterization of sequence motifs in the transit peptide of Arabidopsis small subunit of rubisco. Plant Physiol. 140, 466–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee D. W., Lee S., Oh Y. J., Hwang I. (2009) Multiple sequence motifs in the rubisco small subunit transit peptide independently contribute to Toc159-dependent import of proteins into chloroplasts. Plant Physiol. 151, 129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee K. H., Kim D. H., Lee S. W., Kim Z. H., Hwang I. (2002) In vivo import experiments in protoplasts reveal the importance of the overall context but not specific amino acid residues of the transit peptide during import into chloroplasts. Mol. Cells 14, 388–397 [PubMed] [Google Scholar]
- 21. Chotewutmontri P., Reddick L. E., McWilliams D. R., Campbell I. M., Bruce B. D. (2012) Differential transit peptide recognition during preprotein binding and translocation into flowering plant plastids. Plant Cell 24, 3040–3059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ivey R. A., 3rd, Subramanian C., Bruce B. D. (2000) Identification of a Hsp70 recognition domain within the rubisco small subunit transit peptide. Plant Physiol. 122, 1289–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rial D. V., Arakaki A. K., Ceccarelli E. A. (2000) Interaction of the targeting sequence of chloroplast precursors with Hsp70 molecular chaperones. Eur. J. Biochem. 267, 6239–6248 [DOI] [PubMed] [Google Scholar]
- 24. Zhang X. P., Glaser E. (2002) Interaction of plant mitochondrial and chloroplast signal peptides with the Hsp70 molecular chaperone. Trends Plant Sci. 7, 14–21 [DOI] [PubMed] [Google Scholar]
- 25. Theg S. M., Bauerle C., Olsen L. J., Selman B. R., Keegstra K. (1989) Internal Atp is the only energy requirement for the translocation of precursor proteins across chloroplastic membranes. J. Biol. Chem. 264, 6730–6736 [PubMed] [Google Scholar]
- 26. Marshall J. S., DeRocher A. E., Keegstra K., Vierling E. (1990) Identification of heat shock protein hsp70 homologues in chloroplasts. Proc. Natl. Acad. Sci. U.S.A. 87, 374–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schnell D. J., Kessler F., Blobel G. (1994) Isolation of components of the chloroplast protein import machinery. Science 266, 1007–1012 [DOI] [PubMed] [Google Scholar]
- 28. Shi L. X., Theg S. M. (2010) A stromal heat shock protein 70 system functions in protein import into chloroplasts in the moss physcomitrella patens. Plant Cell 22, 205–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Su P. H., Li H. M. (2010) Stromal Hsp70 is important for protein translocation into pea and arabidopsis chloroplasts. Plant Cell 22, 1516–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Keegstra K., Cline K. (1999) Protein import and routing systems of chloroplasts. Plant Cell 11, 557–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fourie A. M., Sambrook J. F., Gething M. J. (1994) Common and divergent peptide binding specificities of hsp70 molecular chaperones. J. Biol. Chem. 269, 30470–30478 [PubMed] [Google Scholar]
- 32. Nelson B. K., Cai X., Nebenführ A. (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51, 1126–1136 [DOI] [PubMed] [Google Scholar]
- 33. Reddick L. E., Chotewutmontri P., Crenshaw W., Dave A., Vaughn M., Bruce B. D. (2008) Nano-scale characterization of the dynamics of the chloroplast Toc translocon. Methods Cell Biol. 90, 365–398 [DOI] [PubMed] [Google Scholar]
- 34. Dabney-Smith C., van Den Wijngaard P. W., Treece Y., Vredenberg W. J., Bruce B. D. (1999) The C terminus of a chloroplast precursor modulates its interaction with the translocation apparatus and PIRAC. J. Biol. Chem. 274, 32351–32359 [DOI] [PubMed] [Google Scholar]
- 35. Klein R. R., Salvucci M. E. (1992) Photoaffinity labeling of mature and precursor forms of the small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase after expression in Escherichia coli. Plant Physiol. 98, 546–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Olsen L. J., Keegstra K. (1992) The binding of precursor proteins to chloroplasts requires nucleoside triphosphates in the intermembrane space. J. Biol. Chem. 267, 433–439 [PubMed] [Google Scholar]
- 37. Bösl B., Grimminger V., Walter S. (2005) Substrate binding to the molecular chaperone Hsp104 and its regulation by nucleotides. J. Biol. Chem. 280, 38170–38176 [DOI] [PubMed] [Google Scholar]
- 38. Emanuelsson O., Nielsen H., Brunak S., von Heijne G. (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016 [DOI] [PubMed] [Google Scholar]
- 39. Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Miller S., Janin J., Lesk A. M., Chothia C. (1987) Interior and surface of monomeric proteins. J. Mol. Biol. 196, 641–656 [DOI] [PubMed] [Google Scholar]
- 41. Gragerov A., Zeng L., Zhao X., Burkholder W., Gottesman M. E. (1994) Specificity of DnaK-peptide binding. J. Mol. Biol. 235, 848–854 [DOI] [PubMed] [Google Scholar]
- 42. Wilkins M. R., Gasteiger E., Bairoch A., Sanchez J. C., Williams K. L., Appel R. D., Hochstrasser D. F. (1999) Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 112, 531–552 [DOI] [PubMed] [Google Scholar]
- 43. Kyte J., Doolittle R. F. (1982) A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132 [DOI] [PubMed] [Google Scholar]
- 44. Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. (2004) WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Henikoff S. (1996) Scores for sequence searches and alignments. Curr. Opin. Struct. Biol. 6, 353–360 [DOI] [PubMed] [Google Scholar]
- 46. Hertz G. Z., Stormo G. D. (1999) Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics 15, 563–577 [DOI] [PubMed] [Google Scholar]
- 47. Nielsen H., Engelbrecht J., Brunak S., von Heijne G. (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10, 1–6 [DOI] [PubMed] [Google Scholar]
- 48. Horton P., Park K. J., Obayashi T., Fujita N., Harada H., Adams-Collier C. J., Nakai K. (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35, W585–W587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Briesemeister S., Rahnenführer J., Kohlbacher O. (2010) Going from where to why–interpretable prediction of protein subcellular localization. Bioinformatics 26, 1232–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhu X., Zhao X., Burkholder W. F., Gragerov A., Ogata C. M., Gottesman M. E., Hendrickson W. A. (1996) Structural analysis of substrate binding by the molecular chaperone DnaK. Science 272, 1606–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Notredame C., Higgins D. G., Heringa J. (2000) T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302, 205–217 [DOI] [PubMed] [Google Scholar]
- 52. Arnold K., Bordoli L., Kopp J., Schwede T. (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 53. von Heijne G., Hirai T., Klösgen R., Steppuhn J., Bruce B., Keegstra K., Herrmann R. (1991) CHLPEP—A database of chloroplast transit peptides. Plant Mol. Biol. Rep. 9, 104–126 [Google Scholar]
- 54. Rüdiger S., Germeroth L., Schneider-Mergener J., Bukau B. (1997) Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J. 16, 1501–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Klee E. W., Ellis L. B. (2005) Evaluating eukaryotic secreted protein prediction. BMC Bioinformatics 6, 256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Marshall J. S., Keegstra K. (1992) Isolation and characterization of a cDNA clone encoding the major hsp70 of the pea chloroplastic stroma. Plant Physiol. 100, 1048–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Li H. M., Teng Y. S. (2013) Transit peptide design and plastid import regulation. Trends Plant Sci. 18, 360–366 [DOI] [PubMed] [Google Scholar]
- 58. Teng Y. S., Chan P. T., Li H. M. (2012) Differential age-dependent import regulation by signal peptides. PLoS Biol. 10, e1001416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rüdiger S., Buchberger A., Bukau B. (1997) Interaction of Hsp70 chaperones with substrates. Nat. Struct. Biol. 4, 342–349 [DOI] [PubMed] [Google Scholar]
- 60. Killian J. A., von Heijne G. (2000) How proteins adapt to a membrane-water interface. Trends Biochem. Sci. 25, 429–434 [DOI] [PubMed] [Google Scholar]
- 61. de Planque M. R., Bonev B. B., Demmers J. A., Greathouse D. V., Koeppe R. E., 2nd, Separovic F., Watts A., Killian J. A. (2003) Interfacial anchor properties of tryptophan residues in transmembrane peptides can dominate over hydrophobic matching effects in peptide-lipid interactions. Biochemistry 42, 5341–5348 [DOI] [PubMed] [Google Scholar]
- 62. Li H. M., Chiu C. C. (2010) Protein transport into chloroplasts. Annu. Rev. Plant Biol. 61, 157–180 [DOI] [PubMed] [Google Scholar]
- 63. Shi L. X., Theg S. M. (2013) The chloroplast protein import system: from algae to trees. Biochim. Biophys. Acta 1833, 314–331 [DOI] [PubMed] [Google Scholar]
- 64. Jarvis P. (2008) Targeting of nucleus-encoded proteins to chloroplasts in plants. New Phytol. 179, 257–285 [DOI] [PubMed] [Google Scholar]
- 65. Soll J., Schleiff E. (2004) Protein import into chloroplasts. Nat. Rev. Mol. Cell Biol. 5, 198–208 [DOI] [PubMed] [Google Scholar]
- 66. Hinnah S. C., Wagner R., Sveshnikova N., Harrer R., Soll J. (2002) The chloroplast protein import channel Toc75: pore properties and interaction with transit peptides. Biophys. J. 83, 899–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Bionda T., Tillmann B., Simm S., Beilstein K., Ruprecht M., Schleiff E. (2010) Chloroplast import signals: the length requirement for translocation in vitro and in vivo. J. Mol. Biol. 402, 510–523 [DOI] [PubMed] [Google Scholar]
- 68. Ruprecht M., Bionda T., Sato T., Sommer M. S., Endo T., Schleiff E. (2010) On the impact of precursor unfolding during protein import into chloroplasts. Mol. Plant 3, 499–508 [DOI] [PubMed] [Google Scholar]
- 69. Langer T. (2000) AAA proteases: cellular machines for degrading membrane proteins. Trends Biochem. Sci. 25, 247–251 [DOI] [PubMed] [Google Scholar]
- 70. Shanklin J., DeWitt N. D., Flanagan J. M. (1995) The stroma of higher plant plastids contain ClpP and ClpC, functional homologs of Escherichia coli ClpP and ClpA: an archetypal two-component ATP-dependent protease. Plant Cell 7, 1713–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Olinares P. D., Kim J., van Wijk K. J. (2011) The Clp protease system; a central component of the chloroplast protease network. Biochim. Biophys. Acta 1807, 999–1011 [DOI] [PubMed] [Google Scholar]
- 72. Mizuno S., Nakazaki Y., Yoshida M., Watanabe Y. H. (2012) Orientation of the amino-terminal domain of ClpB affects the disaggregation of the protein. FEBS J. 279, 1474–1484 [DOI] [PubMed] [Google Scholar]
- 73. Akita M., Nielsen E., Keegstra K. (1997) Identification of protein transport complexes in the chloroplastic envelope membranes via chemical cross-linking. J. Cell Biol. 136, 983–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Nielsen E., Akita M., Davila-Aponte J., Keegstra K. (1997) Stable association of chloroplastic precursors with protein translocation complexes that contain proteins from both envelope membranes and a stromal Hsp100 molecular chaperone. EMBO J. 16, 935–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Constan D., Froehlich J. E., Rangarajan S., Keegstra K. (2004) A stromal Hsp100 protein is required for normal chloroplast development and function in Arabidopsis. Plant Physiol. 136, 3605–3615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kovacheva S., Bédard J., Patel R., Dudley P., Twell D., Ríos G., Koncz C., Jarvis P. (2005) In vivo studies on the roles of Tic110, Tic40 and Hsp93 during chloroplast protein import. Plant J. 41, 412–428 [DOI] [PubMed] [Google Scholar]
- 77. Chou M. L., Chu C. C., Chen L. J., Akita M., Li H. M. (2006) Stimulation of transit-peptide release and ATP hydrolysis by a cochaperone during protein import into chloroplasts. J. Cell Biol. 175, 893–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Guo F., Maurizi M. R., Esser L., Xia D. (2002) Crystal structure of ClpA, an Hsp100 chaperone and regulator of ClpAP protease. J. Biol. Chem. 277, 46743–46752 [DOI] [PubMed] [Google Scholar]
- 79. Xia D., Esser L., Singh S. K., Guo F., Maurizi M. R. (2004) Crystallographic investigation of peptide binding sites in the N-domain of the ClpA chaperone. J. Struct. Biol. 146, 166–179 [DOI] [PubMed] [Google Scholar]
- 80. Chu C. C., Li H. M. (2012) The amino-terminal domain of chloroplast Hsp93 is important for its membrane association and functions in vivo. Plant Physiol. 158, 1656–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Shi L. X., Theg S. M. (2011) The motors of protein import into chloroplasts. Plant Signal. Behav. 6, 1397–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Flores-Pérez Ú., Jarvis P. (2013) Molecular chaperone involvement in chloroplast protein import. Biochim. Biophys. Acta 1833, 332–340 [DOI] [PubMed] [Google Scholar]
- 83. Kovacheva S., Bédard J., Wardle A., Patel R., Jarvis P. (2007) Further in vivo studies on the role of the molecular chaperone, Hsp93, in plastid protein import. Plant J. 50, 364–379 [DOI] [PubMed] [Google Scholar]
- 84. Rial D. V., Ottado J., Ceccarelli E. A. (2003) Precursors with altered affinity for Hsp70 in their transit peptides are efficiently imported into chloroplasts. J. Biol. Chem. 278, 46473–46481 [DOI] [PubMed] [Google Scholar]
- 85. Bruch E. M., Rosano G. L., Ceccarelli E. A. (2012) Chloroplastic Hsp100 chaperones ClpC2 and ClpD interact in vitro with a transit peptide only when it is located at the N terminus of a protein. BMC Plant Biol. 12, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Schatz G., Dobberstein B. (1996) Common principles of protein translocation across membranes. Science 271, 1519–1526 [DOI] [PubMed] [Google Scholar]
- 87. Schmitt M., Neupert W., Langer T. (1995) Hsp78, a Clp homolog within mitochondria, can substitute for chaperone functions of Mt-Hsp70. EMBO J. 14, 3434–3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Rosenzweig R., Moradi S., Zarrine-Afsar A., Glover J. R., Kay L. E. (2013) Unraveling the mechanism of protein disaggregation through a ClpB-DnaK interaction. Science 339, 1080–1083 [DOI] [PubMed] [Google Scholar]
- 89. Seyffer F., Kummer E., Oguchi Y., Winkler J., Kumar M., Zahn R., Sourjik V., Bukau B., Mogk A. (2012) Hsp70 proteins bind Hsp100 regulatory M domains to activate AAA+ disaggregase at aggregate surfaces. Nat. Struct. Mol. Biol. 19, 1347–1355 [DOI] [PubMed] [Google Scholar]
- 90. Inoue H., Li M., Schnell D. J. (2013) An essential role for chloroplast heat shock protein 90 (Hsp90C) in protein import into chloroplasts. Proc. Natl. Acad. Sci. U.S.A. 110, 3173–3178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. America T., Hageman J., Guéra A., Rook F., Archer K., Keegstra K., Weisbeek P. (1994) Methotrexate does not block import of a DHFR fusion protein into chloroplasts. Plant Mol. Biol. 24, 283–294 [DOI] [PubMed] [Google Scholar]
- 92. Herman C., Prakash S., Lu C. Z., Matouschek A., Gross C. A. (2003) Lack of a robust unfoldase activity confers a unique level of substrate specificity to the universal AAA protease FtsH. Mol. Cell 11, 659–669 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.