Background: Glycosyltransferase inhibitors have important applications in therapeutics and as chemical biology tools.
Results: The human β1,4-galactosyltransferase 7 enzyme active site was mapped by modeling, mutagenesis, and in vitro/in cellulo assays, and novel inhibitors were synthesized.
Conclusion: An efficient inhibitor of β1,4-galactosyltransferase 7 and glycosaminoglycan synthesis was obtained.
Significance: This inhibitory molecule can be exploited to investigate glycosaminoglycan biology and modulate glycosaminoglycan synthesis in therapeutics.
Keywords: Enzyme Inhibitor, Enzyme Kinetics, Glycosaminoglycan, Glycosyltransferase, Proteoglycan Synthesis, Site-directed Mutagenesis
Abstract
Among glycosaminoglycan (GAG) biosynthetic enzymes, the human β1,4-galactosyltransferase 7 (hβ4GalT7) is characterized by its unique capacity to take over xyloside derivatives linked to a hydrophobic aglycone as substrates and/or inhibitors. This glycosyltransferase is thus a prime target for the development of regulators of GAG synthesis in therapeutics. Here, we report the structure-guided design of hβ4GalT7 inhibitors. By combining molecular modeling, in vitro mutagenesis, and kinetic measurements, and in cellulo analysis of GAG anabolism and decorin glycosylation, we mapped the organization of the acceptor binding pocket, in complex with 4-methylumbelliferone-xylopyranoside as prototype substrate. We show that its organization is governed, on one side, by three tyrosine residues, Tyr194, Tyr196, and Tyr199, which create a hydrophobic environment and provide stacking interactions with both xylopyranoside and aglycone rings. On the opposite side, a hydrogen-bond network is established between the charged amino acids Asp228, Asp229, and Arg226, and the hydroxyl groups of xylose. We identified two key structural features, i.e. the strategic position of Tyr194 forming stacking interactions with the aglycone, and the hydrogen bond between the His195 nitrogen backbone and the carbonyl group of the coumarinyl molecule to develop a tight binder of hβ4GalT7. This led to the synthesis of 4-deoxy-4-fluoroxylose linked to 4-methylumbelliferone that inhibited hβ4GalT7 activity in vitro with a Ki 10 times lower than the Km value and efficiently impaired GAG synthesis in a cell assay. This study provides a valuable probe for the investigation of GAG biology and opens avenues toward the development of bioactive compounds to correct GAG synthesis disorders implicated in different types of malignancies.
Introduction
Glycosaminoglycans (GAGs)2 are linear heteropolysaccharide chains covalently attached to the core protein of a variety of proteoglycans (PGs). Because of their high structural diversity and their anionic characteristics, GAGs interact with a network of cellular and extracellular mediators including cytokines and chemokines, enzymes and enzyme inhibitors, matrix proteins, and membrane receptors (1). There is currently great emphasis on the crucial roles of GAGs in numerous physiological events including cell differentiation, proliferation and migration (2), and its pathological aspects, such as tumor formation, progression, and metastasis (3). Furthermore, because PGs are ubiquitously expressed in extracellular matrices and on cell surfaces of virtually every tissue, they are also involved in the normal and pathological functions of the cardiovascular and osteoarticular system (4), in amyloid disorders (5) and in axonal de- and regeneration (6). GAG biosynthesis is initiated by the formation of a tetrasaccharide linkage region (GlcAβ1–3Galβ1–3Galβ1–4Xylβ1-O-) covalently linked to serine residues of the PG core protein (7). This tetrasaccharide acts as a primer for the elongation of major GAG chains, i.e. chondroitin/dermatan sulfate or heparin/heparan sulfate, which polymerization involves the coordinated activities of chondroitin-sulfate synthases and heparan-sulfate synthases (exostosins, EXT), respectively (8, 9). Mature GAG chains are finally produced by the modifications of their constitutive disaccharide units catalyzed by epimerases and sulfotransferases, which considerably increase their structural and functional diversity (10, 11).
The human xylosylprotein β1,4-galactosyltransferase (EC 2.4.1.1337, hβ4GalT7) catalyzes the transfer of the first Gal residue of the tetrasaccharide linkage from the activated sugar UDP-galactose (UDP-Gal) onto Xyl residues attached to the PG core protein (12). Because all GAGs share the same stem core tetrasaccharide, β4GalT7 is a central enzyme in GAG biosynthesis. Indeed, hβ4GalT7 mutations have been associated with a rare genetic condition, the progeroid form of Ehlers-Danlos syndrome (EDS), a group of connective tissue disorders characterized by a major deficiency in PG synthesis. As a consequence of GAG defect, EDS patients exhibit motor development delay, and musculoskeletal malformations, hypermobile joints, and wound healing defaults (13). Patients gene sequencing revealed the presence of missense mutations leading to L206P, A186D (14, 15), and R270C substitutions (16) in the catalytic domain, resulting in a partially or totally inactive enzyme. Recently, we showed that R270C replacement reduced affinity toward the xyloside acceptor and strongly affected GAG chains formation in β4GalT7-deficient CHOpgsB-618 cells (17). There is currently no effective therapy for treating EDS patients.
Interestingly, the biosynthesis of GAGs can be manipulated by simple xylosides carrying a hydrophobic aglycone, which act as substrates and/or inhibitors of hβ4GalT7. Xyloside analogs have been shown to efficiently induce GAG synthesis bypassing the natural Xyl-substituted core protein of PGs for several decades (18, 19). The xyloside-primed GAG chains are usually excreted and show interesting biological functions such as activation of fibroblast growth factor (FGF) signaling (20, 21), antithrombotic (22), tissue regenerating (23), anti-angiogenic (24) and anti-proliferative properties (25, 26). In addition, several groups have synthesized a series of xyloside analogs as potential inhibitors of GAG synthesis. Such compounds would represent highly valuable chemical biology tools to probe the functions of GAGs in cell systems and model organisms and as a starting point toward the development of pharmaceuticals, in particular anti-tumor agents. Recently, Garud et al. (27) and Tsuzuki et al. (28) used click chemistry to generate libraries of 4-deoxy-4-fluorotriazole analogs comprising a set of hydrophobic molecules appended to the anomeric carbon of the xyloside. Siegbahn et al. (29, 30) developed a collection of naphthyl and benzyl xylosides substituted on different positions of the Xyl moiety. These studies led to the discovery of promising xyloside-derived inhibitors of GAG synthesis when screened in cell models.
However, until recently, the development of substrates and inhibitors of β4GalT7 has been mostly limited to the synthesis of libraries of analog compounds and their testing in cell assays. Toward the rational design of hβ4GalT7 inhibitors, we have been involved in structure-activity relationship studies of the recombinant human enzyme for several years and identified critical active site amino acids implicated in catalysis and/or substrate binding (17, 31, 32). We previously investigated the importance of conserved 163DVD165 and 221FWGWRGEDDE230 motifs in the organization of the catalytic domain. Our data have highlighted the crucial role of Trp224 in substrate recognition and suggested a catalytic role for Asp228 (31). These findings were in accordance with the structural data from the recently solved crystal structure of the catalytic domain of Drosophila melanogaster dβ4GalT7 (33) and the human enzyme (34).
In the current study, we developed a structure-guided approach for the design of xyloside inhibitors of hβ4GalT7 that were tested on its galactosyltransferase activity in vitro and on GAG biosynthesis in cell assays. We explored the organization of the acceptor binding pocket, specifically probing the functional and structural contribution of a set of residues located in the vicinity of the catalytic center, and highlighted the crucial role of three tyrosine residues, i.e. Tyr194, Tyr196, and Tyr199, in the architecture of the acceptor substrate binding site and the creation of a hydrophobic environment. Based on these and previous findings, we synthesized compounds that incorporate critical structural elements both on the xylopyranoside and on the aglycone moieties to tightly bind the acceptor site of hβ4GalT7. This work revealed that the 4-deoxy-4-fluoro-Xyl linked to 4-methylumbelliferone (4-MU) strongly inhibited hβ4GalT7 activity in vitro and efficiently impaired GAG synthesis in a cell context. Such a compound will be a valuable tool for the exploration of GAG and PG synthesis and opens avenues toward the development of bioactive oligosaccharide structures for GAG biosynthesis regulation in a number of diseases implicating disorders of GAG synthesis.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
4-Methylumbelliferyl-β-d-xylopyranoside (4-MUX), UDP-α-d-Gal (UDP-Gal), and anti-goat IgG (whole molecule) peroxidase-conjugated antibody were provided from Sigma. Anti-Myc antibodies were from Invitrogen and anti-mouse IgG-peroxidase antibodies were purchased from Cell Signaling, whereas anti-decorin antibodies were from R&D Systems. Na2[35S]SO4 was from PerkinElmer Life Sciences. Cell culture medium was purchased from Invitrogen and restriction enzymes, T4 DNA ligase, and peptide N-glycosidase F from New England Biolabs. The eukaryotic expression vector pcDNA3.1(+) and competent One Shot® Top 10 Escherichia coli cells were provided by Invitrogen and the bacterial expression vector pET-41a(+) and E. coli BL21(DE3) cells were from Novagen-EMD Chemicals. The QuikChange site-directed mutagenesis kit was from Stratagene and the transfection agent ExGen 500 from Euromedex.
Chemical Synthesis
Naphthyl-4-deoxy-β-d-xylopyranoside (4H-Xyl-NP) (29) was obtained after protection of the 2,3 position of naphthyl-β-d-xylopyranoside by isopropylidene acetal followed by radical deoxygenation and deprotection. 4-Methylumbelliferyl-4-deoxy-β-d-xylopyranoside (4H-Xyl-MU) and 4-methylumbelliferyl-4-fluoro-β-d-xylopyranoside (4F-Xyl-MU) were synthesized from the reported starting material 4-methylumbelliferyl-2,3-di-O-benzoyl-β-d-xylopyranoside (35) by radical deoxygenation or stereocontrolled 4-fluorination followed by final deprotection (data not shown).
Molecular Modeling of the hβ4GalT7 Active Site in the Presence of 4-MUX and UDP-Gal
The crystal structure of hβ4GalT7 bound to UDP and to the manganese ion (PDB code 4IRQ) was used as template (34). The crystal structure of dβ4GalT7 (PDB code 4M4K), an inactive mutant (D211N) of dβ4GalT7 in complex with UDP-Gal, and xylobiose was superposed to the human enzyme structure, which was straightforward considering their strong sequence similarity (58% overall identity). Due to crystallization conditions, a Tris molecule is bound within the active site of the hβ4GalT7. When retrieved, it frees space within the cavity that can thus accommodate the Gal moiety. The coordinates of the Gal molecule from the dβ4GalT7 complex were merged to the UDP moiety of hβ4GalT7. This did not generate any steric clash within the active site. The resulting complex was then prepared using the Protein Preparation Wizard tool of the Schrödinger Suite (Schrödinger LLC, New York), with default settings (36). All water molecules were retrieved, except the one that coordinates the manganese ion. The hydrogen atoms were added to the protein and the ligand, ascribing a pH of 7.0. The histidine residues were treated as neutral. The selection of histidine enantiomers and the orientation of the asparagine and glutamine side chains were performed so as to maximize the hydrogen bond network. The partial atomic charges derived from the OPLS-2005 force field (37) were assigned to all ligand and protein atoms. Finally, an all-atom energy minimization with a 0.3 Å heavy-atom root mean square deviation criteria for termination was performed using the Impref module of Impact and OPLS-2005 (38). The 4-MUX ligand was prepared using the ligprep module (Schrödinger Release 2014–22014). The docking program Glide was used in Standard Precision mode, with OPLS-2005, to run rigid-receptor docking calculations (39, 40). The shape and physicochemical properties of the binding site were mapped onto a cubic grid with dimensions of 20 Å3 centered on the xylobiose. During the docking calculations, the parameters for van der Waals radii were scaled by 0.80 for receptor atoms with partial charges less than 0.15e. Ring conformational sampling was not allowed to maintain the 4C1 conformation of the Xyl ring, and no constraint was introduced. A maximum of 100 poses were retained and ranked according to the GlideScore scoring function. The best-docked pose of the 4-MUX ligand showed a root mean square deviation on the Xyl ring heavy atoms of 0.5 Å with the crystallographic xylobiose ligand, thus validating the docking protocol able to recover the position of this moiety.
Expression Vector Construction
The hβ4GalT7 sequence (GenBank® nucleotide sequence accession number NM_007255) was cloned by PCR amplification from a placenta cDNA library (Clontech), as previously described (41). For bacterial expression, a truncated form of hβ4GalT7 was expressed as a fusion protein with glutathione S-transferase (GST). The sequence lacking the codons of the first 60 N-terminal amino acids was amplified from the full-length cDNA and subcloned into NcoI and NotI sites of pET-41a(+) to produce plasmid pET-β4GalT7 (31). For the heterologous expression of hβ4GalT7 in mammalian cell lines, the full-length cDNA sequence was modified by PCR at the 5′ end to include a KpnI site and a Kozak consensus sequence, and at the 3′ end to include a sequence encoding a Myc tag and an XbaI site to be then subcloned into the KpnI-XbaI sites of the eukaryotic expression vector pcDNA3.1(+) to produce pcDNA-β4GalT7 as previously described (31). Mutations were constructed using the QuikChange site-directed mutagenesis kit, employing pcDNA-β4GalT7 or pET-β4GalT7 as template. Mutants were systematically checked by double strand sequencing. The human decorin cDNA sequence (GenBank accession number NM_001920.3) was cloned by PCR amplification from a placenta cDNA library (Clontech). For heterologous expression in eukaryotic cells, the full-length cDNA sequence was modified by PCR to include an AflII site, a Kozak consensus sequence at the 5′ end, a sequence encoding a His5 tag, and an XhoI site at the 3′ end. This sequence was subcloned into pcDNA3.1(+) to produce pcDNA-decorinHis as previously described (31).
Expression and Purification of the Soluble Form of hβ4GalT7
A single colony of E. coli BL21(DE3) cells transformed with the pET-β4GalT7 plasmid was cultured overnight at 37 °C in Luria broth (LB) medium containing 50 μg/ml of kanamycin. The overnight culture was transferred into fresh LB medium (1:100 dilution), supplemented with 50 μg/ml of kanamycin, and incubated at 37 °C until the A600 value reached 0.6–0.8. Expression of hβ4GalT7 was induced by addition of 1 mm isopropyl β-d-thiogalactopyranoside to the cell suspension, which was then incubated overnight at 20 °C under continuous shaking (200 rpm). The bacterial cells were then harvested by centrifugation at 7,000 × g for 10 min at 4 °C. The pellet was resuspended in Lysis buffer (50 mm sodium phosphate, 1 mm phenymethylsulfonyl fluoride, 1 mm EDTA, and 5% (v/v) glycerol, pH 7.4) supplemented with protease inhibitor mixture tablets (1 tablet/12 ml; Roche Diagnostics) and Benzonase® Nuclease (250 units/10 ml, Sigma). The suspended cells were then sonicated for 8 cycles of 30 s, at 30% power (Badelin Sonoplus GM70) with a 20-s interval on ice between each cycle. Soluble proteins were collected from the supernatant after centrifugation for 25 min at 12,000 × g and clarification by filtration (0.2 μm Supor® Membrane; PALL-Life Science). 10 ml of clarified extracts were applied onto a 1-ml glutathione-Sepharose High Performance column (GSTrap HP; GE Healthcare) connected to an AKTA prime plus instrument (GE Healthcare). Protein was eluted as 1-ml fractions using 50 mm Tris-HCl, pH 8.0, containing 10 mm reduced glutathione buffer. Protein purity of the eluted fractions was evaluated by 12% (w/v) SDS-PAGE analysis, followed by staining with Coomassie Brilliant Blue. Fractions containing the pure protein were used to determine the kinetic parameters of the enzyme. The same procedure was used for purification of the mutants. Protein concentration was measured using Quant-iTTM assay kit and QubitTM spectrofluorimeter.
Determination of the in Vitro Kinetic Parameters of hβ4GalT7
The kinetic parameters kcat and Km toward 4-MUX and UDP-Gal were determined as described (31). Briefly, 0.2 μg of purified wild-type or mutated GST-hβ4GalT7 were incubated for 30 min at 37 °C in a 100 mm sodium cacodylate buffer, pH 7.0, 10 mm MnCl2, with concentrations from 0 to 5 mm 4-MUX in the presence of fixed 1 mm UDP-Gal to determine the apparent Km toward 4-MUX, and with concentrations from 0 to 5 mm UDP-Gal in the presence of fixed 5 mm 4-MUX to determine the apparent Km toward UDP-Gal. The incubation mixture was then centrifuged at 10,000 × g for 10 min at 4 °C. The supernatant was analyzed by high performance liquid chromatography (HPLC) with a reverse phase C18 column (xBridge, 4.6 × 150 mm, 5 μm, Waters) using Waters equipment (Alliance Waters e2695) coupled to a UV detector (Shimadzu SPD-10A). Kinetic parameters were determined by nonlinear least squares regression analysis of the data fitted to the Michaelis-Menten rate equation using the curve-fitter program of Sigmaplot 9.0 (Erkraft, Germany).
In Vitro Competition Assays of hβ4GalT7 Activity by C4-modified Xylosides
The in vitro inhibition assays of the wild-type GST-hβ4GalT7 were carried out using 0.2 μg of purified protein incubated for 30 min at 37 °C in a 100 mm sodium cacodylate buffer, pH 7.0, 10 mm MnCl2, with 0.5 mm 4-MUX and 1 mm UDP-Gal, in the presence of concentrations from 0 to 5 mm 4H-Xyl-NP, 4H-Xyl-MU, or 4F-Xyl-MU. Quantification of the reaction product was carried out by HPLC, as described above. The enzyme activities were reported as a function of the logarithmic values of inhibitor concentration. IC50 values were determined by fitting the experimental dose-response curves using the curve-fitter program of Sigmaplot 9.0 (Erkraft, Germany). Ki values were calculated from IC50 values according to the Cheng-Prusoff's equation (42, 43).
In Cellulo Analysis of GAG Chains Biosynthesis by Na2[35SO4] Incorporation
GAG chains biosynthesis using 4-MUX as primer substrate was determined with CHOpgsB-618 cells (American Type Culture Collection). Cells were cultured in Dulbecco's modified Eagle's medium/F-12 (DMEM/F-12) (1:1), supplemented with 10% fetal bovine serum (Dutscher), penicillin (100 units/ml)/streptomycin (100 mg/ml), and 1 mm glutamine, then transfected with the wild-type or mutant pcDNA-β4GalT7-Myc plasmid or with the empty pcDNA3.1 vector at 70% cell confluence. Transfected cells were then incubated in low sulfate medium (Fisher) supplemented with 10 μCi/ml of Na2[35SO4] in the presence of 0.5 or 10 μm 4-MUX for 16 h. For GAG chain isolation 1 ml of culture medium was applied to a G-50 column (GE Healthcare) to separate radiolabeled GAG chains from the non-incorporated Na2[35SO4] and radiolabeling was quantified by scintillation counting. In parallel, the hβ4GalT7 expression level was checked by Western blotting using a primary anti-Myc (1/5,000) and a secondary anti-mouse antibody (1/10,000). To test the inhibitory potency of C4-modified xylosides, the molecules were added at 0 to 100 μm concentration together with 4-MUX (5 μm) for 16 h prior to isolation and quantification of radiolabeled GAGs. To test the cytotoxicity of xyloside inhibitors in CHOpgsB-618 cells expressing the wild-type hβ4GalT7, cells were seeded at 150,000 cells/well in 12-well plates, and incubated for 48 h at 37 °C in the presence of 0 to 400 μm inhibitor, or 4-MUX as a control. The ratio of viable cells upon the total number of cells was determined using the cell counter TC20 (Bio-Rad) in the presence of a vital marker (trypan blue).
In Cellulo Analysis of Decorin Core Protein Glycosylation
CHOpgs-B618 cells stably transfected with pcDNA-decorinHis encoding the human decorin core protein (31, 44) were transiently transfected with pcDNA3.1 or with recombinant vector encoding either the wild-type or mutated hβ4GalT7-Myc as described above. 48 Hours following transfection, the cell medium was collected, concentrated by centrifugation at 4 °C for 15 min at 3000 × g, using the Amicon Ultracell 30 MWCO concentrating system (Merck Millipore, Germany), and submitted to SDS-PAGE (25 μg of protein/well). The glycosylation level of the decorin core protein was monitored by immunoblot using a 1/5,000 dilution of primary polyclonal anti-human decorin antibody (VWR) and a 1/10,000 dilution of secondary anti-goat antibody coupled to horseradish peroxidase (Sigma), then quantified using ImageJ software. Briefly, the level of decorin glycosylation was expressed as relative band intensity by normalizing the band intensity value for the glycosylated form upon the total intensity value for the bands corresponding to the glycosylated and non-glycosylated forms of decorin core protein. The expression level of the decorin core protein in pcDNA3.1-transfected cells was used as the negative control and served as a loading control. On the other hand, the level of glycosylated decorin in cells expressing wild-type hβ4GalT7 was used as positive control for decorin glycosylation.
RESULTS
Molecular Modeling of the hβ4GalT7 Acceptor Binding Site
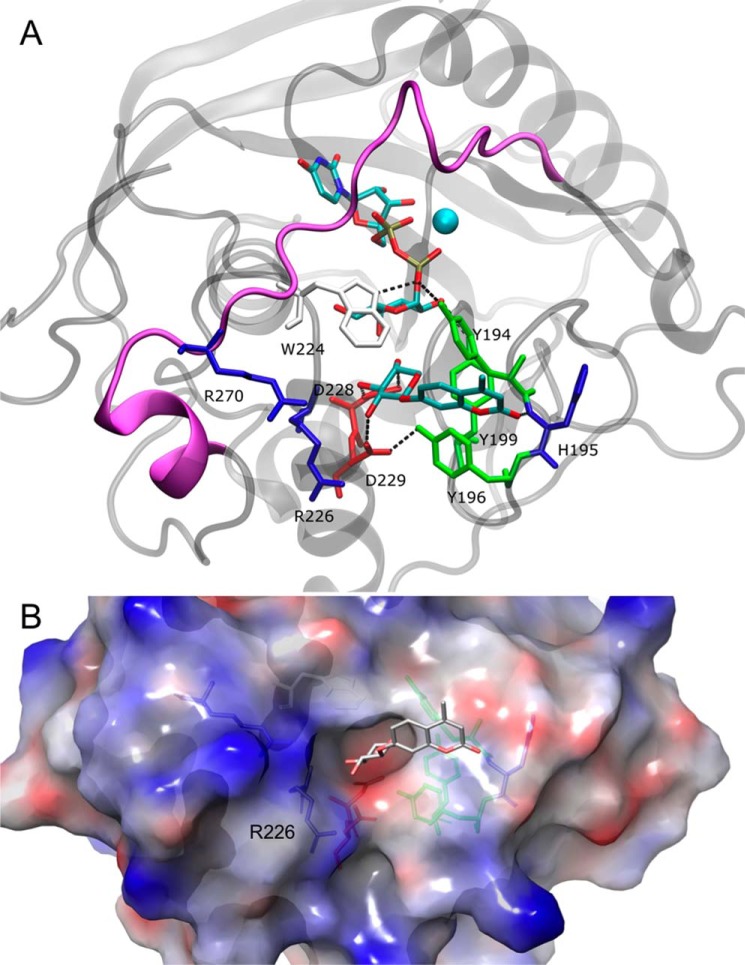
In the present study, we aimed to identify amino acids important for structural organization of the hβ4GalT7 acceptor substrate binding site. We took advantage of the recent crystal structure of hβ4GalT7 in complex with UDP (34) to build a molecular model of this enzyme in complex with both the sugar donor UDP-Gal and the acceptor 4-MUX (Fig. 1A). The modeled structure is in a closed conformation, considered to be the catalytically competent form, and the hydrogen bond network around the UDP moiety is fully conserved. We first examined the position of a series of tyrosine residues, i.e. Tyr194, Tyr196, and Tyr199, that were suggested to be involved in binding of the xylobiose in the dβ4GalT7 structure (33). Our computational analysis indicates that Tyr194 stabilizes both the donor and acceptor substrates location by establishing a hydrogen bond with a β-phosphate oxygen of UDP-Gal and a π-stacking interaction with the 4-methylumbelliferyl moiety, respectively (Fig. 1A). Residue Tyr196 is not hydrogen bonded to the substrates but to the side chain of residue Asp229, allowing its second carboxylic oxygen to be suitably oriented to form a hydrogen bond with the O2 atom of Xyl. The spatial orientation of Tyr199 inside the substrate binding pocket allows the formation of a hydrogen bond between its side chain hydroxyl and the O2 atom of the Gal moiety of UDP-Gal. Altogether, residues Tyr194, Tyr196, and Tyr199 form a strongly hydrophobic cluster that is required for correct binding of the substrates.
FIGURE 1.
Molecular modeling of the hβ4GalT7 structure. A, view of the active site of hβ4GalT7 in complex with donor (UDP-Gal) and acceptor (4-MUX) substrates. The protein α-carbon trace is represented as gray ribbons; the purple ribbon corresponds to the protein backbone that is not seen in the open conformation. B, surface representation of the acceptor binding site, in the same orientation as in A. The electrostatic potential is mapped onto the protein surface and is colored from red (negative) to blue (positive).
Analysis of the His195 position, a conserved amino acid located between the two active site Tyr194 and Tyr196 residues, shows no hydrogen bond involving its side chain. However, the backbone nitrogen atom of this residue is hydrogen bonded with the CO group of 4-MUX (Fig. 1A). As illustrated in Fig. 1B, Arg226 is located on the surface of the acceptor binding site contributing to an amphipathic entry door with the aromatic residues. In our model, there is no hydrogen bond involving the side chain of Arg226. Instead, its backbone nitrogen atom is hydrogen bonded with the O3 atom of the Xyl moiety of 4-MUX (Fig. 1A).
The structural impact of Arg270 on enzyme activity, in the context of EDS was also addressed. The model structure of hβ4GalT7 reveals that Arg270 belongs to the flexible loop (261–284) that moves upon donor substrate binding, thus creating the acceptor substrate binding site (Fig. 1A). This conformational change leads to the closed and catalytically competent conformation of the active site. However, the crystal structure of the human enzyme (34), as well as our own model in complex with both the donor and acceptor substrates do not highlight specific interactions established by this residue, although its close location to the surface of the active site has to be underlined (Fig. 1B).
Kinetic Properties of the Human Recombinant hβ4GalT7 Mutants Expressed in E. coli
To assess the functional importance of the residues of the acceptor binding site highlighted by our model, we carried out point mutagenesis and analyzed the consequences of conservative and non-conservative mutations on the kinetic parameters of hβ4GalT7 expressed and purified from recombinant E. coli cells. The wild-type enzyme and engineered mutants were produced as truncated fusion proteins lacking the 60 N-terminal amino acids (including the transmembrane domain and part of the stem region) linked to GST and purified by affinity chromatography (data not shown). This led to 1.0 to 2.5 mg of pure protein per liter of culture for wild-type and mutant hβ4GalT7. Kinetic assays were performed using 4-MUX as acceptor substrate, which allowed quantification of the transfer reaction product by UV detection coupled to HPLC. The kcat and Km values of the wild-type enzyme toward UDP-Gal and 4-MUX shown in Table 1 were in agreement with previous work (17, 31).
TABLE 1.
Kinetic parameters of wild-type and mutant GST-β4GalT7
Kinetic parameters towards donor (UDP-Gal) and acceptor (4-MUX) substrates were determined as described under “Experimental Procedures.” The results are the mean values of three independent determinations ± S.D. on assays performed in duplicate.
| Enzyme | kcat | UDP-Gal |
4-MUX |
||
|---|---|---|---|---|---|
| Km | kcat/Km | Km | kcat/Km | ||
| min−1 | mm | min−1·mm−1 | mm | min−1·mm−1 | |
| GST-β4GalT7 | 90.5 ± 2.3 | 0.22 ± 0.02 | 425 | 0.35 ± 0.02 | 250 |
| Y194A | NDa | ND | ND | ||
| Y194F | ND | ND | ND | ||
| H195A | 115.9 ± 9.7b | 0.40 ± 0.02b | 291 | 0.64 ± 0.02b | 180 |
| H195Q | 97.4 ± 2.4b | 0.33 ± 0.02b | 295 | 0.55 ± 0.02b | 177 |
| H195R | 89.6 ± 1.3 | 0.32 ± 0.02b | 295 | 0.35 ± 0.03 | 242 |
| Y196A | ND | ND | ND | ||
| Y196F | 30 ± 1.1b | 0.34 ± 0.06b | 88 | 1.06 ± 0.06b | 28 |
| Y199A | ND | ND | ND | ||
| Y199F | 72.3 ± 5.8b | 0.32 ± 0.02b | 243 | 0.59 ± 0.06b | 113 |
| R226A | 53.6 ± 2.0b | 0.34 ± 0.03b | 171 | 0.44 ± 0.05b | 112 |
| R226K | 81.1 ± 2.9b | 0.29 ± 0.02b | 282 | 0.46 ± 0.01b | 175 |
| R270A | 46.4 ± 0.2b | 0.27 ± 0.01b | 184 | 0.60 ± 0.02b | 72 |
| R270K | 48.7 ± 1.5b | 0.37 ± 0.02b | 139 | 0.54 ± 0.07b | 85 |
a ND indicates that no kinetic constant could be determined using excess acceptor or donor substrate.
b The results were analyzed with Student's t test and considered as significant when p < 0.05.
Substitution of Tyr194 by alanine led to an inactive enzyme, and its conservative substitution by phenylalanine did not restore the galactosyltransferase activity of hβ4GalT7 (Table 1), indicating a critical role of this residue and, importantly, of the hydroxyl group of the tyrosine side chain. The mutation of Tyr196 to alanine totally abolished enzyme activity, whereas replacement of this residue by phenylalanine led to a slightly active enzyme. The Y196F mutation did not impair enzyme affinity toward the donor substrate to a major extent but this mutant presented a lower affinity toward 4-MUX with a Km value about 3-fold that of the wild-type enzyme (Table 1). As observed in the case of Tyr194 and Tyr196, the non-conservative mutation Y199A led to a total loss of enzyme activity. However, similarly with what was observed for Tyr196, substitution of Tyr199 by phenylalanine led to an active hβ4GalT7 enzyme with Km values toward UDP-Gal and 4-MUX and a kcat value only weakly affected compared with the wild-type enzyme. These data suggest that the aromatic ring of phenylalanine at position 199 is sufficient to support xyloside binding and activity.
Substitution of His195 by alanine, glutamine, or arginine was carried out. The Km values of all three mutants toward UDP-Gal and 4-MUX were mostly comparable with those of the wild-type enzyme, indicating that these mutations had no major effect upon hβ4GalT7 affinity toward its substrates. Moreover, the substitutions at position 195 did not affect the rate of reaction transfer, as indicated by the kcat values that were essentially unchanged (Table 1). Altogether, these results indicate that the side chain of His195 does not play a critical role in xyloside binding and hβ4GalT7 catalytic activity. Substitution of Arg226 by alanine or lysine did not impair the affinity toward the substrates with Km values for UDP-Gal and 4-MUX, which were in the same range to that of the wild-type enzyme, and produced a moderate decrease (about 2-fold) of the catalytic constant value (Table 1).
The Arg270 residue is mutated to cysteine in the progeroid form of the EDS syndrome, and we previously showed that this mutation led to a significant decrease in hβ4GalT7 activity, mainly due to a reduced affinity toward 4-MUX (about 10-fold, see Ref. 17). To ascertain the contribution of this residue in hβ4GalT7 activity and xyloside binding, we performed kinetic assays following the conservative R270K and non-conservative R270A mutations. The kcat values for both mutants were about two times lower than that found for the wild-type enzyme and the Km value toward UDP-Gal was almost unaffected (Table 1).
Effect of Tyr194, Tyr196, Tyr199, His195, Arg226, and Arg270 Mutations on the Galactosyltransferase Activity of hβ4GalT7 Toward 4-MUX in Cellulo
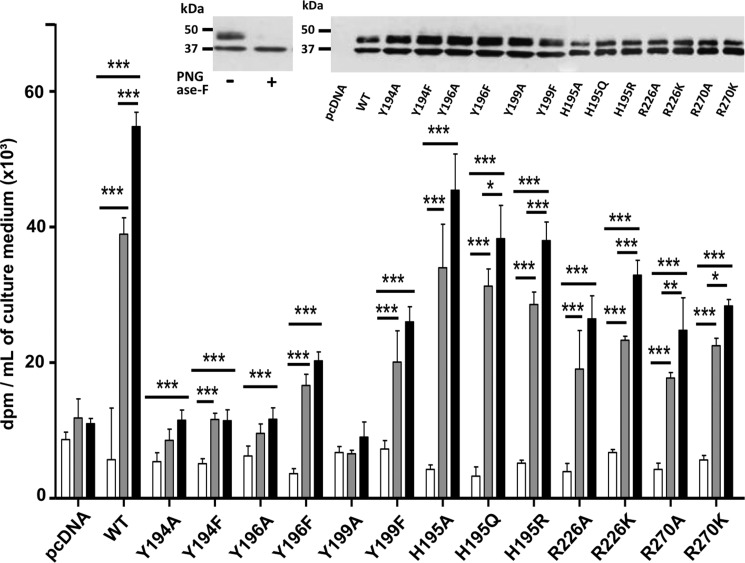
To check the importance of the selected residues on the hβ4GalT7 function in a cellular context, we designed an experimental procedure involving CHOpgsB-618 cells transfected with hβ4GalT7 cDNA encoding either the wild-type or mutant enzymes fused to a Myc tag sequence at their C-terminal end, as described under “Experimental Procedures.” CHOpgsB-618 cells expressing the recombinant enzymes were tested for in cellulo galactosyltransferase activity in the absence or presence of 4-MUX as the exogenous acceptor substrate. The expression level of enzymes was checked by immunoblot analysis using ImageJ software. As shown in Fig. 2, an additional upper band at ∼39 kDa was observed (Fig. 2, inset). This ∼39-kDa band can be attributed to the N-glycosylated form of the enzyme as it disappears upon peptide N-glycosidase F digestion (Fig. 2, inset, left panel). Both bands were taken into account to quantify the total enzyme expression level. The results indicate that all mutants considered in these experiments were expressed at a similar level to that of the wild-type protein (Fig. 2, inset, right panel). As shown in Fig. 2, the GAG synthesis level in cells expressing wild-type hβ4GalT7 was about 7- and 9.5-fold higher in the presence of 5 and 10 μm 4-MUX, respectively, than in the absence of acceptor substrate, indicating that CHOpgsB-618 cells expressing hβ4GalT7 are able to prime efficiently GAG chains synthesis from 4-MUX, in agreement with previous studies (17, 31).
FIGURE 2.
Effect of wild-type and mutated hβ4GalT7 expression on GAG chains primed from 4-MUX in CHOpgsB-618 cells. Cells were transiently transfected with wild-type (WT) or mutated hβ4GalT7 cDNA or with empty vector (pcDNA), and GAG chains synthesis was quantified by scintillation counting following Na2[35SO42−] incorporation, using 0 (white bars), 5 (gray bars), and 10 μm (black bars) 4-MUX. Immunoblot analyses of the protein expression level in CHOpgsB-618 cells transfected with the vector coding for the wild-type or mutated hβ4GalT7 are shown as the inset. The enzyme was identified at the band of ∼35 kDa, whereas the upper band corresponding to ∼39 kDa band could be attributed to the N-glycosylated enzyme as demonstrated by its disappearance upon addition of peptide N-glycosidase F (PNGase F) (left panel). Both bands intensities were used to quantify the total protein expression level (ImageJ software). The immunoblot analysis indicates that the mutated enzymes were all expressed at a comparable level to that of the wild-type hβ4GalT7 (right panel). Data are mean ± S.E. of three independent experiments performed in triplicate. Statistical analysis was carried out by the Student's t test with *, p < 0.05; **, p < 0.01; and ***, p < 0.001 versus GAG synthesis in the absence of 4-MUX.
As expected, cells expressing either Y194A or Y194F mutant in the presence of 4-MUX showed GAG synthesis levels comparable with those obtained with cells transfected with empty vector (Fig. 2). These in cellulo assays confirm that any mutation affecting the Tyr194 position leads to a total loss of hβ4GalT7 activity. Substitution of Tyr196 to alanine dramatically reduced the GAG synthesis rate whose level in the presence of 4-MUX was comparable with that obtained with cells transfected with empty vector. This is consistent with the loss of enzymatic activity observed in the in vitro assays (Table 1). By contrast, the conservative mutation Y196F allowed GAG chain priming from 4-MUX. However, the GAG expression level reached with this mutant was about 2–3-fold lower than that of the wild-type enzyme, at 4-MUX at 5 and 10 μm concentrations, respectively (Fig. 2). This result is consistent with the drastic decrease of the kcat/Km value toward 4-MUX found for the purified Y196F mutant. Comparable results were obtained with cells expressing hβ4GalT7 whose sequence is mutated on the Tyr199 position. Indeed, cells expressing the Y199A mutant were unable to synthesize GAG chains from 4-MUX, whereas cells expressing Y199F showed GAG chain synthesis at a level about half of that observed with cells expressing wild-type enzyme (Fig. 2). These results are in line with the reduced efficiency exhibited in vitro by the enzyme substituted on the Tyr199 position (Table 1). Altogether, these results demonstrate that both conservative and non-conservative mutations affecting Tyr194, Tyr196, or Tyr199 significantly impaired GAG chains biosynthesis in a cellular context, in line with in vitro data (Table 1).
In the presence of 4-MUX, the GAG synthesis rate in cells expressing H195A, H195Q, and H195R mutants was moderately reduced, i.e. 10 to 15% lower than that of cells expressing the wild-type enzyme (Fig. 2). These results indicate that the side chain of this residue does not influence galactosyltransferase activity of hβ4GalT7 in the context of 4-MUX-primed GAG chains in eukaryotic cells, corroborating the findings that none of the mutations of His195 significantly affect in vitro activity (Table 1). The level of [35SO42−] incorporation in the presence of 4-MUX in cells expressing R226A was about 2 times lower than that of the wild-type enzyme (Fig. 2). In addition, replacement of Arg226 by lysine slightly increased the GAG expression level compared with alanine substitution, reaching about 60% that obtained with the wild-type, at 5 and 10 μm 4-MUX. Corroborating in vitro kinetic parameters, these cellular assays indicate that modification of the side chain of Arg226 produces minor effects on galactosyltransferase activity.
We finally examined the impact of mutations of the Arg270 residue upon GAG synthesis in eukaryotic cells. We observed that the GAG synthesis rate in cells expressing the R270A mutant was about 55% lower than that of the wild-type enzyme, at 5 and 10 μm 4-MUX concentration (Fig. 2). The GAG synthesis level of the conservative mutant R270K was also about 2-fold reduced compared with the wild-type (Fig. 2). These results confirm that mutations of Arg270 significantly affect the capacity of hβ4GalT7 to synthesize GAG chains from 4-MUX in a cellular context.
Effect of Tyr194, Tyr196, Tyr199, His195, Arg226, and Arg270 Mutations on the Ability of hβ4GalT7 to Initiate the Glycosylation of the Decorin PG in Cellulo
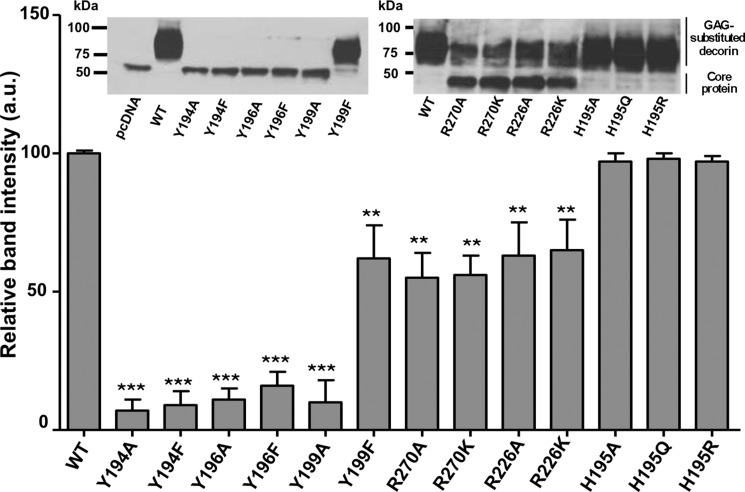
We next determined whether the mutations would affect GAG chain formation on the core protein of decorin, used as a model PG (31). To this aim, CHOpgsB-618 cells were engineered to stably express the recombinant human decorin, and were transiently transfected with a pcDNA3.1 vector encoding the wild-type or mutant forms of hβ4GalT7. This allowed monitoring of the GAG substitution of the secreted PG by Western blot analysis (Fig. 3, inset, left and right panels). The rates of decorin PG glycosylation in cells expressing wild-type or mutant hβ4GalT7 were determined as described under “Experimental Procedures,” then reported onto the histogram (Fig. 3). Results showed that non-conservative mutations of the tyrosine residues at positions 194, 196, and 199 as well as mutations of Tyr194 and Tyr196 to phenylalanine fully abolished the glycosylation of decorin (Fig. 3). These data were consistent with the drop of GAG chains primed from 4-MUX in cells expressing hβ4GalT7 mutated at these positions (Fig. 2). However, the substitution of Tyr196 to phenylalanine induced a more dramatic effect on the glycosylation of decorin than on the in vitro or in cellulo activity toward 4-MUX. Furthermore, results shown in Fig. 3 indicate that conservative mutation Y199F allowed recovery up to 70% of the decorin glycosylation level compared with cells expressing the wild-type hβ4GalT7.
FIGURE 3.
Effect of wild-type and mutated hβ4GalT7 expression on decorin core protein glycosylation in CHOpgsB-618 cells. Cells stably expressing the human recombinant decorin core protein were transfected with the recombinant vector encoding either the wild-type (WT) or mutated hβ4GalT7. The decorin glycosylation level was monitored by immunoblot then quantified using ImageJ software, as described under “Experimental Procedures.” Immunoblot analyses of the decorin core protein glycanation level are shown as inserts for CHOpgsB-618 cells transfected with empty (pcDNA) or with the recombinant vector coding for WT, Y194A, Y194F, Y196A, Y196F, Y199A, or Y199F hβ4GalT7 (left panel), and for WT, R270A, R270K, R226A, R226K, H195A, H195Q, or H195R hβ4GalT7 (right panel). The band observed at a molecular mass of ∼35 kDa can be attributed to the decorin core protein, whereas the wide upper band corresponding to a molecular mass ≥75 kDa corresponds to the glycosylated decorin. Data are mean ± S.E. from three independent experiments performed in triplicate. Statistical analysis was carried out by the Student's t test with **, p < 0.01 and ***, p < 0.001 versus decorin glycosylation in cells expressing the wild-type hβ4GalT7.
We also assessed the role of the His195 residue in the glycosylation process of decorin. The results shown in Fig. 3 indicate that none of the mutations, H195A, H195Q, or H195R, significantly affected the level of decorin glycosylation. These data are consistent with the results obtained on the in vitro and in cellulo activity of the enzyme toward 4-MUX. Together, mutagenesis experiments indicate that modification of the amino acid side chain at position 195 did not greatly affect xyloside binding and galactosyltransferase activity of hβ4GalT7. Investigation of the effect of the Arg226 mutation upon the ability of CHOpgsB-618 cells to glycosylate decorin showed that the glycosylation level reached with cells expressing R226A or R226K was about 65% of that obtained with cells expressing the wild-type enzyme. These results were consistent with the in vitro and in cellulo GAG chain synthesis assays (Fig. 3).
Because we aimed to better understand the molecular basis of the EDS syndrome, it was important to further investigate the impact of mutations affecting the Arg270 position upon the decorin glycosylation. The decorin glycosylation level reached with cells expressing either R270A or R270K mutant was about half of that obtained with cells expressing the wild-type hβ4GalT7, consistent with in vitro and in cellulo galactosyltransferase assays (Fig. 3).
Xyloside Inhibitors Design and in Vitro and in Cellulo Competition Assays
We next took advantage of the knowledge gained from our investigation of the organization of the acceptor substrate binding site to synthesize and test xyloside analogs as potential inhibitors of hβ4GalT7. To this end, in vitro competition assays were performed as described under “Experimental Procedures.” The specific activity as a function of the logarithm values of the inhibitor concentrations are reported in Fig. 4B and data fitted to the logistic equation provided IC50 values as reported in Table 2.
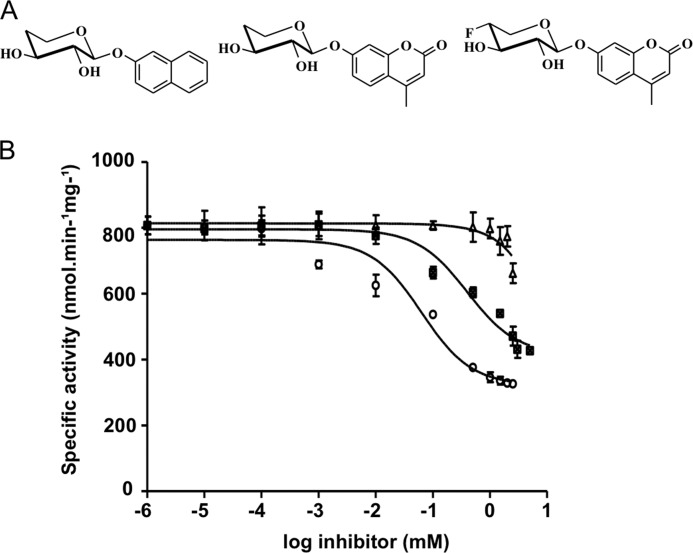
FIGURE 4.
Inhibitory effect of C4-modified xylosides on hβ4GalT7 activity. A, chemical structures of the xyloside analogs synthesized and tested as inhibitors. From left to right: 4H-Xyl-NP, 4H-Xyl-MU, and 4F-Xyl-MU. B, inhibition assays using purified recombinant wild-type hβ4GalT7 in the presence of fixed 4-MUX (0.5 mm) and UDP-Gal (1 mm). Activities are presented as function of the logarithm of increasing inhibitor concentrations (0–5 mm); 4H-Xyl-NP (▴), 4H-Xyl-MU (■), and 4F-Xyl-MU (○). Results are the mean ± S.E. of three independent determinations on assays performed in duplicate.
TABLE 2.
Kinetic inhibition parameters of hβ4GalT7 with C4-modified xylosides
IC50 and Ki values are the values of three independent experiments mean ± S.D. on assays performed in duplicate.
| Xylosides | IC50 | Ki |
|---|---|---|
| mm | ||
| 4H-Xyl-NP | NDa | ND |
| 4H-Xyl-MU | 1.28 ± 0.22 | 0.53 ± 0.10 |
| 4F-Xyl-MU | 0.06 ± 0.02 | 0.03 ± 0.01 |
a ND, not determined.
Because the C4-position is critical for both binding and transfer of the Gal residue from UDP-Gal onto the xyloside acceptor, we first synthesized a 4-deoxy derivative of 4-MUX (4H-Xyl-MU, Fig. 4A) and tested this compound as inhibitor of hβ4GalT7 in vitro. 4H-Xyl-MU was able to inhibit up to 50% of the initial activity at a 2 mm concentration (Fig. 4B), with an IC50 value of about 1 mm and a Ki value of about 0.5 mm (Table 2). To test whether hydrogen bond formation between 4-MUX and the protein via His195 is important for the inhibitory potency, we synthesized 4H-Xyl-NP, which the aglycone structure is unable to establish such an interaction, and compared its inhibitory effect to 4H-Xyl-MU. This compound produced a decrease of hβ4GalT7 activity toward 4-MUX less than 25% at the highest concentration (Fig. 4B) that did not allow determining IC50 and Ki values. These results clearly indicated that 4H-Xyl-NP is a weak inhibitor of hβ4GalT7. We next substituted the equatorial hydrogen of the C4 atom of the Xyl moiety by a fluorine atom, closer to oxygen in terms of electronegativity and predicted to fit the active site in terms of steric hindrance. 4F-Xyl-MU led up to 60% inhibition of the hβ4GalT7 activity at the highest concentration (Fig. 4B), with an IC50 of 0.06 mm and a Ki of 0.03 mm. The inhibition constant for this compound is more than 10 times lower than that reached for the deoxy analog (Table 2).
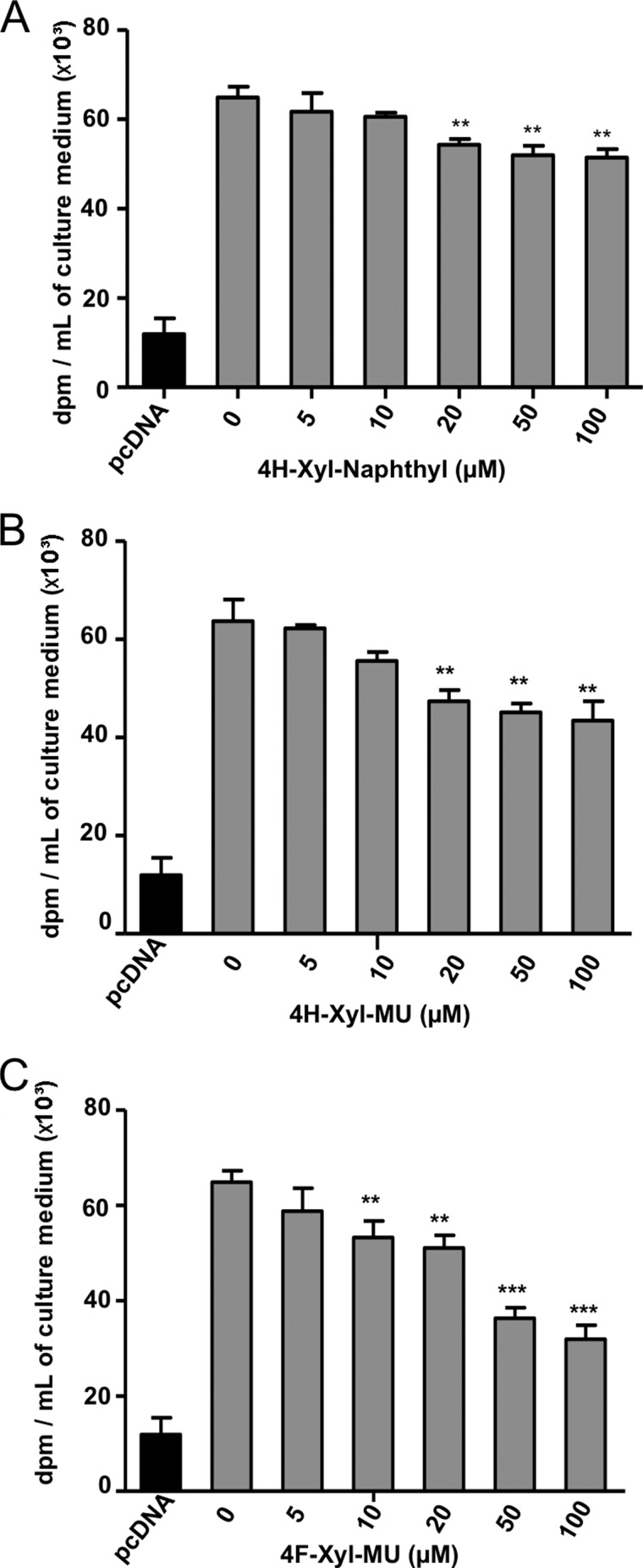
To complement the in vitro assay, we assessed the ability of the synthesized xyloside derivatives to inhibit GAG chain biosynthesis in cellulo. Addition of 4H-Xyl-NP produced a moderate but significant 20% decrease of GAG chain synthesis in CHOpgsB-618 cells, when used at 50 and 100 μm (Fig. 5A). This correlated with the weak in vitro inhibition level obtained with this compound. The compound 4H-Xyl-MU allowed a larger inhibition of GAG chain synthesis, with up to 30% reduction of the GAG synthesis rate at similar concentrations (Fig. 5B). The best inhibitory effect was observed when performed in the presence of 4F-Xyl-MU leading to up to 50% inhibition of the initial GAG chain synthesis rate at 100 μm concentration (Fig. 5C). Interestingly, preliminary results indicated that 400 μm 4F-Xyl-MU inhibited the initial decorin glycosylation rate by about 50%, without affecting the viability of CHOpgsB-618 cells (data not shown). Altogether, the latter data confirmed that this fluorinated compound should be considered as a promising non-cytotoxic xyloside-based inhibitor of hβ4GalT7.
FIGURE 5.
Inhibitory effect of C4-modified xylosides upon 4-MUX-primed GAG chains synthesis in CHOpgsB-618 cells expressing the recombinant wild-type hβ4GalT7. CHOpgsB-618 cells transiently transfected with wild-type hβ4GalT7 cDNA were incubated with 5 μm 4-MUX and Na2[35SO42−], in the presence of 4H-Xyl-NP (panel A), 4H-Xyl-MU (panel B), and 4F-Xyl-MU (panel C). The GAG expression level in cells transfected by the empty pcDNA vector was taken as negative control. Results are the mean ± S.E. of three independent experiments performed in triplicate. Statistical analysis was carried out using the Student's t test with **, p < 0.01 and ***, p < 0.001 versus GAG synthesis rate in the absence of inhibitor.
DISCUSSION
β4GalT7 is a unique enzyme in the GAG biosynthetic pathway with regard to its capacity to use exogenous xyloside molecules as substrates and/or inhibitors that can efficiently modulate GAG synthesis in vitro and in vivo (19, 20, 45). This glycosyltransferase is also central in the GAG synthesis process because the formation of the tetrasaccharide linker is a prerequisite to the polymerization of both heparan sulfate and chondroitin/dermatan sulfate chains. The human enzyme thus represents a prime target for the design of effectors of GAG synthesis as drugs to correct GAG disorders associated with numerous malignant conditions such as genetic diseases and cancer. To meet this challenge, we pioneered structure-function studies of the recombinant hβ4GalT7. We previously carried out structural, thermodynamic, and phylogenetic investigations that identified key amino acid residues mainly implicated in the recognition and binding of the donor substrate (31, 46). We also provided insight into the molecular basis of the GAG defects characterizing rare forms of EDS syndrome (17, 47). In the present work, to develop xyloside compounds that will specifically target the hβ4GalT7 activity for a therapeutic purpose, we explored the architecture of the acceptor substrate binding site. To this aim, we combined functional investigations including site-directed mutagenesis, kinetic analyses, in vitro and in cellulo evaluation of galactosyltransferase activity and GAG synthesis, and a computational approach. This allowed mapping the acceptor binding site and to design and synthesize a potent xyloside-based inhibitor of GAG synthesis.
We first targeted a set of three tyrosine residues, Tyr194, Tyr196, and Tyr199, as well as His195 belonging to the same conserved motif, that occupy a strategic position surrounding the xyloside acceptor substrate (Ref. 34 and the present data). Our mutational analysis led to a remarkable observation because alanine substitution of each of these tyrosine completely abolished hβ4GalT7 activity. The tyrosine-alanine mutants (i) were devoid of galactosyltransferase activity in vitro, (ii) were unable to prime GAG synthesis from 4-MUX in cellulo, and (iii) did not promote decorin glycosylation, thus supporting a prominent function of this set of aromatic residues. Mutating Tyr194, Tyr196, and Tyr199 with phenylalanine revealed that the substitution differently affected hβ4GalT7 activity depending on the position. Noteworthy, the presence of the hydroxyl group of Tyr194 was indispensible because the conservative Y194F mutant completely lacked in vitro or in cellulo galactosyltransferase activity toward 4-MUX and was unable to sustain glycosylation of decorin. This observation is likely to be explained by a functionally important interaction between the hydroxyl group of this tyrosine and the β-phosphoryl group of UDP-Gal that was observed in all structures and models of β4GalT7 (Ref. 34 and this report, see Fig. 1). Most importantly, our computational model and experimental data suggested that the critical role of Tyr194 also arises from a stacking interaction between the aromatic ring of this residue and the 4-methylumbelliferyl moiety of 4-MUX. Altogether, these data indicate that Tyr194 occupies a strategic location in the catalytic center and interacts with both the donor nucleotide and the aglycone group of 4-MUX. In the case of Tyr196, the presence of the hydroxyl group of the tyrosine residue was also a major structural element because its replacement by phenylalanine only slightly restored the activity toward 4-MUX in vitro and in cellulo. The Y196F mutant did not sustain decorin glycosylation, also supporting an important role of this residue in the glycosylation of endogenous proteoglycans. Our model provides a molecular explanation to these results, because it shows that Tyr196 is not directly involved in the binding of the acceptor substrate but rather forms a hydrogen bond between its hydroxyl group and Asp229, this latter residue establishing an important interaction with O2 of the Xyl moiety. This supports the idea that interactions with the hydroxyl in the C2 position control a strict geometry via both Asp229 and Tyr196, in agreement with the physiologically important regulatory role of Xyl-phosphate substitution in position 2 on GAG synthesis (41, 48). Furthermore, our model suggests that this residue is part of the cavity floor in agreement with structural data indicating that the acceptor substrate xylobiose is located in a hydrophobic binding pocket formed by Tyr177, Tyr179, and Trp207 in the Drosophila structure, and by Tyr194, Tyr196, and Trp224 in hβ4GalT7. The present data complements our previous findings demonstrating that Trp224 is a crucial active site residue (31). Differently to the preceding studied tyrosines, hβ4GalT7 tolerated well the substitution of tyrosine to phenylalanine at position 199 leading to a mutant that was active toward 4-MUX in vitro and was able to prime GAG synthesis from 4-MUX and onto the decorin core protein. Consistently, Tyr199 is substituted by a phenylalanine in the Drosophila enzyme, suggesting that the presence of a hydrophobic aromatic ring is sufficient at this position. Further analysis of our molecular model predicts hydrogen bond formation between the tyrosine hydroxyl group of Tyr199 and O2 of the Gal moiety of UDP-Gal. However, no significant change in the Km value toward UDP-Gal was observed for the Y199F mutant, indicating that this interaction may not play a critical functional role in nucleotide binding. The location of Tyr199 favors a role as contributor to the hydrophobic surrounding of the acceptor substrate binding pocket together with Tyr194, Tyr196, and Trp224 residues when hβ4GalT7 is in its closed conformation (34).
Investigation of the structural role of His195 led to the most interesting findings for the design of efficient substrates and inhibitors of hβ4GalT7. We predicted that the nitrogen atom of the peptide backbone of this residue forms a hydrogen bond with the carbonyl group of the coumarin moiety of 4-MUX. This is in full agreement with our mutational study that showed no major effect upon changing the side chain of the histidine residue at this position by alanine, glutamine, or arginine substitution on the hβ4GalT7 activity monitored in vitro and in cellulo. However, the functional importance of an interaction between the His195 backbone and 4-MUX was clearly emphasized by the stronger inhibitory effect of 4-deoxy-Xyl-MU compared with 4-deoxy-Xyl-NP. We also found that 4-MUX was used as a substrate with a better affinity than Xyl-NP (data not shown). Noteworthy, among a series of naphthyl xylosides, Siegbahn et al. (29) showed that 6-hydroxy-naphthyl-Xyl was able to prime GAG chains more efficiently than any other unsubstituted derivative in breast cancer cell lines. Altogether, this gives strong evidence that His195 through its backbone provides an important structural element for efficient binding of 4-MUX derivatives, and offers a molecular explanation for the superiority of 4-MUX synthesized in this study over naphthyl and benzyl-substituted xylosides previously reported in the literature (25, 30).
We also discovered a unique active site basic residue, i.e. Arg226. Interestingly, this residue is located between the aromatic-rich 221FWGWG225 sequence, containing Trp224 that interacts with the β-phosphate O-atom of the donor substrate, and the 227EDDE230 sequence containing acidic residues that are involved in Xyl binding and in the transfer reaction (31). Our functional analysis showed that modifying the side chain of Arg226 by site-directed mutagenesis did not affect enzyme affinity toward acceptor or donor substrate. This is in line with the computational analysis indicating that the nitrogen atom of the peptide backbone of Arg226, but not its side chain, interacts with the O3 atom of the Xyl moiety. Fig. 1, A and B, clearly shows that this residue, together with Trp224, are brought close to the aromatic triad in the closed conformation of hβ4GalT7.
We also investigated the role of Arg270, which substitution by a cysteine residue is implicated in the progeroid form of EDS (49). Our previous studies revealed that this genetic mutation dropped in vitro hβ4GalT7 activity and impaired GAG chains synthesis in CHO-pgsB618 cells (17). These effects were suggested to be due to a loss of hydrogen bonding between the lateral chain of Arg270 and the hydroxyl group of a serine residue from the PG core protein (17). This idea was supported by later molecular modeling indicating that Arg270 borders the catalytic site of hβ4GalT7 in the closed conformation (Ref. 34 and this work, see Fig. 1A). However, the precise mechanism by which Arg270 modulates in vitro and ex vivo hβ4GalT7 activities remains unclear. Indeed, current crystal structures and molecular models do not point to a specific role of this residue in catalysis or substrate binding, consistent with kinetic data showing that mutations of Arg270 in alanine and lysine moderately affect hβ4GalT7 activity and affinity, and the observation that in Drosophila, the corresponding position is occupied by a lysine residue. The Arg270 residue is located in the flexible loop (261–284) that moves upon donor substrate binding, thus creating the acceptor substrate binding site. This conformational change leads to the closed and catalytically competent conformation of the active site. It thus can be expected that any mutations affecting the loop movement would impair the transfer reaction. However, why the substitution of Arg270 by a cysteine residue that causes the progeroid form of EDS patients, produces more deleterious consequences than alanine or lysine mutations requires further investigation of the conformational modifications operating during the catalytic cycle.
Our current and previous functional and computational approaches provide a detailed cartography of the hβ4GalT7 acceptor substrate binding pocket for the rational design of xyloside-based inhibitors (31). We show that the active site organization is governed, on one side, by a series of aromatic amino acids comprising three Tyr residues, i.e. Tyr194, Tyr196, and Tyr199, which together with Trp224 create a hydrophobic environment and provide stacking interactions essential to the binding of both the xylosyl and aglycone parts of the acceptor substrate. On the opposite side of the site, it involves a network of hydrogen-bonding interactions between three charged amino acids, i.e. Asp228, Asp229, and Arg226, and the hydroxyl groups of the Xyl moiety and other active site residues. Until now, most studies aiming to inhibit cellular and extracellular GAG synthesis have been targeted to the synthesis and testing of xyloside derivatives acting as substrates of β4GalT7 thus reducing the glycosylation of endogenous proteoglycan core proteins (19). This approach successfully provided promising pharmacological agents, in particular anti-tumor compounds (45, 50). However, because such molecules behave both as exogenous primers of GAG synthesis and inhibitors of endogenous GAG formation, deciphering their mechanism of action remains challenging. With the perspective of designing xyloside derivatives that selectively act as inhibitors of GAG formation, we opted for C4-modified analogs, whose modification at the C4 position prevents the catalytic transfer, and first synthesized deoxy derivatives. In addition, we took advantage of the information gained from our structural and mutational analyses. We considered two key elements of the aglycone binding, i.e. the strategic position of Tyr194 that forms stacking interactions with the aglycone part of the acceptor substrate as well as the interaction between His195 N-backbone and the carbonyl group of the coumarinyl moiety. In agreement with our prediction, our results clearly show that the 4-deoxy-xyloside appended to 4-MU was superior to the naphthyl-substituted molecule, as indicated by in vitro and in cellulo studies, supporting the idea that the hydrogen bond between His195 and the carbonyl group of the coumaryl group is crucial. Interestingly, Tsuzuki et al. (28) found that among triazole xyloside derivatives generated by click chemistry bearing various aromatic and nonaromatic aglycones, the p-nitrophenyl analog was the best inhibitor of PG synthesis when screened in endothelial cells. Although detailed docking of these triazole derivatives should be performed, it is tempting to speculate that the formation of a hydrogen bond interaction between the aglycone nitro group and His195 enhances the inhibitory potential compared with the other substituted benzyl derivatives. Furthermore, we show here that the 4-deoxy-4-fluorinated 4-MUX was superior to the unsubstituted 4-deoxy analog, indicating that addition of an electronegative atom at this position is an important element in the design of potent inhibitors. The 4-hydroxyl group is involved in two hydrogen bonds with the carboxyl group of Asp228 and the 4-hydroxyl group of Gal, respectively. The replacement of the hydrogen donor hydroxyl group by a fluorine atom that is larger than hydrogen and which van der Waals radius and electronegativity are closer to oxygen, at the C4 position, enhanced binding interactions with hβ4GalT7. This corroborated previous studies showing that several fluorinated xylosides act as “good” substrates or inhibitors of GAG synthesis (18, 29, 30). In the same way, fluorinated thrombin inhibitors showed improved factor Xa binding (51). Further docking calculations of fluoro-substituted xylosides are underway to assess the mechanism underlying the improved interactive properties upon fluorine incorporation.
In summary, we developed a powerful approach for the design of xyloside inhibitors that specifically target hβ4GalT7. By integrating structural elements important for the binding of both the Xyl moiety and the hydrophobic aglycone, we synthesized a xyloside-based inhibitor of hβ4GalT7. We generated a compound that both impact in vitro galactosyltransferase activity and affect GAG synthesis in cells, opening promising pharmacological applications. This molecule also represents a valuable chemical biology tool to explore the biological effects of GAGs.
Acknowledgments
Valérie Gisclard is sincerely acknowledged for her profound implication and Anne Robert is gratefully acknowledged for excellent technical assistance.
This work was supported by Agence Nationale pour la Recherche (ANR) Meca-GT Grant ANR-13-BSV8-0011-01, ANR GAG-Sorting Grant ANR-13-JS07-0004-01, Labex SynOrg Grant ANR-11-LABX-0029, International Associated Laboratory CNRS-Université de Lorraine-University of Dundee, SFGEN), Région Lorraine-FEDER (Glyco-Fluo) grant, grants from the Région Lorraine (to N. R. and S. F. G.), and the AMSEDgenetique association.
- GAG
- glycosaminoglycan
- EDS
- Ehlers-Danlos syndrome
- 4-MU
- 4-methylumbelliferone
- NP
- naphthyl
- PG
- proteoglycan
- Xyl
- xylose
- hβ4GalT7
- β1,4-galactosyltransferase 7.
REFERENCES
- 1. Bernfield M., Götte M., Park P. W., Reizes O., Fitzgerald M. L., Lincecum J., Zako M. (1999) Functions of cell surface heparan sulfate proteoglycans. Annu. Rev. Biochem. 68, 729–777 [DOI] [PubMed] [Google Scholar]
- 2. Clark S. J., Bishop P. N., Day A. J. (2013) The proteoglycan glycomatrix: a sugar microenvironment essential for complement regulation. Front. Immunol. 4, 412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Afratis N., Gialeli C., Nikitovic D., Tsegenidis T., Karousou E., Theocharis A. D., Pavão M. S., Tzanakakis G. N., Karamanos N. K. (2012) Glycosaminoglycans: key players in cancer cell biology and treatment. FEBS J. 279, 1177–1197 [DOI] [PubMed] [Google Scholar]
- 4. Halper J. (2014) Proteoglycans and diseases of soft tissues. Adv. Exp. Med. Biol. 802, 49–58 [DOI] [PubMed] [Google Scholar]
- 5. Wang P., Ding K. (2014) Proteoglycans and glycosaminoglycans in misfolded proteins formation in Alzheimer's disease. Protein Pept. Lett. 21, 1048–1056 [DOI] [PubMed] [Google Scholar]
- 6. Kadomatsu K., Sakamoto K. (2014) Mechanisms of axon regeneration and its inhibition: roles of sulfated glycans. Arch. Biochem. Biophys. 558, 36–41 [DOI] [PubMed] [Google Scholar]
- 7. Sugahara K., Kitagawa H. (2000) Recent advances in the study of the biosynthesis and functions of sulfated glycosaminoglycans. Curr. Opin. Struct. Biol. 10, 518–527 [DOI] [PubMed] [Google Scholar]
- 8. Yada T., Gotoh M., Sato T., Shionyu M., Go M., Kaseyama H., Iwasaki H., Kikuchi N., Kwon Y. D., Togayachi A., Kudo T., Watanabe H., Narimatsu H., Kimata K. (2003) Chondroitin sulfate synthase-2: molecular cloning and characterization of a novel human glycosyltransferase homologous to chondroitin sulfate glucuronyltransferase, which has dual enzymatic activities. J. Biol. Chem. 278, 30235–30247 [DOI] [PubMed] [Google Scholar]
- 9. Lind T., Tufaro F., McCormick C., Lindahl U., Lidholt K. (1998) The putative tumor suppressors EXT1 and EXT2 are glycosyltransferases required for the biosynthesis of heparan sulfate. J. Biol. Chem. 273, 26265–26268 [DOI] [PubMed] [Google Scholar]
- 10. Sheng J., Xu Y., Dulaney S. B., Huang X., Liu J. (2012) Uncovering biphasic catalytic mode of C5-epimerase in heparan sulfate biosynthesis. J. Biol. Chem. 287, 20996–21002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Honke K., Taniguchi N. (2002) Sulfotransferases and sulfated oligosaccharides. Med. Res. Rev. 22, 637–654 [DOI] [PubMed] [Google Scholar]
- 12. Almeida R., Levery S. B., Mandel U., Kresse H., Schwientek T., Bennett E. P., Clausen H. (1999) Cloning and expression of a proteoglycan UDP-galactose: β-xylose β1,4-galactosyltransferase I: a seventh member of the human β4-galactosyltransferase gene family. J. Biol. Chem. 274, 26165–26171 [DOI] [PubMed] [Google Scholar]
- 13. Götte M., Spillmann D., Yip G. W., Versteeg E., Echtermeyer F. G., van Kuppevelt T. H., Kiesel L. (2008) Changes in heparan sulfate are associated with delayed wound repair, altered cell migration, adhesion and contractility in the galactosyltransferase I (β4GalT-7) deficient form of Ehlers-Danlos syndrome. Hum. Mol. Genet. 17, 996–1009 [DOI] [PubMed] [Google Scholar]
- 14. Okajima T., Yoshida K., Kondo T., Furukawa K. (1999) Human homolog of Caenorhabditis elegans sqv-3 gene is galactosyltransferase I involved in the biosynthesis of the glycosaminoglycan-protein linkage region of proteoglycans. J. Biol. Chem. 274, 22915–22918 [DOI] [PubMed] [Google Scholar]
- 15. Furukawa K., Okajima T. (2002) Galactosyltransferase I is a gene responsible for progeroid variant of Ehlers-Danlos syndrome: molecular cloning and identification of mutations. Biochim. Biophys. Acta 1573, 377–381 [DOI] [PubMed] [Google Scholar]
- 16. Götte M., Kresse H. (2005) Defective glycosaminoglycan substitution of decorin in a patient with progeroid syndrome is a direct consequence of two point mutations in the galactosyltransferase I (β4GalT-7) gene. Biochem. Genet. 43, 65–77 [DOI] [PubMed] [Google Scholar]
- 17. Bui C., Talhaoui I., Chabel M., Mulliert G., Coughtrie M. W., Ouzzine M., Fournel-Gigleux S. (2010) Molecular characterization of β1,4-galactosyltransferase 7 genetic mutations linked to the progeroid form of Ehlers-Danlos syndrome (EDS). FEBS Lett. 584, 3962–3968 [DOI] [PubMed] [Google Scholar]
- 18. Lugemwa F. N., Sarkar A. K., Esko J. D. (1996) Unusual β-d-xylosides that prime glycosaminoglycans in animal cells. J. Biol. Chem. 271, 19159–19165 [DOI] [PubMed] [Google Scholar]
- 19. Esko J. D., Weinke J. L., Taylor W. H., Ekborg G., Rodén L., Anantharamaiah G., Gawish A. (1987) Inhibition of chondroitin and heparan sulfate biosynthesis in Chinese hamster ovary cell mutants defective in galactosyltransferase I. J. Biol. Chem. 262, 12189–12195 [PubMed] [Google Scholar]
- 20. Muto J., Naidu N. N., Yamasaki K., Pineau N., Breton L., Gallo R. L. (2011) Exogenous addition of a C-xylopyranoside derivative stimulates keratinocyte dermatan sulfate synthesis and promotes migration. PloS One 6, e25480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nguyen T. K., Tran V. M., Sorna V., Eriksson I., Kojima A., Koketsu M., Loganathan D., Kjellén L., Dorsky R. I., Chien C. B., Kuberan B. (2013) Dimerized glycosaminoglycan chains increase FGF signaling during zebrafish development. ACS Chem. Biol. 8, 939–948 [DOI] [PubMed] [Google Scholar]
- 22. Martin N. B., Masson P., Sepulchre C., Theveniaux J., Millet J., Bellamy F. (1996) Pharmacologic and biochemical profiles of new venous antithrombotic β-d-xyloside derivatives: potential antiathero/thrombotic drugs. Semin. Thromb. Hemost. 22, 247–254 [DOI] [PubMed] [Google Scholar]
- 23. Vassal-Stermann E., Duranton A., Black A. F., Azadiguian G., Demaude J., Lortat-Jacob H., Breton L., Vivès R. R. (2012) A new C-xyloside induces modifications of GAG expression, structure and functional properties. PloS One 7, e47933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Raman K., Ninomiya M., Nguyen T. K., Tsuzuki Y., Koketsu M., Kuberan B. (2011) Novel glycosaminoglycan biosynthetic inhibitors affect tumor-associated angiogenesis. Biochem. Biophys. Res. Commun. 404, 86–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mani K., Havsmark B., Persson S., Kaneda Y., Yamamoto H., Sakurai K., Ashikari S., Habuchi H., Suzuki S., Kimata K., Malmström A., Westergren-Thorsson G., Fransson L. A. (1998) Heparan/chondroitin/dermatan sulfate primer 2-(6-hydroxynaphthyl)-O-β-d-xylopyranoside preferentially inhibits growth of transformed cells. Cancer Res. 58, 1099–1104 [PubMed] [Google Scholar]
- 26. Kolset S. O., Sakurai K., Ivhed I., Overvatn A., Suzuki S. (1990) The effect of β-d-xylosides on the proliferation and proteoglycan biosynthesis of monoblastic U-937 cells. Biochem. J. 265, 637–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Garud D. R., Tran V. M., Victor X. V., Koketsu M., Kuberan B. (2008) Inhibition of heparan sulfate and chondroitin sulfate proteoglycan biosynthesis. J. Biol. Chem. 283, 28881–28887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tsuzuki Y., Nguyen T. K., Garud D. R., Kuberan B., Koketsu M. (2010) 4-Deoxy-4-fluoro-xyloside derivatives as inhibitors of glycosaminoglycan biosynthesis. Bioorg. Med. Chem. Lett. 20, 7269–7273 [DOI] [PubMed] [Google Scholar]
- 29. Siegbahn A., Aili U., Ochocinska A., Olofsson M., Rönnols J., Mani K., Widmalm G., Ellervik U. (2011) Synthesis, conformation and biology of naphthoxylosides. Bioorg. Med. Chem. 19, 4114–4126 [DOI] [PubMed] [Google Scholar]
- 30. Siegbahn A., Manner S., Persson A., Tykesson E., Holmqvist K., Ochocinska A., Rönnols J., Sundin A., Mani K., Westergren-Thorsson G., Widmalmc G., Ellervik U. (2014) Rules for priming and inhibition of glycosaminoglycan biosynthesis: probing the β4GalT7 active site. Chem. Sci. 5, 3501–3508 [Google Scholar]
- 31. Talhaoui I., Bui C., Oriol R., Mulliert G., Gulberti S., Netter P., Coughtrie M. W., Ouzzine M., Fournel-Gigleux S. (2010) Identification of key functional residues in the active site of human β1,4-galactosyltransferase 7: a major enzyme in the glycosaminoglycan synthesis pathway. J. Biol. Chem. 285, 37342–37358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rahuel-Clermont S., Daligault F., Piet M. H., Gulberti S., Netter P., Branlant G., Magdalou J., Lattard V. (2010) Biochemical and thermodynamic characterization of mutated β1,4-galactosyltransferase 7 involved in the progeroid form of the Ehlers-Danlos syndrome. Biochem. J. 432, 303–311 [DOI] [PubMed] [Google Scholar]
- 33. Ramakrishnan B., Qasba P. K. (2010) Crystal structure of the catalytic domain of Drosophila β1,4-galactosyltransferase-7. J. Biol. Chem. 285, 15619–15626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tsutsui Y., Ramakrishnan B., Qasba P. K. (2013) Crystal structures of β1,4-galactosyltransferase 7 enzyme reveal conformational changes and substrate binding. J. Biol. Chem. 288, 31963–31970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Eneyskaya E. V., Ivanen D. R., Shabalin K. A., Kulminskaya A. A., Backinowsky L. V., Brumer, Iii. H., Neustroev K. N. (2005) Chemo-enzymatic synthesis of 4-methylumbelliferyl β-(1→4)-d-xylooligosides: new substrates for β-d-xylanase assays. Org. Biomol. Chem. 3, 146–151 [DOI] [PubMed] [Google Scholar]
- 36. Sastry G. M., Adzhigirey M., Day T., Annabhimoju R., Sherman W. (2013) Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 27, 221–234 [DOI] [PubMed] [Google Scholar]
- 37. Kaminski G. A., Friesner R. A., Tirado-Rives J., Jorgensen W. L. (2001) Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. J. Phys. Chem. B 105, 6474–6487 [Google Scholar]
- 38. Banks J. L., Beard H. S., Cao Y., Cho A. E., Damm W., Farid R., Felts A. K., Halgren T. A., Mainz D. T., Maple J. R., Murphy R., Philipp D. M., Repasky M. P., Zhang L. Y., Berne B. J., Friesner R. A., Gallicchio E., Levy R. M. (2005) Integrated modeling program, applied chemical theory (IMPACT). J. Comput. Chem. 26, 1752–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Friesner R. A., Banks J. L., Murphy R. B., Halgren T. A., Klicic J. J., Mainz D. T., Repasky M. P., Knoll E. H., Shelley M., Perry J. K., Shaw D. E., Francis P., Shenkin P. S. (2004) Glide: a new approach for rapid, accurate docking and scoring: 1. method and assessment of docking accuracy. J. Med. Chem. 47, 1739–1749 [DOI] [PubMed] [Google Scholar]
- 40. Repasky M. P., Shelley M., Friesner R. A. (2007) Flexible ligand docking with Glide. Curr. Protoc. Bioinformatics Chapter 8, Unit 8.12 [DOI] [PubMed] [Google Scholar]
- 41. Gulberti S., Lattard V., Fondeur M., Jacquinet J. C., Mulliert G., Netter P., Magdalou J., Ouzzine M., Fournel-Gigleux S. (2005) Phosphorylation and sulfation of oligosaccharide substrates critically influence the activity of human β1,4-galactosyltransferase 7 (GalT-I) and β1,3-glucuronosyltransferase I (GlcAT-I) involved in the biosynthesis of the glycosaminoglycan-protein linkage region of proteoglycans. J. Biol. Chem. 280, 1417–1425 [DOI] [PubMed] [Google Scholar]
- 42. Cheng Y., Prusoff W. H. (1973) Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50% inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 22, 3099–3108 [DOI] [PubMed] [Google Scholar]
- 43. Lazareno S., Birdsall N. J. (1993) Estimation of competitive antagonist affinity from functional inhibition curves using the Gaddum, Schild and Cheng-Prusoff equations. Br. J. Pharmacol. 109, 1110–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fournel-Gigleux S., Jackson M. R., Wooster R., Burchell B. (1989) Expression of a human liver cDNA encoding a UDP-glucuronosyltransferase catalysing the glucuronidation of hyodeoxycholic acid in cell culture. FEBS Lett. 243, 119–122 [DOI] [PubMed] [Google Scholar]
- 45. Belting M., Borsig L., Fuster M. M., Brown J. R., Persson L., Fransson L. A., Esko J. D. (2002) Tumor attenuation by combined heparan sulfate and polyamine depletion. Proc. Natl. Acad. Sci. U.S.A. 99, 371–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Daligault F., Rahuel-Clermont S., Gulberti S., Cung M. T., Branlant G., Netter P., Magdalou J., Lattard V. (2009) Thermodynamic insights into the structural basis governing the donor substrate recognition by human β1,4-galactosyltransferase 7. Biochem. J. 418, 605–614 [DOI] [PubMed] [Google Scholar]
- 47. Malfait F., Kariminejad A., Van Damme T., Gauche C., Syx D., Merhi-Soussi F., Gulberti S., Symoens S., Vanhauwaert S., Willaert A., Bozorgmehr B., Kariminejad M. H., Ebrahimiadib N., Hausser I., Huysseune A., Fournel-Gigleux S., De Paepe A. (2013) Defective initiation of glycosaminoglycan synthesis due to B3GALT6 mutations causes a pleiotropic Ehlers-Danlos-syndrome-like connective tissue disorder. Am. J. Hum. Genet. 92, 935–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ueno M., Yamada S., Zako M., Bernfield M., Sugahara K. (2001) Structural characterization of heparan sulfate and chondroitin sulfate of syndecan-1 purified from normal murine mammary gland epithelial cells: common phosphorylation of xylose and differential sulfation of galactose in the protein linkage region tetrasaccharide sequence. J. Biol. Chem. 276, 29134–29140 [DOI] [PubMed] [Google Scholar]
- 49. Seidler D. G., Faiyaz-Ul-Haque M., Hansen U., Yip G. W., Zaidi S. H., Teebi A. S., Kiesel L., Götte M. (2006) Defective glycosylation of decorin and biglycan, altered collagen structure, and abnormal phenotype of the skin fibroblasts of an Ehlers-Danlos syndrome patient carrying the novel Arg270Cys substitution in galactosyltransferase I (β4GalT-7). J. Mol. Med. 84, 583–594 [DOI] [PubMed] [Google Scholar]
- 50. Mani K., Belting M., Ellervik U., Falk N., Svensson G., Sandgren S., Cheng F., Fransson L. A. (2004) Tumor attenuation by 2(6-hydroxynaphthyl)-β-d-xylopyranoside requires priming of heparan sulfate and nuclear targeting of the products. Glycobiology 14, 387–397 [DOI] [PubMed] [Google Scholar]
- 51. Hagmann W. K. (2008) The many roles for fluorine in medicinal chemistry. J. Med. Chem. 51, 4359–4369 [DOI] [PubMed] [Google Scholar]