Background: Pseudomonas aeruginosa utilizes extracellular heme as a source of iron on infection of the host.
Results: Both PhuR and HasR are required for efficient utilization of heme by P. aeruginosa.
Conclusion: Non-redundant outer membrane receptors allow for heme acquisition across a range of physiological conditions.
Significance: PhuR and HasR have complimentary roles in heme acquisition and utilization.
Keywords: Cell Surface Receptor, Heme, Iron Metabolism, Mass Spectrometry (MS), Membrane Transport
Abstract
Pseudomonas aeruginosa PAO1 encodes two outer membrane receptors, PhuR (Pseudomonas heme uptake) and HasR (heme assimilation system). The HasR and PhuR receptors have distinct heme coordinating ligands and substrate specificities. HasR is encoded in an operon with a secreted hemophore, HasAp. In contrast the non-hemophore-dependent PhuR is encoded within an operon along with proteins required for heme translocation into the cytoplasm. Herein we report on the contributions of the HasR and PhuR receptors to heme uptake and utilization. Employing bacterial genetics and isotopic [13C]heme labeling studies we have shown both PhuR and HasR are required for optimal heme utilization. However, the unique His-Tyr-ligated PhuR plays a major role in the acquisition of heme. In contrast the HasR receptor plays a primary role in the sensing of extracellular heme and a supplementary role in heme uptake. We propose PhuR and HasR represent non-redundant heme receptors, capable of accessing heme across a wide range of physiological conditions on colonization of the host.
Introduction
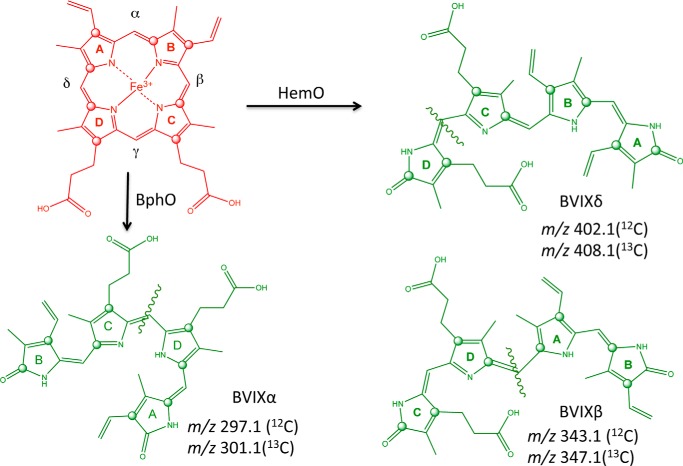
Iron is an essential micronutrient for the survival and virulence of Gram-negative pathogens (1–4). Many pathogens including the opportunistic pathogen Pseudomonas aeruginosa encode systems that utilize the host heme containing proteins as a source of iron (5, 6). P. aeruginosa encodes two heme uptake systems the Pseudomonas heme uptake (phu)2 and the heme assimilation systems (has). The phu system encodes a TonB-dependent outer membrane (OM) receptor (PhuR) that transports heme to the periplasm, where a soluble heme-binding protein (PhuT) interacts with an ATP-dependent permease (ABC-transporter) (PhuUV), which then translocates heme into the cytoplasm. Once internalized heme is sequestered by the cytoplasmic heme-binding protein PhuS. In a series of in vitro studies we have shown that the cytoplasmic heme-binding protein PhuS forms a specific protein complex with the iron-regulated heme oxygenase, HemO (7–9). Heme is transferred to HemO for further degradation with the release of iron, CO, and biliverdin IX (BVIX) δ- and β-isomers (10, 11). P. aeruginosa encodes a second non-iron-regulated heme oxygenase, BphO directly upstream of the phytochrome two-component sensor kinase, BphP (12, 13). BphO catalyzes the degradation of heme to BVIXα that then acts as a ligand for the sensor kinase, BphP. Furthermore, in contrast to HemO the cytoplasmic heme-binding protein, PhuS, does not interact with the non-iron-regulated heme oxygenase BphO (7). In keeping with the specific interaction of PhuS with HemO [13C]heme isotopic labeling studies confirmed in wild type PAO1 extracellular heme is primarily metabolized to [13C]BVIXδ and [13C]BVIXβ (14).
In contrast to the phu operon, the has operon encodes a soluble secreted hemophore (HasA) that scavenges heme and transfers it to the TonB-dependent outer membrane receptor (HasR) (15–17). The HasA protein coordinates heme through His-32 and Tyr-75 with picomolar affinity (18, 19). Recent kinetic and spectroscopic studies of the P. aeruginosa HasA protein have shown that heme acquisition from methemoglobin is relatively passive, occurring at a rate similar to that of heme dissociation (20). Therefore, the capture of dissociated heme by HasA and its reported nanomolar binding affinity to the HasR receptor, would allow the bacteria to access a wider range of heme concentrations within the host environment (21). Sequence alignment of all TonB-dependent OM heme receptors in combination with site-directed mutagenesis of the Yersinia entercolitica HemR, identified the highly conserved His residues located in the predicted N-terminal plug and conserved FRAP/PNPNL loop as being required for heme uptake (22). Spectroscopic and structural characterization of HasR of Serratia marcescens and ShuA of Shigella dysenteriae have confirmed heme to be coordinated through a His on the face of the N-terminal plug occluding the membrane spanning β-barrel, and a corresponding His on the extracellular FRAP/PNPNL loop (Fig. 1) (23–25). Interestingly, the majority of heme receptors so far characterized, both hemophore-dependent and independent, have the characteristic bis-His coordination. An exception to the bis-His coordination was the recently reported five coordinate Tyr ligation in the HmbR receptor of Neisseria meningitidis (26). Recent spectroscopic characterization of the PhuR receptor by our laboratory revealed heme is coordinated through His-124 of the N-terminal plug and Tyr-519 of the FRAP/PNPL loop.3 Interestingly, His-Tyr heme coordination is a common motif in the lipid anchored surface exposed heme receptors of Gram-positive pathogens such as Staphylococcus aureus (27–29), the soluble periplasmic heme-binding proteins (30, 31) and the HasA secreted hemophores of Gram-negative pathogens (18, 19, 32).
FIGURE 1.
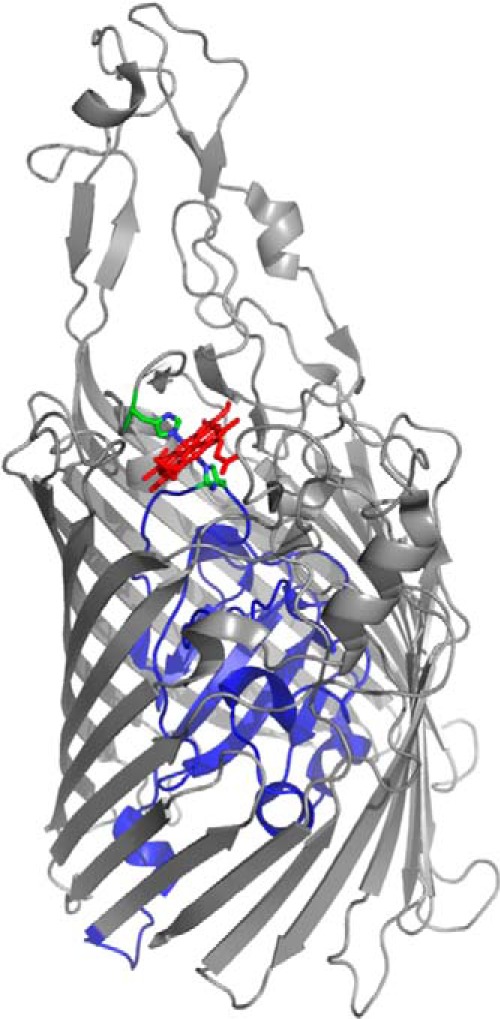
Structure of holo-HasR showing the N-terminal plug (blue) and the β-barrel (gray). The heme coordinating residues His-86 (plug) and His-420 (extracellular loop, L7) are shown as sticks. The image was generated from Protein Data Bank file 3CSL in PyMOL Graphics (49).
Several Gram-negative pathogens have been reported to have multiple heme uptake systems including S. marcescens (33), S. dysenteriae (34), V. cholera (35), Y. pestis (36), and N. meningitidis (37). In a S. marcescens heme auxotroph in vivo studies have shown the hasR and the non-hemophore hemR genes are differentially regulated by heme and iron (38). The HasA-HasR system was reported to access heme at much lower concentrations than HemR, but only under extreme iron limitation. The authors concluded the HemR receptor is a lower affinity receptor expressed at higher concentrations of iron than the high affinity hemophore-dependent HasR. Hence the HasR and HemR systems are optimized for heme acquisition across a wide spectrum of physiological iron gradients. The loss of HasA significantly reduced the range of heme concentrations accessible by HasR alone. In addition the ability to extract heme from the high affinity heme-binding protein hemopexin required the presence of HasA. The authors proposed the differential regulation and spectrum of accessible substrates represented non-redundant uptake systems, and further hypothesized the high affinity has system was absolutely required for vertebrate infection.
In contrast recent transcriptome analyses of P. aeruginosa longitudinal cystic fibrosis lung isolates suggested PhuR is the major heme receptor in clinical infection (39). Utilizing isotopic-labeling studies we have recently shown extracellular heme utilization is dependent on up-regulation of the phu system in clinical isolates (40). We hypothesized the recent His-Tyr heme coordination in PhuR and the emergence of this motif in high affinity heme systems may provide a mechanism to acquire heme across a range of physiological concentrations. The studies herein address the contributions of the hemophore-dependent HasR and PhuR to heme uptake in P. aeruginosa through isotopic [13C]heme labeling studies.
EXPERIMENTAL PROCEDURES
Bacterial Strains
All bacterial strains utilized in this study are listed in Table 1. P. aeruginosa PAO1 and deletion strains were stored as glycerol stocks in LB at −80 °C. P. aeruginosa strains were freshly streaked and maintained on Pseudomonas isolation agar (BD Biosciences) before transferring to liquid culture medium. PAO1 ΔphuR::gmΔhasR::tc was grown in the presence of 50 μg/ml of gentamicin and 100 μg/ml of tetracycline to maintain the respective antibiotic resistance cassettes. P. aeruginosa strains were cultured in Luria-Bertani (LB) (American Bioanalytical) broth. In experiments requiring iron limitation a single colony of P. aeruginosa from freshly streaked Pseudomonas isolation agar plates was used to inoculate LB media (20 ml) and grown overnight at 37 °C with shaking. The following day the cells were used to inoculate 20 ml of fresh LB medium and grown for a further 12 h. The cells were then used to inoculate M9 minimal medium to a final A600 of 0.1 and grown for 12 h to render the cells iron deficient.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain | Description | Source or reference |
|---|---|---|
| E. coli | ||
| DH5α | F′, araD (lac-proAB) rpsL ϕ80dlacZ DM15 hasd R17 | Stratagene |
| S17-1 | pro thi hsdR_ Tpr Smr; chromosome::RP4–2 Tc::Mu-Km::Tn7/λpir | 50 |
| P. aeruginosa | ||
| PAO1 | Wild type | |
| PAO1 ΔphuR::gmFRT | PAO1 with 0.4 kb of phuR replaced by GmR cassette harboring flanking FLP recombinase sites | Oglesby-Sherrouse |
| PAO1 ΔhasR::tcFRT | PAO1 with 1.5 kb of hasR replaced by TcR cassette harboring flanking FLP recombinase sites | Oglesby-Sherrouse |
| PAO1 ΔphuR::gmΔhasR::tc | PAO1 with 0.4 kb of phuR replaced by GmR and 1.5kb of hasR replaced by TcR cassettes harboring flanking FLP recombinase sites | Oglesby-Sherrouse |
| PAO1 ΔphuR | PAO1 with 1.1 kb of phuR gene removed by flip recombinase | This study |
| PAO1 ΔhasR | PAO1 with 1.5 kb of hasR gene removed by flip recombinase | This study |
| Plasmids | ||
| pFLP2 | AmpR; source of FLP recombinase | 51 |
| pMRL2 | AmpR; pET-11a derivate harboring a water-soluble domain of the rat liver cytochrome b5 | 41 |
Construction of the Unmarked PAO1 ΔphuR and ΔhasR Deletion Strains
The unmarked deletion mutants were constructed using flippase (FLP) recombinase encoded on the pFLP2 plasmid. Briefly, the Escherichia coli S17 donor strain was used to transfer the pFLP2 plasmid into either the ΔphuR::gmFRT or ΔhasR::tcFRT marked strains. Loss of the resistance cassette was confirmed by lack of growth on LB-gentamicin or LB-tetracycline, respectively. Sensitive colonies were selected and further streaked on LB, 5% sucrose to remove the pFLP2 plasmid. Loss of the antibiotic cassette was confirmed by PCR and DNA sequencing (Eurofins MWG Operon). The ΔphuR::gmFRT, ΔhasR::tcFRT, and ΔphuR::gmΔhasR::tcFRT strains were a gift from Professor Mandy Oglesby-Sherrouse.
Growth Curves of PAO1 Wild Type and Deletion Strains
Iron-deficient cultures were used to inoculate 50 ml of fresh M9 minimal media to an A600 of 0.08 with or without 0.5 μm heme, 5 μm heme, or 100 μm FeCl3. Cultures were grown an additional 8 h, and cells were harvested and supernatants were stored at −80 °C. All supplements were filtered using a 0.20-μm filter (Corning). Growth curves were generated using a Bioscreen C MBR (Growth Curves USA) with continuous shaking at 37 °C and the A600 was recorded after every 30 min over a 24-h time period.
Preparation and Isolation of [12C]- and [13C]Heme
Unlabeled [12C]δ-aminolevulinic acid (ALA) was purchased from Sigma. Labeled [4-13C]δ-ALA was purchased from Cambridge Isotope Laboratories. δ-ALA or [4-13C]δ-ALA was used as a biosynthetic precursor for the production of [12C]- or [13C]heme, respectively. Expression of cytochrome b5 in the presence of δ-ALA induces heme biosynthesis in E. coli where the produced cytochrome b5 captures and acts as a reservoir for the synthesized heme (41). [13C]Heme was prepared as previously reported (14, 42). Briefly, mitochondrial rat cytochrome b5 was expressed in E. coli BL21(DE3) cells in M9 minimal medium with the following supplements: 2 mm MgSO4, 100 μm CaCl2, 150 nm (NH4)6Mo7O24, 40 μm FeSO4 (acidified with 1 n HCl), 17 μm EDTA, 3 μm CuSO4, 2 μm Co(NO3)2, 7.6 μm ZnSO4, 9.4 μm Na2B4O7·10H2O, and 1 mm thiamine. Following lysis and centrifugation the cellular lysate was loaded onto a Q-Sepharose anion exchange column (2.5 × 10 cm) equilibrated in 50 mm Tris-HCl (pH 7.4), 5 mm EDTA, 50 mm NaCl. The column was washed with the same buffer followed by an additional wash with 50 mm Tris-HCl (pH 7.4), 5 mm EDTA, 125 mm NaCl (5 column volumes). Cytochrome b5 was eluted in 50 mm Tris-HCl (pH 7.4), 5 mm EDTA, 250 mm NaCl and dialyzed overnight in 50 mm Tris-HCl (pH 7.4), 5 mm EDTA, 50 mm NaCl. Heme extraction from cytochrome b5 was performed by the acid/butanone method as previously described (43). Heme concentrations were determined by the pyridine hemochrome assay (44). Heme stocks were prepared immediately prior to use by dissolving in 0.1 n NaOH and buffered to pH 7.4 with 1 m Tris-HCl and the final concentration was determined by pyridine hemochrome (pH 7.4). The labeling pattern of biosynthesized heme using [4-13C]δ-ALA is shown in Scheme 1.
SCHEME 1.
MS/MS fragmentation patterns of the BVIX isomers. The 13C labeling pattern is marked by circles. m/z for the major fragments derived from [12C]- and [13C]heme are shown.
Extraction of BVIX Isomers from P. aeruginosa Supernatants
Analysis of the BVIX isomers was performed as previously reported (14, 42). PAO1 WT and mutant strains were grown as described above and supplemented with either 0.5 or 5 μm 13C-labeled heme. 50-ml cultures were allowed to grow for 8 h at 37 °C in 250-ml baffled flasks with shaking and harvested by centrifugation. Supernatants were acidified to pH ∼ 3 with 10% trifluoroacetic acid (TFA) and loaded over a C18 Sep-Pak column and purified as previously reported (42). Purified BVIX isomers were air dried and stored for up to 1 week at −80 °C prior to LC-MS/MS analysis.
LC-MS/MS Analysis of BVIX Isomers
Samples were analyzed as previously described (42). Briefly, samples were resuspended in 10 μl of dimethyl sulfoxide and diluted to 40 μl with the mobile phase (acetone: 20 mm formic acid (50:50, v/v)) and filtered through a 0.45-μm PTFE syringe. The BVIX isomers were separated and analyzed by LC-MS/MS on a Waters TQD triple quadrupole mass spectrometer with an AQUITY H-Class UPLC over a reverse phase Phenomenex Ultracarb 5 μm ODS analytical column (4.6 × 250 mm) with a flow rate of 0.6 ml/min. The BVIX isomers were detected at 377 nm. Fragmentation patterns of the parent ions were analyzed using multiple reaction monitoring. The source temperature was set to 150 °C, the capillary voltage to 3.30 kV, and the cone voltage to 72 V. The collision energy was set to 34 V for BVIXα, 30 V for BVIXδ, and 36 V for BVIXβ, respectively.
PAO1 RNA Extraction and Purification
PAO1 WT and mutant strains were grown as described above in M9 minimal medium and supplemented with or without 0.5 μm heme, 5 μm heme, or 100 μm FeCl3 as a positive control for ferric uptake regulator-mediated iron regulation. All conditions were performed in triplicate. 1-ml aliquots were taken from cultures at the 8-h time point and cells corrected for differences in A600 were harvested by centrifugation at 12,000 × g for 10 min. Total RNA was extracted from the cell pellets using the RNeasy mini kit (Qiagen). Residual DNA bound to the column was removed by treatment with RNase-free DNase I (Qiagen) for 30 min and RNA was eluted in 40 μl of RNase-free water. Eluted RNA was further treated for DNA contamination with RNase-free DNase I (New England Biolabs) for 2–3 h at room temperature. RNA samples were precipitated overnight at −20 °C by the addition of 5 μl of 3 m sodium acetate (pH 5.5) and 150 μl of 100% ethanol. Precipitated RNA was centrifuged 13,000 × g for 30 min at 4 °C, washed with ice-cold 100% ethanol, and resuspended in 50 μl of RNase-free water. RNA concentrations were determined by measuring the absorbance at 260 nm on a NanoDrop 2000c spectrophotometer (Thermo Scientific) and adjusted to 50 ng/μl.
Q-RT-PCR mRNA Studies
cDNA was generated from 50 ng of total RNA using the ImPromII Reverse Transcription System (Promega). Briefly, 1 μl of RNA (50 ng/μl) was added to 1 μl of Random Primer (0.5 μg/μl) and adjusted to 5 μl with RNase-free water. Samples were incubated 65 °C for 10 min and cooled on ice. Master mix (4 μl of 5 × Buffer, 4 μl of MgCl2, 1 μl of dNTP, 1 μl of reverse transcriptase, 5 μl of RNase-free water) was added to the priming reaction and incubated at 25 °C for 5 min, 42 °C for 1 h, and 70 °C for 15 min. Q-PCR were performed using a Light Cycler 480 (Roche Applied Science) using the TaqMan Gene Expression Master Mix (Life Technologies). ΔΔCT values were calculated, with the relative amounts of cDNA being normalized by dividing the expression values by the relative amounts of the constitutively expressed omlA gene. All primers and probes used for qPCR studies are listed in Table 2.
TABLE 2.
Primers and probes used in this study
| Primers and probes | Sequence |
|---|---|
| omlA Probe | 5′-/56-FAM/CCC GAC GCC AAG TGC GGT TT/3BHQ_1/-3′ |
| omlA F primer | 5′-ACG CAG GAC ATG ATA GAC CAG TT-3′ |
| omlA R primer | 5′-TCG TTG AAG AAC AGG CTG ACG-3′ |
| phuR Probe | 5′-/56-FAM/CTA CGC GCA GAC CCA CCG CAA C/3BHQ_1/-3′ |
| phuR F primer | 5′-TGA CCA ACG ACT TCT TCA GC-3′ |
| phuR R primer | 5′-CTT TAC GAT GTC CGG ATC GAC-3′ |
| hasR Probe | 5′-/FAM/CTG GCC TAC GGG CAG CTC TCC TA/3BHQ_1/-3′ |
| hasR F primer | 5′-CGT GGC GTC GAG TAC CAG-3′ |
| hasR R primer | 5′-GGT CTT CGA ACA GAA GTC GTT G-3′ |
| phuS Probe | 5′-/56-FAM/CTT TCG GCC GCC GCT TCG A/3BHQ_1/-3′ |
| phuS F primer | 5′-TGC CGA CGA ACA CCA TGA-3′ |
| phuS R primer | 5′-TGG CGA CCT GGC GAA A-3′ |
| hemO Probe | 5′-/56-FAM/TTC GTC GCC/ZEN/GCC CAG TAC CTC TTC CAG CAT/3IABkFQ/-3′ |
| hemO F primer | 5′-TGG TGA AGA GCA AGG AAC CCT TC-3′ |
| hemO R primer | 5′-TTC GTT GCG ATA AAG AGG CTC CA-3′ |
| hasA Probe | 5′-/56-FAM/TCG ACC CGA GCC TGT/3BHQ_1/-3′ |
| hasA F primer | 5′-ATC GAC GCG CTG CTG AA-3′ |
| hasA R primer | 5′-TGG TCG AAG GTG GAG TTG ATC-3′ |
SDS-PAGE and Western Blot Analysis
1-ml aliquots of PAO1 WT and mutant cultures were harvested at the 8-h time point as described for the isolation of total mRNA. Cell pellets were resuspended in 100 μl/A600 of 1.0 in Bugbuster (Novagen). Cells were incubated at room temperature for 30 min with occasional agitation to ensure complete cell lysis and then boiled for 10 min. Total protein concentrations were determined using the Bio-Rad RCDC assay. 30 μg of total protein in 2× SDS loading buffer was run on 10% SDS-PAGE for PhuR and HasR detection and 12% SDS-PAGE for PhuS and HemO detection. Proteins were transferred by electrophoresis to a PVDF membrane (Millipore). Membranes were blocked with blocking buffer (5% (w/v) skim milk in Tris-buffered saline (TBS) with 0.2% (v/v) Tween 20), washed, and probed with a 1:1000 dilution of anti-PhuR or anti-HasR primary antibody and a 1:1000 dilution of anti-PhuS or anti-HemO primary antibody in 1% (w/v) skim milk in TBS with 0.2% (v/v) Tween 20. The membrane was then washed and probed with goat anti-rabbit immunoglobin G conjugated to horseradish peroxidase (KPL) at a dilution of 1:10,000 in 1% (w/v) skim milk in TBS with 0.2% (v/v) Tween 20. Proteins were visualized and enhanced by chemiluminescence detection using the Super-signal chemiluminescence kit (Pierce) on a Fluorochem HD2 imager (ProteinSimple). All antibodies were generated commercially from purified protein supplied to Covance Custom Antibodies.
RESULTS
Growth Phenotypes of the P. aeruginosa PAO1 Wild Type, ΔphuR, ΔhasR, and ΔphuR::gmΔhasR::tc Strains
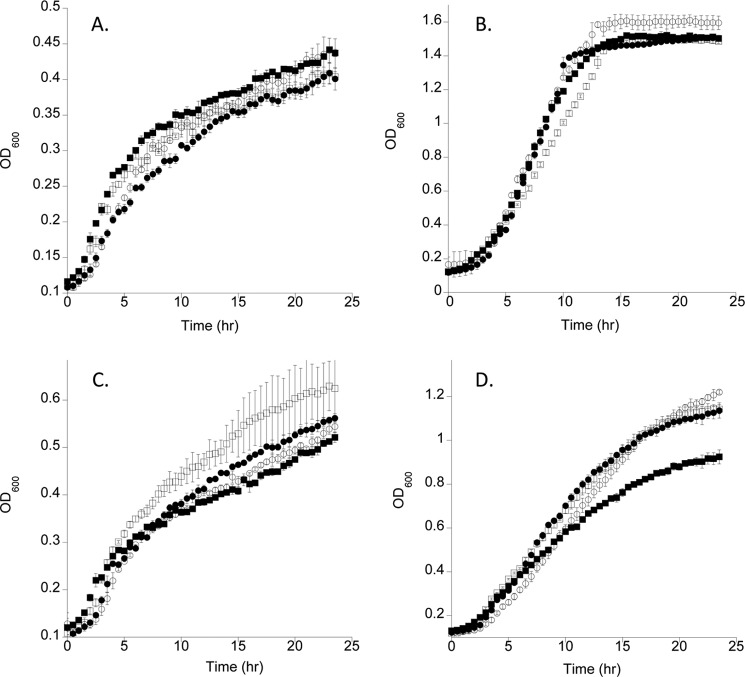
The growth phenotypes of the PAO1 mutant strains were analyzed under low iron conditions with or without heme or iron. All strains grew similarly under iron-restricted conditions or when supplemented with 100 μm FeCl3 (Fig. 2, A and B). On supplementation with 0.5 μm heme all strains grew similarly although the ΔhasR strain consistently grew at a slightly faster rate (Fig. 2C). At 5 μm heme the wild type and mutant strains retaining either the HasR or PhuR receptor grew at similar rates. However, not surprisingly when grown at higher heme concentrations the ΔphuR::gmΔhasR::tc strain in the absence of both OM heme receptors showed a slow growth phenotype (Fig. 2D).
FIGURE 2.
Growth curves of the P. aeruginosa WT and deletion strains supplemented with heme or iron. A, M9 media no iron. PAO1 wild type (open circles), ΔphuR (closed circles), ΔhasR (open squares), and ΔphuR::gmΔhasR::tc (closed squares). B, as in A supplemented with 100 μm FeCl3. C, supplemented with 0.5 μm heme. D, supplemented with 5 μm heme. Growth curves were generated using a Bioscreen C MBR (Growth Curves USA) with continuous shaking at 37 °C and the A600 was recorded after every 30 min over a 24-h time period. Values represent the standard deviation of three separate experiments.
Efficient Extracellular Heme Uptake and Utilization Requires the Concerted Action of the PhuR and HasR OM Receptors
Previous LC-MS studies have utilized isotopic [13C]heme labeling studies to determine BVIX derived from the degradation of extracellular versus intracellular heme (14, 42). Furthermore, tandem ESI-MS/MS analysis can distinguish BVIXδ or -β by the action of HemO versus the BVIXα from cleavage by BphO. By monitoring the [13C]heme metabolite profile in combination with mRNA and protein levels of the Phu and Has systems we have evaluated the contribution of each receptor to extracellular heme utilization. We tested the ability of PAO1 WT, ΔphuR, ΔhasR, and the ΔphuR::gmΔhasR::tc mutants to utilize extracellular [13C]heme at both 0.5 and 5 μm concentrations. Samples for metabolite, mRNA, and protein expression analysis were taken at 8 h. The steady state mRNA and protein levels for HasA, HasR, PhuR, the cytoplasmic heme-binding protein PhuS, and iron-regulated heme oxygenase HemO were measured by qRT-PCR and Western blot.
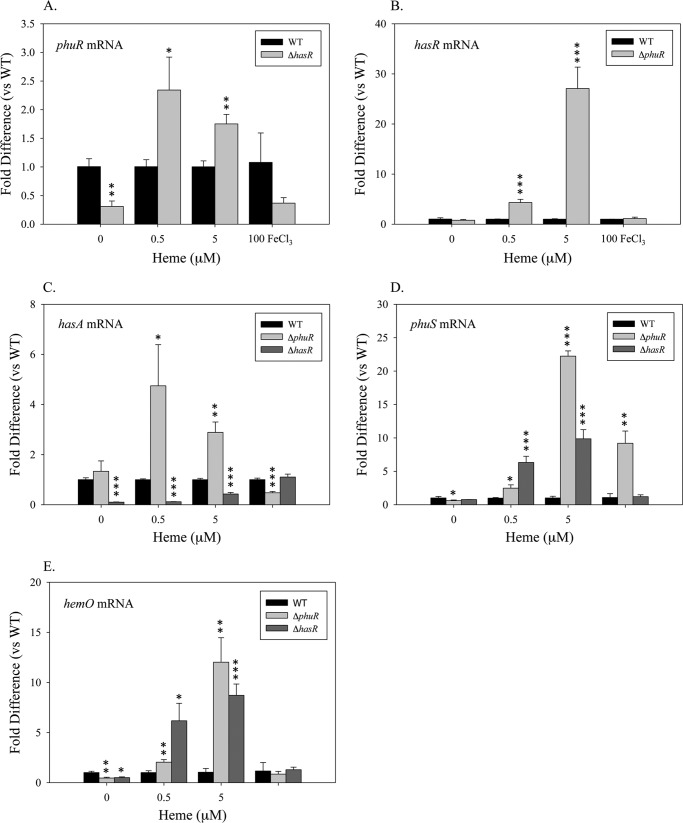
Although the ΔhasR strain in the absence of iron or heme shows a statistically relevant 2-fold decrease in steady state phuR mRNA levels when compared with PAO1 WT (Fig. 3A), this does not translate to increased protein levels (Fig. 4A). When supplemented with 0.5 or 5 μm heme the ΔhasR strain, when compared with wild type PAO1, shows a 2-fold increase in the steady state mRNA levels of phuR (Fig. 3A). Again whereas statistically relevant no apparent change in PhuR protein levels is observed between the ΔhasR strain and PAO1 WT (Fig. 4A). In contrast supplementation with 0.5 or 5 μm heme leads to a 5–10-fold increase in steady state mRNA levels of phuS and hemO (Fig. 3, D and E). The increase in mRNA levels is consistent with an increase in PhuS and HemO protein when compared with wild type PAO1 (Fig. 4B).
FIGURE 3.
mRNA levels of the heme utilization system in the ΔphurR and ΔhasR mutants compared with PAO1 WT. RNA isolated from the indicated strains at the 8-h time point supplemented with either heme or iron was quantified by qRT-PCR as described under “Experimental Procedures.” mRNA levels of phuR (A), hasR (B), hasA (C), phuS (D), and hemO (E). mRNA values represent the standard deviation from at least three experiments performed in triplicate. The indicated p values were normalized to mRNA levels of the PAO1 under the same conditions where *, p < 0.05; **, p < 0.005; or ***, p < 0.001.
FIGURE 4.
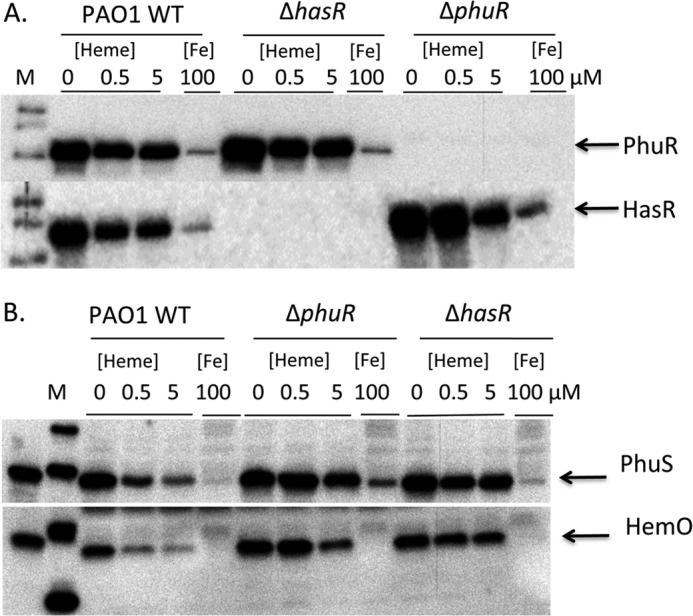
Expression of the heme utilization proteins in the PAO1 WT, ΔphuR, and ΔhasR strains. A, Western blot analysis of PhuR (upper panel) and HasR (lower panel) at the 8-h time point in P. aeruginosa strains as indicated. B, Western blot analysis of PhuS (upper panel) and HemO (lower panel) at the 8-h time point in P. aeruginosa strains as described under “Experimental Procedures.”
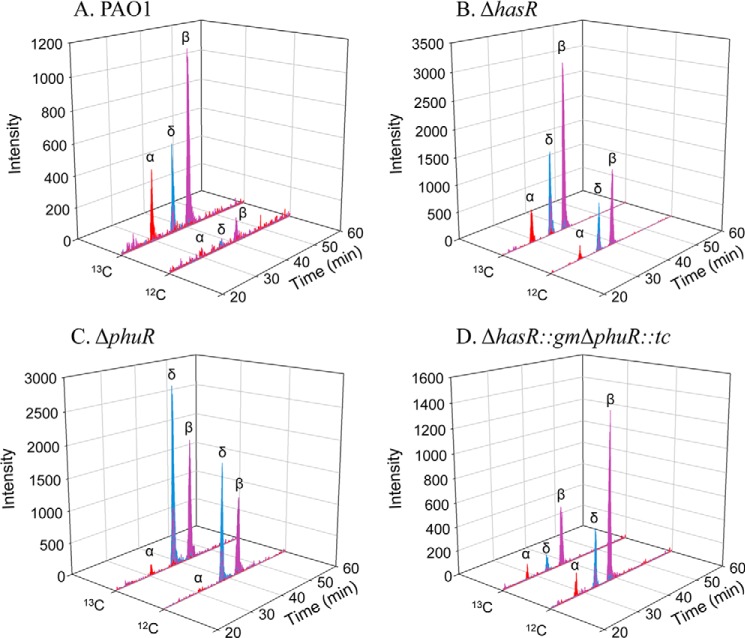
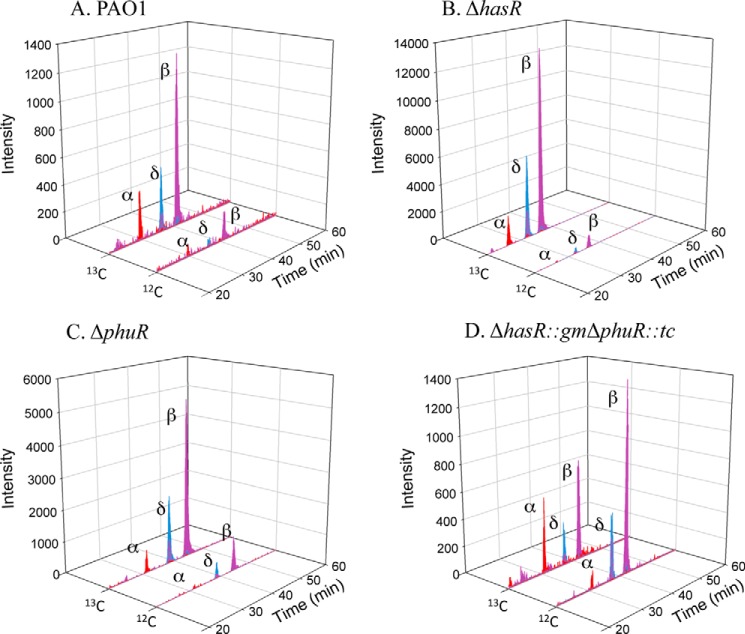
LC-MS/MS of the extracted BVIX metabolites from PAO1 WT supplemented with 0.5 μm heme showed the majority of heme is degraded through HemO to [13C]BVIXβ and -δ, as judged by peaks corresponding to fragment ions at m/z 347.1 and 408.1, respectively (Fig. 5A). A smaller fraction of [13C]BVIXα (m/z 301.1) is produced as a result of extracellular heme being “shunted” through BphO. However, no significant metabolism of intracellular [12C]heme was observed (Fig. 5A). At the higher 5 μm [13C]heme concentration a similar profile to that at 0.5 μm was observed (Fig. 6A).
FIGURE 5.
BVIX isomer fragmentation patterns for PAO1 WT and mutant strains supplemented with 0.5 μm heme. A, LC-MS/MS fragmentation pattern of the eluting isomers in PAO1 wild type. B, as in A for the ΔhasR strain. C, as in A for the ΔphuR strain. D, as in A for the ΔphuR::gmΔhasR::tc strain. LC-MS/MS with multiple reaction monitoring was performed as described under “Experimental Procedures.”
FIGURE 6.
BVIX isomer fragmentation patterns for PAO1 WT and mutant strains supplemented with 5 μm heme. A, LC-MS/MS fragmentation pattern of the eluting isomers in PAO1 wild type. B, as in A for the ΔhasR strain. C, as in A for the ΔphuR strain. D, as in A for the ΔphuR::gmΔhasR::tc strain. LC-MS/MS was performed as described under “Experimental Procedures” with multiple reaction monitoring.
[13C]Heme isotopic labeling studies of the ΔhasR strain supplemented with 0.5 μm heme produced a similar BVIX metabolite profile to that of PAO1 WT (Fig. 5B). However, in the absence of HasR a significant increase in the HemO-catalyzed degradation of intracellular [12C]heme was observed at 0.5 μm heme (Fig. 5B). The overall increase in heme metabolism correlates with up-regulation of the intracellular heme chaperone, PhuS, and heme oxygenase, HemO protein levels compared with the PAO1 WT strain (Fig. 4B). At the higher 5 μm heme concentration the ΔhasR strain shows a 10-fold increase in [13C]BVIXβ and -δ metabolites over that of PAO1 WT (Fig. 6B). Taken together the up-regulation of PhuS and HemO together with the turnover of intracellular biosynthesized [12C]heme indicates that on loss of HasR the cells are less efficient at utilizing heme. However, the significant increase in overall levels of [13C]BVIX metabolites on loss of HasR may be less a result of inefficient heme uptake but more an inability to sense and regulate heme uptake.
In contrast, deletion of the phuR gene results in a 5- and 30-fold increase in hasR steady state mRNA levels when compared with PAO1 WT at 0.5 and 5 μm heme, respectively (Fig. 3B). The increase in hasR mRNA levels in the presence of 0.5 and 5 μm heme is accompanied by a 5- to 2-fold increase in hasA mRNA, respectively (Fig. 3C). Western blot analysis of HasR protein levels is consistent with the increase in mRNA levels (Fig. 4A). The significant increase in mRNA and HasR protein levels at 0.5 μm heme indicates HasR is less efficient than PhuR in acquiring heme (Figs. 3B and 4A). This is further supported by the fact that compared with PAO1 WT at 0.5 μm heme, the increase in HasR protein along with the elevated PhuS and HemO proteins, the BVIXδ and -β derived from intracellular heme approximates that from extracellular heme uptake (Fig. 5C). The flux of [12C]heme through HemO can be suppressed on increasing the extracellular heme levels supporting the fact that HasR is less efficient at utilizing heme in the absence of PhuR (Fig. 6C). The significant up-regulation of HasR in the ΔphuR mutant suggests the Phu system plays a major role in heme acquisition in P. aeruginosa.
Interestingly, the 2:1 ratio of BVIXβ to -δ observed for the wild type and ΔhasR strain is reversed in the ΔphuR strain at the lower 0.5 μm heme concentration (Fig. 5C). It has previously been shown that the BVIXβ and -δ ratio is determined by a rotation of the heme around the α/γ axis (10, 45). Therefore, the BVIXβ to -δ ratio is determined by how the heme is “handed off” to HemO and suggests heme taken up via either PhuR or HasR may be delivered to PhuS in alternate orientations. The physiological relevance if any of the switch in the ratio of BVIXβ to -δ has not been determined. Furthermore, although it has always been assumed heme acquired through the HasA-HasR system required the intracellular components of the Phu system for translocation into the cytoplasm, our current studies are the first to link heme uptake via HasR to the Phu cytoplasmic heme uptake proteins.
PhuR and HasR Are the Primary Extracellular Heme Receptors in P. aeruginosa
We further determined if PhuR and HasR are the primary contributors to extracellular heme uptake in the ΔphuR::gmΔhasR::tc strain. On analysis of the [13C]BVIX metabolites at 0.5 μm heme concentration a decrease in [13C]BVIX metabolites correlates with an increase in intracellular [12C]heme being shunted through HemO (Fig. 5D). At the higher heme concentration increased background levels of [13C]BVIX metabolites were observed, presumably as a result of heme uptake via less efficient transporters or nonspecific diffusion across the membrane (Fig. 5D). Taken together the data confirms the PhuR and HasR OM receptors are primarily responsible for the acquisition of extracellular heme.
Heme-dependent Regulation of the Outer Membrane Receptors
As expected in the presence of 100 μm FeCl3 all strains show a significant decrease in the protein expression of the OM receptors, presumably by ferric uptake regulator-mediated repression (Fig. 4A). However, in the ΔphuR deletion the ferric uptake regulator-dependent repression of phuS is disrupted (Figs. 3D and 4B). We have noted a similar effect on the ferric uptake regulator-mediated repression of phuR on deletion of phuS.4 This loss of regulation is due to as yet unidentified transcriptional or translational regulatory elements upstream of the currently defined promoter (46). We are re-evaluating the heme and iron regulation of the phu operon through transcriptional and translational fusions. Interestingly, in WT PAO1 the PhuR and HasR receptors, as well as PhuS, are constitutively expressed at a low level in the presence of 100 μg/ml of FeCl3 (Fig. 4, A and B). In contrast the expression of HemO is more tightly regulated by iron (Fig. 4B). This would suggest that on encountering heme in the environment the bacteria are “primed” for heme uptake. Furthermore, the data are consistent with previous metabolism studies from our laboratory showing extracellular heme uptake, despite the expression of the heme uptake proteins, is dependent on the presence of a catalytically active HemO (42).
DISCUSSION
It is not possible to extrapolate heme binding affinities of the isolated receptors to in vivo function given the fact that the stability of any particular heme-ligated state will be dependent on its interaction with the TonB energy transducing system. Furthermore, heme binding to the isolated receptors may introduce heterogeneity not observed during active transport. In keeping with this reasoning we have recently shown that the isolated P. aeruginosa HasR and PhuR receptors can bind free heme but are not capable of stripping heme from hemoglobin or holo-HasAp.3 However, incubation of E. coli cultures expressing HasR or PhuR with either heme or hemoglobin increased the heme content of the purified receptor. This is in contrast to previous reports where the purified S. marcescens HasR receptor was reported to strip heme from holo-HasA (21). Additionally, the ShuA receptor of S. dysenteriae was shown to accept heme from methemoglobin in lipid bicelles but not free heme (23) and the recently characterized N. meningitidis HmbR receptor was unable to be reconstituted with heme or hemoglobin following purification (26). Therefore given the variation in the heme-binding properties of the detergent-solubilized receptors we have employed a combination of bacterial genetics and 13C-isotopic heme labeling to assess the contributions of the OM hemophore-dependent HasR and PhuR receptors to heme acquisition in P. aeruginosa.
Deletion of hasR does not lead to a significant increase in PhuR protein levels (Fig. 4A), nor does the loss of HasR affect the cells ability to efficiently acquire extracellular heme (Fig. 4B). However, whereas PAO1 WT heme uptake and metabolism leads to down-regulation of intracellular heme utilization proteins PhuS and HemO, the hasR mutant is unable to down-regulate the flux of heme. The increase in steady state levels of the intracellular heme utilization proteins PhuS and HemO most likely accounts for the “shunting” of intracellular biosynthesized heme through HemO at the lower heme concentration. The inability to regulate intracellular heme utilization proteins is not ferric uptake regulator-dependent as in the presence of iron, the steady state mRNA levels of phuS and hemO are down-regulated to the same degree as the wild type strain (Fig. 3, D and E) and consistent with decreased protein expression (Fig. 4B).
The P. aeruginosa HasR receptor shows high sequence homology with HasR of S. marcescens (46–48). The S. marcescens HasR mediates signal transduction through the extra cytoplasmic function σ and anti-σ factors HasI and HasS, respectively (47). In S. marcescens binding of the holo-HasA to HasR inactivates the anti-σ factor HasS and activates the σ factor HasI allowing transcription of the hasAR operon itself. It was further shown that transcription of hasS was autoregulated by HasI in a ferric uptake regulator-independent manner. Therefore, as HasS is subject to a heme-dependent signaling cascade, HasI regulation of hasS allows for expression and accumulation of inactive HasS. Extracellular heme levels that decrease HasR expression can be rapidly fine-tuned to the fluctuating extracellular heme concentrations. Therefore, as heme acquired through HasR requires the periplasmic and cytoplasmic components of the Phu uptake system we hypothesize some “cross-talk” exists between the hemophore signaling cascade and the phu operon.
In contrast to the ΔhasR mutant, deletion of phuR results in a significant up-regulation of HasR to compensate for loss of PhuR. It is not clear if PhuR represents a higher affinity receptor, however, it is interesting that the His-Tyr heme ligation found in PhuR is an emerging motif in the high affinity lipid anchored heme receptors of Gram-positive organisms (27–29), the soluble periplasmic heme-binding proteins (30, 31), and the HasA-secreted hemophores of Gram-negative pathogens (18, 19, 32). We propose P. aeruginosa has exploited the higher affinity His-Tyr ligation for the soluble extracellular HasA “heme sensor” and the high capacity PhuR receptor to rapidly respond and transport heme across a wide range of physiological concentrations within the host.
Furthermore, loss of PhuR is accompanied by an increase in intracellular [12C]heme metabolism, presumably due to decreased efficiency of HasR coupled with the increased steady state levels of the PhuS and HemO proteins. In contrast to the ΔhasR mutant we attribute the elevated levels of PhuS protein to the disruption in ferric uptake regulator-regulated repression of phuS. Specifically, in contrast to PAO1 WT addition of 100 μm FeCl3 to the ΔphuR strain does not suppress PhuS protein levels (Fig. 4B). We have observed a similar disruption in ferric uptake regulator regulation of phuR on deletion of the phuS gene.4 Interestingly, we also observe that increased PhuS protein levels in the presence of heme in both the ΔhasR and ΔphuR strains compared with PAO1 WT is coupled to an increase in HemO (Fig. 4B). This regulatory link between phuS and hemO is heme dependent given the fact that in the presence of iron hemO transcription is suppressed in the ΔphuR strain to the same degree as in PAO1 wild type. We are currently investigating the heme-dependent regulatory link between phuS and hemO.
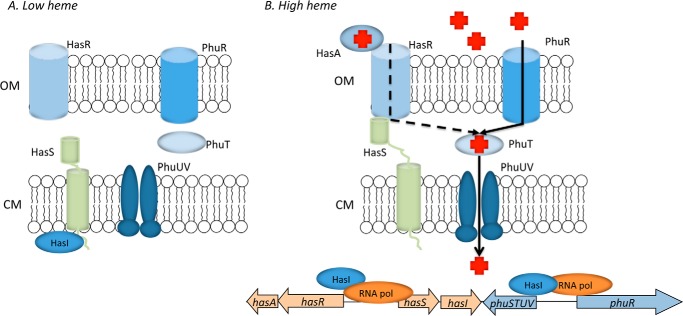
Taken together the present data confirms that although PhuR is the major facilitator of heme uptake, both the Has and Phu systems are required for efficient utilization of heme in P. aeruginosa. Previous studies in S. marcescens have suggested that the hemophore-dependent HasR is the high affinity receptor system with HemR, representing a lower affinity system (38). However, the present data suggests a testable hypothesis whereby the HasR receptor has a lower capacity for heme uptake but acts as a high affinity heme sensor of extracellular heme through its interaction with HasA (Fig. 7). Activation of the HasA-HasR signaling cascade on sensing extracellular heme leads to further up-regulation of the HasA-HasR system, which on heme uptake directly or indirectly up-regulates the high capacity PhuR receptor, the major facilitator of extracellular heme acquisition. The signaling cascade also positively regulates the levels of the anti-σ factor HasS, which accumulates in an inactive state. As extracellular heme levels decrease HasS inactivates the σ-factor, HasI, and down-regulates the heme acquisition systems. Such a fine-tuning mechanism would allow the cell to rapidly respond to external heme levels in a ferric uptake regulator-independent manner. Although beyond the scope of the current studies we are further investigating the heme-dependent regulation of the phu and has systems. Finally, further support for PhuR being the primary receptor for heme acquisition in P. aeruginosa comes from recent studies of clinical isolates where mutations within the phuR promoter, leading to increased PhuR expression, conferred a growth advantage in the presence of hemoglobin. Furthermore, this evolution within the host toward increased reliance on heme via up-regulation of PhuR coincides with loss of alternate iron-scavenging systems (39). Therefore, in acute infection the HasA-HasR system may play a more significant role in iron acquisition, however, on chronic infection bacteria adapt to utilize heme as an iron source through the high capacity Phu system.
FIGURE 7.
Proposed model for the P. aeruginosa heme cell surface signaling system. A, in the absence of heme HasR binds the HasI σ-factor inhibiting its activity. B, in the presence of heme HasA binds to HasR initiating TonB-dependent import of heme into the cell and signaling displacement of the anti-σ factor, freeing HasI for increased expression of the heme uptake systems. Positive regulation of HasS allows inactive HasS to accumulate so that as extracellular heme levels decrease, HasI can be inactivated to rapidly down-regulate the Phu and Has heme uptake systems.
Acknowledgments
We thank Professor Mandy Oglesby-Sherrouse for the PAO1 transposon mutants and Professor Mario Rivera for the kind gift of the cytochrome b5 expression plasmid.
This work was supported, in whole or in part, by National Institutes of Health Grant AI 102883 (to A. W.).
A. D. Smith, A. R. Modi, S. Sun, J. H. Dawson, and A. Wilks, unpublished data.
S. Mourino-Lopez and A. Wilks, unpublished data.
- phu
- Pseudomonas heme uptake
- has
- heme assimilation system
- HemO
- heme oxygenase
- OM
- outer membrane
- ALA
- aminolevulinic acid
- qRT
- quantitative RT.
REFERENCES
- 1. Wilks A., Barker K. D. (2011) Mechanism of heme uptake and utilization in bacterial pathogens in Handbook of Porphyrin Science (Kadish K. M., Smith K. M., Guilard R., eds) 1st Ed., pp. 357–398, World Scientific, Singapore [Google Scholar]
- 2. Wilks A., Burkhard K. A. (2007) Heme and virulence: how bacterial pathogens regulate, transport and utilize heme. Nat. Prod. Rep. 24, 511–522 [DOI] [PubMed] [Google Scholar]
- 3. Contreras H., Chim N., Credali A., Goulding C. W. (2014) Heme uptake in bacterial pathogens. Curr. Opin. Chem. Biol. 19, 34–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cornelis P. (2010) Iron uptake and metabolism in pseudomonads. Appl. Microbiol. Biotechnol. 86, 1637–1645 [DOI] [PubMed] [Google Scholar]
- 5. Ochsner U. A., Vasil M. L. (1996) Gene repression by the ferric uptake regulator in Pseudomonas aeruginosa: cycle selection of iron-regulated genes. Proc. Natl. Acad. Sci. U.S.A. 93, 4409–4414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cornelis P., Dingemans J. (2013) Pseudomonas aeruginosa adapts its iron uptake strategies in function of the type of infections. Front. Cell. Infect. Microbiol. 3, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lansky I. B., Lukat-Rodgers G. S., Block D., Rodgers K. R., Ratliff M., Wilks A. (2006) The cytoplasmic heme-binding protein (PhuS) from the heme uptake system of Pseudomonas aeruginosa is an intracellular heme-trafficking protein to the δ-regioselective heme oxygenase. J. Biol. Chem. 281, 13652–13662 [DOI] [PubMed] [Google Scholar]
- 8. O'Neill M. J., Bhakta M. N., Fleming K. G., Wilks A. (2012) Induced fit on heme binding to the Pseudomonas aeruginosa cytoplasmic protein (PhuS) drives interaction with heme oxygenase (HemO). Proc. Natl. Acad. Sci. U.S.A. 109, 5639–5644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Block D. R., Lukat-Rodgers G. S., Rodgers K. R., Wilks A., Bhakta M. N., Lansky I. B. (2007) Identification of two heme-binding sites in the cytoplasmic heme-trafficking protein PhuS from Pseudomonas aeruginosa and their relevance to function. Biochemistry 46, 14391–14402 [DOI] [PubMed] [Google Scholar]
- 10. Friedman J., Lad L., Li H., Wilks A., Poulos T. L. (2004) Structural basis for novel δ-regioselective heme oxygenation in the opportunistic pathogen Pseudomonas aeruginosa. Biochemistry 43, 5239–5245 [DOI] [PubMed] [Google Scholar]
- 11. Ratliff M., Zhu W., Deshmukh R., Wilks A., Stojiljkovic I. (2001) Homologues of neisserial heme oxygenase in Gram-negative bacteria: degradation of heme by the product of the pigA gene of Pseudomonas aeruginosa. J. Bacteriol. 183, 6394–6403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wegele R., Tasler R., Zeng Y., Rivera M., Frankenberg-Dinkel N. (2004) The heme oxygenase(s)-phytochrome system of Pseudomonas aeruginosa. J. Biol. Chem. 279, 45791–45802 [DOI] [PubMed] [Google Scholar]
- 13. Barkovits K., Harms A., Benkartek C., Smart J. L., Frankenberg-Dinkel N. (2008) Expression of the phytochrome operon in Pseudomonas aeruginosa is dependent on the alternative σ factor RpoS. FEMS Microbiol. Lett. 280, 160–168 [DOI] [PubMed] [Google Scholar]
- 14. Barker K. D., Barkovits K., Wilks A. (2012) Metabolic flux of extracellular heme uptake in Pseudomonas aeruginosa is driven by the iron-regulated heme oxygenase (HemO). J. Biol. Chem. 287, 18342–18350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Létoffé S., Delepelaire P., Wandersman C. (1996) Protein secretion in Gram-negative bacteria: assembly of the three components of ABC protein-mediated exporters is ordered and promoted by substrate binding. EMBO J. 15, 5804–5811 [PMC free article] [PubMed] [Google Scholar]
- 16. Létoffé S., Delepelaire P., Wandersman C. (2004) Free and hemophore-bound heme acquisitions through the outer membrane receptor HasR have different requirements for the TonB-ExbB-ExbD complex. J. Bacteriol. 186, 4067–4074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cescau S., Cwerman H., Létoffé S., Delepelaire P., Wandersman C., Biville F. (2007) Heme acquisition by hemophores. Biometals 20, 603–613 [DOI] [PubMed] [Google Scholar]
- 18. Deniau C., Gilli R., Izadi-Pruneyre N., Létoffé S., Delepierre M., Wandersman C., Briand C., Lecroisey A. (2003) Thermodynamics of heme binding to the HasA(SM) hemophore: effect of mutations at three key residues for heme uptake. Biochemistry 42, 10627–10633 [DOI] [PubMed] [Google Scholar]
- 19. Jepkorir G., Rodríguez J. C., Rui H., Im W., Lovell S., Battaile K. P., Alontaga A. Y., Yukl E. T., Moënne-Loccoz P., Rivera M. (2010) Structural, NMR spectroscopic, and computational investigation of hemin loading in the hemophore HasAp from Pseudomonas aeruginosa. J. Am. Chem. Soc. 132, 9857–9872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yukl E. T., Jepkorir G., Alontaga A. Y., Pautsch L., Rodriguez J. C., Rivera M., Moënne-Loccoz P. (2010) Kinetic and spectroscopic studies of hemin acquisition in the hemophore HasAp from Pseudomonas aeruginosa. Biochemistry 49, 6646–6654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Izadi-Pruneyre N., Huché F., Lukat-Rodgers G. S., Lecroisey A., Gilli R., Rodgers K. R., Wandersman C., Delepelaire P. (2006) The heme transfer from the soluble HasA hemophore to its membrane-bound receptor HasR is driven by protein-protein interaction from a high to a lower affinity binding site. J. Biol. Chem. 281, 25541–25550 [DOI] [PubMed] [Google Scholar]
- 22. Bracken C. S., Baer M. T., Abdur-Rashid A., Helms W., Stojiljkovic I. (1999) Use of heme-protein complexes by the Yersinia enterocolitica HemR receptor: histidine residues are essential for receptor function. J. Bacteriol. 181, 6063–6072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Burkhard K. A., Wilks A. (2007) Characterization of the outer membrane receptor ShuA from the heme uptake system of Shigella dysenteriae: substrate specificity and identification of the heme protein ligands. J. Biol. Chem. 282, 15126–15136 [DOI] [PubMed] [Google Scholar]
- 24. Cobessi D., Meksem A., Brillet K. (2010) Structure of the heme/hemoglobin outer membrane receptor ShuA from Shigella dysenteriae: heme binding by an induced fit mechanism. Proteins 78, 286–294 [DOI] [PubMed] [Google Scholar]
- 25. Krieg S., Huché F., Diederichs K., Izadi-Pruneyre N., Lecroisey A., Wandersman C., Delepelaire P., Welte W. (2009) Heme uptake across the outer membrane as revealed by crystal structures of the receptor-hemophore complex. Proc. Natl. Acad. Sci. U.S.A. 106, 1045–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mokry D. Z., Nadia-Albete A., Johnson M. K., Lukat-Rodgers G. S., Rodgers K. R., Lanzilotta W. N. (2014) Spectroscopic evidence for a 5-coordinate oxygenic ligated high spin ferric heme moiety in the Neisseria meningitidis hemoglobin binding receptor. Biochim. Biophys. Acta 1840, 3058–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Grigg J. C., Mao C. X., Murphy M. E. (2011) Iron-coordinating tyrosine is a key determinant of NEAT domain heme transfer. J. Mol. Biol. 413, 684–698 [DOI] [PubMed] [Google Scholar]
- 28. Sharp K. H., Schneider S., Cockayne A., Paoli M. (2007) Crystal structure of the heme-IsdC complex, the central conduit of the Isd iron/heme uptake system in Staphylococcus aureus. J. Biol. Chem. 282, 10625–10631 [DOI] [PubMed] [Google Scholar]
- 29. Gaudin C. F., Grigg J. C., Arrieta A. L., Murphy M. E. (2011) Unique heme-iron coordination by the hemoglobin receptor IsdB of Staphylococcus aureus. Biochemistry 50, 5443–5452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Eakanunkul S., Lukat-Rodgers G. S., Sumithran S., Ghosh A., Rodgers K. R., Dawson J. H., Wilks A. (2005) Characterization of the periplasmic heme-binding protein ShuT from the heme uptake system of Shigella dysenteriae. Biochemistry 44, 13179–13191 [DOI] [PubMed] [Google Scholar]
- 31. Ho W. W., Li H., Eakanunkul S., Tong Y., Wilks A., Guo M., Poulos T. L. (2007) Holo- and apo-bound structures of bacterial periplasmic heme-binding proteins. J. Biol. Chem. 282, 35796–35802 [DOI] [PubMed] [Google Scholar]
- 32. Czjzek M., Létoffé S., Wandersman C., Delepierre M., Lecroisey A., Izadi-Pruneyre N. (2007) The crystal structure of the secreted dimeric form of the hemophore HasA reveals a domain swapping with an exchanged heme ligand. J. Mol. Biol. 365, 1176–1186 [DOI] [PubMed] [Google Scholar]
- 33. Ghigo J. M., Létoffé S., Wandersman C. (1997) A new type of hemophore-dependent heme acquisition system of Serratia marcescens reconstituted in Escherichia coli. J. Bacteriol. 179, 3572–3579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Payne S. M., Wyckoff E. E., Murphy E. R., Oglesby A. G., Boulette M. L., Davies N. M. (2006) Iron and pathogenesis of Shigella: iron acquisition in the intracellular environment. Biometals 19, 173–180 [DOI] [PubMed] [Google Scholar]
- 35. Henderson D. P., Payne S. M. (1994) Characterization of the Vibrio cholerae outer membrane heme transport protein HutA: sequence of the gene, regulation of expression, and homology to the family of TonB-dependent proteins. J. Bacteriol. 176, 3269–3277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rossi M. S., Fetherston J. D., Létoffé S., Carniel E., Perry R. D., Ghigo J. M. (2001) Identification and characterization of the hemophore-dependent heme acquisition system of Yersinia pestis. Infect. Immun. 69, 6707–6717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Stojiljkovic I., Hwa V., de Saint Martin L., O'Gaora P., Nassif X., Heffron F., So M. (1995) The Neisseria meningitidis haemoglobin receptor: its role in iron utilization and virulence. Mol. Microbiol. 15, 531–541 [DOI] [PubMed] [Google Scholar]
- 38. Benevides-Matos N., Biville F. (2010) The Hem and Has haem uptake systems in Serratia marcescens. Microbiology 156, 1749–1757 [DOI] [PubMed] [Google Scholar]
- 39. Marvig R. L., Damkiær S., Khademi S. M., Markussen T. M., Molin S., Jelsbak L. (2014) Within-host evolution of Pseudomonas aeruginosa reveals adaptation toward iron acquisition from hemoglobin. mBio 5, e00966–00914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nguyen A. T., O'Neill M. J., Watts A. M., Robson C. L., Lamont I. L., Wilks A., Oglesby-Sherrouse A. G. (2014) Adaptation of iron homeostasis pathways by a Pseudomonas aeruginosa pyoverdine mutant in the cystic fibrosis lung. J. Bacteriol. 196, 2265–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rivera M., Walker F. A. (1995) Biosynthetic preparation of isotopically labeled heme. Anal. Biochem. 230, 295–302 [DOI] [PubMed] [Google Scholar]
- 42. O'Neill M. J., Wilks A. (2013) The P. aeruginosa heme binding protein PhuS is a heme oxygenase titratable regulator of heme uptake. ACS Chem. Biol. 8, 1794–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Teale F. W. (1959) Cleavage of the haem-protein link by acid methylethylketone. Biochim. Biophys. Acta 35, 543. [DOI] [PubMed] [Google Scholar]
- 44. Fuhrop J. H., Smith K. M. (eds) (1975) Porphyrins and Metalloporphyrins, pp. 804–807, Elsevier, Amsterdam [Google Scholar]
- 45. Caignan G. A., Deshmukh R., Wilks A., Zeng Y., Huang H. W., Moënne-Loccoz P., Bunce R. A., Eastman M. A., Rivera M. (2002) Oxidation of heme to β- and δ-biliverdin by Pseudomonas aeruginosa heme oxygenase as a consequence of an unusual seating of the heme. J. Am. Chem. Soc. 124, 14879–14892 [DOI] [PubMed] [Google Scholar]
- 46. Ochsner U. A., Johnson Z., Vasil M. L. (2000) Genetics and regulation of two distinct haem-uptake systems, phu and has, in Pseudomonas aeruginosa. Microbiology 146, 185–198 [DOI] [PubMed] [Google Scholar]
- 47. Biville F., Cwerman H., Létoffé S., Rossi M. S., Drouet V., Ghigo J. M., Wandersman C. (2004) Haemophore-mediated signalling in Serratia marcescens: a new mode of regulation for an extra cytoplasmic function (ECF) σ factor involved in haem acquisition. Mol. Microbiol. 53, 1267–1277 [DOI] [PubMed] [Google Scholar]
- 48. Rossi M. S., Paquelin A., Ghigo J. M., Wandersman C. (2003) Haemophore-mediated signal transduction across the bacterial cell envelope in Serratia marcescens: the inducer and the transported substrate are different molecules. Mol. Microbiol. 48, 1467–1480 [DOI] [PubMed] [Google Scholar]
- 49. Schrodinger L. (2012) The PyMOL Molecular Graphics System, 1.5.0.4 Ed., Schrodinger LLC, New York [Google Scholar]
- 50. de Lorenzo V., Timmis K. N. (1994) Analysis and construction of stable phenotypes in Gram-negative bacteria with Tn5- and Tn-10-derived minitransposons. Methods Enzymol. 235, 386–405 [DOI] [PubMed] [Google Scholar]
- 51. Hoang T. T., Karkhoff-Schweizer R. R., Kutchma A. J., Schweizer H. (1998) A broad-host range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences. Application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212, 77–86 [DOI] [PubMed] [Google Scholar]