Background: To date, few glycosaminoglycan (GAG) endosulfatases have been identified.
Results: A novel chondroitin sulfate/dermatan sulfate (CS/DS) endosulfatase was identified for the first time from a marine bacterium.
Conclusion: The endosulfatase has low homology to the conventional GAG sulfatases and can specifically remove 4-O-sulfate from CS/DS chains.
Significance: The endosulfatase will be a useful tool for CS/DS-related research and applications.
Keywords: Chondroitin Sulfate, Dermatan Sulfate, Glycosaminoglycan, Oligosaccharide, Polysaccharide, Endosulfatase, Marine Bacterium, Sulfated Polysaccharide
Abstract
Sulfatases are potentially useful tools for structure-function studies of glycosaminoglycans (GAGs). To date, various GAG exosulfatases have been identified in eukaryotes and prokaryotes. However, endosulfatases that act on GAGs have rarely been reported. Recently, a novel HA and CS lyase (HCLase) was identified for the first time from a marine bacterium (Han, W., Wang, W., Zhao, M., Sugahara, K., and Li, F. (2014) J. Biol. Chem. 289, 27886–27898). In this study, a putative sulfatase gene, closely linked to the hclase gene in the genome, was recombinantly expressed and characterized in detail. The recombinant protein showed a specific N-acetylgalactosamine-4-O-sulfatase activity that removes 4-O-sulfate from both disaccharides and polysaccharides of chondroitin sulfate (CS)/dermatan sulfate (DS), suggesting that this sulfatase represents a novel endosulfatase. The novel endosulfatase exhibited maximal reaction rate in a phosphate buffer (pH 8.0) at 30 °C and effectively removed 17–65% of 4-O-sulfates from various CS and DS and thus significantly inhibited the interactions of CS and DS with a positively supercharged fluorescent protein. Moreover, this endosulfatase significantly promoted the digestion of CS by HCLase, suggesting that it enhances the digestion of CS/DS by the bacterium. Therefore, this endosulfatase is a potential tool for use in CS/DS-related studies and applications.
Introduction
Chondroitin sulfates (CS)2/dermatan sulfates (DS) are the major classes of glycosaminoglycans (GAGs) and are ubiquitously expressed on cell surfaces, in extracellular matrices, and in basement membrane in the animal kingdom. CS/DS not only play structural roles in connective tissues, such as cartilage and bone, but also participate in various important physiological and pathological processes, such as development (1), neuronal growth (2–5), inflammation (6), tumor progression (7, 8), and infection (9–12), by interacting with a large number of growth factors, growth factor receptors, and other target proteins (13, 14). Various functions of CS/DS have been attributed to their variety of structures.
CS/DS chains are composed of repeating disaccharide units consisted of d-glucuronic acid (GlcUA)/l-iduronic acid (IdoUA) and N-acetyl-d-galactosamine (GalNAc). CS chains are initially synthesized on the common GAG-protein linkage tetrasaccharide sequence (-GlcUA-Gal-Gal-Xyl-), which is attached to specific serine residues of the respective core proteins in endoplasmic reticulum/Golgi compartments. After or while CS chains are polymerized by the actions of various enzyme complexes, some GlcUA residues are epimerized into IdoUA by glucuronyl C5 epimerase, thus generating DS chains comprising repeated disaccharide units of -IdoUA-GalNAc- (15, 16). CS/DS chains are further subjected to differential sulfation by various specific sulfotransferases at C-2 of GlcUA/IdoUA and/or C-4 and/or C-6 of GalNAc to yield remarkable structural diversity (17). CS and DS chains are often detected as co-polymeric hybrid structures (CS/DS) and are usually periodically distributed in a cell/tissue-specific manner (18, 19). These modifications introduce enormous microheterogeneity within the CS/DS chains that usually play key roles in various biological functions through interacting with various CS- and/or DS-binding proteins, such as heparin-binding growth factors and cytokines (20). However, the structural complexity and diversity of CS/DS chains present a huge challenge for elucidating their structure-function relationships.
Accumulating evidence shows that the sulfation of CS/DS chains is not random and that specific sulfation patterns are crucial to the interaction with various target proteins (21). Hence, the investigation of sulfation patterns is an important focus of studies on CS/DS structure-activity relationships. Sulfatases are a large enzyme family and can hydrolyze sulfate esters in a wide range of substrates from small steroids to complex cell surface carbohydrates, such as GAGs (22). Undoubtedly, sulfatases, which selectively remove sulfate groups from hexuronic acid residues or hexamine residues, will be useful tools for the structure-function analysis of GAGs. However, most of the GAG sulfatases that have been identified thus far from eukaryotes and prokaryotes are exosulfatases, which act only on sulfated saccharide residues at the end of GAG chains, in particular on short oligosaccharides, such as disaccharides, which are generated by the digestion of GAGs by GAG-degrading enzymes (23); thus, major limitations hinder their application. More recently, a CS/DS 4-O-endosulfatase has been identified for the first time in B. thetaiotaomicron (24); however, its biochemical properties remain to be characterized in detail.
Recently, we identified a novel GAG lyase (HCLase) from a newly isolated marine bacterium Vibrio sp. FC509 (25). The unique features exhibited by HCLase prompted us to search for more GAG-related enzyme tools from marine bacteria. In this study, a putative sulfatase closely linked to the hclase gene of Vibrio sp. FC509 has been expressed in a recombinant form in Escherichia coli, and the recombinant protein exhibits very efficient endosulfatase activity that specifically removes 4-O-sulfates from both CS/DS disaccharides and polysaccharides. The presence of this endosulfatase significantly increases the rate of CS/DS degradation by HCLase. Furthermore, the treatment of CS/DS with this endosulfatase significantly inhibits the interaction of CS/DS chains with a positively charged green fluorescent protein (GFP). Taken together, these findings demonstrate that this novel sulfatase is a very useful tool for structural and functional studies of CS/DS chains.
EXPERIMENTAL PROCEDURES
Materials
The strains and plasmids used in this study are listed in Table 1. SDS, PrimeSTARTM HS DNA polymerases, restriction endonucleases, and other genetic engineering enzymes were purchased from Takara Inc. (Dalian, China). Standard CS unsaturated disaccharides were purchased from Iduron (Manchester, UK). CS-C from shark cartilage, CS-E from squid cartilage, and DS from porcine skin were obtained from Seikagaku Corp. (Tokyo, Japan). Avidin from egg white, 2-aminobenzamide (2-AB), cyanoborohydride (NaBH3CN), CS-A from bovine trachea, and CSase ABC (EC 4.2.2.4) were obtained from Sigma. All other chemicals and reagents were of the highest quality available. CS-H was purified from hagfish notochord (26).
TABLE 1.
Bacterial strains, plasmids, and primers used for sequencing
Restriction enzyme sites are underlined. Kanr, kanamycin-resistant. Cmr, cefamandole-resistant.
| Strains and plasmids | Description | Sources or References |
|---|---|---|
| Strains | ||
| Vibrio sp. FC509 | A GAG-degrading marine bacterium (patented as CGMCC 8913) | (25) |
| E. coli BL21(DE3) | F-, ompT, hsdSB (rB−, mB−), dcm, gal, λ (DE3), pLysS, Cmr | Novogen |
| Plasmids | ||
| pET30a | Expression vector; Kanr | Invitrogen |
| pE30a-endosulfatase | pET30a, carrying an amplified KpnI-BamHI fragment encoding the recombinant protein of 4-O-endosulfatase, fused with a His6 tag at the N terminus | This study |
| Sequencing primers | ||
| 4-O-Endosulfatase-F | 5′-GGGTACCGCAACCATGGTGACTGCTGGCTG-3′ | |
| 4-O-Endosulfatase-R | 5′-GGGATCCTTAGGTATAGGCTTTAGCCACGCTGG-3′ | |
Sequence Analyses of Genes and Proteins of Chondroitin Sulfatases
Promoter motifs of the 5′-flanking DNA region upstream to the open reading frame (ORF) were identified using Primer Premier version 5.0 (PREMIER Biosoft International, Palo Alto, CA) and the Promoter 2.0 Prediction Server. The G + C content (G + C%) of the ORF was calculated using Bio-Edit version 7.0.5.3.
An online similarity search of the protein sequence was performed using the BLASTp algorithm. Secretion signal peptides and their types were identified using the SignalP 4.0 server and the LipoP 1.0 server, respectively. The molecular mass of the protein was estimated using the peptide mass tool on the ExPASy server of the Swiss Institute of Bioinformatics. Sequence alignment and phylogenetic analysis were performed using MEGA version 5.05. Protein modules and domains were identified using the Simple Modular Architecture Research Tool, the Pfam database (SMART), and the Carbohydrate-Active Enzyme (CAZy) database.
Heterologous Expression of the 4-O-Sulfatase Gene
To express 4-O-endosulfatase in E. coli strains, the full-length gene of 4-O-endosulfatase was amplified using primer pairs (as listed in Table 1) and high-fidelity PrimeSTARTM HS DNA polymerases (Takara, Dalian, China). Primer pairs with restriction enzyme sites (underlined in Table 1) were designed according to the inserting site sequences of the expression plasmid pET-30a(+) (Invitrogen). Gel-recovered PCR products were cloned into the expression vector. The expression plasmid (pE30a-endosulfatase), which was constructed from pET-30a(+), was transformed into E. coli BL21(DE3) cells. The integrity of the nucleotide sequence of the constructed plasmid was confirmed by DNA sequencing.
E. coli cells harboring the expression vector (pE30a-endosulfatase) were initially cultured in 100 ml of LB broth at 37 °C. When the cell density reached an A600 of 0.8–1.0, the broth was supplemented with the inducer isopropyl 1-thio-β-d-galactopyranoside (final concentration 0.05 mm) to initiate the expression of targeting protein. After continuous cultivation for an additional 24 h at 16 °C, the cells were harvested by centrifugation at 6,000 × g for 15 min, washed twice using ice-cold buffer A (50 mm Tris-HCl, 150 mm NaCl, pH 8.0), resuspended in buffer A, and disrupted by sonication (50 repetitions, 5 s) in an ice-cold environment. After centrifugation at 15,000 × g for 30 min, the supernatant was collected for further purification of the soluble targeting protein.
Purification of Recombinant Protein 4-O-Endosulfatase
To purify the 4-O-endosulfatase protein, the supernatant containing the soluble native enzyme was loaded onto a column packed with nickel-SepharoseTM 6 Fast Flow resin (GE Healthcare); then the column was washed with buffer A containing 50 mm imidazole to remove impurities, and 4-O-endosulfatase was finally eluted from the nickel-nitrilotriacetic acid column using a concentration gradient of imidazole from 50 to 250 mm. Fractions containing 4-O-endosulfatase were concentrated using an Amicon Ultra 0.5-ml 10K unit (Millipore) and then loaded onto a SuperdexTM 200 10/300 GL column and eluted with buffer A at a flow rate of 0.4 ml/min. The absorbance of the eluates was monitored at 280 nm, and the largest peak was collected. The purity of 4-O-endosulfatase was analyzed using SDS-PAGE according to Sambrook and Russell (27). Coomassie Brilliant Blue R-250 was used to stain the proteins in the gels. Protein concentrations were determined using the Folin-Lowry method (28).
Screening for 4-O-Endosulfatase Activity with Unsaturated Disaccharides
To determine the substrate specificity of 4-O-endosulfatase, various unsaturated CS/DS disaccharides, including Δ4,5HexUAα1–3GalNAc(4S) (ΔA), Δ4,5HexUAα1–3GalNAc(6S) (ΔC), Δ4,5HexUA(2S)α1–3GalNAc(6S) (ΔD), Δ4,5HexUAα1–3GalNAc(4S,6S) (ΔE), and Δ4,5HexUA(2S) α1–3GalNAc(4S,6S) (ΔT) were individually dissolved in deionized water to prepare stock solutions (300 pmol/μl), where Δ4,5HexUA, 2S, 4S, and 6S represent 4,5-unsaturated uronic acid, 2-O-sulfate, 4-O-sulfate, and 6-O-sulfate, respectively. Each stock solution (10 μl) was mixed with 20 μl of 250 mm NaH2PO4-Na2HPO4 buffer (pH 7.0), 60 μl of water, and 10 μl of appropriately diluted enzyme (2 μg/μl); the solutions were then incubated at 37 °C for 12 h. Enzyme-treated disaccharide samples were heated in boiling water for 10 min and then cooled in ice-cold water for 10 min. After centrifugation at 15,000 × g for 15 min, the supernatants were collected and labeled with 2-AB in the presence of sodium cyanoborohydride, as described by Bigge et al. (29). Free 2-AB was removed by extraction with chloroform. All these preparations were individually analyzed by anion exchange HPLC on a YMC-Pack PA-G column (YMC-Pack PA, Kyoto, Japan) eluted with a linear gradient from 16 to 460 mm NaH2PO4 over 60 min at a flow rate of 1.0 ml/min at room temperature. The eluates were monitored by measuring absorbance using a fluorescent detector at excitation and emission wavelengths of 330 and 420 nm, respectively. Disaccharides were identified and quantified by comparison with CS-derived authentic unsaturated disaccharides (30).
Biochemical Characterization of 4-O-Endosulfatase
To determine the optimal pH for the rate of the recombinant enzyme, an aliquot of ΔA unit (30 nmol) was digested with 0.1 μg of the enzyme in buffers with different pH values, including 50 mm NaAc-HAc buffer (pH 5.0–6.0), 50 mm NaH2PO4-Na2HPO4 buffer (pH 6.0–8.0), and 50 mm Tris-HCl buffer (pH 7.0–10.0) in a total volume of 100 μl at 37 °C for 30 min. After the optimum pH was determined, the effect of temperature on 4-O-endosulfatase rate was examined in 50 mm NaH2PO4-Na2HPO4, pH 8.0, at temperatures from 0 to 70 °C for 30 min. Furthermore, the effects of metal ions or chelating agent (5 mm) on 4-O-endosulfatase rate were investigated in 50 mm NaH2PO4-Na2HPO4, pH 8.0, at 30 °C. To determine the thermostability of 4-O-endosulfatase, the enzyme (in 50 mm NaH2PO4-Na2HPO4 buffer, pH 8.0) was preincubated for 0–12 h at 30 °C, and the residual activity was determined under the optimum conditions (i.e. 50 mm NaH2PO4-Na2HPO4, pH 8.0, at 30 °C). The half-life of the enzyme at the optimal temperature was calculated by drawing a semi-log plot. All reactions were performed in triplicate.
Degradation of Polysaccharides by 4-O-Endosulfatase
To investigate the activity of the novel 4-O-endosulfatase toward CS polysaccharides, CS or DS (500 μg) was treated with 4-O-endosulfatase (50 μg) for over 72 h at 30 °C in a volume of 1 ml with the addition of 50 μg of fresh enzyme every 24 h two times. The resulting products were loaded onto a DEAE-Sephadex column (16 × 100 mm), which had been pre-equilibrated with 50 mm NaH2PO4. After the column was washed with the equilibration buffer, CS, which bound to the column, was eluted with a linear gradient of NaCl from 0 to 2 m in the equilibration buffer over a period of 160 min at a flow rate of 0.5 ml/min. The elutes were collected at 2-min intervals and were analyzed using the carbazole reaction (31).
Additionally, CS-A, DS, CS-E, or CS-H (5 μg) was exhaustively digested by the novel 4-O-endosulfatase (0.5 μg) for 72 h at 30 °C in 20 μl with the addition of 0.5 μg of fresh enzyme every 24 h two times and immediately boiled for 10 min. The resulting products were then digested with CSase ABC (32) and labeled with 2-AB. The 2-AB-labeled digests were analyzed by anion exchange HPLC on a YMC-Pack PA-G column. The resulting disaccharides were identified and quantified by comparison with CS-derived authentic unsaturated disaccharides.
The Activity Assay of 4-O-Endosulfatase
Briefly, 4-O-endosulfatase (2 μg) was added to 50 nmol of disaccharides or 25 μg of polysaccharides (CA-A, CS-E, DS, and CS-H) in 50 mm NaH2PO4-Na2HPO4 buffer at pH 8.0 in a total volume of 500 μl. Each reaction mixture was incubated at 30 °C. At various time intervals (up to 30 min), 10-μl aliquots were withdrawn in duplicate, boiled for 10 min, and then cooled in ice-cold water for 10 min. In the case of polysaccharides, the digests were further digested using CSase ABC (5 mIU/reaction) for 2 h and then cooled in ice-cold water for 10 min. After centrifugation at 15,000 × g for 15 min, the supernatant fluid of each digest was collected and analyzed using anion exchange HPLC on a YMC-Pack PA-G column. One unit of enzyme was defined as the amount of enzyme required to produce 1 μmol of free sulfate per minute. The specific activity of crude protein and protein eluates from the nickel column was measured as described above.
Effect of 4-O-Endosulfatase on the Digestion of CS by rHCLase
To investigate whether the desulfation of 4-O-endosulfatase promotes the digestion of CS, the activities of rHCLase against CS-A and CS-E in the absence or presence of 4-O-endosulfatase were measured according to the method reported by Yamagata et al. (33). Briefly, rHCLase (2 ng) alone or with 4-O-endosulfatase (50 μg) was added to 1 mg/ml CS-A or CS-E in 50 mm NaH2PO4-Na2HPO4 at pH 8.0 in a total volume of 1 ml, and the reaction mixture was incubated at 30 °C. At various time intervals (up to 10 min), 100-μl aliquots were withdrawn in duplicate, boiled for 10 min, and then cooled in ice-cold water for 10 min. After centrifugation at 15,000 × g for 15 min, the supernatant was collected, diluted 5-fold, and analyzed based on the absorbance at 232 nm. One unit of enzyme was defined as the amount of enzyme required to produce 1 μmol of unsaturated carbon bonds per minute.
Interaction Assay of Supercharged GFP with CS/DS Treated with 4-O-Endosulfatase
A supercharged GFP with a net charge of +36 (ScGFP) was recombinantly expressed and purified as described previously (34). The ScGFP, as a model protein with positive charge, was used to evaluate the protein-binding activity of CS/DS treated with or not treated with 4-O-endosulfatase. CS-A, CS-E, and DS were individually partially biotinylated (assuming a few percent of the total carboxyl groups) at the carboxyl groups of their GlcUA/IdoUA residues (35). Biotinylated CS or DS (50 μg) was treated with 4-O-endosulfatase or inactive 4-O-endosulfatase (30 μg) for 72 h at 30 °C in a volume of 50 μl; then the enzyme was denatured by heating at 100 °C for 10 min and removed by centrifugation at 15,000 × g for 15 min. The supernatant containing biotinylated CS or DS (1 μg/μl) was collected for the following assay. Briefly, a Corning 96-well black plate was coated with avidin (0.5 μg/well) at 4 °C overnight and then blocked with 1% BSA in PBS. After washing with PBS, the avidin-coated wells were incubated with enzyme-treated or -untreated biotinylated CS or DS (0.5 μg/well) for 1 h at room temperature. After the wells were washed twice with PBS, 50 μl of ScGFP (0.1 μg) in 10 mm Tris-HCl (pH 7.0) containing 100 mm NaCl was added to each well, and the plates were then incubated for 30 min at room temperature. Finally, the ScGFP solution was discarded, and the wells were washed three times with PBS before the fluorescence intensity of each well was measured using an EnSpire® multimode plate reader (PerkinElmer Life Sciences).
RESULTS
Information Relating to the 4-O-Endosulfatase Gene and Protein Sequences
A putative CS/DS sulfatase gene (GenBankTM number KP123433) encoding 4-O-endosulfatase was 1,566 bp in length and contained a GC content of 47.5%. The predicted 4-O-sulfatase protein (4-O-endosulfatase) comprised 521 amino acid residues.
The molecular weight of the 4-O-endosulfatase protein was 59.5. The isoelectric point (pI) was 6.21. A BLASTp search showed that among elucidated sulfatases, 4-O-endosulfatase shared the highest sequence identity (38%) with sulfatase BT_3349 from Bacteroides thetaiotaomicron (24). CaZy and SMART analyses show that the 4-O-endosulfatase contains an N-terminal signal peptide and a sulfatase module.
Heterologous Expression of 4-O-Endosulfatase in E. coli
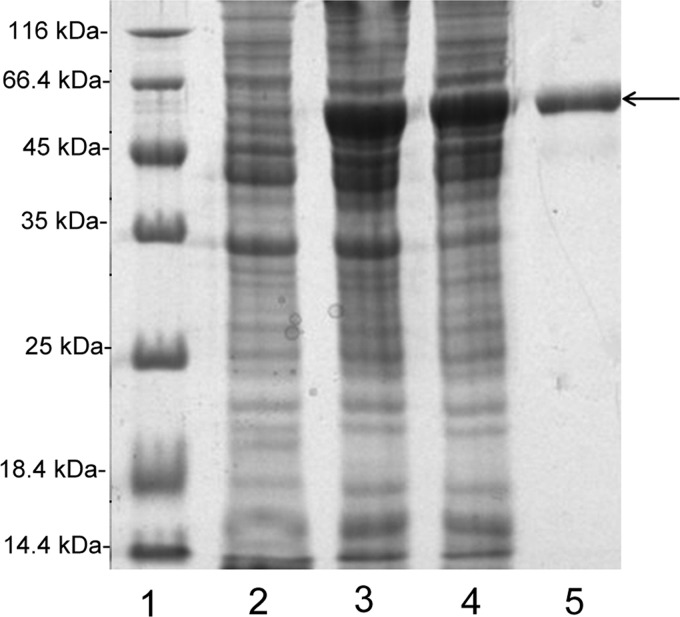
The full-length sequence of the 4-O-endosulfatase ORF was amplified directly from the genomic DNA of Vibrio sp. FC509. The PCR product was recovered and cloned into the pET-30a(+) vector following a T7 promoter. In this 4-O-endosulfatase expression vector (pE30a-endosulfatase), a His6 tag was added at the N terminus of the recombinant protein. SDS-PAGE analysis indicated that BL21(DE3) cells harboring the pE30a-endosulfatase plasmid yielded soluble products (∼100 mg/liter) of the correct molecular mass (i.e. 60 kDa).
The crude enzyme was extracted from cultures of the host cells harboring pE30a-endosulfatase by sonication and centrifugation. The recombinant enzyme (4-O-endosulfatase) was further purified by nickel-nitrilotriacetic acid affinity chromatography and gel filtration chromatography. As shown in Table 2, compared with the crude bacterial lysate, the specific activities of the purified enzyme against ΔA and CS-A were increased by 2.5 and 2.44 times, respectively. SDS-PAGE analysis showed that the purified 4-O-endosulfatase protein was more than 95% pure, and the initial concentration was 2 mg/ml (Fig. 1).
TABLE 2.
Purification of recombinant 4-O-endosulfatase
Results are the means ± S.D. for at least two experiments.
| Total protein | On ΔA |
On CS-A |
|||||
|---|---|---|---|---|---|---|---|
| Total activity | Specific activity | Yield | Total activity | Specific activity | Yield | ||
| mg | units | milliunits/mg | % | units | milliunits/mg | % | |
| Crude protein | 40.1 ± 5.1 | 80.5 | 2008 ± 84 | 100 | 13.5 | 336 ± 23 | 100 |
| Elution from Ni2+ column | 12.0 ± 2.4 | 50.2 | 4185 ± 143 | 62.3 | 8.1 | 675 ± 56 | 60.0 |
| Elution from gel filtration | 7.2 ± 1.3 | 36.1 | 5020 ± 186 | 44.8 | 5.9 | 820 ± 63 | 43.7 |
FIGURE 1.
Purification of recombinant 4-O-endosulfatase from E. coli by Ni2+ chelation chromatography and subsequent gel filtration. Enzyme purity was assessed following each fractionation step by SDS-PAGE using 13.2% polyacrylamide gels and Coomassie Brilliant Blue staining. Lane 1, unstained protein molecular weight Marker SM 0431 (Thermo); lane 2, uninduced cell lysate; lane 3, induced cell lysate; lane 4, the supernatant fluid of the induced cell lysate; lane 5, purified recombinant 4-O-endosulfatase. Molecular weight markers (Mr) and their corresponding masses are indicated.
Specific Activity of Recombinant Sulfatase toward Disaccharides
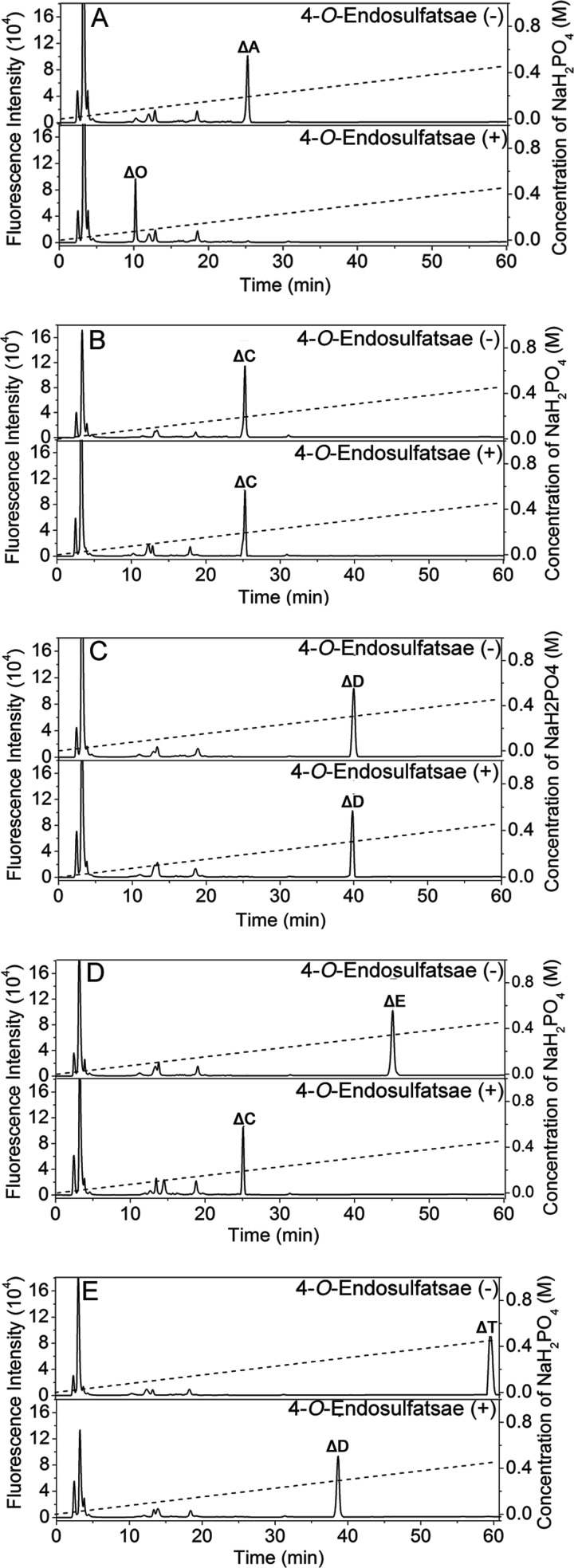
To investigate the specific activity of the recombinant protein, five types of unsaturated CS disaccharides (ΔA, ΔC, ΔD, ΔE, and ΔT) with different sulfation patterns were individually treated with or without 4-O-endosulfatase. As shown in Fig. 2, treatment with the recombinant protein completely transformed ΔA (Fig. 2A, top), ΔE (Fig. 2D, top), and ΔT (Fig. 2E, top) to ΔO (Fig. 2A, bottom), ΔC (Fig. 2D, bottom), and ΔD (Fig. 2E, bottom), respectively, but did not affect ΔC (Fig. 2B) and ΔD (Fig. 2C). These results showed that the sulfatase was able to specifically hydrolyze the 4-O-sulfate group at the GalNAc residues of disaccharides; thus, the sulfatase was designated as 4-O-sulfatase.
FIGURE 2.
Analysis of the final products of the digestion of CS/DS disaccharides by 4-O-endosulfatase. Unsaturated CS/DS disaccharides ΔA (A), ΔC (B), ΔD (C), ΔE (D), and ΔT (E) were exhaustively digested without (top) or with (bottom) 4-O-endosulfatase, labeled with 2-AB, and then analyzed by anion exchange HPLC as described under “Experimental Procedures.” The elution positions of the following standard oligosaccharides are indicated: ΔO, ΔC, ΔA, ΔD, ΔE, and ΔT.
Enzymatic Characteristics of 4-O-Endosulfatase
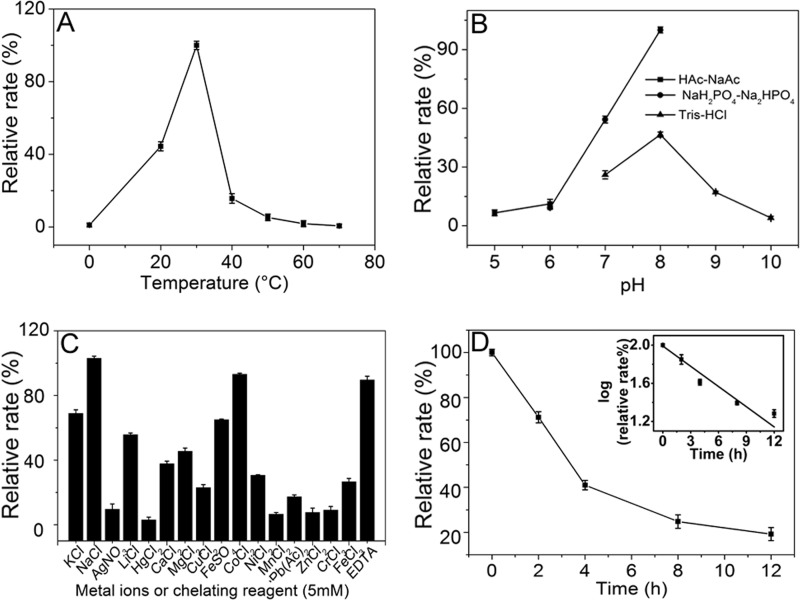
After achieving the recombinant expression and purification of the 4-O-endosulfatase and demonstrating its high specificity for the 4-O-sulfate of GalNAc, the reaction conditions required for optimal enzyme activity were determined. The parameters obtained included optimum pH, temperature, and metal ion dependence. Using ΔA as a substrate, 4-O-endosulfatase exhibited the maximal rate at 30 °C, and its rate rapidly decreased at temperatures lower or higher than 30 °C, suggesting that this enzyme is temperature-sensitive, especially in the high temperature region from 40 to 70 °C (Fig. 3A). The effects of pH on the reaction rate of 4-O-endosulfatase were investigated at the optimum temperature 30 °C, and the results showed that the optimum pH was 8.0, whereas the rate of enzyme was much higher in 50 mm NaH2PO4-Na2HPO4 buffer than in 50 mm Tris-Cl buffer (Fig. 3B). Moreover, the enzyme also exhibited pH sensitivity. As shown in Fig. 3B, the activity was completely abolished at the outlying pH values of 5.0 and 10.0. To determine the effects of metal ions on the enzyme rate, various metal ions were added to the basic reaction buffer (50 mm NaH2PO4-Na2HPO4, pH 8.0), and the reaction rate of 4-O-endosulfatase was measured at 30 °C. As shown in Fig. 3C, no metal ion exhibited a significant enhancing effect; however, most tested metal ions, such as Ag+, Hg2+, Mn2+, Zn2+, and Cr3+ (but not Na+), strongly inhibited the enzyme activity. Additionally, the chelating regent EDTA exhibited no significant effect.
FIGURE 3.
Biochemical reaction conditions for recombinant 4-O-endosulfatase. A, effect of temperature. The enzyme activities of 4-O-endosulfatase were measured using the ΔA as a substrate in 50 mm NaH2PO4-Na2HPO4 buffer, pH 8.0, at various temperatures for 30 min. The data are shown as percentages of the activity obtained at 30 °C (100%). B, effect of pH. The activities of 4-O-endosulfatase against the ΔA were measured in buffers with pH values from 5 to 10 at 30 °C for 30 min. The data are shown as percentages of the activity obtained in the NaH2PO4-Na2HPO4 buffer at pH 8.0 (100%). C, effect of metal ions. The activities of 4-O-endosulfatase against the ΔA were measured in NaH2PO4-Na2HPO4 buffer (pH 8.0) containing a 5 mm concentration of various metal ions at 30 °C for 30 min. The data are shown as percentages of the activity obtained in the buffer without the tested metal ions. D, to determine the thermostability of 4-O-endosulfatase, the enzyme in 50 mm NaH2PO4-Na2HPO4 buffer (pH 8.0) was preincubated for 0–12 h at the optimum temperature, and the residual activity was determined in the optimum conditions; to calculate the half-life of the enzyme, a semi-log curve was plotted (inset). The data are shown as the activity relative to that of untreated 4-O-endosulfatase. Shown are mean values of triplicates ± S.D. (error bars).
Thus, the optimum conditions for 4-O-endosulfatase were determined to be 50 mm NaH2PO4-Na2HPO4 (pH 8.0) at 30 °C. The thermostability of this enzyme was assayed under the optimum conditions; the rate of 4-O-endosulfatase gradually declined from 100 to 20% during 12 h, indicating mild stability. The calculated half-life at the optimal temperature is 4.1 h according to the semi-log curve (Fig. 3D, inset).
Degradation of Polysaccharides by 4-O-Endosulfatase
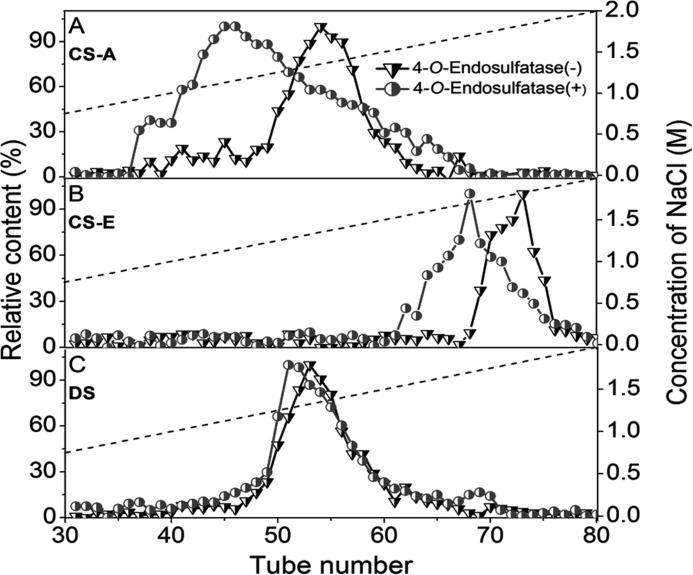
To explore whether the novel 4-O-endosulfatase acts on CS/DS polysaccharides, 4-O-sulfate-enriched CS/DS polysaccharides (including CS-A, which is rich in the monosulfated disaccharide GluUAβ1–3GalNAc(4S) (36); CS-E, which is rich in GluUAβ1–3GalNAc(4S,6S) (36); and DS, which is rich in IdoUAβ1–3GalNAc(4S) (36)) were individually used as substrates for enzyme digestion. The digests were analyzed by anion exchange chromatography on a DEAE-Sephadex column, and the results showed that all samples treated with enzyme were eluted at lower salt concentrations than the corresponding enzyme-untreated parent polysaccharides (Fig. 4), suggesting that the enzyme-treated samples had less negative charge; this finding indicates that some sulfate groups were removed from the polysaccharide chains by the 4-O-endosulfatase. Among the three test samples, CS-A was most strongly affected by treatment with this sulfatase, suggesting that the internal structure of CS-A chains, such as the dominant GlcUAβ1–3GalNAc(4S), is most suitable for the enzyme action.
FIGURE 4.
Digestion of polysaccharides by 4-O-endosulfatase. The digestion of CS-A (A), CS-E (B), and DS (C) by 4-O-endosulfatase was evaluated over 72 h at 30 °C in a volume of 1 ml; then each digest was loaded on a DEAE-Sephadex column (16 × 100 mm), which was pre-equilibrated with 50 mm NaH2PO4. CS chains were eluted with a linear gradient of NaCl from 0 to 2 m in 50 mm NaH2PO4. CS samples were collected at 2-min intervals and quantified using the carbazole reaction.
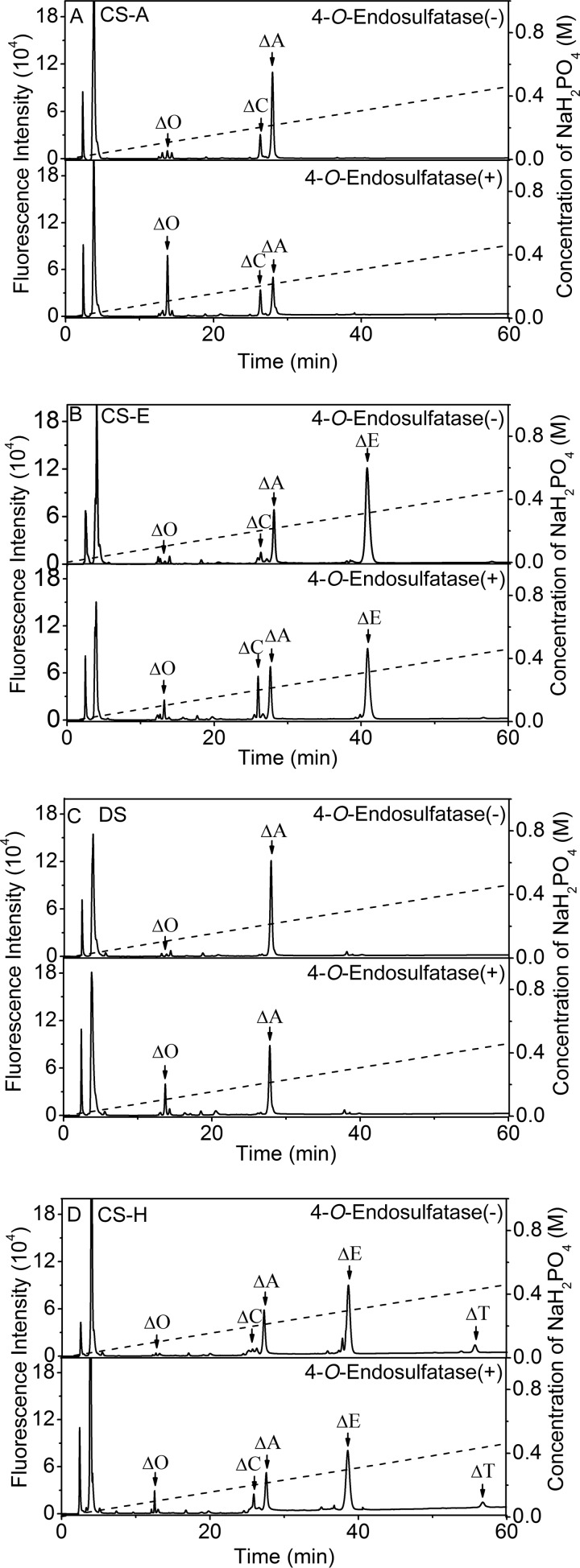
To further confirm the desulfation of polysaccharides by 4-O-endosulfatase, CS/DS samples, including CS-A, CS-E, DS, and CS-H (which is rich in GlcUAβ1–3GalNAc(4S,6S) and IdoUAβ1–3GalNAc(4S,6S) (26)), were exhaustively treated by the enzyme and then were digested with CSase ABC and labeled with 2-AB for disaccharide composition analysis by anion exchange HPLC as described under “Experimental Procedures.” Consistent with the results obtained from the above described assay for polysaccharides using anion exchange chromatography, CS-A was the best substrate for the 4-O-endosulfatase; more than 65% of GlcUAβ1–3GalNAc(4S) was transformed into the nonsulfated disaccharide GlcUAβ1–3GalNAc by the specific desulfation catalyzed by the enzyme (Fig. 5A and Table 3). In contrast, only 27% of the IdoUAβ1–3GalNAc(4S) in DS, which is a DS counterpart of GlcUA in CS-A, was converted to IdoUAβ1–3GalNAc (Fig. 5C and Table 3), indicating that the IdoUA attached to GalNAc(4S) in DS exerts a more negative impact on the action of 4-O-endosulfatase than the GlcUA in CS. In the case of CS-E, 20% of GlcUAβ1–3GalNAc(4S,6S) and 22% of GlcUAβ1–3GalNAc(4S) were converted to GlcUAβ1–3GalNAc(6S) and GlcUAβ1–3GalNAc, respectively (Fig. 5B and Table 3); the low conversion rate of GlcUAβ1–3GalNAc(4S,6S) suggests that the 6-O-sulfation of GalNAc inhibits the activity of 4-O-endosulfatase. The above findings were confirmed by the digestion of CS-H containing IdoUAβ1–3GalNAc(4S,6S); only 17% of HexUAβ1–3GalNAc(4S,6S) was transformed to HexUAβ1–3GalNAc(6S) (Fig. 5D and Table 3).
FIGURE 5.
Disaccharide analysis of CS and DS polysaccharides digested with 4-O-endosulfatase. CS-A (A), CS-E (B), DS (C), or CS-H (D) was exhaustively digested without (top) or with (bottom) 4-O-endosulfatase and then further digested with CSase ABC; the products were analyzed by anion exchange HPLC as described under “Experimental Procedures.” The elution positions of the following standard disaccharides are indicated: ΔO, ΔC, ΔA, ΔE, and ΔT.
TABLE 3.
Disaccharide analysis of polysaccharides treated with 4-O-endosulfatase
Results are the means ±S.D. for at least two experiments.
| Unsaturated disaccharide | CS-A |
CS-E |
DS |
CS-H |
||||
|---|---|---|---|---|---|---|---|---|
| Untreated | Treated | Untreated | Treated | Untreated | Treated | Untreated | Treated | |
| mol %a | ||||||||
| ΔO | 5.9 ± 0.4 | 49.5 ± 1.1 | 0.6 ± 0.1 | 6.6 ± 0.1 | 1 ± 0.1 | 28.1 ± 0.4 | 0.5 ± 0.1 | 6.2 ± 0.2 |
| ΔA | 66.9 ± 0.8 | 23 ± 0.4 | 22.2 ± 0.3 | 17.4 ± 0.3 | 99 ± 0.1 | 71.8 ± 0.4 | 11.7 ± 0.4 | 6.6 ± 0.1 |
| ΔC | 27.2 ± 0.7 | 27.5 ± 0.3 | 12.8 ± 0.2 | 24.4 ± 0.4 | 19.6 ± 0.2 | 30.8 ± 0.4 | ||
| ΔE | 64.4 ± 1.0 | 51.6 ± 0.7 | 64.3 ± 0.4 | 53.2 ± 0.3 | ||||
| ΔT | 3.9 ± 0.1 | 3.2 ± 0.1 | ||||||
| S/unitsb | 0.94 | 0.50 | 1.64 | 1.45 | 0.99 | 0.72 | 1.72 | 1.53 |
a Molar proportion.
b Molar ratio of sulfate to disaccharide.
The Activity of 4-O-Endosulfatase
Under the optimum conditions, the specific activity of 4-O-endosulfatase was calculated using various CS/DS disaccharides and polysaccharides. The specific activities of 4-O-endosulfatase against the CS/DS-derived unsaturated disaccharides ΔA, ΔE, and ΔT were 5,020, 1,210, and 800 milliunits/mg of protein, respectively (one unit of enzyme was defined as the amount of enzyme required to produce 1 μmol of free sulfate/min) (Table 4). In contrast, specific activities toward polysaccharides CS-A, CS-E, and DS were 820, 390, and 610 milliunits/mg of protein, respectively; these values were lower than those obtained with the disaccharides (Table 4).
TABLE 4.
Activity analysis for 4-O-endosulfatase
| Substrate | 4-O-Endosulfatase | |
|---|---|---|
| milliunits/mga | ||
| Disaccharide | ΔA | 5,020 ± 186b |
| ΔE | 1,210 ± 75 | |
| ΔT | 800 ± 87 | |
| Polysaccharide | CS-A | 820 ± 63 |
| DS | 610 ± 71 | |
| CS-E | 390 ± 45 |
a One unit of enzyme was defined as the amount of enzyme required to produce 1 μmol of free sulfate/min.
b Results are the means ± S.D. for at least two experiments.
Effect of 4-O-Endosulfatase on the Digestion of CS/DS by rHCLase
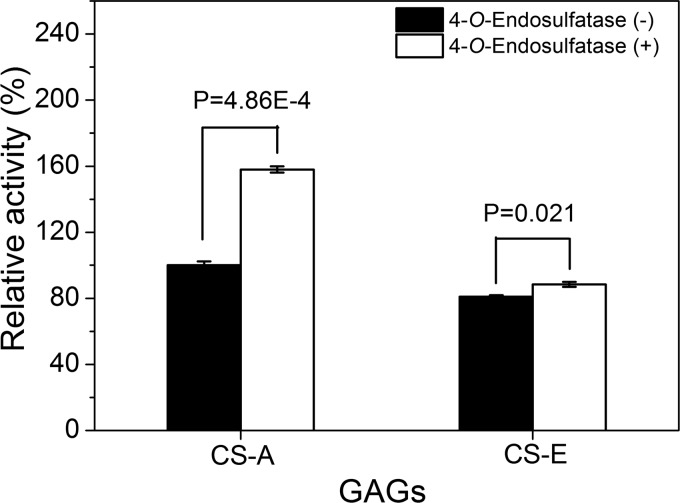
In our previous study, we found that the GAG lyase rHCLase was more active against nonsulfated and low sulfated GAGs, such as hyaluronan (HA), chondroitin, and CS-A, than against more highly sulfated CS chains (25); thus, we speculate that 4-O-endosulfatase, which is closely linked to HCLase in the genome, might promote the digestion of CS by HCLase via the prior desulfation of substrates. To investigate this hypothesis, 4-O-endosulfatase and HCLase were combined and used to digest CS samples. The presence of 4-O-endosulfatase significantly promoted the ability of HCLase to digest CS-A and CS-E (Fig. 6). Specifically, the activities toward CS-A and CS-E were increased by 60 and 10%, respectively (Fig. 6); these values are consistent with the desulfation activities of 4-O-endosulfatase against the two substrates.
FIGURE 6.
The activity of 4-O-endosulfatase toward rHCLase. rHCLase (2 ng) alone or with 4-O-endosulfatase (50 μg) was added to 1 mg/ml CS-A or CS-E in 50 mm NaH2PO4-Na2HPO4 at pH 8.0 in a total volume of 1 ml, and the reaction mixture was incubated at 30 °C. The activity of rHCLase against CS-A or CS-E was determined as described under “Experimental Procedures.” The data are shown as the activity relative to CS-A treated with rHCLase only. The p values are shown to indicate a significant difference from the control. Shown are mean values of triplicates ± S.D. (error bars).
Interaction of ScGFP with CS and DS Treated with 4-O-Endosulfatase
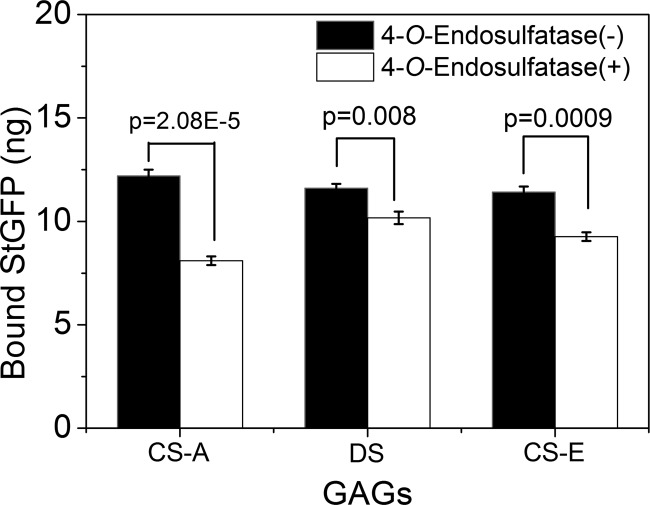
The unique ability of 4-O-endosulfatase to specifically remove 4-O-sulfate groups from CS/DS polysaccharides renders it of potential use as a tool for structure-function studies of CS/DS. As an example, we used ScGFP as a model protein with a positive charge to investigate the interactions of proteins with CS/DS-treated with 4-O-endosulfatase. The interaction analysis was carried out using a simple, rapid, and inexpensive assay of ScGFP binding to immobilized CS/DS on an avidin-coated 96-well plate, as described under “Experimental Procedures.” As shown in Fig. 7, treatment with 4-O-endosulfatase significantly reduced the binding activity of CS-A, CS-E, and DS to ScGFP, by 31.2, 15.9, and 12.4%, respectively. Thus, the novel 4-O-endosulfatase is a very useful tool for structural and functional studies of CS/DS.
FIGURE 7.
Effects of the 4-O-endosulfatase treatment on the binding of CS and DS preparations with positively supercharged GFP. CS-A, CS-E, or DS was exhaustively digested with 4-O-endosulfatase or inactive enzyme, and the binding capacity of each treated polysaccharide preparation was analyzed using ScGFP as described under “Experimental Procedures.” The data are shown as percentages of the untreated CS-A (100%). The p values are shown to indicate a significant difference from the control. Shown are mean values of triplicates ± S.D. (error bars).
DISCUSSION
Sulfation by various sulfotransferases is an important postpolymerization modification that introduces high structural diversity to CS/DS chains. However, details of the mechanism underlying the desulfation of CS/DS chains by sulfatases have rarely been investigated because CS/DS endosulfatases, which can remove sulfate from internal positions of CS/DS chains, were not identified until recently, when a novel CS/DS 4-O-endosulfatase was found in B. thetaiotaomicron (24) (however, several CS/DS exosulfatases have been reported previously (33, 37, 38)). In the present study, a novel CS/DS 4-O-endosulfatase was identified from a marine bacterium, Vibrio sp. FC509. This enzyme shares very low homology with other identified sulfatases but shares 38% identity with the sulfatase from B. thetaiotaomicron, suggesting the existence of a novel CS/DS endosulfatase family. The 4-O-endosulfatase also shared no significant homology (24%) with the human N-acetylgalactosamine-4-O-sulfatase (UniProt ID: P15848). The human N-acetylgalactosamine-4-O-sulfatase is a strict exo-enzyme that only hydrolyzes 4-O-sulfate on the nonreducing end of GAG (39). The 4-O-endosulfatase from Vibrio acts like the 4-O-endosulfatase from B. thetaiotaomicron, both of which can efficiently remove 4-O-sulfate groups from CS/DS disaccharides to polysaccharides. These findings open a new line of research regarding the discovery of specific CS/DS endosulfatases, which might play important roles in various metabolic processes and regulate various biological functions of CS/DS chains; these sulfatases might prove useful as tools for investigating the structure-function relationship of CS/DS.
In this study, the recombinantly expressed sulfatase exhibited specific CS/DS 4-O-sulfatase activity toward both disaccharides and polysaccharides, similar to the findings for the 4-O-endosulfatase from B. thetaiotaomicron. Currently, few data are available regarding the biochemical characterization of CS/DS sulfatases, including the sulfatase from B. thetaiotaomicron. In this study, the effects of temperature, pH, and metal ions on the novel 4-O-endosulfatase were investigated in detail, and the optimal conditions were determined. The enzyme exhibited maximal activity in phosphate buffer (pH 8.0) at 30 °C. Interestingly, however, no observation was made regarding the activation of the enzyme by metal cations, as found for other sulfatases (22). In addition, this enzyme exhibited much higher activity in phosphate buffer than in Tris-HCl buffer at the optimum pH of 8.0, unlike two well known CS exosulfatases from Proteus vulgaris (33). These unique properties suggest that this CS/DS endosulfatase might possess a novel catalytic mechanism. Hence, further investigation of this enzyme to determine its mechanism of action is required.
Although the novel 4-O-endosulfatase from a marine bacterium exhibited very strict specificity for the 4-O-sulfate groups of galactosamine, it acted on various 4-O-sulfate-containing CS/DS substrates from disaccharides to polysaccharides with various sulfation patterns. This sulfatase exhibited the highest activity against monosulfated ΔA (5,020 milliunits/mg); in contrast, the degradation rates of disulfated ΔE and trisulfated ΔT were only 24 and 16% of the rate for the ΔA unit, respectively, suggesting that the sulfation of other hydroxy groups exerts inhibitory effects on the activity of this enzyme. The activity of this sulfatase toward polysaccharides was lower than that for disaccharides and was higher for CS-A (820 milliunits/mg) than for CS-E (390 milliunits/mg) or DS (610 milliunits/mg). CS-E from squid cartilage contains more than 60% disulfated E unit GlcUAβ1–3GalNAc(4S,6S) (36), which might explain the lower activity of the sulfatase against this material, as found for disaccharides. In contrast, DS from porcine skin contains more than 90% monosulfated iA unit IdoUAα1–3GalNAc(4S) (36) and should represent an optimum substrate; however, the lower enzyme activity toward the iA unit than toward the A unit suggests that the attachment of a uronic acid residue to the 4-O-sulfated galactosamine affects the enzyme activity and that GlcUA in CS is a more suitable substrate than IdoUA. In addition, other structural features in CS/DS chains should also affect the activity of this enzyme. In an exhaustive degradation of the best substrate, CS-A, little more than 65% of GlcUAβ1–3GalNAc(4S) was converted to GlcUAβ1–3GalNAc, indicating that ∼35% of GlcUAβ1–3GalNAc(4S) might contain structures or sequences near the A unit that inhibit the action of 4-O-endosulfatase. Notably, the bovine trachea-derived CS-A contains 27.2% 6-O-sulfate-containing C unit GlcUAβ1–3GalNAc(6S) and 5.9% nonsulfated O unit GlcUAβ1–3GalNAc (Table 3), which may be involved in such inhibiting structures. This phenomenon is more prominent in the digestion of more complex CS/DS, such as CS-E and CS-H. Further investigation of the resistant structures in CS/DS chains will help to reveal the mechanism of action of this enzyme.
Vibrio sp. FC509 is a polysaccharide-degrading marine bacterium isolated from costal sediments (25). It can grow rapidly by utilizing multiple GAG chains as the sole carbon sources. The studies suggest that Vibrio sp. FC509 contains an efficient GAG-degrading system not yet explored in any Vibrio or marine bacteria. In the genome of Vibrio sp. FC509, the 4-O-endosulfatase is closely linked to an HA/CS lyase, HCLase, as reported in our recent work (25). Although the HCLase exhibits very high activity for HA, chondroitin, and various types of CS isoforms, it notably prefers nonsulfated substrates, such as HA and chondroitin. Considering that 4-O-endosulfatase can remove sulfate groups from CS/DS polysaccharides and that both enzymes exhibit similar optimal reaction conditions, these related enzymes might work together in the metabolism of CS/DS by bacteria. To study this, we investigated the effect of the endosulfatase on the digestion of CS/DS by HCLase; the results clearly showed that the simultaneous action of the endosulfatase and HCLase significantly accelerated the rate of degradation of various CS chains. A similar phenomenon was described in a study on the 4-O-endosulfatase from B. thetaiotaomicron (24). Taken together, these new findings will help to reveal the biological role of these endosulfatases in the degradation and utilization of host CS/DS by bacteria, which will improve the conventional understanding of CS/DS metabolism in bacteria.
Undoubtedly, endosulfatases, which can effectively and specifically remove sulfate groups from GAG chains, are very useful tools for structure-function studies of sulfated GAGs. Together with the sulfatase reported here, however, only three GAG endosulfatases have yet been identified from animals (40) and bacteria (24). The roles of HS 6-O-endosulfatase in animals have been widely and actively studied in biological processes from embryo development to tumorigenesis (41–43). However, this enzyme has rarely been used as a tool for structure-function studies of HS in vitro, which might be due to its low activity and instability. The 4-O-endosulfatase isolated from a marine bacterium has a comparable or higher activity and stability than previously identified GAG sulfatases (24, 38, 40) under optimum conditions. To evaluate the biological value of this endosulfatase for studying the structure-function relationships of CS/DS, a highly positively charged GFP was used as a model protein that was targeted by CS/DS chains. The results of this experiment showed that treatment with this enzyme significantly weakened the interactions of CS and DS with the ScGFP, suggesting that this enzyme is a very useful tool for CS/DS-related studies.
Similar to the first marine-derived GAG lyase, HCLase (25), the first CS/DS sulfatase identified from the same marine bacterium possesses several unique features, in particular the rare endosulfatase activity. This reminds us that the ocean represents a repository of novel enzymes that can be applied to GAG-related basic research and medical applications.
This work was supported by Major State Basic Research Development Program of China Grant 2012CB822102, National High Technology Research and Development Program of China Grant 2012AA021504, Shandong Province Science and Technology Development Plan Grant 2013GSF12106, and National Natural Science Foundation of China Grant 31300664. This study was also supported in part by Grant-in-aid for Challenging Exploratory Research 25670018 from the Japan Society for the Promotion of Science.
- CS
- chondroitin sulfate(s)
- DS
- dermatan sulfate(s)
- HA
- hyaluronan
- GAG
- glycosaminoglycan
- GalNAc
- N-acetyl-d-galactosamine
- IdoUA
- l-iduronic acid
- GlcUA
- d-glucuronic acid
- HexUA
- hexuronic acid
- Δ4,5HexUA
- Δ4,5-unsaturated hexuronic acid
- 2S
- 4S, and 6S, 2-O-sulfate, 4-O-sulfate, and 6-O-sulfate, respectively
- ΔO
- ΔA, ΔC, ΔD, ΔE, and ΔT units, Δ4,5HexUAα1–3GalNAc, Δ4,5HexUAα1–3GalNAc(4S), Δ4,5HexUAα1–3GalNAc(6S), Δ4,5HexUA(2S) α1–3GalNAc(6S), Δ4,5HexUAα1–3GalNAc(4S,6S), and Δ4,5HexUA(2S) α1–3GalNAc(4S,6S), respectively
- CSase
- chondroitinase
- 2-AB
- 2-aminobenzamide
- ScGFP
- supercharged GFP
- HCLase
- HA and CS lyase.
REFERENCES
- 1. Klüppel M., Wight T. N., Chan C., Hinek A., Wrana J. L. (2005) Maintenance of chondroitin sulfation balance by chondroitin-4-sulfotransferase 1 is required for chondrocyte development and growth factor signaling during cartilage morphogenesis. Development 132, 3989–4003 [DOI] [PubMed] [Google Scholar]
- 2. Faissner A., Clement A., Lochter A., Streit A., Mandl C., Schachner M. (1994) Isolation of a neural chondroitin sulfate proteoglycan with neurite outgrowth promoting properties. J. Cell Biol. 126, 783–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Clement A. M., Nadanaka S., Masayama K., Mandl C., Sugahara K., Faissner A. (1998) The DSD-1 carbohydrate epitope depends on sulfation, correlates with chondroitin sulfate D motifs, and is sufficient to promote neurite outgrowth. J. Biol. Chem. 273, 28444–28453 [DOI] [PubMed] [Google Scholar]
- 4. Lafont F., Rouget M., Triller A., Prochiantz A., Rousselet A. (1992) In vitro control of neuronal polarity by glycosaminoglycans. Development 114, 17–29 [DOI] [PubMed] [Google Scholar]
- 5. Sugahara K., Mikami T. (2007) Chondroitin/dermatan sulfate in the central nervous system. Curr. Opin. Struct. Biol. 17, 536–545 [DOI] [PubMed] [Google Scholar]
- 6. Taylor K. R., Gallo R. L. (2006) Glycosaminoglycans and their proteoglycans: host-associated molecular patterns for initiation and modulation of inflammation. FASEB J. 20, 9–22 [DOI] [PubMed] [Google Scholar]
- 7. Li F., Ten Dam G. B., Murugan S., Yamada S., Hashiguchi T., Mizumoto S., Oguri K., Okayama M., van Kuppevelt T. H., Sugahara K. (2008) Involvement of highly sulfated chondroitin sulfate in the metastasis of the Lewis lung carcinoma cells. J. Biol. Chem. 283, 34294–34304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mizumoto S., Takahashi J., Sugahara K. (2012) Receptor for advanced glycation end products (RAGE) functions as receptor for specific sulfated glycosaminoglycans, and anti-RAGE antibody or sulfated glycosaminoglycans delivered in vivo inhibit pulmonary metastasis of tumor cells. J. Biol. Chem. 287, 18985–18994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hsiao J. C., Chung C. S., Chang W. (1999) Vaccinia virus envelope D8L protein binds to cell surface chondroitin sulfate and mediates the adsorption of intracellular mature virions to cells. J. Virol. 73, 8750–8761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Williams R. K., Straus S. E. (1997) Specificity and affinity of binding of herpes simplex virus type 2 glycoprotein B to glycosaminoglycans. J. Virol. 71, 1375–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bergefall K., Trybala E., Johansson M., Uyama T., Naito S., Yamada S., Kitagawa H., Sugahara K., Bergström T. (2005) Chondroitin sulfate characterized by the E-disaccharide unit is a potent inhibitor of herpes simplex virus infectivity and provides the virus binding sites on gro2C cells. J. Biol. Chem. 280, 32193–32199 [DOI] [PubMed] [Google Scholar]
- 12. Uyama T., Ishida M., Izumikawa T., Trybala E., Tufaro F., Bergström T., Sugahara K., Kitagawa H. (2006) Chondroitin 4-O-sulfotransferase-1 regulates E disaccharide expression of chondroitin sulfate required for herpes simplex virus infectivity. J. Biol. Chem. 281, 38668–38674 [DOI] [PubMed] [Google Scholar]
- 13. Trowbridge J. M., Rudisill J. A., Ron D., Gallo R. L. (2002) Dermatan sulfate binds and potentiates activity of keratinocyte growth factor (FGF-7). J. Biol. Chem. 277, 42815–42820 [DOI] [PubMed] [Google Scholar]
- 14. Maeda N., Fukazawa N., Hata T. (2006) The binding of chondroitin sulfate to pleiotrophin/heparin-binding growth-associated molecule is regulated by chain length and oversulfated structures. J. Biol. Chem. 281, 4894–4902 [DOI] [PubMed] [Google Scholar]
- 15. Silbert J. E., Sugumaran G. (2002) Biosynthesis of chondroitin/dermatan sulfate. IUBMB Life 54, 177–186 [DOI] [PubMed] [Google Scholar]
- 16. Maccarana M., Olander B., Malmström J., Tiedemann K., Aebersold R., Lindahl U., Li J. P., Malmström A. (2006) Biosynthesis of dermatan sulfate: chondroitin-glucuronate C5-epimerase is identical to SART2. J. Biol. Chem. 281, 11560–11568 [DOI] [PubMed] [Google Scholar]
- 17. Kusche-Gullberg M., Kjellén L. (2003) Sulfotransferases in glycosaminoglycan biosynthesis. Curr. Opin. Struct. Biol. 13, 605–611 [DOI] [PubMed] [Google Scholar]
- 18. Izumikawa T., Kitagawa H., Mizuguchi S., Nomura K. H., Nomura K., Tamura J., Gengyo-Ando K., Mitani S., Sugahara K. (2004) Nematode chondroitin polymerizing factor showing cell-/organ-specific expression is indispensable for chondroitin synthesis and embryonic cell division. J. Biol. Chem. 279, 53755–53761 [DOI] [PubMed] [Google Scholar]
- 19. Cheng F., Heinegård D., Malmström A., Schmidtchen A., Yoshida K., Fransson L. A. (1994) Patterns of uronosyl epimerization and 4-/6-O-sulphation in chondroitin/dermatan sulphate from decorin and biglycan of various bovine tissues. Glycobiology 4, 685–696 [DOI] [PubMed] [Google Scholar]
- 20. Sugahara K., Mikami T., Uyama T., Mizuguchi S., Nomura K., Kitagawa H. (2003) Recent advances in the structural biology of chondroitin sulfate and dermatan sulfate. Curr. Opin. Struct. Biol. 13, 612–620 [DOI] [PubMed] [Google Scholar]
- 21. Mikami T., Kitagawa H. (2013) Biosynthesis and function of chondroitin sulfate. Biochim. Biophys. Acta 1830, 4719–4733 [DOI] [PubMed] [Google Scholar]
- 22. Hanson S. R., Best M. D., Wong C. H. (2004) Sulfatases: structure, mechanism, biological activity, inhibition, and synthetic utility. Angew. Chem. Int. Ed. Engl. 43, 5736–5763 [DOI] [PubMed] [Google Scholar]
- 23. Sugahara K., Kojima T. (1996) Specificity studies of bacterial sulfatases by means of structurally defined sulfated oligosaccharides isolated from shark cartilage chondroitin sulfate D. Eur. J. Biochem. 239, 865–870 [DOI] [PubMed] [Google Scholar]
- 24. Ulmer J. E., Vilén E. M., Namburi R. B., Benjdia A., Beneteau J., Malleron A., Bonnaffé D., Driguez P.-A., Descroix K., Lassalle G., Le Narvor C., Sandström C., Spillmann D., Berteau O. (2014) Characterization of glycosaminoglycan (GAG) sulfatases from the human gut symbiont Bacteroides thetaiotaomicron reveals the first GAG-specific bacterial endosulfatase. J. Biol. Chem. 289, 24289–24303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Han W., Wang W., Zhao M., Sugahara K., Li F. (2014) A novel eliminase from a marine bacterium that degrades hyaluronan and chondroitin sulfate. J. Biol. Chem. 289, 27886–27898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nandini C. D., Mikami T., Ohta M., Itoh N., Akiyama-Nambu F., Sugahara K. (2004) Structural and functional characterization of oversulfated chondroitin sulfate/dermatan sulfate hybrid chains from the notochord of hagfish: neuritogenic and binding activities for growth factors and neurotrophic factors. J. Biol. Chem. 279, 50799–50809 [DOI] [PubMed] [Google Scholar]
- 27. Sambrook J., Russell D. W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Ed., pp. A8.40–A8.47, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 28. Bramhall S., Noack N., Wu M., Loewenberg J. R. (1969) A simple colorimetric method for determination of protein. Anal. Biochem. 31, 146–148 [DOI] [PubMed] [Google Scholar]
- 29. Bigge J. C., Patel T. P., Bruce J. A., Goulding P. N., Charles S. M., Parekh R. B. (1995) Nonselective and efficient fluorescent labeling of glycans using 2-amino benzamide and anthranilic acid. Anal. Biochem. 230, 229–238 [DOI] [PubMed] [Google Scholar]
- 30. Sugahara K., Yamada S. (2000) Structure and function of oversulfated chondroitin sulfate variants: unique sulfation patterns and neuroregulatory activities. Trends Glycosci. Glycotechnol. 12, 321–349 [Google Scholar]
- 31. Bitter T., Muir H. M. (1962) A modified uronic acid carbazole reaction. Anal. Biochem. 4, 330–334 [DOI] [PubMed] [Google Scholar]
- 32. Saito H., Yamagata T., Suzuki S. (1968) Enzymatic methods for the determination of small quantities of isomeric chondroitin sulfates. J. Biol. Chem. 243, 1536–1542 [PubMed] [Google Scholar]
- 33. Yamagata T., Saito H., Habuchi O., Suzuki S. (1968) Purification and properties of bacterial chondroitinases and chondrosulfatases. J. Biol. Chem. 243, 1523–1535 [PubMed] [Google Scholar]
- 34. Lawrence M. S., Phillips K. J., Liu D. R. (2007) Supercharging proteins can impart unusual resilience. J. Am. Chem. Soc. 129, 10110–10112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Deepa S. S., Umehara Y., Higashiyama S., Itoh N., Sugahara K. (2002) Specific molecular interactions of oversulfated chondroitin sulfate E with various heparin-binding growth factors: implications as a physiological binding partner in the brain and other tissues. J. Biol. Chem. 277, 43707–43716 [DOI] [PubMed] [Google Scholar]
- 36. Li F., Yamada S., Basappa, Shetty A. K., Sugiura M., Sugahara K. (2008) Determination of iduronic acid and glucuronic acid in sulfated chondroitin/dermatan hybrid chains by 1H-nuclear magnetic resonance spectroscopy. Glycoconj. J. 25, 603–610 [DOI] [PubMed] [Google Scholar]
- 37. Bielicki J., Fuller M., Guo X. H., Morris C. P., Hopewood J. J., Anson D. S. (1995) Expression, purification and characterization of recombinant human N-acetylgalactosamine-6-sulphatase. Biochem. J. 311, 333–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shaklee P. N., Glaser J. H., Conrad H. E. (1985) A sulfatase specific for glucuronic acid 2-sulfate residues in glycosaminoglycans. J. Biol. Chem. 260, 9146–9149 [PubMed] [Google Scholar]
- 39. Bond C. S., Clements P. R., Ashby S. J., Collyer C. A., Harrop S. J., Hopwood J. J., Guss J. M. (1997) Structure of a human lysosomal sulfatase. Structure 5, 277–289 [DOI] [PubMed] [Google Scholar]
- 40. Morimoto-Tomita M., Uchimura K., Werb Z., Hemmerich S., Rosen S. D. (2002) Cloning and characterization of two extracellular heparin-degrading endosulfatases in mice and humans. J. Biol. Chem. 277, 49175–49185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dhoot G. K., Gustafsson M. K., Ai X., Sun W., Standiford D. M., Emerson C. P., Jr. (2001) Regulation of Wnt signaling and embryo patterning by an extracellular sulfatase. Science 293, 1663–1666 [DOI] [PubMed] [Google Scholar]
- 42. Seffouh A., Milz F., Przybylski C., Laguri C., Oosterhof A., Bourcier S., Sadir R., Dutkowski E., Daniel R., van Kuppevelt T. H., Dierks T., Lortat-Jacob H., Vivès R. R. (2013) HSulf sulfatases catalyze processive and oriented 6-O-desulfation of heparan sulfate that differentially regulates fibroblast growth factor activity. FASEB J. 27, 2431–2439 [DOI] [PubMed] [Google Scholar]
- 43. Narita K., Chien J., Mullany S. A., Staub J., Qian X., Lingle W. L., Shridhar V. (2007) Loss of HSulf-1 expression enhances autocrine signaling mediated by amphiregulin in breast cancer. J. Biol. Chem. 282, 14413–14420 [DOI] [PubMed] [Google Scholar]