Background: No investigation on daptomycin production at the transcriptional regulatory level has been reported.
Results: The autoregulator AtrA directly regulates daptomycin gene cluster expression, and atrA is the transcriptional target of AdpA.
Conclusion: The AtrA-mediated transcriptional signaling pathway directly regulates daptomycin production.
Significance: We reveal for the first time the transcriptional regulatory mechanism of daptomycin production for its potential rational genetic engineering.
Keywords: Antibiotics, Bacterial Genetics, Gene Regulation, Protein-DNA Interaction, Secondary Metabolism
Abstract
Daptomycin is a cyclic lipopeptide antibiotic produced by Streptomyces roseosporus. To reveal the transcriptional regulatory mechanism of daptomycin biosynthesis, we used the biotinylated dptE promoter (dptEp) as a probe to affinity isolate the dptEp-interactive protein AtrA, a TetR family transcriptional regulator, from the proteome of mycelia. AtrA bound directly to dptEp to positively regulate gene cluster expression and daptomycin production. Meanwhile, both ΔatrA and ΔadpA mutants showed bald phenotype and null production of daptomycin. AdpA positively regulated atrA expression by direct interaction with atrA promoter (atrAp), and removal of ArpA in S. roseosporus, a homolog of the A-factor receptor, resulted in accelerated morphological development and increased daptomycin production, suggesting that atrA was the target of AdpA to mediate the A-factor signaling pathway. Furthermore, AtrA was positively autoregulated by binding to its own promoter atrAp. Thus, for the first time at the transcriptional level, we have identified an autoregulator, AtrA, that directly mediates the A-factor signaling pathway to regulate the proper production of daptomycin.
Introduction
Daptomycin is a cyclic lipopeptide antibiotic against infection caused by a broad spectrum of Gram-positive bacterial pathogens such as Staphylococcus aureus and Enterococcus species (1, 2). It has been reported that daptomycin is synthesized by a non-ribosomal peptide synthetase in Streptomyces roseosporus NRRL 11379 (3). Because of its clinical importance, many efforts have been made to improve daptomycin production, including random mutagenesis (4), metabolic engineering (5, 6), and ribosomal engineering by mutations based on resistance to antibiotics (7, 8). Nevertheless, no direct evidence has been reported on the transcriptional regulation of the daptomycin gene cluster expression, which might hinder targeted genetic engineering based on the rational designing of the transcriptional circuit to improve daptomycin production.
Secondary metabolism development including antibiotic production in Streptomyces is transcriptionally regulated by extracellular stimuli, intracellular signaling pathways, and their cross-talks (9). AdpA is one of the key pleiotropic transcriptional regulators in Streptomyces griseus, and is widely found in Streptomyces to control morphological development and secondary metabolism (10–13). adpA is under the transcriptional control of a repressor (such as ArpA in S. griseus), which dissociates from the adpA promoter after binding to the growth phase-dependent γ-butyrolactone molecules, thus causing derepression of adpA expression (11, 14). Once activated, AdpA binds to the highly conserved binding elements (5′-TGGCSNGWWY-3′) on the promoters of its regulons, which are involved in various cell developmental programs (11, 15, 16).
In the daptomycin industrial producer S. roseosporus SW0702, we have characterized the DeoR-type regulator DptR2 as required for daptomycin production but not for gene cluster expression (17). To further reveal the transcriptional regulatory mechanism of daptomycin production, we applied affinity purification with biotinylated dptEp as a probe to isolate a TetR-type transcriptional regulator, AtrA. In Streptomyces coelicolor, AtrA binds to the coding regions of actII-orf4 and actII-orf3 to positively regulate actinorhodin production, but it is not required for undecylprodigiosin or calcium-dependent antibiotic production or for morphological development (18). In S. griseus, AtrA binds to the strR promoter synergistically along with AdpA to conditionally regulate streptomycin production (19). However, AveI, the homolog of AtrA in Streptomyces avermitilis, negatively regulates avermectin biosynthesis and does not bind to the aveR promoter (20).
Here we have provided evidence that AtrA could positively regulate daptomycin production by directly binding to the gene cluster promoter; atrA was one of AdpA transcriptional targets to mediate the conserved A-factor signaling pathway. We also have suggested the positive feedback regulation of atrA expression for the proper production of daptomycin. This is the first report to provide direct evidence at the transcriptional regulatory level for daptomycin production and also suggests that an AtrA-mediated A-factor signaling pathway is required for daptomycin production.
EXPERIMENTAL PROCEDURES
Strains and Media
S. roseosporus SW0702 is an industrial daptomycin producer (17). ΔatrA, ΔadpA, and ΔarpA mutants are the in-frame deletion strains of S. roseosporus SW0702. Escherichia coli strain TG1 was the host for routine plasmid subcloning. BL21(DE3) was used for protein expression and ET12456/pUZ8002 for conjugation of plasmids into Streptomyces.
E. coli cells were cultured in LB medium for plasmid preparation and protein expression. Solid R5 medium was used for morphological development and sporulation of Streptomyces cells, and MSF medium was used for conjugation (21). For daptomycin production, tryptic soy broth plus 5% PEG 6000 was the seed medium, and YEME (0.3% yeast extract, 0.3% malt extract, 0.5% tryptone, 4% glucose) medium was used for fermentation (17).
Plasmid Construction
Plasmids and primers were listed in Table 1 and Table 2, respectively. Primer pairs 1 + 2 and 3 + 4 were used to amplify the right and left homologous regions of atrA, respectively, and cloned into the HindIII/BamHI and BamHI/EcoRI sites of pKC1139 (22), respectively, to give rise to pKC1139-atrA. Plasmids pKC1139-adpA and pKC1139-arpA were constructed by cloning homologous fragments into the HindIII/BamHI and BamHI/EcoRI sites of pKC1139. Primers 5 + 6 and 7 + 8 were for adpA, and primers 9 + 10 and 11 + 12 were for arpA. The complementation DNA fragments for atrA, adpA, and arpA were amplified with primer pairs 13 + 14, 15 + 16, and 17 + 18, respectively, and cloned into the HindIII/BamHI site of pSET152 (22) to create the complementation plasmids pSET152-atrA, pSET152-adpA, and pSET152-arpA, respectively. dptEp and atrAp were obtained with primers 19 + 20 and 21 + 22, respectively, and cloned into pTA2 after deoxyadenylic acid (dA) addition with Taq polymerase. Both promoters were digested with BamHI and ligated into the BamHI site of pIPP1 (23) to get pIPP1-dptEp and pIPP1-atrAp, respectively. BamHI-digested dptEp was ligated into the BamHI site of pUC18 to generate pUC18-dptEp. atrA and adpA were amplified with primer pairs 24 + 25 and 26 + 27 and cloned into the BamHI/HindIII site of pET32a to obtain the expression plasmids pET32a-atrA and pET32a-adpA, respectively. All PCR were performed with KOD-Plus-Neo (Toyobo Co.) from the genomic DNA of S. roseosporus SW0702.
TABLE 1.
Plasmids used in this study
| Plasmid | Description | References |
|---|---|---|
| pKC1139 | Temperature-sensitive vector for gene knock-out | (22) |
| pKC1139-atrA | atrA in-frame deletion plasmid | This study |
| pKC1139-adpA | adpA in-frame deletion plasmid | This study |
| pKC1139-arpA | arpA in-frame deletion plasmid | This study |
| pSET152 | Integrative plasmid for complementation in Streptomyces | (22) |
| pSET152-atrA | atrA gene complementation plasmid | This study |
| pSET152-adpA | adpA gene complementation plasmid | This study |
| pSET152-arpA | arpA gene complementation plasmid | This study |
| pTA2 | T vector | Toyobo Co. |
| pTA2-dptEp | dptE promoter (dptEp) cloned in pTA2 vector | This study |
| pTA2-atrAp | atrA promoter (atrAp) cloned in pTA2 vector | This study |
| pUC18 | Cloning vector | Novagen |
| pUC18-dptEp | dptEp cloned in BamHI site of pUC18 vector | This study |
| pET32a | E. coli expression vector | Novagen |
| pET32a-atrA | atrA ORF cloned in BamHI/HindIII site of pET32a | This study |
| pET32a-adpA | adpA ORF cloned in BamHI/HindIII site of pET32a | This study |
| pIPP1 | Promoter probing vector with xylE reporter | (23) |
| pIPP1-dptEp | dptEp cloned in BamHI site of pIPP1 | This study |
| pIPP1-atrAp | atrAp cloned in BamHI site of pIPP1 | This study |
TABLE 2.
Primers used in this study
Restriction sites are underlined.
| No. | Sequence |
|---|---|
| 1 | TGTTGGATCCCTGGAGGGACTGCGGTC |
| 2 | CTAAGAATTCGAGATACGCCTGCTTGAG |
| 3 | TATCAAGCTTGACGAAGACGAAGGTGAG |
| 4 | TACTAGATCTCGAGCTCATGACGCCCAG |
| 5 | TACTAAGCTTCTGTTCGCGTACCACAC |
| 6 | TACTGGATCCGGCGGAGTCCTGGCTC |
| 7 | TACTGGATCCCAGCGGAGTGCCCCGTAG |
| 8 | TACTGAATTCCGTCATGGAACACCAGC |
| 9 | ACTTCAAGCTTGATGCCGTTGGTGGACCC |
| 10 | ACTTAGGATCCAGCCTGCTTCGCCATTTC |
| 11 | ACTTAGGATCCGCTGCCTGATCCTCGACC |
| 12 | ACTTAGAATTCCTCCTCGGCACGCTGTTC |
| 13 | ACTTAGGATCCACGACGAGAACGCGCAC |
| 14 | TACTAAGCTTGCAGCAGGAAGAGCCGCT |
| 15 | ACTTAGGATCCTGTCTCGGCGAAGAGAGTC |
| 16 | TACTAAGCTTCTACGGGGCACTCCGCTG |
| 17 | TACTAGATCTGAAACAACGGATGTGAATC |
| 18 | TACTAAGCTTGAGGTGGACTTCATCGTTC |
| 19 | GGATCCCCTCCCCACCACCTGC |
| 20 | GGATCCGGCAGGCACTCCCCCTCC |
| 21 | GGATCCGGAGCCGAGGCGTCCTG |
| 22 | GGATCCCCGCCGACGACGAGAAC |
| 23 | FAM-GCCAGGGTTTTCCCAGTCACGA |
| 24 | TACTGGATCCGTGGGTGCGCTGGGCGTC |
| 25 | TACTAAGCTTCGCCGGCCGTGACCGCAG |
| 26 | AATAGGATCCATGAGCCAGGACTCCGCC |
| 27 | TACTAAGCTTCTACGGGGCACTCCGCTG |
| 28 | Biotin-GCCAGGGTTTTCCCAGTCACGA |
| 29 | GAGCGGATAACAATTTCACACAGG |
Strain Construction
atrA, adpA, and arpA were knocked out by in-frame deletion in S. roseosporus SW0702 (17). pKC1139-based knock-out plasmids were introduced into S. roseosporus SW0702 by conjugation. The conjugates were streaked on the solid R5 medium with apramycin at 37 °C to get the single crossover strains. They were patched on the R5 medium at 37 °C for two rounds of sporulation. Spores were then diluted on the R5 medium into single colonies. The mutants were identified from the apramycin-sensitive clones by PCR and Southern blot (data not shown).
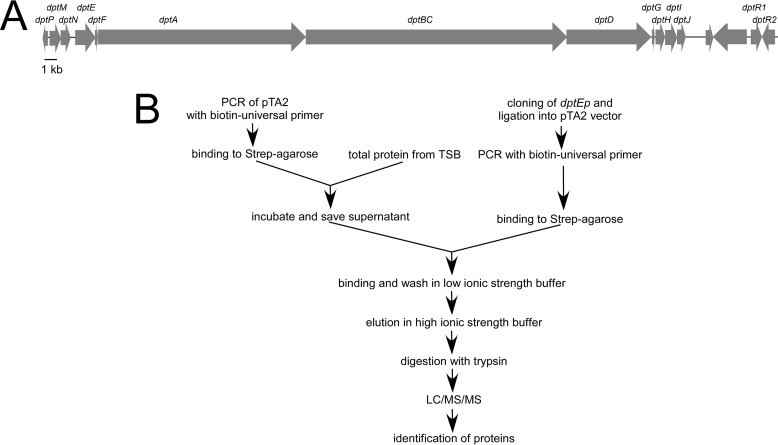
Affinity Isolation of dptEp-interactive Proteins
The biotinylated vector and vector-dptEp were prepared by PCR from pUC18 and pUC18-dptEp, respectively, with universal primers 28 + 29 and gel-purified. About 500 ng of biotinylated DNA was incubated with streptavidin-agarose (Prozyme) and washed twice with the binding buffer (20 mm Tris, pH 8.0, 50 mm NaCl, 1 mm EDTA, and 10% glycerol) to eliminate the unbound probes and save the pellet. S. roseosporus SW0702 cells were cultured in tryptic soy broth medium to the logarithmic phase, collected, sonicated in the binding buffer, and centrifuged at 4 °C for 15 min to collect the total lysate. The proteome was incubated initially with vector-streptavidin-agarose at room temperature for 30 min in the presence of 500 μg of sheared sperm DNA to prevent the nonspecific binding of proteins. The mixture was centrifuged briefly, and the supernatant was incubated with dptEp-streptavidin-agarose. After binding at room temperature for 60 min, the beads were spun down and washed with the binding buffer three times, and the binding proteins were eluted with high ionic buffer (binding buffer plus 1 m NaCl). The eluted protein was buffer-exchanged with 100 mm NH4HCO3, pH 8.0 on a 10-kDa cut-off YM-10 column (Millipore). Then it was incubated with 10 mm DTT at 56 °C for 30 min and cooled down to room temperature followed by incubation with 20 mm iodoacetamide in the dark for 30 min. The protein mixture was digested with 100 ng of trypsin at 37 °C overnight, vacuum-dried, and subjected to LC/MS/MS to identify the peptide sequences.
Protein Expression and Purification from E. coli
BL21(DE3) containing expression plasmids for AtrA and AdpA, respectively, was cultured in LB medium at 37 °C to OD = 0.5 and induced with 0.1 mm IPTG at 16 °C for 16 h. Cells were collected and disrupted in buffer A (50 mm Tris, pH 8.0, 500 mm NaCl, and 10 mm imidazole) by sonication. Proteins expressed in soluble form were purified on Ni2+-nitrilotriacetic acid resin as described by the manufacturer (Qiagen) and eluted in buffer A plus 500 mm imidazole. Proteins were ultra-filtered on a 10-kDa cut-off YM-3 column (Millipore) with 20 mm Tris buffer, pH 7.5.
Electrophoretic Mobility Shift Assay (EMSA)
5′-Biotin-labeled DNA probes including vector, vector-dptEp, and vector-atrAp were prepared by high fidelity PCR from plasmids pTA2, pTA2-dptEp, and pTA2-atrAp, respectively, with universal primers 28 + 29. These probes were gel-purified and eluted in sterile water. EMSA was demonstrated as described previously with 1 ng of probes and an increasing amount of protein (24).
DNase I Footprinting Assay
The DNase I footprinting assays were demonstrated as described previously (24). 5′-FAM-labeled dptEp and atrAp were prepared by PCR from plasmids pTA2-dptEp and pTA2-atrAp, respectively, with universal primers 29 + 23 and then gel-purified. About 50 ng of probes were incubated with various amounts of protein, and 0.01 unit of DNase I was added for partial digestion.
HPLC Analysis of Daptomycin
Daptomycin was produced in S. roseosporus as described previously (17). After fermentation, the culture was mixed with an equal volume of methanol and centrifuged. The supernatant was analyzed by HPLC on a reverse phase column (Phenomenex, 250 × 4.6 mm, C18) with UV detection at 366 nm at a flow rate of 0.1 ml/min. The solutions used were solution A (50 mm Na2HPO4, pH 3.15 ± 0.05) and solution B (100% acetonitrile). HPLC was run using the following procedure: 0–10 min, A:B = 20:50%; 10–40 min, A:B = 50:80%; 40–50 min, A:B = 80:95%. Pure daptomycin was used as a standard.
Catechol 2,3-Dioxygenase Activity Assay
The catechol 2,3-dioxygenase activity assays were demonstrated as described previously with minor modifications (25). Streptomyces cells were harvested, washed with 20 mm potassium phosphate, pH 7.2, and resuspended in 1 ml of sample buffer (100 mm potassium phosphate, pH 7.5, 10% acetone). Cells were lysed by sonication, and cell debris was removed by centrifugation for 15 min at 4 °C. The assay buffer (100 mm potassium phosphate, pH 7.5, 1 mm catechol) was preincubated at 37 °C for 1 min, and the reactions were initiated by the addition of 100 μl of cell extract to 900 μl of assay buffer. The optical density at 375 nm was measured over time (0, 1, 2, and 3 min, respectively). Catechol dioxygenase activity was calculated as the change rate in the optical density at 375 nm and converted to milliunits/min/mg of total protein.
RESULTS
Identification of AtrA as a dptEp-interactive Regulator
Two pathway-specific regulators, DptR1 and DptR2, were found when the daptomycin gene cluster was initially sequenced in S. roseosporus NRRL 11379 (3). We have reported that the industrial daptomycin producer S. roseosporus SW0702 contains the daptomycin gene cluster in high identity with that from S. roseosporus NRRL 11379 (17) (Fig. 1A). The whole gene cluster was reported to be transcribed as a large mRNA from the promoter of dptE (dptEp) (3). To further reveal its transcriptional regulatory mechanism, we used the biotinylated dptEp DNA fragment as a probe to isolate regulators that might interact with dptEp directly from the total lysate of mycelia (Fig. 1B). In total, 82 proteins were identified by LC/MS/MS, including the transcription machinery (RNA polymerase and accessary proteins), several transcriptional regulators with DNA-binding domains, and other proteins that might interact with these DNA-binding proteins (data not shown).
FIGURE 1.
Isolation of dptEp-interactive proteins. A, gene cluster of daptomycin in S. roseosporus SW0702. The homologous genes were labeled on each open reading frame. B, strategy for affinity isolation of dptEp-interactive proteins from S. roseosporus SW0702 proteome.
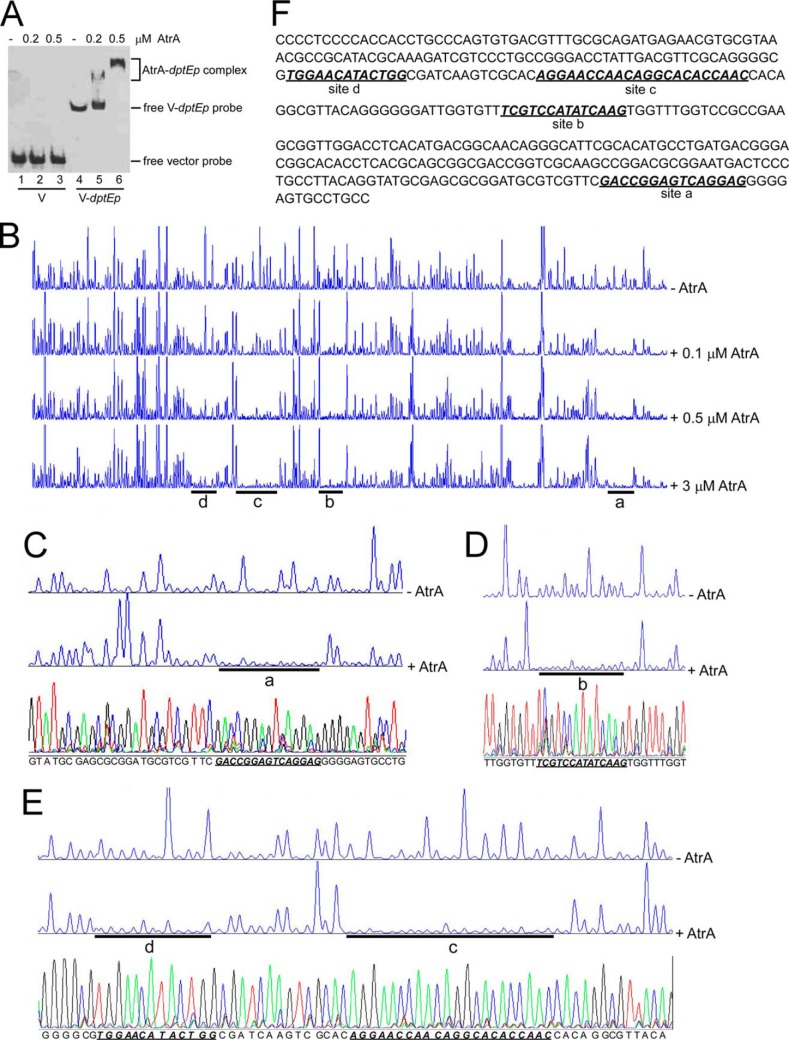
Among these identified regulators, AtrA is a TetR-type transcriptional regulator with a DNA-binding domain at its N terminus, and protein alignment showed that it had 86, 70, and 67% identity with its homologs from S. griseus, S. avermitilis, and S. coelicolor, respectively, with 100% identity in the DNA-binding domains (data not shown). AtrA was expressed and purified from E. coli. EMSA showed that AtrA specifically bound to dptEp (Fig. 2A), which confirmed our screening strategy. Four AtrA-binding sites on dptEp were determined by DNase I footprinting assays (Fig. 2B). The site closest to the start codon (ATG) of dptE (Fig. 2B, site a) had the same binding pattern, despite the concentration of AtrA, suggesting that a small amount of AtrA could fully occupy this site. On other sites, however, the protection patterns became more significant with the increased amount of AtrA. Farther away from dptE ATG, more AtrA was needed to completely protect these sites (Fig. 2B, site b to d), suggesting gradient binding affinity with AtrA along with dptEp. Moreover, deletion of all four sites on dptEp could abrogate binding of AtrA on dptEp, but removal of each of the binding sites could not apparently alter the binding affinity of AtrA to dptEp (data not shown), further supporting multiple interactive sites on dptEp for AtrA.
FIGURE 2.
Binding of AtrA to the dptE promoter (dptEp). A, EMSA of AtrA binding to dptEp. Biotin-labeled dptEp (V-dptEp) was used as a probe, and biotin-labeled void vector (V) was the negative control. B, DNase I footprinting assays of AtrA binding sites on dptEp. FAM-labeled dptEp was used as a probe with gradient concentrations of AtrA. The protected regions were labeled as site a, b, c, and d, respectively. C–E, binding sequence determination of AtrA on dptEp by DNase I footprinting assays for site a (C), site b (D), and sites c and d (E). F, the binding sequences of AtrA on dptEp as determined by DNase I footprinting assays. The four binding sites are shown in bold italics and are underlined.
After alignment with the DNA sequencing data (Fig. 2, C–E), the binding sequence of these sites was determined (Fig. 2F). However, the four AtrA-binding sites had no consensus or apparent palindrome sequence, which is often recognized by TetR-type regulators (26). S. coelicolor AtrA has been shown to bind to the coding region of actII-orf4, and these sites do not show significant conserved sequences but have imperfect inverted repeats of the hexanucleotide GGAAT(G/C) sequence (18). Also an imperfect inverted repeat (GGAGGGNNNCGTTCC) was identified in S. griseus, and mutation of this repeat abolishes S. griseus AtrA binding to strRp (19). Consistent with the 100% identity of the AtrA DNA-binding domain from three Streptomyces species, we found two GGAG motifs on site a, a GGAC motif on site b, and a GGAA motif on sites c and d, respectively. However, no apparent inverted repeat was observed on any of the sites. Nevertheless, our results suggested that AtrA is a transcriptional regulator that binds directly to the promoter of the daptomycin gene cluster.
AtrA Is Required for dptEp Activity and Daptomycin Production
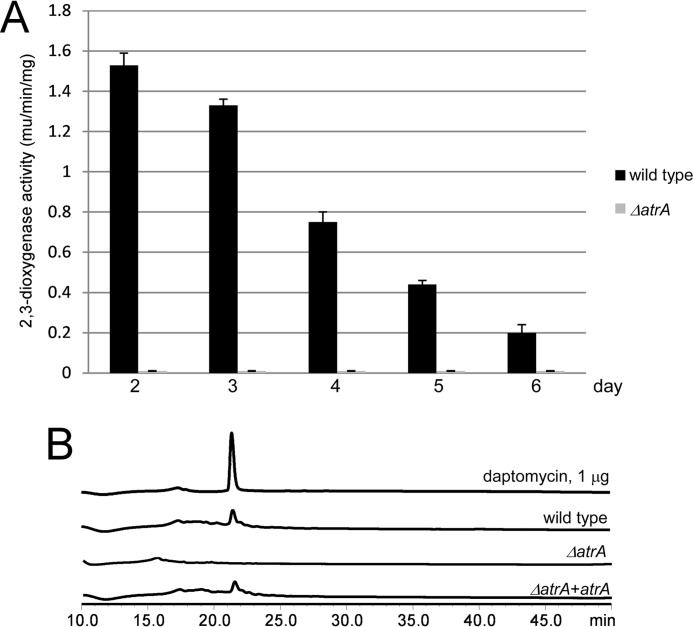
The xylE reporter assays were used to investigate the effects of AtrA on dptEp activity. atrA was knocked out by in-frame deletion. S. roseosporus had the highest daptomycin productivity, between 3 and 4 days, and subsequently decreased (17). However, the highest dptEp activity was observed at day 2 and then dropped continuously during daptomycin production. But the ΔatrA mutant showed null dioxygenase activities on all the days examined (Fig. 3A). Consistent with these results, HPLC assays showed that the ΔatrA mutant did not produce daptomycin, whereas reintroduction of atrA rescued daptomycin production (Fig. 3B). These results suggested that AtrA can bind directly to the daptomycin gene cluster promoter to positively regulate daptomycin production.
FIGURE 3.
AtrA is required for dptEp activity and daptomycin production. A, xylE reporter assays for dptEp activity. The wild type and the ΔatrA mutant were transformed with pIPP1-dptEp (dptEp-xylE) and cultured in YEME medium. Cells were collected and disrupted by sonication every day, and the 2,3-dioxygenase activity was measured. B, HPLC assay of daptomycin production. The wild type and the ΔatrA mutant were cultured in YEME medium, and the daptomycin production was measured on day 5. The purified daptomycin was used as a standard.
Both AtrA and AdpA Are Required for Daptomycin Production and Morphological Development
The ΔatrA mutant in S. coelicolor does not show a significant difference in morphological development from the wild type (18). However, the phenotype of the ΔatrA mutant in S. roseosporus was quite different from the wild type in that this mutant grew only in substrate mycelia on the R5 medium, whereas the wild type and the complementation strain underwent morphological development with aerial mycelia (Fig. 4A) and spores (data not shown). This phenotype was reminiscent of the ΔadpA mutant, which also shows the bald phenotype from many Streptomyces species, including S. coelicolor, S. griseus, etc. (11, 12).
FIGURE 4.
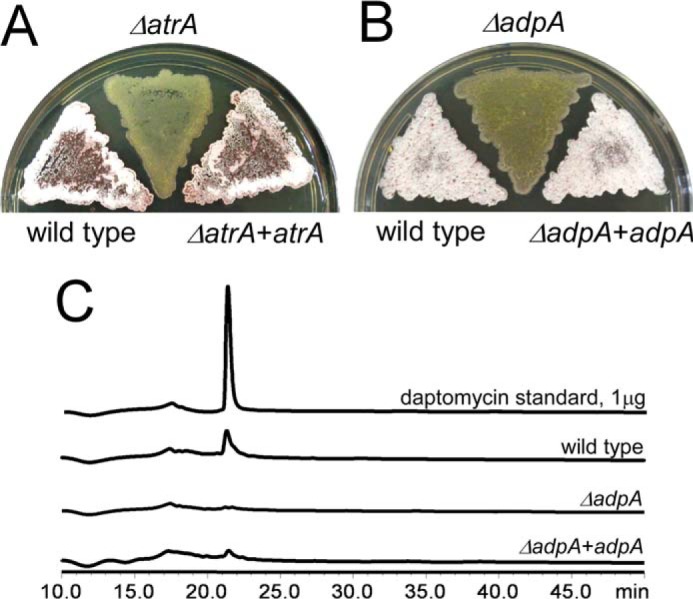
AdpA is required for morphological development and daptomycin production. Mycelia of the wild type, the ΔatrA mutant, and the complementation strain (A) and the wild type, the ΔadpA mutant, and the complementation strain (B) were collected from tryptic soy broth medium, spread on the R5 medium for 3 days, and photographed. C, AdpA is required for daptomycin production. The wild type and the ΔadpA mutant were cultured in YEME medium, and daptomycin production was measured on day 5. The purified daptomycin was used as the standard.
S. roseosporus AdpA was identified based on a protein BLAST with S. griseus AdpA as a query against the S. roseosporus SW0702 genome sequence. It displayed 97 and 84% identity with its homologs in S. griseus and S. coelicolor, respectively, and the DNA-binding domain also showed 100% identity (data not shown), suggesting that conserved functions of AdpA in S. roseosporus. adpA was disrupted by in-frame deletion. The ΔadpA mutant constantly grew in substrate mycelia, as expected (Fig. 4B), and did not produce daptomycin (Fig. 4C). Moreover, after 3 days on the R5 medium, red pigment production was observed in wild type and complementation strains but not in the ΔatrA or ΔadpA mutant (data not shown). These results suggested that both AtrA and AdpA are positive regulators of secondary metabolism (especially daptomycin production) and morphological development.
AdpA Positively Regulates atrA Expression
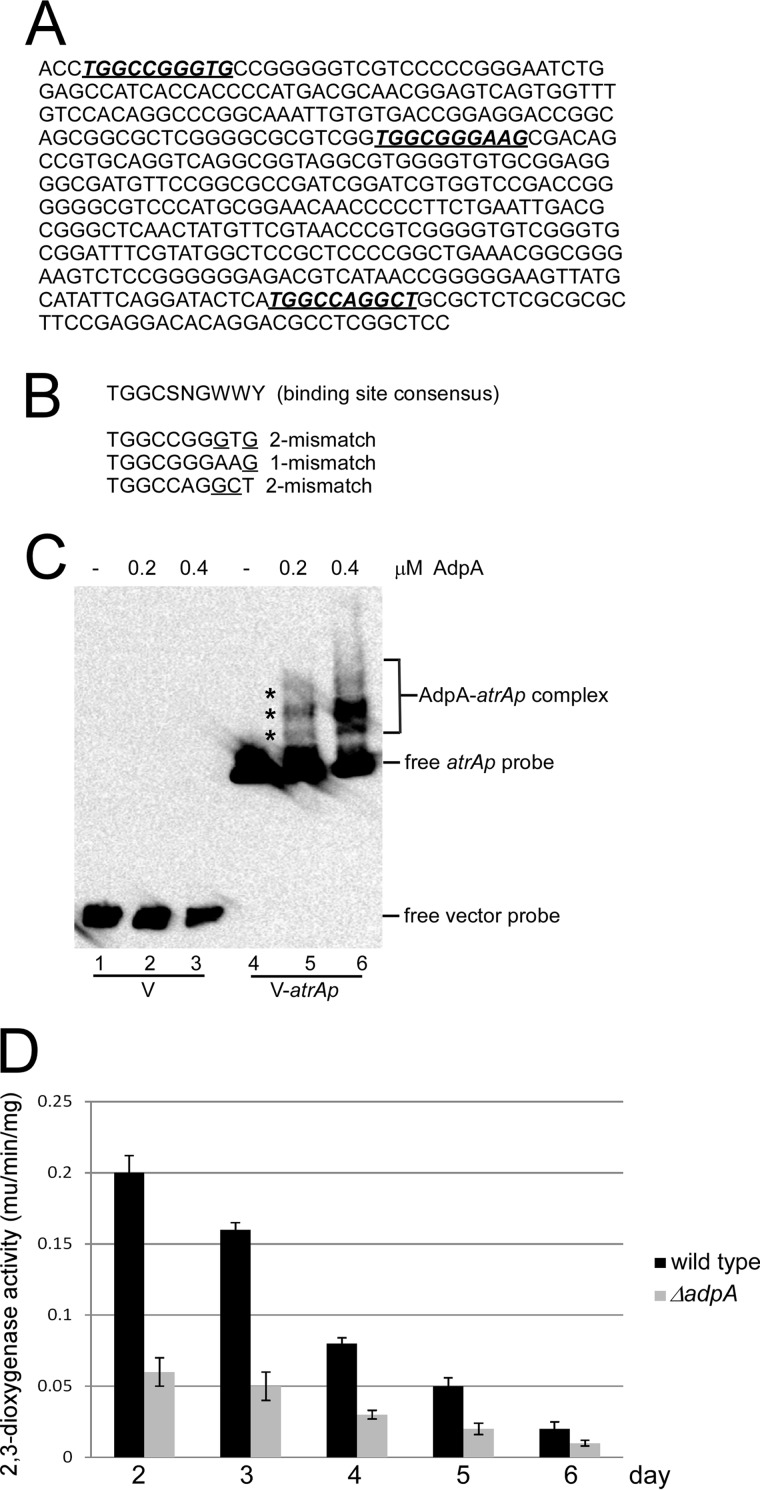
AdpA is a pleiotropic regulator that binds to the promoters of a variety of regulons with a consensus motif TGGCSNGWWY (S, G, or C; W, A, or T; Y, T, or C; N, any nucleotide) (15). Three potential binding motifs were identified on the atrA promoter (atrAp) of S. roseosporus (Fig. 5A), although one or two mismatches were still found (Fig. 5B). However, these motifs, even with two or three mismatches, can still be recognized by AdpA, as exemplified by the wblA promoter in S. coelicolor (13), suggesting that atrA might also be one of the AdpA targets in S. roseosporus. AdpA was expressed and purified from E. coli. Consistent with the in silico prediction, EMSA showed that AdpA could specifically bind to atrAp, and three shifted bands were observed (Fig. 5C).
FIGURE 5.
Binding of AdpA to atrA promoter (atrAp). A, DNA sequence of atrAp. The deduced AdpA binding sites are shown in bold italics and underlined. B, alignment of deduced AdpA binding sites on atrAp with the binding consensus. One or two mismatched nucleotides are indicated. C, EMSA of AdpA binding to atrAp. Biotin-labeled atrAp (V-atrAp) was used as a probe, and biotin-labeled void vector (V) was the negative control. D, AdpA positively regulates atrAp activity. The wild type and the ΔadpA mutant were transformed with pIPP1-atrAp (atrAp-xylE) and cultured in YEME medium. Cells were collected and disrupted by sonication every day, and 2,3-dioxygenase activity was measured. Triplicate independent experiments were demonstrated, and S.D. bars are shown.
Moreover, xylE reporter assays indicated that atrAp activities decreased during daptomycin production in wild type (Fig. 5D), which was coincident with dptEp activities. Deletion of adpA caused a 2–4-fold decrease in atrAp activities (Fig. 5D). These results suggest that AdpA is required for daptomycin production and morphological development by directly regulating atrA expression.
Deletion of arpA Causes Accelerated Morphological Development and Increased Daptomycin Production
The two positive regulators AtrA and AdpA in S. roseosporus showed the highest homology to their counterparts in S. griseus (data not shown), where A-factor-triggered morphological development and secondary metabolism have been extensively studied. The A-factor binds to its receptor protein, ArpA, and releases it from the adpA promoter (adpAp), thus derepressing adpA expression to switch on the developmental programs (11). S. roseosporus ArpA was also identified from the genome sequence by a protein BLAST with S. griseus ArpA. It showed 95% identity with its counterpart in S. griseus and 100% identity in the DNA-binding domain (data not shown). Meanwhile, the binding sequence on adpAp also showed 100% identity in S. roseosporus and S. griseus (data not shown), suggesting that ArpA in S. roseosporus must function via the same mechanism as in S. griseus. This prompted us to construct a strain for a high yield of daptomycin, because deletion of arpA in S. griseus can enhance streptomycin production and accelerate morphological development (27). arpA was also deleted in S. roseosporus by in-frame knock-out. Consistent with this hypothesis, the ΔarpA mutant showed earlier development of aerial mycelia compared with the wild type (Fig. 6A) and about 2.5-fold higher production of daptomycin (Fig. 6B). These data suggested that an A-factor-motivated signaling pathway is conserved in S. roseosporus and that removal of the A-factor receptor can promote daptomycin production and morphological development.
FIGURE 6.
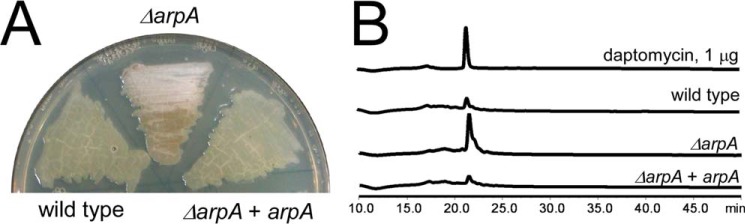
ArpA negatively regulates morphological development and daptomycin production. A, spores of the wild type, the ΔarpA mutant, and the complementation strain were patched on the R5 medium for 30 h and photographed. B, ArpA negatively regulates daptomycin production. The wild type and the ΔarpA mutant were cultured in YEME medium, and daptomycin production was measured on day 5. The purified daptomycin was used as a standard.
Autoregulation of atrA Expression during Daptomycin Production
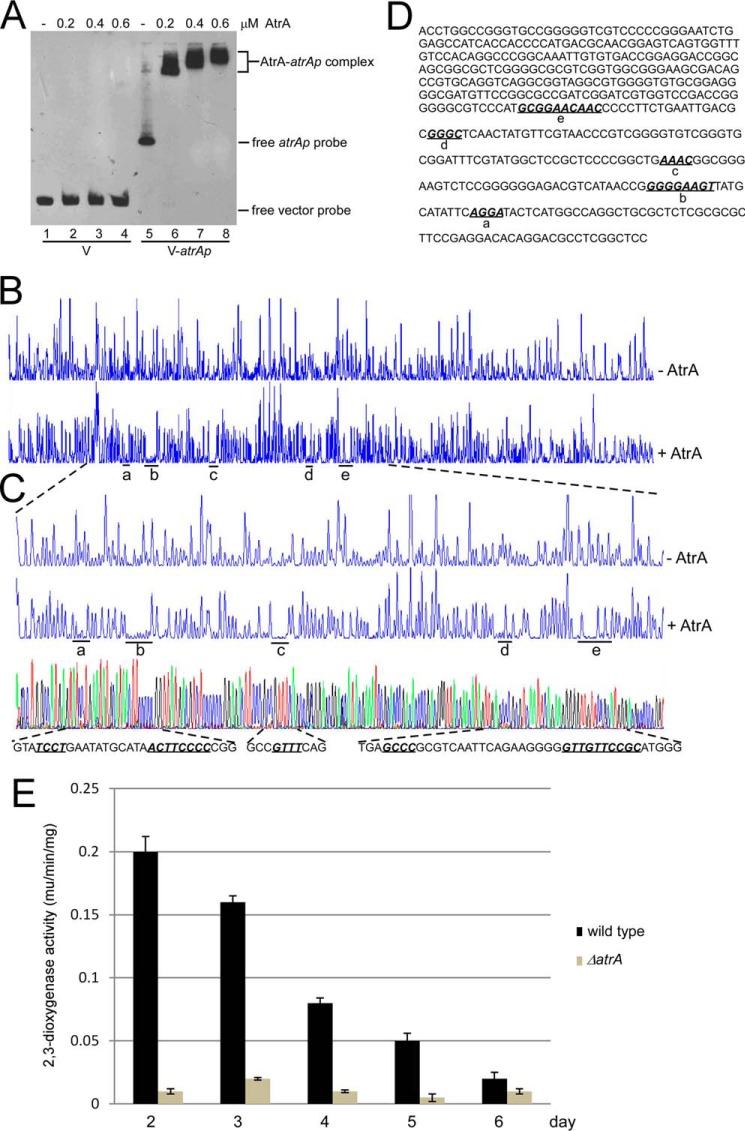
AtrA was predicted to be a TetR-type transcriptional regulator, which often binds to its own promoter (26). EMSA showed that AtrA could bind directly to atrAp at a low concentration (0.2 μm or about 100 ng), but no shifted band was observed even with more AtrA in the control DNA (Fig. 7A), suggesting AtrA could specifically bind to atrAp at high affinity. Meanwhile, five binding sites were determined by DNase I footprinting assays (Fig. 7B), and the sequences were also determined (Fig. 7C). These binding sites were all very small (4, 8, or 10 bp), and no consensus sequence was found (Fig. 7D). Moreover, deletion of all five sites abolished AtrA-atrAp interaction, whereas removal of each site did not affect AtrA binding to atrAp (data not shown), also supporting the idea of multiple interactive sites of AtrA on its own promoter. Nevertheless, we still observed a GGA motif in site a and a GGAA(G/C) motif in sites b and e (Fig. 7D). Although no GGA motif was found in site c or d (Fig. 7D), these sites were rich in purine, suggesting a more complex binding pattern of AtrA on its own promoter. These results, to some extent, were consistent with the previous report that in S. coelicolor, even though AtrA-atrAp interact at high affinity, no AtrA-binding sites could be determined on atrAp by DNase I footprinting assays (28).
FIGURE 7.
Binding of AtrA to its own promoter (atrAp). A, AtrA binds to atrAp. Biotin-labeled atrAp (V-atrAp) was used as a probe, and biotin-labeled void vector (V) was the negative control. Gradient concentrations of AtrA were added for EMSA. B, DNase I footprinting assay of AtrA binding sites on atrAp. FAM-labeled atrAp was used as the probe with 1 μm AtrA. The protected regions were labeled as a–e, respectively. C, binding sequence determination of AtrA on atrAp by DNase I footprinting assays. D, the binding sequences of AtrA on atrAp as determined by DNase I footprinting assays. The five binding sites are shown in bold italics and are underlined. E, AtrA positively regulates atrAp activity. The wild type and the ΔatrA mutant were transformed with pIPP1-atrAp (atrAp-xylE) and cultured in YEME medium. Cells were collected and disrupted by sonication every day, and 2,3-dioxygenase activity was measured. The data were from three independent experiments. S.D. bars are shown.
Based on xylE reporter assays, atrAp activities in the ΔatrA mutant was only about 1/20th that of the wild type on day 2 and persisted at a lower level than wild type in the following days. However, atrAp activities in the wild type also kept dropping (Fig. 7E), suggesting that atrA is positively autoregulated.
DISCUSSION
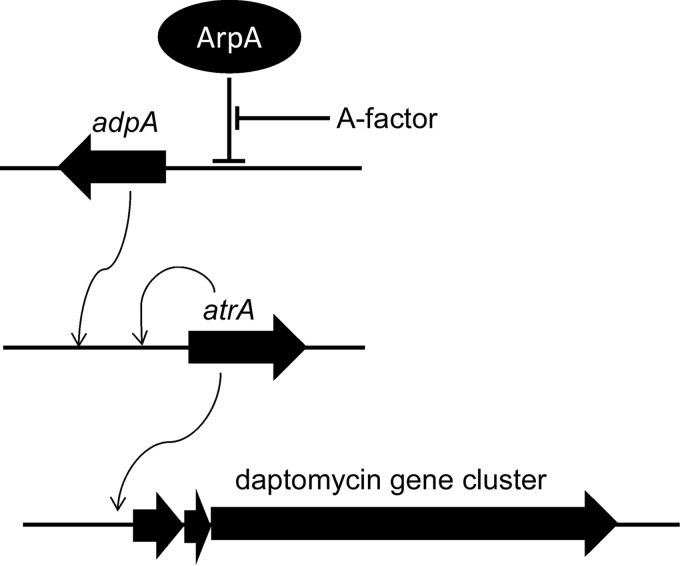
Here, we used the biotinylated dptEp to isolate AtrA as a positive regulator for daptomycin production by directly binding to the gene cluster promoter. In other Streptomyces species, AtrA can also positively regulate streptomycin and actinorhodin production in S. griseus and S. coelicolor, respectively, by direct binding to strRp and the flanking regions of actII-orf4p, respectively (18, 19), but not the promoter of the gene clusters. Meanwhile, AtrA negatively regulates avermectin production in S. avermitilis without binding to aveRp (20), suggesting diverse and complex regulatory patterns of AtrA in Streptomyces.
AtrA was required for morphological development and secondary metabolism in S. roseosporus, suggesting that AtrA is also a pleiotropic regulator as well as AdpA, the key regulator in the A-factor-induced signaling pathway. In S. griseus, AtrA and AdpA synergistically bind to strRp to positively regulate streptomycin production (19). Here in S. roseosporus, we demonstrated that AdpA and AtrA bind to different sites of atrAp, as determined by bioinformatics analysis and DNase I footprinting assays, respectively (Figs. 5 and 7). We also proved that atrA expression was under the direct control of AdpA, thus establishing the signaling pathway from extracellular stimulation of the potential A-factor to daptomycin gene cluster expression and also, for the first time, providing direct evidence about the transcriptional regulation of daptomycin production (Fig. 8). Based on these observations, we propose that targeted genetic engineering by deletion of arpA results in enhanced daptomycin production. Although this industrial strain showed the highest daptomycin production at about day 4, gene cluster expression decreased continuously. The decreasing dptEp activity might be explained by the decreased expression of atrA, as AtrA bound directly to atrAp, and positively regulated its activity, thus composing a positive feedback regulatory pattern of atrA expression by itself. We observed that adpA expression also declined during daptomycin production (data not shown), which also might contribute to the decreased expression of atrA, because AdpA positively regulates atrA expression directly. This ingenious regulation of atrA expression might ensure proper gene cluster expression and daptomycin production.
FIGURE 8.
A proposed model of transcriptional regulation by AtrA for daptomycin production. The TetR-type regulator AtrA directly regulates daptomycin gene cluster expression; atrA expression is autoregulated and under the direct regulation of AdpA.
AtrA protein from all three Streptomyces species had 100% identity in their DNA-binding domains, suggesting the same binding pattern among the three. Although imperfect inverted repeats were reported in S. coelicolor and S. griseus, no apparent palindromic sequence was observed in S. roseosporus dptEp or atrAp, suggesting the atypical binding pattern of this TetR-type transcriptional regulator. However, most binding sites of AtrA in S. roseosporus contained GGA motifs and were rich in purine. These observations suggested complex binding mechanism of AtrA. Therefore it might not be possible to map precisely the universal conserved binding motifs of AtrA on DNA. However, it might be possible that the adjacent DNA sequence environment or higher DNA architecture conformation is also critical for AtrA recognition and binding. We may need more experimental information, such as chromatin immunoprecipitation (ChIP) assays or AtrA-DNA co-crystallization, to establish this binding model.
Acknowledgments
We are grateful to Professor George H. Jones, Department of Biology, Emory University, for providing the xylE reporter plasmid pIPP1. We also thank the reviewers for comments made during the revision process that led to an enhanced discussion of the results.
Footnotes
This work was supported by Grants 2012CB721005 from the National Basic Research Program of China (973 Program), 31370103 from the National Natural Science Foundation of China, 2012AA02A706 from the National High Technology Research and Development Program of China (863 Program), and LZ12C01001 from the Key Program of Zhejiang Provincial Natural Science Foundation of China and by fellowships from the New Star Project from Zhejiang University. The nucleotide sequence(s) reported in this paper has been submitted to the DDBJ/GenBankTM/EBI Data Bank with accession number(s) KM091937, KM091938, KM091939, and KM091940.
REFERENCES
- 1. Arbeit R. D., Maki D., Tally F. P., Campanaro E., Eisenstein B. I. (2004) The safety and efficacy of daptomycin for the treatment of complicated skin and skin-structure infections. Clin. Infect. Dis. 38, 1673–1681 [DOI] [PubMed] [Google Scholar]
- 2. Sader H. S., Flamm R. K., Farrell D. J., Jones R. N. (2013) Daptomycin activity against uncommonly isolated streptococcal and other Gram-positive species groups. Antimicrob. Agents Chemother. 57, 6378–6380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Miao V., Coëffet-Legal M. F., Brian P., Brost R., Penn J., Whiting A., Martin S., Ford R., Parr I., Bouchard M., Silva C. J., Wrigley S. K., Baltz R. H. (2005) Daptomycin biosynthesis in Streptomyces roseosporus: cloning and analysis of the gene cluster and revision of peptide stereochemistry. Microbiology 151, 1507–1523 [DOI] [PubMed] [Google Scholar]
- 4. Yu G., Jia X., Wen J., Lu W., Wang G., Caiyin Q., Chen Y. (2011) Strain improvement of Streptomyces roseosporus for daptomycin production by rational screening of He-Ne laser and NTG-induced mutants and kinetic modeling. Appl Biochem. Biotechnol. 163, 729–743 [DOI] [PubMed] [Google Scholar]
- 5. Huang D., Wen J., Wang G., Yu G., Jia X., Chen Y. (2012) In silico aided metabolic engineering of Streptomyces roseosporus for daptomycin yield improvement. Appl. Microbiol. Biotechnol. 94, 637–649 [DOI] [PubMed] [Google Scholar]
- 6. Huang D., Jia X., Wen J., Wang G., Yu G., Caiyin Q., Chen Y. (2011) Metabolic flux analysis and principal nodes identification for daptomycin production improvement by Streptomyces roseosporus. Appl. Biochem. Biotechnol. 165, 1725–1739 [DOI] [PubMed] [Google Scholar]
- 7. Li L., Ma T., Liu Q., Huang Y., Hu C., Liao G. (2013) Improvement of daptomycin production in Streptomyces roseosporus through the acquisition of pleuromutilin resistance. Biomed. Res. Int. 2013, 479742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang L., Zhao Y., Liu Q., Huang Y., Hu C., Liao G. (2012) Improvement of A21978C production in Streptomyces roseosporus by reporter-guided rpsL mutation selection. J. Appl. Microbiol. 112, 1095–1101 [DOI] [PubMed] [Google Scholar]
- 9. Liu G., Chater K. F., Chandra G., Niu G., Tan H. (2013) Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol. Mol. Biol. Rev. 77, 112–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pan Y., Liu G., Yang H., Tian Y., Tan H. (2009) The pleiotropic regulator AdpA-L directly controls the pathway-specific activator of nikkomycin biosynthesis in Streptomyces ansochromogenes. Mol. Microbiol. 72, 710–723 [DOI] [PubMed] [Google Scholar]
- 11. Ohnishi Y., Yamazaki H., Kato J. Y., Tomono A., Horinouchi S. (2005) AdpA, a central transcriptional regulator in the A-factor regulatory cascade that leads to morphological development and secondary metabolism in Streptomyces griseus. Biosci. Biotechnol. Biochem. 69, 431–439 [DOI] [PubMed] [Google Scholar]
- 12. Wolanski M., Donczew R., Kois-Ostrowska A., Masiewicz P., Jakimowicz D., Zakrzewska-Czerwinska J. (2011) The level of AdpA directly affects expression of developmental genes in Streptomyces coelicolor. J. Bacteriol. 193, 6358–6365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lee H. N., Kim J. S., Kim P., Lee H. S., Kim E. S. (2013) Repression of antibiotic downregulator WblA by AdpA in Streptomyces coelicolor. Appl. Environ. Microbiol. 79, 4159–4163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang Y. H., Song E., Kim J. N., Lee B. R., Kim E. J., Park S. H., Kim W. S., Park H. Y., Jeon J. M., Rajesh T., Kim Y. G., Kim B. G. (2012) Characterization of a new ScbR-like γ-butyrolactone binding regulator (SlbR) in Streptomyces coelicolor. Appl. Microbiol. Biotechnol. 96, 113–121 [DOI] [PubMed] [Google Scholar]
- 15. Higo A., Hara H., Horinouchi S., Ohnishi Y. (2012) Genome-wide distribution of AdpA, a global regulator for secondary metabolism and morphological differentiation in Streptomyces, revealed the extent and complexity of the AdpA regulatory network. DNA Res. 19, 259–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yamazaki H., Tomono A., Ohnishi Y., Horinouchi S. (2004) DNA-binding specificity of AdpA, a transcriptional activator in the A-factor regulatory cascade in Streptomyces griseus. Mol. Microbiol. 53, 555–572 [DOI] [PubMed] [Google Scholar]
- 17. Wang F., Ren N. N., Luo S., Chen X. X., Mao X. M., Li Y. Q. (2014) DptR2, a DeoR-type auto-regulator, is required for daptomycin production in Streptomyces roseosporus. Gene 544, 208–215 [DOI] [PubMed] [Google Scholar]
- 18. Uguru G. C., Stephens K. E., Stead J. A., Towle J. E., Baumberg S., McDowall K. J. (2005) Transcriptional activation of the pathway-specific regulator of the actinorhodin biosynthetic genes in Streptomyces coelicolor. Mol. Microbiol. 58, 131–150 [DOI] [PubMed] [Google Scholar]
- 19. Hirano S., Tanaka K., Ohnishi Y., Horinouchi S. (2008) Conditionally positive effect of the TetR-family transcriptional regulator AtrA on streptomycin production by Streptomyces griseus. Microbiology 154, 905–914 [DOI] [PubMed] [Google Scholar]
- 20. Chen L., Lu Y., Chen J., Zhang W., Shu D., Qin Z., Yang S., Jiang W. (2008) Characterization of a negative regulator AveI for avermectin biosynthesis in Streptomyces avermitilis NRRL8165. Appl. Microbiol. Biotechnol. 80, 277–286 [DOI] [PubMed] [Google Scholar]
- 21. Kieser T., Bibb M. J., Butter M. J., Chater K. F., Hopwood D. A. (2000) Practical Streptomyces Genetics, pp. 125–160, John Innes Foundation, Norwich, United Kingdom [Google Scholar]
- 22. Bierman M., Logan R., O'Brien K., Seno E. T., Rao R. N., Schoner B. E. (1992) Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116, 43–49 [DOI] [PubMed] [Google Scholar]
- 23. Jones G. H. (2011) Integrative, xylE-based promoter probe vectors for use in Streptomyces. Plasmid 65, 219–225 [DOI] [PubMed] [Google Scholar]
- 24. Mao X. M., Sun Z. H., Liang B. R., Wang Z. B., Feng W. H., Huang F. L., Li Y. Q. (2013) Positive feedback regulation of stgR expression for secondary metabolism in Streptomyces coelicolor. J. Bacteriol. 195, 2072–2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ingram C., Brawner M., Youngman P., Westpheling J. (1989) xylE functions as an efficient reporter gene in Streptomyces spp: use for the study of galP1, a catabolite-controlled promoter. J. Bacteriol. 171, 6617–6624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ramos J. L., Martínez-Bueno M., Molina-Henares A. J., Terán W., Watanabe K., Zhang X., Gallegos M. T., Brennan R., Tobes R. (2005) The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 69, 326–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Onaka H., Ando N., Nihira T., Yamada Y., Beppu T., Horinouchi S. (1995) Cloning and characterization of the A-factor receptor gene from Streptomyces griseus. J. Bacteriol. 177, 6083–6092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ahn S. K., Cuthbertson L., Nodwell J. R. (2012) Genome context as a predictive tool for identifying regulatory targets of the TetR family transcriptional regulators. PLoS One 7, e50562. [DOI] [PMC free article] [PubMed] [Google Scholar]