Abstract
Chicken immune responses to human proteins are often more robust than rodent responses because of the phylogenetic relationship between the different species. For discovery of a diverse panel of unique therapeutic antibody candidates, chickens therefore represent an attractive host for human-derived targets. Recent advances in monoclonal antibody technology, specifically new methods for the molecular cloning of antibody genes directly from primary B cells, has ushered in a new era of generating monoclonal antibodies from non-traditional host animals that were previously inaccessible through hybridoma technology. However, such monoclonals still require post-discovery humanization in order to be developed as therapeutics. To obviate the need for humanization, a modified strain of chickens could be engineered to express a human-sequence immunoglobulin variable region repertoire. Here, human variable genes introduced into the chicken immunoglobulin loci through gene targeting were evaluated for their ability to be recognized and diversified by the native chicken recombination machinery that is present in the B-lineage cell line DT40. After expansion in culture the DT40 population accumulated genetic mutants that were detected via deep sequencing. Bioinformatic analysis revealed that the human targeted constructs are performing as expected in the cell culture system, and provide a measure of confidence that they will be functional in transgenic animals.
Keywords: gene conversion, somatic hypermutation, DT40, antibody repertoire, chicken B cells, human antibodies, deep sequencing
Introduction
Historically, therapeutic monoclonal antibodies have been derived from immunized mice and phage display technologies. However, antigens that are conserved throughout mammalian evolution are typically weakly or non-antigenic in mice. In some cases, the failure to elicit an immune response in mice has been obviated by immunizing chickens (1–3). Early attempts to use chicken-derived antibodies were thwarted by the lack of technology to derive monoclonal antibodies from non-murine animals. A fusion partner for chicken B cells was identified to create an avian version of the classical murine hybridoma technology (4) although it has not gained wide usage and phage display has been used more frequently to isolate chicken monoclonals (5–11). We developed technology to isolate antigen-specific monoclonal antibodies from immunized chickens using a single lymphocyte screening and recovery method, the gel-encapsulated microenvironment (GEM) assay (see US Patents 8030095 and 841517382). The GEM assay involves placing a single antibody-secreting lymphocyte in proximity with reporters (which can be cells or beads). The secreted antibody diffuses locally within the GEM and has the opportunity to bind to the reporters. Bound antibody can be detected either directly through the use of a secondary antibody or by eliciting a response in the reporter that generates a visual signal. Each GEM may contain multiple types of reporters which can be differentiated from each other based on color. Selected GEMs are isolated and antibody genes are amplified through RT PCR and cloned into a mammalian expression vector, usually in scFv format.
The advantage of producing antibodies to conserved epitopes in chickens prompted the development of humanization protocols to obviate the immune response in patients to the avian V regions of chimeric antibodies (7, 8). An alternative approach to eliminating the anti-animal response in patients is to engineer the animal to produce human immunoglobulins (12). We are currently creating a line of chickens that will produce antibodies with fully human V regions. Human V regions will be recovered from these birds using GEMs. We will then combine the human V segments with human constant regions to produce fully human antibodies with therapeutic potential. The human V region sequences have been designed to replace the equivalent chicken coding regions while leaving most of the endogenous IgH and IgL regulatory sequences intact.
Diversification of chicken immunoglobulin genes is achieved through gene conversion (GC) and somatic hypermutation (SHM) (13). In humans, diversification is achieved through V(D)J recombination and SHM. Because of the phylogenetic distance between humans and chickens and the known differences in the mechanism of diversity generation, it was prudent to evaluate the genetically modified V regions in vitro before investing in the much longer timeline to produce genetically modified birds.
A preliminary evaluation of expression and diversification of human immunoglobulin V regions in DT40 cells was previously reported (14). Briefly, chicken VL and VH loci were knocked out in DT40 and replaced with human VK (VK3-15) and VH (VH3-23) genes. To achieve GC of human genes in chicken B cells, human pseudogene arrays were inserted upstream of the functional human VK and VH regions. The sequences of the VK and VH functional genes served as the starting template for the design of the human pseudogenes. Proper expression of chimeric IgM comprises human variable regions and chicken constant regions were shown. Sanger-based sequencing of selected DT40 genetic variants confirmed that the human pseudogene arrays contributed to the generation of diversity through GC at both the Igl and Igh loci. Although these data showed that engineered pseudogene arrays contribute to human antibody sequences in chicken B cells, a more thorough repertoire analysis was not possible as only a relatively small number of events were analyzed.
Here, we have used next-generation sequencing methods to study much more comprehensively the repertoire generated by a long-term, non-selected culture of DT40 cells harboring targeted human V genes, analyzing well over 1 million sequences for each of the heavy and light chains. We are now able to show that the engineered locus can produce a diverse pool of human antibody sequences in chicken B cells.
Materials and Methods
Culture of chicken DT40 cells carrying human V genes
A derivative of the chicken B cell line DT40 was made in which the chicken immunoglobulin variable regions were replaced with human variable regions in both the IgL and IgH loci (14). In both loci, the active functional allele was targeted, thereby switching the cells from expressing normal chicken surface IgM to the expression of chimeric IgM, consisting of human variable regions and chicken constant regions. A derivative of DT40, cell line 1208-1, was produced by serial transfection with knockout constructs followed by site-specific insertion of constructs for the expression of human V regions. To take advantage of the GC machinery in DT40, upstream arrays of human-sequence pseudogenes were included in the transgenes to provide the donor sequences for mutating the single functional human kappa (HuVK) and human heavy chain (HuVH) regions (Figure 1). Pseudogene arrays were synthesized by Bio Basic (Markham, ON, Canada). These pseudogenes were based on the sequences of the functional HuVK and HuVH regions, with diversity incorporated into the complementarity determining regions (CDR), and in some cases, the framework regions as well (Figure 2). The pseudogenes were thus designed de novo and not based on the endogenous pseudogenes found in the human genomic heavy and light chain loci. We refer to the HuVK pseudogenes as the SynVK array and the HuVH pseudogenes as the SynVH array. Diversity in the SynVK array was derived from human EST sequences, whereas the SynVH array was made by scanning substitution of CDR positions with tyrosine, tryptophan, or serine residues. Furthermore, additional AID hotspots (nucleotides WRC/GYW) were incorporated into the SynVK-C construct, as silent changes. In the 1208-1 cell line, construct SynVH-B was inserted at the heavy chain locus, followed by insertion of the SynVK-C construct at the light chain locus. The sequences of the pseudogene arrays are shown in Figure 2.
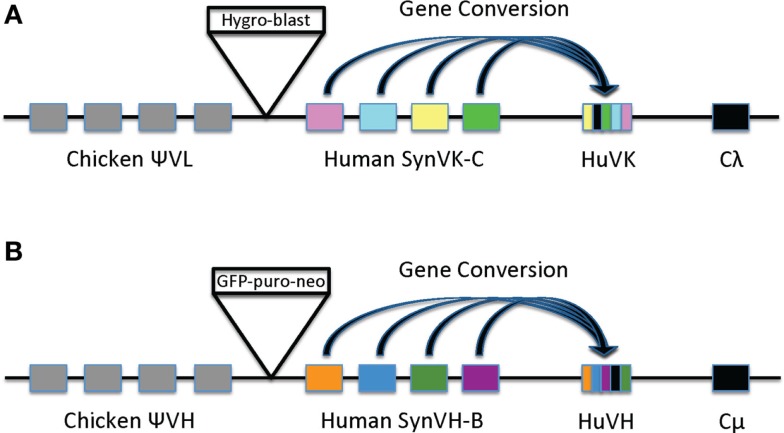
Figure 1.
Diagrams of light chain (A) and heavy chain (B) loci in cell line 1208-1. (A) In the light chain, the endogenous rearranged chicken VL and its promoter in DT40 was replaced by an array of human SynVK-C pseudogenes and a rearranged functional HuVK gene driven by the chicken VL promoter. The chicken Ψ VL pseudogene array, constant region (Cλ), J–C intron, and 3′ flanking DNA are intact. A β-actin-hygromycin, β-actin- blasticidin resistance cassette (box labeled Hygro-blast) was placed between the chicken and human pseudogene arrays as part of the transfection process. (B) In the heavy chain, the endogenous rearranged chicken VH and 350 bp of its promoter region were replaced by the SynVH-B human pseudogene array, the chicken VH promoter, and a rearranged functional human VH gene. The upstream chicken Ψ VH pseudogene array, the chicken JH-Cμ intron, and constant regions are intact. A β-actin-EGFP, β-actin-puromycin, β-actin-neomycin selectable marker cassette (box labeled GFP-puro-neo) was placed between the chicken and human pseudogene arrays as part of the transfection process. Gene conversion in both heavy and light chains is depicted as blocks of sequences (colored blocks) being transferred from the pseudogenes to the HuVK and HuVH functional genes.
Figure 2.
Human light and heavy chain pseudogene sequences. (A) Alignment of SynVK pseudogenes in the SynVK-C construct. Top line shows the sequence of the functional HuVK gene that is mutated by the SynVK pseudogenes. The CDRs (boxes; IMGT nomenclature) were derived from human EST databases. Some pseudogenes also contain framework changes derived from the ESTs. At the 3′ end of the pseudogenes, the sequence of the flanking DNA downstream of CDR3 in each pseudogene is shown. This flanking sequence is part of the 100 bp spacer sequence inserted between each pair of pseudogenes. (B) Alignment of the SynVH pseudogenes in the SynVH-B construct. Top line shows the sequence of the functional HuVH gene. The CDRs consist of a tyrosine/tryptophan/serine scan. The framework regions contain no changes.
The 1208-1 cell line was propagated for 10 weeks with both SynVK-C and SynVH-B transgenes to allow mutations to accumulate prior to harvesting genomic DNA for sequencing (additionally, the precursor cell line carrying only the SynVH-B construct was cultured for 3 weeks before transfection of the SynVK-C construct). The culture was expanded to 1.85e8 cells and gDNA was purified by Qiagen DNeasy kit.
Generation of amplicons for sequencing
Purified gDNA from the 1208-1 DT40 cell line was sent for further processing to Genewiz, Inc. The HuVK and HuVH regions were amplified using the primers in Table 1. Amplicons were sequenced by Genewiz, Inc. (South Plainfield, NJ, USA) on the Illumina MiSeq 2x250 platform (Illumina, Inc., San Diego, CA, USA). Raw data files are available online at the NCBI sequence read archive (SRA), project PRJNA275158, accession number SRP055184.
Table 1.
Primers for amplification of HuVK and HuVH sequences from gDNA from 1208-1 cells.
| For HuVH amplicon | Sequence |
|---|---|
| HuVHampf1 (forward) | TCCTTCCCCACAGGTGTC |
| HuVHampr1 (reverse) | GGTTGAAGACTCACCTGAG |
| For HuVK amplicon | |
| HuVKampf1 (forward) | CAGACTGCACCGGAGAAA |
| HuVKampr1 (reverse) | GTCAGCGACTCACGTTTG |
Sequence data analysis
High throughput sequencing reads were analyzed using VDJFasta (http://www.distributedbio.com/vdjfastadocs/), a general antibody repertoire algorithm suitable for interpretation of engineered antibody diversity. In order to control for possible residual chicken content, we used a combination natural chicken and human synthetic segment classification database. In order to analyze the repertoire comprehensively in a manner unbiased by the underlying GC mechanics, we used general profile Hidden–Markov models to identify immunoglobulin content and align sequences in a consistent manner independent of nucleotide composition. Kabat positional annotations were transferred from Hidden–Markov model columns to every aligned sequence in the resulting database, enabling consistent annotation of frameworks and CDR boundaries (15–17).
Results
Read quality analysis
Analysis was performed by processing all reads through VDJFasta. Sequences were assigned closest segments with a probabilistic classifier. All reads were translated into six-frames of translation and analyzed for Ig content by profile-Hidden–Markov model scoring with VDJFasta, with a 1e-10 cutoff for significance. Pass-cutoff frames were aligned using the pHMMs and analyzed for framestate and coverage. Over 96% of reads contained full-length clones, with over 1 million HuVH reads and 2 million HuVK reads available for downstream analysis (Table 2).
Table 2.
Analysis of on-target proportions in data.
| HuVH |
HuVK |
|||
|---|---|---|---|---|
| Reads | Proportion (%) | Reads | Proportion (%) | |
| Total | 1117578 | 100.0 | 2066629 | 100.0 |
| wV | 1115812 | 99.8 | 2029766 | 98.2 |
| wJ | 1093284 | 97.8 | 2014189 | 97.5 |
| wVJ | 1093270 | 97.8 | 2014173 | 97.5 |
| wVJclone | 1074927 | 96.2 | 2002661 | 96.9 |
wV, contains detectable V-segment; wJ, contains detectable J-segment; wVJ, contains both a detectable V and J segment; wVJclone, contains a detectable V and J segment, and a CDR3 that remains in-frame.
Comparison to inserted arrays of V genes
Nucleotide and translated CDRs were extracted from profile Hidden–Markov model alignments, using minimum profile annealing cutoffs to ensure high fidelity CDR capture [see Ref. (15)]. CDRs were compared to a reference database containing all V genes and pseudogenes that were included in the targeted array. Counts of exact match to reference database were stored for all CDRs.
Sequence complexity
The extracted sequences were highly redundant in both the heavy chain and light chain data sets, with the single functional human V gene being seen predominately in its respective group. The non-mutated VK represented 81% of all the full-length sequence reads and the non-mutated VH 57% of the total. These non-mutated sequences are referred to as the “reference” sequences (one for VH and one for VK). For the heavy chain, 9125 unique clones were found at a minimum 2× sequence depth; for the light chain, 7671 unique clones were found. If the sequences are counted at a 1× sequence depth a total of 21,403 unique heavy chains and 33,848 unique light chain genes were seen.
Identifying gene conversion and somatic hypermutation events
Each framework and CDR was analyzed separately, with an exact-match assignment performed to reference synthetic human frameworks designed into the transgenic organism. A control search was also performed with all known native IgL and IgH chicken segments, but they were never encountered in the repertoire. GC events were scored if a sequence found in the VK (Table 3) or VH (Table 4) pool could be traced back to particular pseudogenes present in the array. In the cases where gene converted sequences are shared by multiple pseudogenes, one GC event is counted and all possible donor pseudogenes are indicated. Individual GC events were counted at the 1× sequence depth since it is unlikely that stretches of nucleotides would occur through sequencing or read error. Regardless, all CDR events classified as GC occurred more than twice in the data set. Within each CDR a high proportion of the unique sequences matched perfectly with the reference sequence. Those that deviated from the reference sequence in ways that could not be clearly attributed to GC are labeled as “SHM or fusion” and this category includes single or multiple point mutations as well as possible complex events (i.e., multiple sequential GC). Sequencing errors would be expected to show up in this category, possibly inflating the observed events.
Table 3.
Number of unique SHM and gene conversion sequences within CDRs for light chain.
| CDR1 | Events | CDR2 | Events | CDR3 | Events |
|---|---|---|---|---|---|
| SynVK_ Reference | 28735 | SynVK_ Reference | 17179 | SynVK_ Reference | 13995 |
| SHM or fusion | 3853 | SHM or fusion | 15340 | SHM or fusion | 22630 |
| SynVK20 | 20 | SynVK20/21 | 160 | SynVK23_ CDR3 | 87 |
| SynVK21 | 44 | SynVK22 | 76 | Chicken pseudo | 0 |
| SynVK22 | 39 | SynVK24 | 21 | ||
| SynVK23 | 219 | SynVK26 | 83 | ||
| SynVK24 | 195 | SynVK27 | 11 | ||
| SynVK25 | 121 | SynVK28/29 | 203 | ||
| SynVK26 | 52 | SynVK30/31 | 284 | ||
| SynVK27 | 44 | SynVK32 | 204 | ||
| SynVK28 | 30 | SynVK33 | 129 | ||
| SynVK29 | 49 | SynVK34 | 88 | ||
| SynVK30 | 314 | SynVK35 | 70 | ||
| SynVK31 | 39 | Chicken pseudo | 0 | ||
| SynVK32 | 4 | ||||
| SynVK33 | 15 | ||||
| SynVK34 | 27 | ||||
| SynVK35 | 48 | ||||
| Chicken pseudo | 0 | ||||
| Sum GC | 1260 | Sum GC | 1329 | Sum GC | 87 |
| Total | 33848 | Total | 33848 | Total | 36712a |
aIncludes a small proportion of non-full-length reads.
Table 4.
Number of unique SHM and gene conversion sequences within CDRs for heavy chain.
| CDR1 | Events | CDR2 | Events | CDR3 | Events |
|---|---|---|---|---|---|
| SynVH_ Reference | 19440 | SynVH_ Reference | 5600 | SynVH_ Reference | 19379 |
| SHM or fusion | 647 | SHM or fusion | 15096 | SHM or fusion | 1830 |
| SynVH_21 | 185 | SynVH_21 | 111 | SynVH_22 | 13 |
| SynVH_22 | 120 | SynVH_22 | 61 | SynVH_23 | 11 |
| SynVH_23 | 39 | SynVH_23 | 138 | SynVH_24 | 32 |
| SynVH_24 | 160 | SynVH_24 | 3 | SynVH_25 | 21 |
| SynVH_25 | 96 | SynVH_25 | 15 | SynVH_26 | 20 |
| SynVH_26 | 39 | SynVH_26 | 149 | SynVH_27 | 9 |
| SynVH_28 | 79 | SynVH_27 | 22 | SynVH_28 | 3 |
| SynVH_29 | 38 | SynVH_28 | 28 | SynVH_30 | 4 |
| SynVH_30 | 37 | SynVH_29 | 66 | SynVH_31 | 20 |
| SynVH_31 | 102 | SynVH_30 | 12 | SynVH_32 | 18 |
| SynVH_32 | 101 | SynVH_31 | 19 | SynVH_37 | 43 |
| SynVH_33 | 17 | SynVH_32 | 4 | Chicken pseudo | 0 |
| SynVH_34 | 116 | SynVH_33 | 5 | ||
| SynVH_35 | 5 | SynVH_34 | 48 | ||
| SynVH_36 | 169 | SynVH_35 | 11 | ||
| SynVH_37 | 13 | SynVH_36 | 15 | ||
| Chicken pseudo | 0 | Chicken pseudo | 0 | ||
| Sum GC | 1316 | Sum GC | 707 | Sum GC | 194 |
| Total | 21403 | Total | 21403 | Total | 21403 |
Evaluation of potential contribution of endogenous chicken pseudogenes
The IgL and IgH knockouts were made by deleting portions of the functional VJC and VDJ regions, respectively, by homologous recombination. Since endogenous chicken pseudogenes remain upstream of the inserted human V gene array, they could in principle contribute to repertoire diversity. We specifically checked for such events by creating a library of all known chicken pseudogenes and running the analysis as we did with the library containing our human pseudogenes. Evidence of endogenous pseudogenes participating in GC events was never observed.
Gene conversion and SHM in framework regions
Since some diversity was incorporated into the frameworks of the inserted VK pseudogenes, it was possible to identify GC events as well as SHM events in these regions (Table 5). Fewer GC attributable sequences were found in the VK frameworks as compared to the VK CDRs; however, this may be due simply to the lower framework diversity that was incorporated in the pseudogene design.
Table 5.
Number of unique SHM and gene conversion sequences within VK framework regions.
| FW1 | Events | FW2 | Events | FW3 | Events |
|---|---|---|---|---|---|
| SynVK_ Reference | 26576 | SynVK_ Reference | 17114 | SynVK_ Reference | 22447 |
| SHM or fusion | 7163 | SHM or fusion | 16405 | SHM or fusion | 11235 |
| SynVK22 | 43 | SynVK24 | 94 | SynVK23 | 144 |
| SynVK25 | 41 | SynVK27 | 32 | SynVK25 | 11 |
| SynVK26 | 13 | SynVK28 | 54 | SynVK27 | 10 |
| SynVK27 | 8 | SynVK29 | 24 | SynVK30 | 1 |
| SynVK30 | 1 | SynVK33 | 49 | ||
| SynVK31 | 1 | SynVK34 | 76 | ||
| SynVK33 | 2 | ||||
| Sum GC | 109 | Sum GC | 329 | Sum GC | 166 |
| Total | 33848 | Total | 33848 | Total | 33848 |
Identification of multiple gene conversion events
In some cases, we were able to identify sequences with contributions from two different pseudogenes, and these are termed paired-fusion events (Tables 6–8). Partial GC was analyzed using the parsimonious assumption of single-conversion events within the CDR as a source of non-100% identity match to the SynVH pseudogene segment reference database. Custom software was written to generate all non-redundant fusion events that can emerge between pairwise interactions of the reference database sequences. Paired-fusion events are highly biased in their relative occurrence, as an analysis of the most commonly encountered rearrangements demonstrates. Paired-fusion events were not feasible to determine for SynVK due to the sequence complexity inherent in this array.
Table 6.
Top 10 most common HuVH CDR-H1 paired-fusion events.
| FREQ | |
| 67 | SynVH_22_CDR_1 invades > SynVH_21_CDR_1 at position 2 |
| 21 | SynVH_25_CDR_1 invades > SynVH_21_CDR_1 at position 13 |
| 17 | SynVH_21_CDR_1 invades > SynVH_28_CDR_1 at position 3 |
| 13 | SynVH_22_CDR_1 invades > SynVH_29_CDR_1 at position 1 |
| 12 | SynVH_27ref_CDR_1 invades > SynVH_36_CDR_1 at position 22 |
| 11 | SynVH_21_CDR_1 invades > SynVH_27ref_CDR_1 at position 1 |
| 10 | SynVH_25_CDR_1 invades > SynVH_32_CDR_1 at position 11 |
| 8 | SynVH_23_CDR_1 invades > SynVH_21_CDR_1 at position 7 |
| 7 | SynVH_27ref_CDR_1 invades > SynVH_37_CDR_1 at position 20 |
| 6 | SynVH_36_CDR_1 invades > SynVH_21_CDR_1 at position 23 |
Table 8.
Top 10 most common HuVH CDR-H3 paired-fusion events.
| FREQ | |
| 204 | SynVH_21_CDR_3 invades > SynVH_35_CDR_3 at position 20 |
| 153 | SynVH_21_CDR_3 invades > SynVH_24_CDR_3 at position 10 |
| 39 | SynVH_21_CDR_3 invades > SynVH_36_CDR_3 at position 23 |
| 30 | SynVH_21_CDR_3 invades > SynVH_27_CDR_3 at position 19 |
| 21 | SynVH_37_CDR_3 invades > SynVH_21_CDR_3 at position 14 |
| 13 | SynVH_27_CDR_3 invades > SynVH_21_CDR_3 at position 19 |
| 8 | SynVH_24_CDR_3 invades > SynVH_21_CDR_3 at position 10 |
| 7 | SynVH_36_CDR_3 invades > SynVH_21_CDR_3 at position 23 |
| 7 | SynVH_22_CDR_3 invades > SynVH_21_CDR_3 at position 4 |
| 6 | SynVH_25_CDR_3 invades > SynVH_21_CDR_3 at position 14 |
Table 7.
Top 10 most common HuVH CDR-H2 paired-fusion events.
| FREQ | |
| 4304 | SynVH_26_CDR_2 invades > SynVH_29_CDR_2 at position 13 |
| 1381 | SynVH_21_CDR_2 invades > SynVH_32_CDR_2 at position 10 |
| 562 | SynVH_26_CDR_2 invades > SynVH_29_CDR_2 at position 13 |
| 318 | SynVH_21_CDR_2 invades > SynVH_30_CDR_2 at position 4 |
| 185 | SynVH_28_CDR_2 invades > SynVH_27_CDR_2 at position 19 |
| 115 | SynVH_21_CDR_2 invades > SynVH_32_CDR_2 at position 10 |
| 80 | SynVH_21_CDR_2 invades > SynVH_31_CDR_2 at position 7 |
| 78 | SynVH_23_CDR_2 invades > SynVH_29_CDR_2 at position 13 |
| 49 | SynVH_22_CDR_2 invades > SynVH_29_CDR_2 at position 13 |
| 41 | SynVH_CDR_2 invades > SynVH_21_CDR_2 at position 23 |
Positional variation profiling of the repertoire
Analysis of positional amino acid variation was performed by converting a total alignment of non-redundant amino acid sequences into a positional weight matrix (PWM), with reference residue frequency omitted to emphasize non-reference residue variation (Figure 3). The number of amino acids observed at each position cannot be attributed to GC events including both complete replacements and single paired fusions. These observations suggest that the diversity generated by GC is augmented by SHM.
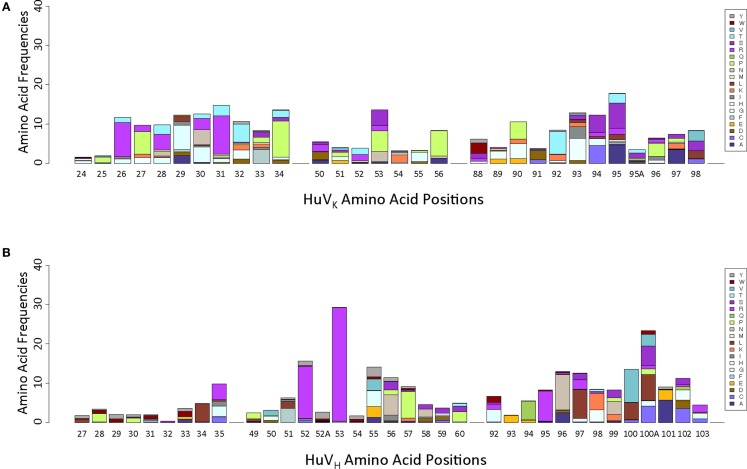
Figure 3.
Positional weight matrices of non-reference residue variation, by Kabat position. (A) HuVK variation, indicated positionally. (B) HuVH variation, indicated positionally.
Discussion
The DT40 cell line has been used extensively to better understand the nature of immunoglobulin diversification in chickens, including the mechanism of GC (18–21). The cell line has also been used to develop chicken antibodies to novel targets using in vitro selection strategies (22, 23). We have inserted human V gene arrays into the chicken immunoglobulin loci of DT40. In principle, the human V genes in DT40 cells could be diversified in vitro to provide an unselected library of immunoglobulin sequences from which antigen-specific antibodies could be extracted. However, most therapeutic antibodies are derived from immunized animals producing affinity-matured, antigen-specific antibodies. In the current context, we have used DT40 cells to provide in vitro proof of concept that arrays of human-derived immunoglobulin gene sequences can be diversified by chicken B cells. Subsequently, these sequences will be introduced into chickens to provide genetically engineered animals that can be immunized to produce affinity-matured, antigen-specific antibodies with therapeutic potential. Thus, our purpose with DT40 is to determine whether targeted synthetic human V gene arrays can be used as a substrate for genetic diversification in chicken cells in a way that mirrors what is known regarding the native chicken immunoglobulin loci. Affirmative data in the DT40 culture system inspires confidence that the effort required on the arduous path to generating a genetically engineered chicken will be rewarded with a transgenic animal that performs as expected.
We have previously shown that our heavy and light chain arrays can be diversified by both GC and SHM in DT40 cells (14). This analysis involved conventional sequencing of a few hundred clones sampled from a large population of DT40 cells. Some examples of expected diversification events were seen, but most events were likely missed at that depth. Next generation sequence technology allows for identification of very rare events in a non-selected, non-biased cellular population. Indeed, we were able to show in the current work that every pseudogene in our array was used by some cell in the population. The finding of paired-fusion events, wherein GC occurs using two different pseudogenes in succession is expected in a fully functional locus. It has been estimated that wild-type chicken B cells undergo 1–2 independent GC events on average during affinity maturation (24).
We were also able to confirm our previous conclusion that for both heavy and light chains, GC is more prevalent in CDR1 and CDR2 than CDR3, which is heavily skewed toward SHM. This finding is also consistent with previously published results (25). Nonetheless, a mix of both templated and non-templated mutations is seen in all CDRs, resulting in a repertoire with amino acid diversity at every CDR position. It will be interesting to see if a similar bias can be seen in the SynV chicken, in which processes of cellular selection may affect the repertoire in a way that is not seen in DT40, which has no selection pressure for surface Ig expression or specificity.
While it is reasonable to use deep sequencing on our DT40 population to determine whether certain types of events have occurred, caution should be used in interpreting the observed frequency results because of the nature of the long-term DT40 culture system. In such a system, clones with particular sequences could have a growth advantage, or alternatively, a particular mutation could occur very early in the expansion of the culture and then subsequent mutations could occur in addition, potentially creating a large number of “unique clones,” which carry the original mutation. For instance, in our VHCDR2 data, we find a very large number of clones bearing a S53R substitution (Figure 3). The high frequency observed could be the result of a true mutational hotspot that mutated many times independently, or simply the result of a single random mutation that occurred early in the expansion of the population. One sequence bearing this substitution is highly redundant in our sample, second only to the starting sequence; this is consistent with the existence of a large subpopulation wherein secondary mutations could have occurred. Further, if S53R is an aberration, it skewed our data to make it appear that SHM in HC CDR2 is extremely high relative to GC, which may not be the correct interpretation.
In summary, the DT40 culture system, coupled with deep sequencing methodologies, is an excellent tool for the functional testing of arrays of synthetic human V genes designed to be diversified and affinity-matured in vivo. Our deep sequencing results confirm that arrays of human V genes can be targeted into the immunoglobulin loci of chicken cells and the host machinery can diversify those genes over time in a manner that recapitulates in vivo GC in the B cells of wild-type chickens. Furthermore, when rare events are included in the analysis, it is clear that even in a relatively small population of cells, all of the introduced pseudogenes are capable of participating in GC, and that codons for non-templated amino acid residues are generated through SHM in every CDR of both light chain and heavy chain. These data support the concept of introducing constructs containing all necessary genetic elements required for diversification in the B cell compartment, contributing to a functionally diverse repertoire of human-sequence antibodies in a transgenic chicken. Once made, this bird will be the most evolutionarily divergent host of any human-Ig transgenic animal currently available, and will be particularly well suited to generating novel antibodies to therapeutic targets that are conserved among mammals.
Conflict of Interest Statement
Crystal Bioscience has an issued patent (US 8592644) regarding the production of human antibodies in transgenic chickens.
References
- 1.Carroll S, Stollar B. Antibodies to calf thymus RNA polymerase II from egg yolks of immunized hens. J Biol Chem (1983) 258:24–6. [PubMed] [Google Scholar]
- 2.Gassmann M, Thommes P, Weiser T, Hubscher U. Efficient production of chicken egg yolk antibodies against a conserved mammalian protein. FASEB J (1990) 4:2528–32. [DOI] [PubMed] [Google Scholar]
- 3.Camenisch G, Tini M, Chilov D, Kvietikova I, Srinivas V, Caro J, et al. General applicability of chicken egg yolk antibodies: the performance of IgY immunoglobulins raised against the hypoxia-inducible factor 1alpha. FASEB J (1999) 13:81–8. [DOI] [PubMed] [Google Scholar]
- 4.Nishinaka S, Suzuki T, Matsuda H, Murata M. A new cell line for the production of chicken monoclonal antibody by hybridoma technology. J Immunol Methods (1991) 139:217–22. 10.1016/0022-1759(91)90191-H [DOI] [PubMed] [Google Scholar]
- 5.Nakamura N, Shimokawa M, Miyamoto K, Hojyo S, Horiuchi H, Furusawa S, et al. Two expression vectors for the phage-displayed chicken monoclonal antibody. J Immunol Methods (2003) 280:157–64. 10.1016/S0022-1759(03)00204-7 [DOI] [PubMed] [Google Scholar]
- 6.van Wyngaardt W, Malatji T, Mashau C, Fehrsen J, Jordaan F, Miltiadou D, et al. A large semi-synthetic single-chain Fv phage display library based on chicken immunoglobulin genes. BMC Biotechnol (2004) 4:6. 10.1186/1472-6750-4-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsurushita N, Park M, Pakabunto K, Ong K, Avdalovic A, Fu H, et al. Humanization of a chicken anti-IL-12 monoclonal antibody. J Immunol Methods (2004) 295:9–19. 10.1016/j.jim.2004.08.018 [DOI] [PubMed] [Google Scholar]
- 8.Nishibori N, Horiuchi H, Furusawa S, Matsuda H. Humanization of chicken monoclonal antibody using phage-display system. Mol Immunol (2006) 43:634–42. 10.1016/j.molimm.2005.04.002 [DOI] [PubMed] [Google Scholar]
- 9.Miyamoto K, Kimura S, Nakamura N, Yokoyama T, Horiuchi H, Furusawa S, et al. Chicken antibody against a restrictive epitope of prion protein distinguishes normal and abnormal prion proteins. Biologicals (2007) 35:303–8. 10.1016/j.biologicals.2007.01.007 [DOI] [PubMed] [Google Scholar]
- 10.Bowes T, Hanley SA, Liew A, Eglon M, Mashayekhi K, O’Kennedy R, et al. Developing cell-specific antibodies to endothelial progenitor cells using avian immune phage display technology. J Biomol Screen (2011) 16:744–54. 10.1177/1087057111407067 [DOI] [PubMed] [Google Scholar]
- 11.Wu L, Oficjalska K, Lambert M, Fennell BJ, Darmanin-Sheehan A, Ní Shúilleabháin D, et al. Fundamental characteristics of the immunoglobulin VH repertoire of chickens in comparison with those of humans, mice, and camelids. J Immunol (2012) 188:322–33. 10.4049/jimmunol.1102466 [DOI] [PubMed] [Google Scholar]
- 12.Lonberg N. Human antibodies from transgenic animals. Nat Biotechnol (2005) 23:1117–25 10.1038/nbt1135 [DOI] [PubMed] [Google Scholar]
- 13.Ratcliffe M. Antibodies, immunoglobulin genes and the bursa of Fabricius in chicken B cell development. Dev Comp Immunol (2006) 30:101–18. 10.1016/j.dci.2005.06.018 [DOI] [PubMed] [Google Scholar]
- 14.Schusser B, Yi H, Collarini EJ, Izquierdo SM, Harriman WD, Etches RJ, et al. Harnessing gene conversion in chicken B cells to create a human antibody sequence repertoire. PLoS One (2013) 8:e80108. 10.1371/journal.pone.0080108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glanville J, Zhai W, Berka J, Telman D, Huerta G, Mehta GR, et al. Precise determination of the diversity of a combinatorial antibody library gives insight into the human immunoglobulin repertoire. Proc Natl Acad Sci U S A (2009) 106:20216–21. 10.1073/pnas.0909775106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhai W, Glanville J, Fuhrmann M, Mei L, Ni I, Sundar PD, et al. Synthetic antibodies designed on natural sequence landscapes. J Mol Biol (2011) 412:55–71. 10.1016/j.jmb.2011.07.018 [DOI] [PubMed] [Google Scholar]
- 17.Glanville J, Kuo TC, von Büdingen H-C, Guey L, Berka J, Sundar PD, et al. Naive antibody gene-segment frequencies are heritable and unaltered by chronic lymphocyte ablation. Proc Natl Acad Sci U S A (2011) 108:20066–71. 10.1073/pnas.1107498108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buerstedde J, Reynaud C, Humphries E, Olson W, Ewert D, Weill J. Light chain gene conversion continues at high rate in an ALV-induced cell line. EMBO J (1990) 9:921–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harris R, Sale J, Petersen-Mahrt S, Neuberger M. AID is essential for immunoglobulin V gene conversion in a cultured B cell line. Curr Biol (2002) 12:435–8. 10.1016/S0960-9822(02)00717-0 [DOI] [PubMed] [Google Scholar]
- 20.Arakawa H, Hauschild J, Buerstedde J. Requirement of the activation-induced deaminase (AID) gene for immunoglobulin gene conversion. Science (2002) 295:1301–6. 10.1126/science.1067308 [DOI] [PubMed] [Google Scholar]
- 21.Ordinario E, Yabuki M, Larson R. Temporal regulation of Ig gene diversification revealed by single-cell imaging. J Immunol (2009) 183(7):4545–53. 10.4049/jimmunol.0900673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seo H, Hashimoto S, Tsuchiya K, Lin W, Shibata T, Ohta K. An ex vivo method for rapid generation of monoclonal antibodies (ADLib system). Nat Protoc (2006) 1:1502–6. 10.1038/nprot.2006.248 [DOI] [PubMed] [Google Scholar]
- 23.Todo K, Miyake K, Magari M, Kanayama N, Ohmori H. Novel in vitro screening system for monoclonal antibodies using hypermutating chicken B cell library. J Biosci Bioeng (2006) 102:478–81. 10.1263/jbb.102.478 [DOI] [PubMed] [Google Scholar]
- 24.Arakawa H, Furusawa S, Ekino S, Yamagishi H. Immunoglobulin gene hyperconversion ongoing in chicken splenic germinal centers. EMBO J (1996) 15:2540–6. [PMC free article] [PubMed] [Google Scholar]
- 25.McCormack WT, Thompson CB. Chicken IgL variable region gene conversions display pseudogene donor preference and 5” to 3” polarity. Genes Dev (1990) 4:548–58. 10.1101/gad.4.4.548 [DOI] [PubMed] [Google Scholar]