Abstract
In collective resistance, microbial communities are able to survive antibiotic exposures that would be lethal to individual cells. In this review, we explore recent advances in understanding collective resistance in bacteria. The population dynamics of “cheating” in a system with cooperative antibiotic inactivation have been described, providing insight into the demographic factors that determine resistance allele frequency in bacteria. Extensive work has elucidated mechanisms underlying collective resistance in biofilms and addressed questions about the role of cooperation in these structures. Additionally, recent investigations of “bet-hedging” strategies in bacteria have explored the contributions of stochasticity and regulation to bacterial phenotypic heterogeneity and examined the effects of these strategies on community survival.
Introduction
The mechanisms that confer antibiotic resistance in microbes are well understood on the level of the individual cell [1]. However, it is increasingly apparent that the social dynamics of microbes can affect the outcome of antimicrobial chemotherapy, and there has been considerable recent progress in identifying the population-level processes that affect antibiotic response.
In this review, we focus on how a community of bacteria can survive antimicrobial therapy where a single bacterium cannot. For the purposes of this review, we define a microbial “community” as a collection of microorganisms within which different phenotypes and/or genotypes coexist. We discuss recent work in collective resistance in bacteria via cooperative inactivation of antibiotics, where population dynamics within communities are shown to play a role in determining the prevalence of antibiotic resistance genes. Further, we review recent advances in biofilm ecology that demonstrate the importance of spatial structure and cooperation for antibiotic resistance in these structures. Finally, we consider current theories of stochastic and responsive phenotypic diversity, with a focus on signaling within communities and interactions between communities and their environment as factors controlling the emergence of antibiotic-tolerant phenotypes. For each of these topics, we explore the implications for antimicrobial chemotherapy.
Cooperative Resistance
Dense populations of bacteria resist eradication by drug concentrations considerably greater than those required to kill the same population at a lower density [2]in a phenomenon known as the “inoculum effect”. One mechanism for density-dependent antibiotic efficacy is drug inactivation by bacterially expressed resistance enzymes (Figure 1a,b).. The population’s collective capacity to inactivate the drug depends upon several factors, including the number of cells expressing the enzyme conferring resistance. Cooperative resistance is therefore expected to be less effective at small initial population sizes, resulting in separation between the minimum antibiotic concentration required to affect single cells and the canonical MIC (Figure 1c), which is typically measured using relatively high inocula (~106 CFU/mL) [3] [T. Artemova, unpublished].
Figure 1. Enzymatic inactivation of antibiotics allows cooperative resistance.
(a) When a resistance enzyme (blue) is expressed (from a resistance plasmid, red), the amount of antibiotic (green squares) inside the cell is determined by the balance between flux and inactivation (inactivated antibiotic, green 3/4 square). If the intracellular concentration is below some critical threshold at a given external concentration of drug, the cells will survive and grow; otherwise, the cells will die. (b) The outcome of antibiotic treatment depends on cell density and resistance enzyme production. Starting resistant cell density determines whether the population will be able to inactivate enough of the drug to allow re-growth before it is driven to extinction. (c) This results in density-dependent changes in the external concentration of drug that is sufficient to prevent re-growth (called the minimum inhibitory concentration or MIC), characteristic of the inoculum effect. (d) Cooperative inactivation can protect sensitive cells which do not carry the resistance gene, and the population structure after re-growth may be very different from the starting structure due to fitness differences between sensitive cells and cells expressing the resistance enzyme.
Enzymatic inactivation of antibiotic can lower the environmental antibiotic concentration, allowing sensitive cells in that environment to survive (Figure 1d). Indeed, cooperative inactivation of β-lactam antibiotics and protection of sensitive cells occurs with some regularity in natural microbial communities [4]. In these communities, antibiotic-sensitive bacteria can be regarded as “cheaters” which benefit from resistance without incurring the metabolic cost of enzyme production.
Experimental evidence from synthetic microbial systems has demonstrated that, in a community where cooperative resistance alleles exist prior to antibiotic treatment, cheating can drive the population dynamics of the resistance allele [3]. Antibiotic inactivation may be thought of as a common goods problem, where the dynamics of these mixed populations depend on whether resistance enzyme production has a fitness cost. When resistance carries a cost, the system is vulnerable to cheating - sensitive bacteria may grow more quickly than resistant cells once the antibiotic concentration drops beneath their MIC [5], resulting in a decrease in the proportion of resistant cells [3]. However, evolution of compensatory mechanisms can occur rapidly, minimizing the cost of resistance and stabilizing the resistance allele [3,6].
Collective resistance can occur when cells do not express antibiotic-inactivating enzymes. For example, a recent study has demonstrated a mechanism for inoculum effects during aminoglycoside treatment of sensitive Escherichia coli; here, the amount of antibiotic per cell determines whether individual cells grow or die, resulting in density-dependent bistability in the outcome of antibiotic treatment [2]. In this case the cells are acting as “sinks” for the antibiotic, resulting in effective cooperation by load-sharing between cells in the population. Collective resistance in dense bacterial populations is therefore likely to be a ubiquitous feature that will require new measurements, analyses, and treatments.
Biofilm-Associated Resistance
Density-dependent collective resistance is also characteristic of structured environments such as surface-associated biofilms. These structures are ubiquitous in nature, probably representing the most common structured bacterial environment in the natural world, and are notoriously difficult to eradicate in settings from industry to the clinic [7]. Bacteria within biofilms can show remarkable decreases in susceptibility to antibiotics and other toxins, resisting drug concentrations much higher than those required to kill free-living bacteria at comparable densities [8]. Together with cell density, spatial organization and the physical structure itself can all contribute to resistance.
The structure and composition of the biofilm matrix can contribute to resistance. Exopolysaccharide and extracellular DNA in the biofilm matrix can act as a barrier to diffusion, preventing drugs from reaching living cells (Figure 2a) [9]. The effectiveness of this barrier varies between antibiotics – large molecules, positively charged aminoglycosides, and antimicrobial peptides diffuse poorly in biofilms, but quinolones and β-lactams appear to move freely [10,11].
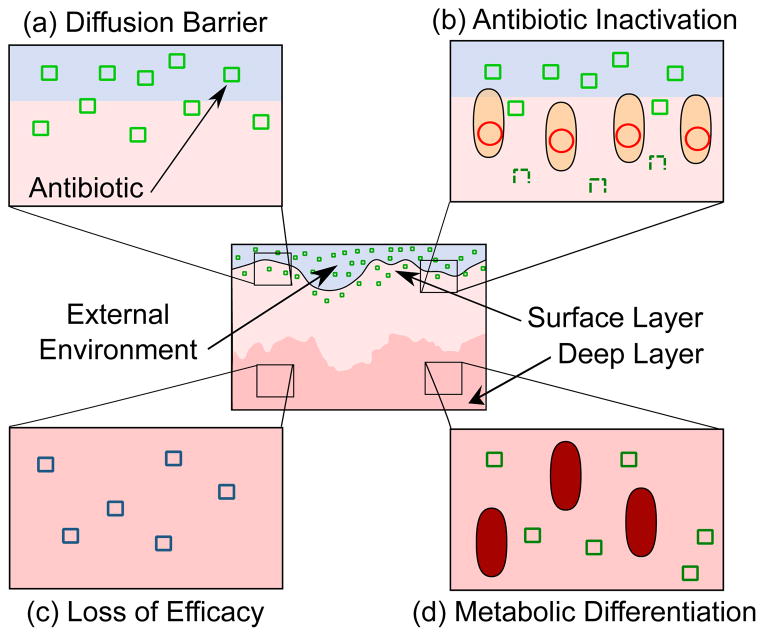
Figure 2. Mechanisms of collective resistance in biofilms.
Within the static structure of a biofilm, multiple mechanisms may contribute to community resistance. (a) The matrix itself may act as a diffusion barrier, preventing the drug (green squares) from reaching its target. (b) Enzymatic inactivation by bacteria near the surface of the film (white layer) can protect sensitive bacteria deeper within the structure. In the deep layers of a biofilm (dark pink layer), diffusion gradients will produce a hostile environment where (c) the drug itself may lose efficacy (blue squares) and (d) the member bacteria of the biofilm may enter altered metabolic states (red cells) inimical to antibiotic action.
For antibiotics that can penetrate the matrix, inactivation by resistance enzymes can produce collective resistance as described in the previous section (Figure 2b) [12]. Enzymatic inactivation of antibiotics in the outer regions of a biofilm can prevent drugs from reaching deeper layers, potentially allowing sensitive bacteria in these regions to survive. Resistance enzyme-expressing bacteria can therefore occupy a spatial niche near the biofilm surface, where concentrations of drugs as well as oxygen and other nutrients will be highest [13,14]. Here, spatial organization can decrease the cost of resistance enzyme production by allowing preferential access to resources, while simultaneously allowing less-resistant bacteria to survive in a detoxified environment. Consistent with this idea, increases in resistance plasmid copy number and hyperexpression of antibiotic-inactivating enzymes occur heterogeneously in biofilm-associated Enterococcus faecalis, increasing resistance in the biofilm as a whole [15].
Antibiotics that do reach the deep layers of a biofilm may be less effective due to conditions generated by bacterial metabolism and diffusion limitation. These processes create gradients in oxygen, cations, and other solutes as well as pH, which can affect the uptake and efficacy of antibiotics [16,17]. (Figure 2c). Moreover, slow or arrested cell growth deep in the biofilm is known to decrease antibiotic susceptibility in biofilms (Figure 2d) [18], and metabolic responses to nutrient limitation may control antibiotic tolerance in growth-arrested cells under these conditions [19,20].
Metabolic cooperation within biofilms can also promote antibiotic resistance. This has been observed in polymicrobial biofilms [21], probably due in part to increased productivity and matrix thickness in these assemblies [22]. Diversity in biofilms may therefore imply a sort of division of labor [23] where mutualistic interactions between taxa produce net gains in total productivity.
As observed previously for cooperative antibiotic degradation, cooperation in biofilms is vulnerable to exploitation by cheating [24]. Biofilm assembly may be thought of as a common goods problem, where membership in the communal structure is advantageous but production and maintenance of the structure is costly. When an excess of cheaters is present, the protective capacity of the collective structure is diminished [24,25], and the entire structure may be lost.
Phenotypic Heterogeneity and Signaling
Collective resistance need not be “cooperative” in a normal sense; instead, phenotypic variation within isogenic bacterial populations can act as a risk-buffering or “bet-hedging” strategy by generating stress-resistant variants [26,27] (Figure 3). There is an observed trade-off between growth rate and robustness of individual cells [27], and stress-resistant “persister” phenotypes show reduced or arrested growth under permissive conditions but survive and re-grow after antibiotic treatment [18]. Importantly, the resulting population of cells restores the original distribution of stress-sensitive and stress-resistant phenotypes, demonstrating that the rare survivors were not genetically resistant to the antibiotic [28]. This diversity allows the community to survive environmental changes that would kill off individual cells, increasing the community’s long-term fitness in a variable environment.
Figure 3. Phenotypic heterogeneity is produced by underlying stochasticity and environmental information.
In a population of genetically identical cells, phenotypic variants emerge as a result of stochastic switching (black cells, left) and responsive switching (red cells, right) after detection of environmental changes or chemical signals (blue hexagons). When the environment becomes stressful, cells that have undergone phenotypic switching are able to endure and thereby gain fitness relative to their undifferentiated clonemates.
Though bet-hedging only requires increased survival of variant phenotypes, generation of stress-tolerant phenotypes can also protect the population as a whole. In the case of Staphylococcus aureus, stochastic differentiation produces reversibly gentamycin-resistant small colony variants (SCVs) which divide slowly but can grow in the presence of antibiotic [29]. Interestingly, the by-products of SCV fermentation alter environmental pH, reducing the efficacy and uptake of gentamycin. As a result, SCV frequency increases initially and then declines in favor of wild-type revertants as the environment becomes more permissive.
In S. aureus, stress-tolerant phenotypes are generated and reversed stochastically, at rates that do not depend on environmental conditions [38]. Stochastic differentiation of phenotypes is common in microbes [30], and the canonical mechanism describes emergence of distinct phenotypes as a consequence of bistability in networks with feedback and inherent noise [31,32]. These phenotypes are not usually heritable [33], but phenotypes of individual cells may in some cases be inherited epigenetically; in Mycobacterium smegmatis, deterministic heterogeneity in elongation rate is a source of physiological variation, and this heterogeneity predictably affects antibiotic response [34].
In other cases, bet-hedging can be regulated by environmental information, including bacterially-produced chemical signals [35] (Figure 3). Differentiation of antibiotic-tolerant phenotypes may be regulated by self-produced signals as observed in Pseudomonas aeruginosa [36] and E. coli [37] and by interspecies communication in polymicrobial communities [38,39]. Additionally, chemical communication by resistant bacteria can directly affect antibiotic response in sensitive cells through induction of protective mechanisms; this has been observed as “charitable” indole signaling in E. coli under antibiotic stress [40]. Environmental information can therefore be used to alter both the diversity of phenotypes in a bet-hedging strategy and the stress tolerance of the entire population.
Conclusions
Community-based resistance presents challenges and opportunities for antimicrobial chemotherapy. We have seen that community-based resistance often relies on cooperation, which can be disrupted by cheating; weakening cooperative links within collectively-resistant communities may therefore reduce their protective effects. In particular, the recalcitrance of biofilms is being addressed from an ecological standpoint in efforts to disrupt these communities and remove the protection of the collective, including methods for interfering with inter-cell signaling [41,42]. Inhibition of microbial cooperation is a promising pathway for treatment regimes that minimize collective resistance; continued discovery of new inhibitors and deployment of these compounds against antibiotic-recalcitrant communities will show how likely this promise is to be realized.
However, signal interference may be less effective when cooperation within assemblies is not the rule. It has been suggested that conflict rather than cooperation should dominate polymicrobial interactions [43], minimizing the effects of internal control. Even where cooperation is the rule, physical conditions within biofilms may limit the distance over which communication is possible [44], raising difficulties in targeting interference for maximum effect. Furthermore, in some cases these structures may not require communication but instead represent emergent properties of collective behavior [45].
It is therefore important to understand the conditions under which signal interference is likely to be useful. Recent work has demonstrated that signaling performs optimally when cell densities are high or diffusion is limited [46], allowing high local concentrations of signal. Signaling under these conditions can be highly informative, particularly when multiple signals are integrated [47]. Chemical communication can therefore minimize costs by restricting cooperative behaviors to conditions where these behaviors are efficient [46]. More work is needed to understand the ecology of microorganisms, including their co-aggregation habits in natural environments, and integrate this information into rules for determining whether signal disruption is likely to be effective against a given community and whether resistance to signal disruption is likely to develop [48].
Few attempts have been made to directly address cooperative inactivation of antibiotics or taxonomic diversity as mechanisms of collective resistance. Current efforts rely heavily on empirical treatment regimes, using a set of antibiotics targeted to the vulnerabilities of individual taxa [49] or otherwise targeting multiple vulnerabilities within a system [50–54] to provide ad hoc means of navigating a landscape of unknown defenses. For cooperatively resistant communities containing resistance alleles and/or multiple taxa, it may be possible to design more precise treatments that disrupt ecological interactions (removing key portions of the community or more broadly reducing diversity) with the aim of reducing overall resistance.
More work is needed to develop therapies for eradication of physiologically heterogeneous bacterial communities. Regulation of phenotypic differentiation has been targeted in attempts to prevent or reverse formation of stress-resistant variants, and there have been promising recent advances targeting bacterial metabolism and signaling to reduce phenotypic tolerance through environmental regulation [55–57]. Additionally, increased understanding of metabolism in phenotypic variants has raised the possibility of treatments that can specifically eradicate these cells [20,56,57], potentially allowing elimination of persistent infections.
It may be possible to address stochastic differentiation in its ecological context. Selective forces are expected to tune rates and regulatory mechanisms of bet-hedging to the expected environmental variation. Random switching is expected to fare well in environments with rare, sudden fluctuations, while reliable environmental signaling is expected to favor regulation [33,35]. Recent work [35] has expanded theories for the evolution of bet-hedging strategies into scenarios incorporating spatial structure and population density; here, success of a strategy depends on the size and composition of the local community, indicating that the ecological context can affect how bet-hedging strategies are evolved. As our understanding of stochastic differentiation increases, it may be possible to estimate current fitness and evolutionary trajectories of bet-hedging strategies in different environments, and ultimately even to design therapies to counter the full range of physiological adaptation.
Highlights.
Bacterial communities survive antimicrobial therapy where individuals cannot.
This effect can be produced by cooperative inactivation of antibiotics.
Biofilms produce community- and drug-dependent collective resistance.
Bet-hedging allows community survival in variable environments.
Collective resistance offers opportunities for new therapeutic approaches.
Acknowledgments
We thank the students and postdocs of the Gore lab for helpful comments on the manuscript, especially B. Deris and Y. Friedman.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hogan D, Kolter R. Why are bacteria refractory to antimicrobials? Curr Opin Microbiol. 2002;5:472–477. doi: 10.1016/s1369-5274(02)00357-0. [DOI] [PubMed] [Google Scholar]
- 2.Tan C, Phillip Smith R, Srimani JK, Riccione KA, Prasada S, Kuehn M, You L. The inoculum effect and band_pass bacterial response to periodic antibiotic treatment. Mol Syst Biol. 2012;8:617. doi: 10.1038/msb.2012.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3**.Yurtsev EA, Chao HX, Datta MS, Artemova T, Gore J. Bacterial cheating drives the population dynamics of cooperative antibiotic resistance plasmids. Mol Syst Biol. 2013;9 doi: 10.1038/msb.2013.39. Using a combination of experiments and modeling, this paper explores the population dynamics between resistant and sensitive bacteria growing in a medium containing the β-lactam antibiotic ampicillin. The paper shows that in the presence of resistant cells, sensitive cells can survive high antibiotic concentrations. The authors provide a simple model that successfully explains the observed dynamics, providing intuition for how inactivation of the antibiotic by resistant cells can result in coexistence between resistant and sensitive cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brook I. The role of beta-lactamase-producing-bacteria in mixed infections. BMC Infect Dis. 2009;9:202. doi: 10.1186/1471-2334-9-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andersson DI, Hughes D. Persistence of antibiotic resistance in bacterial populations. FEMS Microbiol Rev. 2011;35:901–911. doi: 10.1111/j.1574-6976.2011.00289.x. [DOI] [PubMed] [Google Scholar]
- 6.Gullberg E, Cao S, Berg OG, Ilbäck C, Sandegren L, Hughes D, Andersson DI. Selection of Resistant Bacteria at Very Low Antibiotic Concentrations. PLoS Pathog. 2011;7:e1002158. doi: 10.1371/journal.ppat.1002158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drescher K, Shen Y, Bassler BL, Stone HA. Biofilm streamers cause catastrophic disruption of flow with consequences for environmental and medical systems. Proc Natl Acad Sci. 2013;110:4345–4350. doi: 10.1073/pnas.1300321110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8*.Kirby AE, Garner K, Levin BR. The Relative Contributions of Physical Structure and Cell Density to the Antibiotic Susceptibility of Bacteria in Biofilms. Antimicrob Agents Chemother. 2012;56:2967–2975. doi: 10.1128/AAC.06480-11. The authors determine susceptibility of biofilm and planktonic cells of S. aureus and E. coli to several classes of antibiotic using a time-kill method. By assessing the relative contributions of density, physiological state, and biofilm physical structure, the authors discover that the effects of biofilm structure on antibiotic susceptibility differ dramatically between antibiotic classes. The paper outlines broadly applicable protocols for determining the effects of structured habitats on antibiotic susceptibility. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson GG, O’Toole GA. Innate and Induced Resistance Mechanisms of Bacterial Biofilms. In: Romeo T, editor. Bacterial Biofilms. Springer; Berlin Heidelberg: 2008. pp. 85–105. Available from: http://link.springer.com/chapter/10.1007/978-3-540-75418-3_5. [DOI] [PubMed] [Google Scholar]
- 10.Tseng BS, Zhang W, Harrison JJ, Quach TP, Song JL, Penterman J, Singh PK, Chopp DL, Packman AI, Parsek MR. The extracellular matrix protects Pseudomonas aeruginosa biofilms by limiting the penetration of tobramycin. Environ Microbiol. 2013;15:2865–2878. doi: 10.1111/1462-2920.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson L, Horsman SR, Charron-Mazenod L, Turnbull AL, Mulcahy H, Surette MG, Lewenza S. Extracellular DNA-induced antimicrobial peptide resistance in Salmonella enterica serovar Typhimurium. BMC Microbiol. 2013;13:115. doi: 10.1186/1471-2180-13-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Narisawa N, Haruta S, Arai H, Ishii M, Igarashi Y. Coexistence of Antibiotic-Producing and Antibiotic-Sensitive Bacteria in Biofilms Is Mediated by Resistant Bacteria. Appl Environ Microbiol. 2008;74:3887–3894. doi: 10.1128/AEM.02497-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13**.Nadell CD, Bucci V, Drescher K, Levin SA, Bassler BL, Xavier JB. Cutting through the complexity of cell collectives. Proc R Soc B Biol Sci. 2013;280:20122770. doi: 10.1098/rspb.2012.2770. In this review, the authors discuss modeling approaches for understanding social behavior in microbes, highlight recent work that has used these methods to approach problems in nutrient transport and consumption, selective sweeps, and quorum sensing, and provide an example analyzing cooperative enzyme secretion. This review presents background and analytical techniques for models that bridge social evolution, population dynamics, and the physical properties of bacterial communities. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hengzhuang W, Ciofu O, Yang L, Wu H, Song Z, Oliver A, Høiby N. High β-Lactamase Levels Change the Pharmacodynamics of β-Lactam Antibiotics in Pseudomonas aeruginosa Biofilms. Antimicrob Agents Chemother. 2013;57:196–204. doi: 10.1128/AAC.01393-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cook LC, Dunny GM. Effects of Biofilm Growth on Plasmid Copy Number and Expression of Antibiotic Resistance Genes in Enterococcus faecalis. Antimicrob Agents Chemother. 2013;57:1850–1856. doi: 10.1128/AAC.02010-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stewart PS, Franklin MJ. Physiological heterogeneity in biofilms. Nat Rev Microbiol. 2008;6:199–210. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- 17.Thomas J, Linton S, Corum L, Slone W, Okel T, Percival SL. The affect of pH and bacterial phenotypic state on antibiotic efficacy. Int Wound J. 2012;9:428–435. doi: 10.1111/j.1742-481X.2011.00902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wood TK, Knabel SJ, Kwan BW. Bacterial Persister Cell Formation and Dormancy. Appl Environ Microbiol. 2013;79:7116–7121. doi: 10.1128/AEM.02636-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nguyen D, Joshi-Datar A, Lepine F, Bauerle E, Olakanmi O, Beer K, McKay G, Siehnel R, Schafhauser J, Wang Y, et al. Active Starvation Responses Mediate Antibiotic Tolerance in Biofilms and Nutrient-Limited Bacteria. Science. 2011;334:982–986. doi: 10.1126/science.1211037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20**.Amato SM, Fazen CH, Henry TC, Mok WWK, Orman MA, Sandvik EL, Volzing KG, Brynildsen MP. The role of metabolism in bacterial persistence. Microb Physiol Metab. 2014;5:70. doi: 10.3389/fmicb.2014.00070. This review presents current evidence for the importance of metabolism in bacterial persistence, including methods for measuring persister metabolism and metabolic approaches for reverting persisters to a susceptible state. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dalton T, Dowd SE, Wolcott RD, Sun Y, Watters C, Griswold JA, Rumbaugh KP. An In Vivo Polymicrobial Biofilm Wound Infection Model to Study Interspecies Interactions. PLoS ONE. 2011;6:e27317. doi: 10.1371/journal.pone.0027317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brenner K, Arnold FH. Self-Organization, Layered Structure, and Aggregation Enhance Persistence of a Synthetic Biofilm Consortium. PLoS ONE. 2011;6:e16791. doi: 10.1371/journal.pone.0016791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23*.Estrela S, Brown SP. Metabolic and Demographic Feedbacks Shape the Emergent Spatial Structure and Function of Microbial Communities. PLoS Comput Biol. 2013;9 doi: 10.1371/journal.pcbi.1003398. This paper uses an individual-based modeling framework to simulate growth of a two-species cooperative community where each species produces a different common good (food or detoxification). The results indicate that the strength of metabolic interdependence dictates the level of interspecies mixing, cooperation vs. conflict, and community productivity, indicating that spatial and functional relationships within structured communities can reflect the underlying metabolic interdependencies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Popat R, Crusz SA, Messina M, Williams P, West SA, Diggle SP. Quorum-sensing and cheating in bacterial biofilms. Proc R Soc B Biol Sci. 2012;279:4765–4771. doi: 10.1098/rspb.2012.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Billings N, Ramirez Millan M, Caldara M, Rusconi R, Tarasova Y, Stocker R, Ribbeck K. The Extracellular Matrix Component Psl Provides Fast-Acting Antibiotic Defense in Pseudomonas aeruginosa Biofilms. PLoS Pathog. 2013;9:e1003526. doi: 10.1371/journal.ppat.1003526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kester JC, Fortune SM. Persisters and beyond: Mechanisms of phenotypic drug resistance and drug tolerance in bacteria. Crit Rev Biochem Mol Biol. 2014;49:91–101. doi: 10.3109/10409238.2013.869543. [DOI] [PubMed] [Google Scholar]
- 27.Carlquist M, Fernandes RL, Helmark S, Heins A-L, Lundin L, Sørensen SJ, Gernaey KV, Lantz AE. Physiological heterogeneities in microbial populations and implications for physical stress tolerance. Microb Cell Factories. 2012;11:94. doi: 10.1186/1475-2859-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sufya N, Allison DG, Gilbert P. Clonal variation in maximum specific growth rate and susceptibility towards antimicrobials. J Appl Microbiol. 2003;95:1261–1267. doi: 10.1046/j.1365-2672.2003.02079.x. [DOI] [PubMed] [Google Scholar]
- 29.Edwards AM. Phenotype Switching Is a Natural Consequence of Staphylococcus aureus Replication. J Bacteriol. 2012;194:5404–5412. doi: 10.1128/JB.00948-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Balázsi G, van Oudenaarden A, Collins JJ. Cellular Decision Making and Biological Noise: From Microbes to Mammals. Cell. 2011;144:910–925. doi: 10.1016/j.cell.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rotem E, Loinger A, Ronin I, Levin-Reisman I, Gabay C, Shoresh N, Biham O, Balaban NQ. Regulation of phenotypic variability by a threshold-based mechanism underlies bacterial persistence. Proc Natl Acad Sci. 2010;107:12541–12546. doi: 10.1073/pnas.1004333107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32**.Deris JB, Kim M, Zhang Z, Okano H, Hermsen R, Groisman A, Hwa T. The Innate Growth Bistability and Fitness Landscapes of Antibiotic-Resistant Bacteria. Science. 2013;342:1237435. doi: 10.1126/science.1237435. Using bulk and single-cell techniques as well as mathematical modeling to analyze populations of drug-resistant E. coli, the authors demonstrate that feedback between growth rate and expression of drug resistance genes creates heterogeneity in the resistance phenotype of genetically identical cells. Upon exposure to antibiotic the population splits stochastically in two, with many cells appearing susceptible while others grow rapidly. These results contradict the usual assumption that genetically encoded resistance provides uniform protection to cells carrying the gene. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kussell E, Leibler S. Phenotypic Diversity, Population Growth, and Information in Fluctuating Environments. Science. 2005;309:2075–2078. doi: 10.1126/science.1114383. [DOI] [PubMed] [Google Scholar]
- 34.Aldridge BB, Fernandez-Suarez M, Heller D, Ambravaneswaran V, Irimia D, Toner M, Fortune SM. Asymmetry and Aging of Mycobacterial Cells Lead to Variable Growth and Antibiotic Susceptibility. Science. 2012;335:100–104. doi: 10.1126/science.1216166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35**.Arnoldini M, Mostowy R, Bonhoeffer S, Ackermann M. Evolution of stress response in the face of unreliable environmental signals. PLoS Comput Biol. 2012;8:e1002627. doi: 10.1371/journal.pcbi.1002627. The authors use analytical and individual-based models to explore the evolution of stochastic and responsive bet-hedging strategies in fluctuating environments with unreliable signals of transition. The incorporation of spatial heterogeneity in the environment produces density-dependent and frequency-dependent effects on the success of switching strategies, demonstrating the importance of ecological and demographic factors in the evolution of stochastic switch-based differentiation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moker N, Dean CR, Tao J. Pseudomonas aeruginosa increases formation of multidrug tolerant persister cells in response to quorum sensing signaling molecules. J Bacteriol. 2010;192:1946–1955. doi: 10.1128/JB.01231-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vega NM, Allison KR, Khalil AS, Collins JJ. Signaling-mediated bacterial persister formation. Nat Chem Biol. 2012;8:431–433. doi: 10.1038/nchembio.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38*.Vega NM, Allison KR, Samuels AN, Klempner MS, Collins JJ. Salmonella typhimurium intercepts Escherichia coli signaling to enhance antibiotic tolerance. Proc Natl Acad Sci. 2013;110:14420–14425. doi: 10.1073/pnas.1308085110. In this follow-up study to Vega et al. 2012, the authors demonstrate regulation of antibiotic tolerance by inter-cell signaling in a two-species system, where indole signaling produced by E. coli protects non-indole-producing S. typhimurium during antibiotic treatment. Protection is demonstrated in vitro and in a C. elegans host system. This study demonstrates a case of inter-species signaling altering the effects of antibiotic treatment in a host-associated bacterial community. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pereira CS, Thompson JA, Xavier KB. AI-2-mediated signalling in bacteria. FEMS Microbiol Rev. 2013;37:156–181. doi: 10.1111/j.1574-6976.2012.00345.x. [DOI] [PubMed] [Google Scholar]
- 40.Lee HH, Molla MN, Cantor CR, Collins JJ. Bacterial charity work leads to population-wide resistance. Nature. 2010;467:82–85. doi: 10.1038/nature09354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boyle KE, Heilmann S, van Ditmarsch D, Xavier JB. Exploiting social evolution in biofilms. Curr Opin Microbiol. 2013;16:207–212. doi: 10.1016/j.mib.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42*.Böttcher T, Kolodkin-Gal I, Kolter R, Losick R, Clardy J. Synthesis and Activity of Biomimetic Biofilm Disruptors. J Am Chem Soc. 2013;135:2927–2930. doi: 10.1021/ja3120955. The authors develop a biomimetic library based on norspermidine, a known biofilm disruptor in Bacillus subtilis, and identify compounds with increased activity against B. subtilis and S. aureus biofilms. Structure-activity relationships and chemical information are analyzed to determine the structural and chemical properties that distinguish potent biofilm inhibitors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43*.Foster KR, Bell T. Competition, Not Cooperation, Dominates Interactions among Culturable Microbial Species. Curr Biol. 2012;22:1845–1850. doi: 10.1016/j.cub.2012.08.005. The authors compare single-species and pairwise productivities of 72 bacteria strains isolated from a common aquatic environment and determine that in most cases, mixed communities show lower productivity, indicating that the majority of pairwise interactions are competitive. These results are robust to higher-order interactions among larger numbers of species, indicating that antagonistic interactions may dominate in natural bacterial communities. [DOI] [PubMed] [Google Scholar]
- 44**.Drescher K, Nadell CD, Stone HA, Wingreen NS, Bassler BL. Solutions to the public goods dilemma in bacterial biofilms. Curr Biol CB. 2014;24:50–55. doi: 10.1016/j.cub.2013.10.030. This study explores chitin digestion by diffusible chitinases as a public good within Vibrio cholerae biofilms to explain how this cooperative trait is maintained robustly in natural isolates. Diffusion-limited environments (thick biofilms) and high-flow environments (analogous to particles falling through a water column) allowed producers to outcompete cheaters. The observed mechanisms are consistent with an inclusive fitness theory - the distance over which public goods are shared is small, allowing preferential access by producers and their kin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nadell CD, Xavier JB, Foster KR. The sociobiology of biofilms. FEMS Microbiol Rev. 2009;33:206–224. doi: 10.1111/j.1574-6976.2008.00150.x. [DOI] [PubMed] [Google Scholar]
- 46.Mina P, di Bernardo M, Savery NJ, Tsaneva-Atanasova K. Modelling emergence of oscillations in communicating bacteria: a structured approach from one to many cells. J R Soc Interface. 2013;10 doi: 10.1098/rsif.2012.0612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47**.Cornforth DM, Popat R, McNally L, Gurney J, Scott-Phillips TC, Ivens A, Diggle SP, Brown SP. Combinatorial quorum sensing allows bacteria to resolve their social and physical environment. Proc Natl Acad Sci. 2014;201319175 doi: 10.1073/pnas.1319175111. This study addresses the functional role of multiple quorum sensing systems in the bacterium P. aeruginosa, demonstrating that integration of multiple signals with different chemical half-lives allows this bacterium to decode its physical and social environment with high resolution. Furthermore, the authors show that this combinatorial sensing can be used to restrict expression of a costly behavior (production of a secreted enzyme) to high-density, low-mass transfer environments where this behavior is efficient, demonstrating a clear fitness benefit for integration of quorum sensing signals. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48*.García-Contreras R, Maeda T, Wood TK. Resistance to Quorum Quenching Compounds. Appl Environ Microbiol. 2013 doi: 10.1128/AEM.02378-13. In this review, the authors discuss potential problems and limitations of signal interference-based therapies, including the possibility of evolved resistance. The existing literature on quorum-interference treatments is addressed thoroughly. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brook I. Microbiology of polymicrobial abscesses and implications for therapy. J Antimicrob Chemother. 2002;50:805–810. doi: 10.1093/jac/dkg009. [DOI] [PubMed] [Google Scholar]
- 50.Fischbach MA. Combination therapies for combating antimicrobial resistance. Curr Opin Microbiol. 2011;14:519–523. doi: 10.1016/j.mib.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lázár V, Pal Singh G, Spohn R, Nagy I, Horváth B, Hrtyan M, Busa-Fekete R, Bogos B, Méhi O, Csörgő B, et al. Bacterial evolution of antibiotic hypersensitivity. Mol Syst Biol. 2013;9 doi: 10.1038/msb.2013.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bollenbach T, Kishony R. Resolution of Gene Regulatory Conflicts Caused by Combinations of Antibiotics. Mol Cell. 2011;42:413–425. doi: 10.1016/j.molcel.2011.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Torella JP, Chait R, Kishony R. Optimal Drug Synergy in Antimicrobial Treatments. PLoS Comput Biol. 2010;6:e1000796. doi: 10.1371/journal.pcbi.1000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Imamovic L, Sommer MOA. Use of Collateral Sensitivity Networks to Design Drug Cycling Protocols That Avoid Resistance Development. Sci Transl Med. 2013;5:204ra132–204ra132. doi: 10.1126/scitranslmed.3006609. [DOI] [PubMed] [Google Scholar]
- 55.Roy V, Adams BL, Bentley WE. Developing next generation antimicrobials by intercepting AI-2 mediated quorum sensing. Enzyme Microb Technol. 2011;49:113–123. doi: 10.1016/j.enzmictec.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 56.Allison KR, Brynildsen MP, Collins JJ. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature. 2011;473:216–220. doi: 10.1038/nature10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57**.Orman MA, Brynildsen MP. Establishment of a Method To Rapidly Assay Bacterial Persister Metabolism. Antimicrob Agents Chemother. 2013;57:4398–4409. doi: 10.1128/AAC.00372-13. Using a variety of techniques to isolate viable cells after antibiotic treatment, the authors distinguish multiple antibiotic-tolerant phenotypes - viable but nonculturable cells (VBNCs) and persisters - within isogenic cultures of E. coli. The metabolic heterogeneity of antibiotic-tolerant cells is characterized, indicating metabolic differences between susceptible and antibiotic-tolerant E. coli. These results are used to suggest metabolite-based treatments for eradication of physiologically diverse infections. [DOI] [PMC free article] [PubMed] [Google Scholar]