Abstract
Recent discoveries of microRNAs (miRNAs) that control HDL abundance and function have expanded our knowledge of the mechanisms regulating this important lipoprotein subclass. miRNAs have been shown to regulate gene networks that control HDL biogenesis and uptake, as well as discrete steps in the reverse cholesterol transport pathway. Furthermore, HDL itself has been shown to selectively transport miRNAs in health and disease, offering new possibilities of how this lipoprotein may alter gene expression in distal target cells and tissues. Collectively, these discoveries offer new insights into the mechanisms governing HDL metabolism and function, and open new avenues for the development of therapeutics for the treatment of cardiovascular disease.
Keywords: miRNA, lipid metabolism, HDL, post-transcriptional gene regulation
Introduction
Over 60 years ago, an observation was made that would change the way we understood how cholesterol contributes to cardiovascular disease development. In 1951, Barr and colleagues noted that individuals who suffered from atherosclerosis tended to have low levels of plasma α-lipoproteins, now widely known as high-density lipoprotein (HDL)1. He then put forth the notion that measurement of the levels of these lipoproteins would be a valuable tool for assessing an individual's risk of developing atherosclerosis, and even perhaps aid in its early detection. These observations, combined with the findings of the Framingham Study2 set in motion decades of research that would eventually contribute to the “HDL cholesterol hypothesis”, which states that low levels of circulating HDL are a causative factor in the development of cardiovascular disease. Since then, a wealth of epidemiologic studies have demonstrated an inverse correlation between plasma levels of HDL cholesterol and the risk of cardiovascular disease and its thrombotic complications3. This correlation is believed to reflect the ability of HDL particles to remove excess cholesterol from peripheral cells, particularly macrophages in atherosclerotic plaques, for return to the liver. Supporting this, a number of studies in animal models have demonstrated that raising the number of HDL particles by either direct infusion of HDL or by over-expressing apoA-I (a major protein component of HDL) can reduce atherosclerosis progression or promote its regression 4, 5. As a result of these collective observations, HDL has earned the moniker of the “good” cholesterol 6. Yet, recent Mendelian randomization studies have shown that certain single nucleotide polymorphisms that raise plasma HDL cholesterol (HDL-C) levels do not lower the risk of myocardial infarction, challenging the concept that HDL is atheroprotective 7. Compounding these findings, several clinical trials of HDL cholesterol-raising therapeutics, including niacin and inhibitors of cholesterol ester transfer protein (CETP), have failed to show benefit 8, 9. These studies have begun to cast doubt on HDL's atheroprotective functions, and spurred further investigations of the molecular mechanisms regulating levels of plasma HDL and its function. One area of rapid growth in this regard, has been the discovery of microRNAs (miRNA) as potent regulators of the gene pathways controlling HDL genesis, cholesterol efflux and reverse cholesterol transport.
Over the last decade, the import of non-coding RNA as regulators of pre-, co-, and post-transcriptional gene expression has emerged 10. Of the various classes of non-coding RNAs, miRNAs are currently the most widely-studied and have established roles in regulating a myriad of biological processes. The human genome contains over 2,500 unique mature miRNAs11, and these are encoded in both intergenic and intronic regions of the genome. A number of reviews describing the transcription and processing of miRNAs have recently been published12, 13. In their mature form, these small, non-coding RNAs of ∼22 nucleotides bind to partially complementary sites primarily found in the 3′UTR region of target mRNAs, and inhibit gene expression via induction of mRNA degradation or translational repression. Although the founding work on miRNAs was performed in C. elegans14, 15, this mechanism of post-transcriptional gene regulation has been highly conserved throughout evolution. It is now estimated that over 60% of all human genes are regulated by miRNAs16, 17. Studies of miRNA expression and function have demonstrated that miRNAs allow for context-dependent fine-tuning of gene expression18, for example during embryonic development or environmental stresses (eg. starvation). In this regard, a powerful aspect of miRNAs is their ability to simultaneously repress numerous genes to influence the output of biological gene networks18. Although the magnitude of a miRNA's effect on any one gene may be modest (often on the order of 10-30%), the ability of miRNAs to repress multiple genes or steps in a pathway can lead to robust inhibition of the output.
Recent studies in the field of lipid metabolism have identified microRNAs as regulators of plasma levels of lipoproteins19, 20, novel intercellular signaling molecules21, and plasma biomarkers of physiological status22, 23. As described below, multiple genes affecting HDL and the reverse cholesterol transport pathway have been shown to be under control of these small non-coding RNAs, including those affecting HDL biogenesis, cellular cholesterol efflux, selective cholesterol uptake from HDL and bile transport (Figure 1). These studies have revealed how single miRNAs can target multiple components of this pathway, and also identified key genes that are under control of multiple miRNAs. These findings have garnered considerable interest not only for their insight into how cells and tissues regulate HDL function, but also for their potential as targets for miRNA-based therapies.
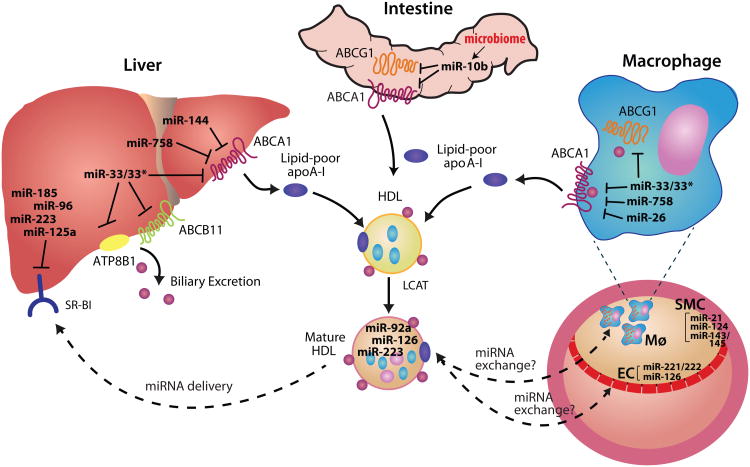
Figure 1. microRNA coordination of HDL homeostasis.
miRNAs have been shown to regulate genes involved in cholesterol efflux and HDL homeostasis in various tissues, including the liver, intestine and macrophage. These microRNAs repress mRNA and/or protein expression of their target genes as indicated. In the liver, ABCA1 is a target of a number of microRNAs that reduce cholesterol efflux to lipid-poor apolipoprotein A-I (apoA-I), which generates nascent HDL. Free cholesterol (FC) on nascent HDL particles is then esterified to cholesteryl esters (CE) by lecithin-cholesterol acyltransferase (LCAT), converting nascent HDL to mature HDL. In peripheral tissues, ABCG1 mediates cholesterol transfer to mature HDL, and is a target of miR-33, as well as miR-10b, whose expression is impacted by the microbiome. Secreted microRNAs, such as miR-223, miR-92a and miR-126 are carried on circulating HDL particles and may mediate extracellular signaling by repressing genes in target tissues. HDL interaction with macrophages and endothelial cells may result in miRNA exchange (ie. pick-up or delivery), although this remains an area of open investigation. Upon its return to the liver, HDL's cholesterol and microRNA cargo is selectively taken up by scavenger receptor B-I (SR-BI), and excess cholesterol is excreted from the liver into the bile. Targeting of SR-BI and the ABC11 and ATP8B1 transporters reduce cholesterol hepatic clearance and/or excretion.
MicroRNAs controlling HDL biogenesis
Nascent HDL is generated in the liver through the efflux of cholesterol and phospholipid across the hepatocyte cell membrane onto newly-synthesized lipid-poor apolipoprotein A-I (apoA-I)3. The ATP-binding cassette transporter A1 (ABCA1) plays a critical role in this process as evidenced by the near-absence of plasma HDL-C in patients with Tangier disease, which results from mutations in the ABCA1 gene24-27. The levels of ABCA1 at the plasma membrane controls the rate of cholesterol efflux to apoA-I, and hepatic-specific deletion of ABCA1 results in an ∼85% loss of total HDL-C28, with ABCA1 in the adipose and intestine contributing to the residual balance29-32. Several miRNAs have recently been identified that target ABCA1 and thus, regulate plasma levels of HDL-C. Among these, miR-33a and miR-33b were the first to be reported 20, 33 and were intriguing because of their genomic context: miR-33a and b are embedded in intronic regions of the SREBF2 and SREBF1 genes, which code for the SREBP2 and SREBP1 transcription factors that control the expression of genes involved in cholesterol and fatty acid synthesis. miR-33a/b are co-regulated with their host genes and act to repress gene programs that oppose SREBP functions, eg. cholesterol efflux and fatty acid oxidation. For example, under low cholesterol conditions that trigger transcription of SREBF2 and the regulation of genes involved in cholesterol synthesis and uptake, co-transcription of miR-33a acts to inhibit cellular cholesterol export by targeting ABCA120, 33-35. The 3′UTRs of mouse and human ABCA1 mRNA harbor 4 miR-33a binding sites, resulting in strong repression of ABCA1 mRNA and protein. Furthermore, consistent with the ability of most miRNAs to mediate pathway regulation, miR-33a/b also target other genes that contribute to cholesterol mobilization and efflux from the cell, including NPC1 and ABCG1 20. The physiological relevance of miR-33 targeting of ABCA1 was initially demonstrated using inhibitors of miR-33, which increased cholesterol efflux from hepatocytes to apoA-I in vitro and raised levels of plasma HDL-C 30% in mice by 25-30% 20, 33. These findings were subsequently confirmed by targeted deletion of miR-33, which resulted in 25 and 40% increases in plasma HDL-C in male and female miR-33 null mice, respectively36.
The two members of the miR-33 family, miR-33a and b, differ by only 2 nucleotides in their mature form, however these nucleotides lie outside the seed region (5′ bases 2 to 8 of the mature miRNA) that dictates target recognition. Thus, miR-33a and b are predicted to target a similar subset of genes, and experiments to date have shown comparable repression of ABCA1 and other known targets by these two isoforms 37. Nonetheless, differences in their genomic context and evolutionary conservation may influence biological outcomes. For example, the abundance of miR-33a and miR-33b is controlled by factors that regulate their host genes, and the amplitude of the induction of SREBF2/1, e.g. levels of SREBP2 mRNA are increased 2-3 fold by sterol depletion, while levels of SREBP1 can be induced over ten times that amount by insulin. Furthermore, the presence of miR-33a within the SREBF2 locus is highly conserved across species, whereas miR-33b is present in primates, but lacking in rodents and lower organisms 38. Such differences would lead to high levels of miR-33b in insulin-resistant states in humans, and thus repression of ABCA1 and plasma HDL, that would not be observed in mice. To determine whether the findings of miR-33 inhibition in mice were translatable to primates, a study was undertaken in African green monkeys using 2′F/MOE anti-miR-33 oligonucleotides designed to inhibit both miR-33a and miR-33b. Anti-miR33 treatment increased hepatic expression of ABCA1 and plasma HDL-C in monkeys fed both a chow diet and a high-carbohydrate diet designed to increase levels of SREBF1 and thus miR-33b39. Notably, these effects of miR-33 inhibition were accompanied by increases in the number of large HDL particles and apoA-1 in the circulation, attributes that have been shown to be atheroprotective in studies of other HDL-raising therapies40, 41. These studies solidified the notion that miR-33 represses hepatic ABCA1 expression and thus, dampens plasma HDL-C levels, in a model highly relevant to humans, and highlighted its potential as a therapeutic target to raise HDL.
Although the majority of miR-33 studies have focused on the 5p strand, a recent report indicates that the miR-33 passenger strand or * strand may also be active in certain cell types or tissues42. Based on the low abundance of passenger strands of many mature miRNAs, these *strands were thought to be degraded. Yet an increasing number of miRNA* sequences with abundant expression have been reported to act as guide miRNAs43, prompting renewed interest in their function. Goedeke et al showed that miR-33a* and miR-33b* accumulate under steady state conditions in various mouse, monkey and human tissues42, and other groups have noted the specific regulation of miR-33a* in endothelial cells subjected to hypoxia44 and in M2-polarized macrophages45. The miR-33a* and miR-33b* strands are highly conserved across species, suggesting a conserved function. Interestingly, miR-33a* and miR-33b* were shown to target a similar subset of lipid metabolism genes (NPC1, CROT, IRS2, SRC3, NFYC, RIP140, ABCA1 indirectly) as their sister strands42, implying that both strands of the miR-33 locus may work in concert to regulate cellular cholesterol metabolism. MiR-33a* and miR-33b* have different seed sequences from miR-33a and miR-33b, and thus are predicted to bind different sites in the 3′UTR of their target genes. This dual targeting of lipid metabolism genes by both strands of the miR-33 duplex would result in strong repression of their targets. Future studies examining the regulation of miR-33 and miR-33* abundance will be important to understand the factors that support their accumulation in the cell. The mechanisms of miRNA strand selection and loading into the RISC complex remain obscure, but are thought to be related to the thermodynamic stability of each strand of the duplex. Interestingly a recent study reported stabilization of strands by their target mRNAs46, suggesting a scenario in which miR-33* may be stabilized to help miR-33 regulate genes involved in cholesterol efflux and/or fatty acid oxidation. The functional effects of miR-33* on HDL and/or triglyceride levels in vivo have yet to be examined, but are likely to be relevant to the design of current therapeutic strategies to inhibit miR-33 for the treatment of atherosclerosis, as targeting of one arm of the duplex would not necessarily inhibit the functionality of the other, and may in fact lead to stabilization of the non-targeted strand.
ABCA1 has an uncommonly long 3′UTR of >3.3kb, rendering it particularly susceptible to post-transcriptional regulation by miRNAs. Indeed, shortly after the discovery of miR-33, other miRNAs were found to repress ABCA1 and cholesterol efflux in vitro, including miR-75847, miR-26 48 and miR-106b 49. Recently, two groups reported that miR-144, an intergenic miRNA present in a bicistronic cluster with miR-451, also targets ABCA1 in the liver and modulates plasma HDL cholesterol levels 50, 51. Interestingly, miR-144 expression is regulated by two members of the nuclear hormone receptor family, the farsenoid X receptor (FXR) and the liver X receptors (LXR)52, 53, providing fine-tuning of ABCA1 expression under specific biological contexts. These ligand-activated transcription factors contribute to the regulation of cholesterol homeostasis through transcriptional regulation of lipid associated genes: FXR controls hepatic sterol and bile acid levels, and LXR controls components of the cellular cholesterol efflux pathway in the liver and macrophages, including ABCA1. FXR induction of miR-144 transcription may channel cholesterol to the bile for excretion by repressing hepatic ABCA1, and thus HDL biogenesis. While this serves to reduce plasma HDL-C levels, this effect of miR-144 may be favorable overall by promoting reverse cholesterol transport, as has been observed with probucol treatment 54. On the other hand, LXR upregulation of miR-144 during cholesterol excess may function as a feedback mechanism to prevent uncontrolled LXR-induced cholesterol efflux through ABCA1. In both studies, anti-miR inhibition of miR-144 in mice resulted in increased hepatic ABCA1 expression and plasma HDL-C levels 50, 51. However, further studies of the effect of miR-144 inhibition on reverse cholesterol transport will be required to determine its impact on biliary cholesterol excretion.
Collectively, the studies of miR-33 and miR-144 have begun to illuminate the intricate network that fine-tunes ABCA1-dependent cholesterol efflux from the liver to regulate plasma HDL-C. It is likely that numerous other microRNAs will be identified to act in concert to regulate ABCA1 expression in the liver, with their individual and combined contributions determined by factors that regulate their expression and abundance under specific conditions. The identification of such metabolic rheostats will no doubt provide new opportunities for therapeutic manipulation of plasma HDL-C levels and reverse cholesterol transport.
MicroRNAs controlling cellular cholesterol mobilization
The efflux of excess cholesterol from peripheral tissues, particularly macrophages in the artery wall29, 31, is essential for maintaining cholesterol homeostasis. At the cellular level, this requires that cholesterol first be mobilized from internal stores via cooperation of the lysosome, lipid droplets, neutral cholesteryl ester hydrolase, and the autophagy machinery55. The final step of cholesterol efflux from the plasma membrane is mediated by ABCA1 to lipid poor ApoA-I (ApoE in the brain) and through the related transporter, ABCG1, to mature HDL particles. This ability of HDL and ApoA-I to act as acceptors of excess cholesterol from cells is thought to be central to their protective functions and this constitutes the first step in the reverse cholesterol transport (RCT) pathway through which HDL ferries cholesterol back to the liver for excretion. Through the coordination of these cholesterol mobilization pathways, net cholesterol balance in the arterial wall is maintained and pro-inflammatory responses by arterial cholesterol-loaded macrophages are reduced. As each of these steps represents potential points of microRNA control, the complexity of microRNA regulation of cholesterol efflux is likely to be much greater than originally anticipated.
As an example, autophagy, which regulates the availability of free cholesterol for efflux56-59 and contributes prominently to macrophage RCT in vivo57, is a complex process that requires multiple sequential membrane remodeling and trafficking events, orchestrated by a small army of autophagy-related gene (ATG) proteins. This pathway has recently been shown to be regulated by a number of miRNAs, including miR-18a, miR-20a, miR-30a, miR-30d, miR-101, miR-106b, miR-132, miR-181a, miR-196, miR-212, miR-221, miR-222, miR-376b, miR-50260-69, which act by targeting ATG proteins (ATG2, ATG4, ATG5, ATG12)62, 63, 65, 68 or their upstream effectors (BECN1, mTOR, ULK1)61, 63, 64, 66, 68. To date, the majority of these miRNAs regulating autophagy have been characterized in different types of cancer (breast cancer,60, 62 hepatocellular carcinoma,67 chronic myelogenous leukemia,69 colon cancer,64, 70 melanomas61), as well as in cardiac hypertrophy,71, 72 Parkinson's disease,73 and Crohn's disease.74, 75 While the role of these miRNAs in regulating lysosomal trafficking of cholesterol has yet to be investigated, they are likely to have a major impact on cholesterol efflux, RCT and HDL function. As the activation of autophagy in macrophages has been shown to suppress foam cell formation and atherogenesis in mice56, therapeutic targeting of microRNAs that limit this pathway may provide new therapeutic targets for enhancing cholesterol flux.
Although macrophage RCT does not significantly alter total plasma HDL-C levels76, 77, it's contribution is critical to atheroprotection. Indeed, Khera et al showed that the efflux capacity of HDL is an independent and robust predictor of atherosclerosis in humans, which is not simply explained by levels of HDL-C in the circulation78. As ABCA1 and ABCG1 control the terminal steps of cholesterol efflux to nascent and mature HDL from extrahepatic cells, microRNAs that target these genes would be predicted to inhibit reverse cholesterol transport. In mice, miR-33 targets both ABCA1 and ABCG1 (ABCG1 is not a target in humans), and miR-33 inhibitors enhance macrophage cholesterol efflux to apoA-1 and HDL in vitro20, 33, 35. Furthermore, parenteral delivery of anti-miR33 oligonucleotides in mice increased RCT from labeled macrophages in vivo79, 80, and directly upregulated ABCA1 in atherosclerotic plaque macrophages to reduce plaque cholesterol content79. Although miR-33 has been the most extensively studied in vivo, several other microRNAs have been shown to regulate ABCA1 in macrophages and other cell types, including miR-758, miR-26, miR-106 and miR-14447-49. Like miR-33, miR-26 also downregulates other genes involved in cholesterol mobilization in addition to ABCA1, such as ADP-ribosylation factor-like 7 (ARL7), an intracellular transport protein that moves cholesterol to the membrane for removal by ABCA148. Expression of miR-26 is suppressed by LXR, and thus would be predicted to be downregulated under conditions of cholesterol excess during which increased levels of ABCA1 would be needed. On the other hand, miR-144 would be induced by LXR to target ABCA1 under similar conditions50, and thus further studies of the temporal expression and abundance of these two microRNAs will be needed to resolve their relative contribution to cholesterol efflux control. Finally, miR-758 and miR-106b have been found to be highly enriched in the brain where ABCA1 plays a key role in effluxing excess cholesterol to apoE, the predominant apolipoprotein in the brain81 49. Notably, ABCA1-dependent cholesterol efflux appears to reduce the accrual of amyloid-β in the brain, and in accordance with this, overexpression of miR-106b in neuronal cells increased the accumulation of amyloid-β in these cells49. Thus, while multiple miRNAs can mediate post-transcriptional regulation of ABCA1, their individual impact on cholesterol efflux and RCT will be influenced by factors such as their relative tissue enrichment, transcriptional regulation, as well as miRNA cooperation and/or competition.
Recent studies have highlighted the complex interplay of dietary nutrients and intestinal microbiota composition in influencing cardiometabolic diseases82, 83. Dietary anthocyanins, such as the cyanidin-3-O-B-glucoside (Cy-3-G) polyphenol commonly found in fruits, berries and red wine, have been associated with reduced risk of cardiovascular disease. This has now been linked in a series of studies to the actions of a Cy-3-G metabolite, protocatechuic acid (PCA), which reduces levels of miR-10b, a newly identified repressor of ABCA1 and ABCG184, 85. Studies in antibiotic-treated and germ-free mice established that dietary Cy-3-G conversion to PCA, and its downstream enhancement of RCT, was dependent on the gut microbiota85. Using physiological concentrations of PCA achieves with Cy-3-G dietary supplementation, the authors showed that PCA treatment of macrophages reduced miR-10b, causing derepression of its target genes ABCA1 and ABCG1, and increasing cholesterol efflux capacity. In Apoe−/− mice treated for 4 weeks with dietary Cy-3-G or PCA, these observed changes in miR-10b, macrophage cholesterol efflux and RCT were associated with a reduction in atherosclerotic plaque size. This study underscores the complex interaction of the gut microbiome with risk factors for cardiovascular disease, and highlights a new mechanism through which miRNAs that regulate cholesterol homeostasis could be modulated. Future studies investigating how other gut-microbiota derived compounds, such as the recently identified plasma metabolite trimethylamine N-oxide (TMAO) that promotes macrophage cholesterol accumulation and atherosclerosis 83, might also alter cholesterol-associated microRNAs will no doubt reveal new mechanistic links between metabolism and host-microbial interactions.
MicroRNAs targeting hepatic HDL uptake and excretion
Transport of HDL-C to the liver for bile acid synthesis and excretion is the final step of RCT and can occur either directly via the scavenger receptor B-I (SR-BI), or after transfer to apolipoprotein B-containing lipoproteins by the cholesteryl ester transfer protein (CETP) present in humans3. MicroRNAs targeting these pathways are just beginning to be explored, and represent exciting new therapeutic targets to influence the route of delivery of HDL's cargo. SR-BI is a plasma membrane glycoprotein structurally similar to CD36 that is most highly expressed in liver and steroidogenic tissues, where it delivers HDL-derived cholesterol for excretion and steroid hormone synthesis. SR-BI-mediated selective uptake of HDL-C is considered a beneficial pathway, as it both increases the rate of delivery of cholesterol to the liver and results in the release of cholesterol-depleted HDL particles that are recycled to further promote cholesterol efflux. The level of SR-BI expression in controlled at the transcriptional level by nuclear hormone receptor transcription factors such as PPARg and LXR, and at the post-transcriptional level by alternative splicing of the mRNA. The additional post-transcriptional control of this pathway by microRNAs was recently demonstrated using small interfering RNA (siRNA) silencing of the miRNA-processing enzymes Drosha and Dicer, which resulted in a marked increase in SR-BI mRNA and protein in HEPG2 cells86. Bioinformatic prediction algorithms, such as Targetscan and MiRanda, indicate that up to 50 microRNAs may target the 3′UTR of human SR-BI. Among these, miR-185, miR-96, and miR-223 were validated as strong repressors of SR-BI mRNA and cell surface expression, and their inhibition in HEPG2 cells increased SR-BI expression and selective HDL-C uptake86. Notably, when two or three of these miRNAs were combined, there was greater repression of SR-BI than that conferred by any single miRNA, suggesting that these miRNAs may coordinately repress SR-BI mRNA by simultaneously binding to different regions of the SR-BI 3′UTR. Interestingly, miR-185, miR-96 and miR-223 have all been reported to regulate genes involved in the proliferation of various tumor cell lines, although changes in proliferation were not observed in the HEPG2 cells used for the study of SR-BI. While the factors regulating expression of these miRNAs have not yet been explored, miR-185 is located within the first intron of a gene of unknown function, C22orf25, whereas miR-96 and miR-223 are intergenic miRNAs encoded on chromosomes 7 and X, respectively. Of these miRNAs, only miR-223 does not have conserved target sites in the rodent SR-BI 3′UTR. An analogous screen of miRNAs targeting the 3′UTR of mouse SR-BI identified miR-125a and miR-455 as potent regulators of SR-BI expression in murine steroidogenic and hepatic cell lines87. Overexpression of these miRNAs reduced both SR-BI-mediated selective HDL uptake and HDL-stimulated progesterone production. The miR-125a binding site is conserved in the human SR-BI 3′UTR, however studies of its function in human cells have yet to be performed. While in vivo studies demonstrating that inhibition of mouse and primate SR-BI targeting microRNAs can increase HDL-C uptake are still lacking, these hold promise as a therapeutic approach to hold to increase RCT through this pathway.
During its journey to the liver, HDL undergoes numerous remodeling events that affect its size and composition. Cholesteryl ester transfer protein (CETP) mediates the exchange of HDL-cholesteryl esters for triglycerides from apoB-containing lipoproteins, thus shifting HDL's cholesterol cargo for uptake by the LDL receptor in the liver. Inhibitors of CETP have been actively pursued by the pharmaceutical industry as a means to raise plasma HDL-C and RCT88, 89. Although no microRNAs have yet been described to target the CETP gene, and its relatively small UTR (<200 nucleotides) is only predicted to contain few miRNA binding sites, it is likely that the expression of this and other enzymes that modulate HDL composition and function, such as lecithin-cholesterol acyltransferase (LCAT), hepatic lipase, and endothelial lipase, will be found to be under microRNA control.
Hepatic cholesterol delivered via either SR-BI or the LDL receptor can be oxygenated, converted into bile acids, and secreted into the intestine via canalicular transporters. While the majority of bile acids are reabsorbed in the intestines, a proportion is eliminated in the feces, thereby ridding the body of excess cholesterol. In addition to its roles in regulating HDL biogenesis and macrophage cholesterol efflux, miR-33 has also been shown to regulate hepatic bile metabolism80. miR-33 targets the 3′UTRs of ABCB11 and ATP8B1, transporters that reside in hepatic canalicular membranes and play essential roles in regulating biliary output (Figure 1). Using locked nucleic acid (LNA)-mediated silencing of miR-33, Allen et al showed that miR-33 inhibition increased sterols in the bile and enhanced reverse cholesterol transport in vivo80. Similar studies in Ldlr-/- mice by Rayner et al using 2′F/MOE oligonucleotide inhibitors of miR-33 noted a step-wise increase in RCT to the serum, liver and feces -30, 50 and 85%, respectively79 – reinforcing the notion that miR-33 coordinates reverse cholesterol transport at multiple levels.
HDL transport of microRNAs: a predictor of functionality?
Although miRNAs act intracellularly, microRNAs have been shown to be exported from both healthy and diseased tissues and cells. Extracellular microRNAs can be transported in membrane-derived vesicles (exosomes and microparticles), on lipoproteins, or bound to proteins like Argonaute2, and these circulating microRNAs are remarkably stable in plasma. As a result, extracellular miRNAs are being studied as novel biomarkers of disease states, including CVD, and distinct circulating miRNA signatures are beginning to be identified in health and disease90. For example, recent studies have identified miRNA-208b and miR-499 as promising biomarkers of acute myocardial infarction, however these remain to be validated in larger populations91, 92. Perhaps more exciting is the recognition that extracellular miRNAs represent a novel class of signaling molecules that may mediate cell-to-cell communication. A key role for such extracellular miRNA signaling was demonstrated in the artery wall between endothelial and smooth muscle cells and was shown to mediate atheroprotection93. Dimmeler et al showed that atheroprotective shear stress regulates the expression of multiple miRNAs in endothelial cells via the transcription factor KLF2, which are exported in extracellular vesicles93. Most prominent among these were miR-143 and miR-145, which can regulate smooth muscle cell phenotype prevent de-differentiation. Indeed, extracellular vesicles derived from KLF2-overexpressing endothelial cells, injected intravenously in Apoe−/− mice for 6 weeks, protected from atherosclerotic lesion formation, and this could be reversed by inhibiting miR-143/145. Endothelial functions such as migration have also been shown to be regulated by such cell-to-cell miRNA-based communication from monocyte-derived (miR-150)94 and apoptotic cell-derived (miR-126)21 miRNAs.
The discovery that HDL can transport miRNAs in the plasma, and stably deliver these to cells for uptake, suggests that HDL may participate in extracellular miRNA signaling 95, 96. Vickers et al demonstrated that several species of RNAs, including miRNAs and tRNA- and RNase P-derived RNA fragments, are carried on plasma HDL. Although miRNAs can also be isolated from LDL, for reasons that are unclear they appear to be more highly enriched on HDL. For example, miR-223 is one of the most abundant miRNAs on both HDL and LDL, yet it is approximately 7-fold higher on HDL (10,000 copies per μg of HDL)96. Other miRNAs with defined roles in vascular biology and inflammation are also present on HDL, albeit at lower levels than miR-223: the endothelial enriched miRNAs, miR-126 and miR-92a, are present at 3,000 copies per μg of HDL, whereas the inflammation-associated miRNAs miR-146a and miR-155, and the metabolically controlled miR-378 are present at fewer than 120 copies/μg of HDL96. Notably, of these microRNAs, only the pro-inflammatory miR-155 was found to be greater quantities on LDL than HDL, a distinction that bears further investigation given the finding that miR-155 is proatherosclerotic. Similar to what has been reported for exosomes and microvesicles, HDL-derived miR-223 was shown to be delivered to cells, including hepatocytes and a kidney cell line overexpressing the HDL receptor SR-BI, which was shown to be required for HDL-mediated delivery of miRNAs to recipient cells95. Notably, HDL-transferred miR-223 reduced target gene expression in the recipient cells, including the miR-223 target SR-BI86, 95. However, a second study of HDL-transported miRNAs in which the Caenorhabditis elegans miRNA, cel-miR-39, was used to track delivery of HDL-derived miRNAs reported that only a small number of cel-miR-39 copies could be detected in recipient cells such as endothelial cells, monocytes and smooth muscle cells96. Thus, further investigation will be need to understand the functional relevance and physiological impact of HDL-derived miRNAs, as well as such basic questions as how such miRNAs are selected for export and associate with HDL.
In addition to its critical role in RCT, HDL can exert anti-inflammatory, anti-oxidant, and anti-thrombotic effects—and these functions appear to vary among individuals. It is thus possible that these functions of HDL may be mediated in part by, or altered by, the subset of microRNAs that it carries. Indeed, the microRNA cargo of HDL has been shown to be altered in both mice and humans by hypercholesterolemia and atherosclerosis95. An analysis of HDL from subjects with familial hypercholesterolemia revealed approximately 22 miRNAs that were significantly altered as compared to HDL from normal subjects. Moreover, HDL from familial hypercholesterolemia subjects was found to impact target gene expression in recipient hepatoma cells, as compared to normal HDL. These findings offer insight into new potential mechanisms by which HDL may mediate pleiotropic effects, however whether microRNAs are responsible for some of the observed variations in HDL's anti-inflammatory, antioxidant, and antithrombotic effects await further testing.
Therapeutic targeting of miRNAs to increase HDL abundance and function
MicroRNA-based therapeutics represent a new class of drugs that hold promise for the treatment of cardiovascular and other diseases. The recent FDA approval of Kynamro (previously known as Mipomersen), a first-in-class antisense oligonucleotide inhibitor that targets apolipoprotein B-100 to reduce LDL cholesterol for the treatment of homozygous familial hypercholesterolemia97, represents a giant leap forward for oligonucleotide-based therapies, including miRNA therapeutics. The first anti-microRNA therapy has yet to reach the clinic, yet anti-sense oligonucleotides against miR-122 (known as Miraversen) have shown efficacy in patients with hepatitis C infection, where the Miraversen-treated group showed prolonged dose-dependent reductions in hepatitis C viral RNA levels without evidence of viral resistance 98. These results have generated considerable excitement for the possibility of using microRNA-based therapies to treat cardiovascular disorders. miR-33 inhibition is thought to be particularly promising as a therapeutic for atherosclerosis, as it would enhance multiple components of the reverse cholesterol transport pathway, including HDL biogenesis, cholesterol efflux from plaque macrophages, and cholesterol excretion to the bile. Indeed preclinical studies of miR-33 inhibition in mice and nonhuman primates for up to 12 weeks showed sustained increases in HDL-C (on the order of 40-50%)39,40. Furthermore, miR-33 deletion or inhibition has now been tested in several different mouse models of atherosclerosis progression and regression. The first study, performed in Ldlr−/− mice with established atherosclerotic plaques, showed that a 4 week regimen of anti-miR-33 treatment led a 40% increase in HDL-C, greater RCT and a marked regression of atherosclerotic lesions79. Characterization of the plaques in anti-miR-33 treated mice revealed 35% reductions in plaque size, lipids and macrophages, and an accompanying increase in plaque collagen content. Of note, the 2′F/MOE modified anti-miR-33 oligonucleotides used in that study were shown to penetrate the plaque, where they accumulated in lesional macrophages to directly upregulate ABCA1 mRNA. Microarray expression profiling of plaque macrophages isolated by laser capture microdissection revealed an overall decrease in inflammatory gene expression, as well as a polarization of macrophages to the reparative M2 macrophage phenotype which has been shown to characterize regressing atherosclerotic plaques99, 100.
The beneficial effects of miR-33 targeting on atherosclerosis were confirmed in miR33−/− mice crossed onto the Apoe−/− background, which showed 20-25% reductions in plaque size and lipid content compared to control Apoe−/− mice after 14 weeks on a 0.15% cholesterol-containing western diet101. Peritoneal macrophages isolated from miR33−/−Apoe−/− mice showed enhanced cholesterol efflux to apoA-I and HDL-C compared to macrophages from Apoe−/− mice, reinforcing the concept that miR-33 targeting enhances RCT at the level of both HDL biogenesis and macrophage cholesterol efflux. However, studies in mice treated with miR-33 inhibitors for 12 weeks during the progression of atherosclerosis have been less clear. One study by Rotlan et al in Ldlr−/− mice fed a western diet (0.3% cholesterol) together with treatment with 2′F/MOE modified anti-miR-33 oligonucleotides showed 20% reductions in both plaque size and macrophage content, and a decrease in miR-33 target genes in the aorta102, while a second study by Marquart et al in Ldlr−/− mice fed a western diet (1.25% cholesterol) together with treatment with an anti-miR33 LNA failed to show any benefit103. Notably, both groups reported that although miR-33 inhibitors increased plasma HDL-C in mice fed a chow diet, this effect was lost when the mice were switched to the atherogenic western diets. This absence of effect of miR-33 inhibition on HDL-C in mice fed a diet enriched in cholesterol may be due to low levels of hepatic miR-33 under these conditions: miR-33 is co-regulated with its host gene SREBF2, whose transcription in the liver is decreased by dietary cholesterol20. However, the reasons for the divergent outcomes of miR-33 inhibition on atherosclerosis are less clear, but may relate to differences in the cholesterol contents of the different western diets (0.3 and 1.25%) or the bioavailability of the miR-33 inhibitors used (i.e. LNA vs. 2′F/MOE). While the ability of the anti-miR33 LNA used by Marquart et al to reach macrophages in the plaque has not been tested, this could potentially account for the efficacy of the 2′F/MOE modified anti-miR-33 in reducing atherosclerosis in the absence of an increase in plasma HDL. The study by Rotlan et al used the same 2′F/MOE modified anti-miR-33 oligonucleotides that had proven to be effective at increasing ABCA1 in plaque macrophages and regressing atherosclerosis in the previous study by Rayner et al, however miR-33 target gene expression in these cells was not evaluated. Future studies will be needed to determine whether anti-miR-33 targeting of plaque macrophages is responsible for its atheroprotective effects in the absence of increased plasma HDL. These have further underscored the importance of studying how miRNAs modulate cholesterol flux through the HDL pathway and not HDL cholesterol alone.
Inhibition of miR-33, as well as other ABCA1-targeting microRNAs, may also prove advantageous in a number of other conditions in which increased cholesterol efflux is thought to be beneficial. For example, to reduce islet cholesterol levels which impair β-cell function and glucose tolerance104, and to reduce the secretion of β-amyloid in the brain which is inversely correlated with ABCA1 -mediated cholesterol efflux to apoE 105. Studies of these approaches are eagerly awaited. Furthermore, although the pre-clinical studies of miR-33 inhibitors in mice and monkeys appear promising, many questions remain to be addressed, such as the effects of long-term suppression of miR-33, and whether compensatory mechanisms may become activated during miR-33 inhibition such as the upregulation of other ABCA1 targeting miRNAs. Finally, as the miRNA networks that regulate hepatic and systemic lipid homeostasis are unraveled, this will no doubt be paralleled by the identification of additional targets for therapeutic intervention.
Summary and Future Directions
Since the original hypothesis that HDL may be cardioprotective, HDL's halo has become somewhat tarnished as a series of HDL-C raising therapies have failed to confer protection from cardiovascular disease 106. This has prompted a reconsideration of the “HDL cholesterol hypothesis” and its evolution into the “HDL flux hypothesis”, owing to the renewed interest in understanding the mechanisms controlling HDL flux and functionality. The discoveries of microRNAs that control HDL biosynthesis, function and uptake, have greatly expanded our understanding of the molecular mechanisms regulating plasma levels of HDL-C and components of the RCT pathway, and have identified new therapeutic targets to regulate HDL flux. The list of microRNAs targeting lipoprotein metabolism pathways continues to grow at a rapid pace, and will no doubt expand to include microRNAs targeting other genes involved in HDL biogenesis (eg. apoAI), remodeling (eg. CETP and LCAT) and functionality. There remains much to understand about how microRNAs contribute to HDL functionality in health and disease. For example, whether dysregulation of miRNA activity contributes to the pathogenesis of cardiovascular disease or HDL dysfunction remains to be ascertained. The prospect that HDL-carried microRNAs contribute to the heterogeneous effects of HDL on endothelial cells, macrophages, and other cell types that influence vascular health, is intriguing and may provide insight into how the protective effects of HDL may be altered in disease or enhanced for therapeutic purposes. As our knowledge of these points of post-transcriptional control of HDL increases, so too will the potential for translating these discoveries from animal models to humans and, eventually, new therapies to treat and prevent cardiovascular disease burden.
Table 1. MicroRNAs targeting the HDL pathway and their regulation.
| miRNA | Target gene(s) | Regulation of miR Expression | Effects in vivo? | Refs |
|---|---|---|---|---|
| miR-33a/b | ABCA1, ABCG1, NPC1, ABCB11, ATP8B1, CROT, CPT1a, HADHB, IRS2, PRKAA1, R1P140, SRC3 | Sterols, LXR ligands, insulin | ↓HDL, ↑atherosclerosis, ↓RCT | 20, 33-35, 39, 79, 80 |
| miR-33* | NPC1, CROT, IRS2, SRC3, NFYC, RIP140 | Sterols, LXR ligands | Not yet tested | 42 |
| miR-758 | ABCA1 | Sterols | Not yet tested | 47 |
| miR-144 | ABCA1 | LXR & FXR ligands | ↓HDL | 50,51 |
| miR-26 | ABCA1.ARL7 | LXR ligands | Not yet tested | 48 |
| miR-106b | ABCA1 | Not known | Not yet tested | 49 |
| miR-10b | ABCA1, ABCG1 | Dietary anthocyanins (i.e.Cy-3-G metabolites) | Reduced expression ↓associated with atherosclerosis, ↑RCT | 84 |
| miR-185 | SR-61 | Not known | Reduced expression associated with ↑SR-BI in Apoe-/- liver | 86 |
| miR-96 | SR-BI | Not known | Reduced expression associated with ↑SR-BI in Apoe-/- liver | 86 |
| miR-223 | SR-BI | Carried on HDL | Reduced expression associated with ↑SR-BI in Apoe-/- liver | 86, 95, 96 |
refers to miR-33 passenger strand
Acknowledgments
Sources of funding: Research on microRNAs in the Moore Lab is supported by the NIH (R01 HL108182). K.J.R. is supported by a CIHR Operating Grant and Salary Award.
Footnotes
Disclosures K.J.M. is a member of the miR-33 Clinical Advisory Board of Regulus Therapeutics. K.J.M. and K.J.R. have a patent application on the use of miR-33 inhibitors for the treatment of atherosclerosis.
References
- 1.Barr DP, Russ EM, Eder HA. Protein-lipid relationships in human plasma. Ii. In atherosclerosis and related conditions. Am J Med. 1951;11:480–493. doi: 10.1016/0002-9343(51)90183-0. [DOI] [PubMed] [Google Scholar]
- 2.Gordon T, Castelli WP, Hjortland MC, Kannel WB, Dawber TR. High density lipoprotein as a protective factor against coronary heart disease. The framingham study. Am J Med. 1977;62:707–714. doi: 10.1016/0002-9343(77)90874-9. [DOI] [PubMed] [Google Scholar]
- 3.Tall AR, Yvan-Charvet L, Terasaka N, Pagler T, Wang N. Hdl, abc transporters, and cholesterol efflux: Implications for the treatment of atherosclerosis. Cell Metab. 2008;7:365–375. doi: 10.1016/j.cmet.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 4.Shah PK. Apolipoprotein a-i/hdl infusion therapy for plaque stabilization-regression: A novel therapeutic approach. Current pharmaceutical design. 2007;13:1031–1038. doi: 10.2174/138161207780487520. [DOI] [PubMed] [Google Scholar]
- 5.Smith JD. Apolipoprotein a-i and its mimetics for the treatment of atherosclerosis. Current opinion in investigational drugs (London, England : 2000) 2010;11:989–996. [PMC free article] [PubMed] [Google Scholar]
- 6.Boden WE. High-density lipoprotein cholesterol as an independent risk factor in cardiovascular disease: Assessing the data from framingham to the veterans affairs high--density lipoprotein intervention trial. The American journal of cardiology. 2000;86:19L–22L. doi: 10.1016/s0002-9149(00)01464-8. [DOI] [PubMed] [Google Scholar]
- 7.Voight BF, Peloso GM, Orho-Melander M, Frikke-Schmidt R, Barbalic M, Jensen MK, Hindy G, Holm H, Ding EL, Johnson T, Schunkert H, Samani NJ, Clarke R, Hopewell JC, Thompson JF, Li M, Thorleifsson G, Newton-Cheh C, Musunuru K, Pirruccello JP, Saleheen D, Chen L, Stewart A, Schillert A, Thorsteinsdottir U, Thorgeirsson G, Anand S, Engert JC, Morgan T, Spertus J, Stoll M, Berger K, Martinelli N, Girelli D, McKeown PP, Patterson CC, Epstein SE, Devaney J, Burnett MS, Mooser V, Ripatti S, Surakka I, Nieminen MS, Sinisalo J, Lokki ML, Perola M, Havulinna A, de Faire U, Gigante B, Ingelsson E, Zeller T, Wild P, de Bakker PI, Klungel OH, Maitland-van der Zee AH, Peters BJ, de Boer A, Grobbee DE, Kamphuisen PW, Deneer VH, Elbers CC, Onland-Moret NC, Hofker MH, Wijmenga C, Verschuren WM, Boer JM, van der Schouw YT, Rasheed A, Frossard P, Demissie S, Willer C, Do R, Ordovas JM, Abecasis GR, Boehnke M, Mohlke KL, Daly MJ, Guiducci C, Burtt NP, Surti A, Gonzalez E, Purcell S, Gabriel S, Marrugat J, Peden J, Erdmann J, Diemert P, Willenborg C, Konig IR, Fischer M, Hengstenberg C, Ziegler A, Buysschaert I, Lambrechts D, Van de Werf F, Fox KA, El Mokhtari NE, Rubin D, Schrezenmeir J, Schreiber S, Schafer A, Danesh J, Blankenberg S, Roberts R, McPherson R, Watkins H, Hall AS, Overvad K, Rimm E, Boerwinkle E, Tybjaerg-Hansen A, Cupples LA, Reilly MP, Melander O, Mannucci PM, Ardissino D, Siscovick D, Elosua R, Stefansson K, O'Donnell CJ, Salomaa V, Rader DJ, Peltonen L, Schwartz SM, Altshuler D, Kathiresan S. Plasma hdl cholesterol and risk of myocardial infarction: A mendelian randomisation study. Lancet. 2012;380:572–580. doi: 10.1016/S0140-6736(12)60312-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boden WE, Probstfield JL, Anderson T, Chaitman BR, Desvignes-Nickens P, Koprowicz K, McBride R, Teo K, Weintraub W. Niacin in patients with low hdl cholesterol levels receiving intensive statin therapy. The New England journal of medicine. 2011;365:2255–2267. doi: 10.1056/NEJMoa1107579. [DOI] [PubMed] [Google Scholar]
- 9.Schwartz GG, Olsson AG, Abt M, Ballantyne CM, Barter PJ, Brumm J, Chaitman BR, Holme IM, Kallend D, Leiter LA, Leitersdorf E, McMurray JJ, Mundl H, Nicholls SJ, Shah PK, Tardif JC, Wright RS. Effects of dalcetrapib in patients with a recent acute coronary syndrome. The New England journal of medicine. 2012;367:2089–2099. doi: 10.1056/NEJMoa1206797. [DOI] [PubMed] [Google Scholar]
- 10.Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, Epstein CB, Frietze S, Harrow J, Kaul R, Khatun J, Lajoie BR, Landt SG, Lee BK, Pauli F, Rosenbloom KR, Sabo P, Safi A, Sanyal A, Shoresh N, Simon JM, Song L, Trinklein ND, Altshuler RC, Birney E, Brown JB, Cheng C, Djebali S, Dong X, Ernst J, Furey TS, Gerstein M, Giardine B, Greven M, Hardison RC, Harris RS, Herrero J, Hoffman MM, Iyer S, Kelllis M, Kheradpour P, Lassmann T, Li Q, Lin X, Marinov GK, Merkel A, Mortazavi A, Parker SC, Reddy TE, Rozowsky J, Schlesinger F, Thurman RE, Wang J, Ward LD, Whitfield TW, Wilder SP, Wu W, Xi HS, Yip KY, Zhuang J, Bernstein BE, Green ED, Gunter C, Snyder M, Pazin MJ, Lowdon RF, Dillon LA, Adams LB, Kelly CJ, Zhang J, Wexler JR, Good PJ, Feingold EA, Crawford GE, Dekker J, Elinitski L, Farnham PJ, Giddings MC, Gingeras TR, Guigo R, Hubbard TJ, Kellis M, Kent WJ, Lieb JD, Margulies EH, Myers RM, Starnatoyannopoulos JA, Tennebaum SA, Weng Z, White KP, Wold B, Yu Y, Wrobel J, Risk BA, Gunawardena HP, Kuiper HC, Maier CW, Xie L, Chen X, Mikkelsen TS, Gillespie S, Goren A, Ram O, Zhang X, Wang L, Issner R, Coyne MJ, Durham T, Ku M, Truong T, Eaton ML, Dobin A, Tanzer A, Lagarde J, Lin W, Xue C, Williams BA, Zaleski C, Roder M, Kokocinski F, Abdelhamid RF, Alioto T, Antoshechkin I, Baer MT, Batut P, Bell I, Bell K, Chakrabortty S, Chrast J, Curado J, Derrien T, Drenkow J, Dumais E, Dumais J, Duttagupta R, Fastuca M, Fejes-Toth K, Ferreira P, Foissac S, Fullwood MJ, Gao H, Gonzalez D, Gordon A, Howald C, Jha S, Johnson R, Kapranov P, King B, Kingswood C, Li G, Luo OJ, Park E, Preall JB, Presaud K, Ribeca P, Robyr D, Ruan X, Sammeth M, Sandu KS, Schaeffer L, See LH, Shahab A, Skancke J, Suzuki AM, Takahashi H, Tilgner H, Trout D, Walters N, Wang H, Hayashizaki Y, Reymond A, Antonarakis SE, Hannon GJ, Ruan Y, Carninci P, Sloan CA, Learned K, Malladi VS, Wong MC, Barber GP, Cline MS, Dreszer TR, Heitner SG, Karolchik D, Kirkup VM, Meyer LR, Long JC, Maddren M, Raney BJ, Grasfeder LL, Giresi PG, Battenhouse A, Sheffield NC, Showers KA, London D, Bhinge AA, Shestak C, Schaner MR, Kim SK, Zhang ZZ, Mieczkowski PA, Mieczkowska JO, Liu Z, McDaniell RM, Ni Y, Rashid NU, Kim MJ, Adar S, Zhang Z, Wang T, Winter D, Keefe D, Iyer VR, Sandhu KS, Zheng M, Wang P, Gertz J, Vielmetter J, Partridge EC, Varley KE, Gasper C, Bansal A, Pepke S, Jain P, Amrhein H, Bowling KM, Anaya M, Cross MK, Muratet MA, Newberry KM, McCue K, Nesmith AS, Fisher-Aylor KI, Pusey B, DeSalvo G, Parker SL, Balasubramanian S, Davis NS, Meadows SK, Eggleston T, Newberry JS, Levy SE, Absher DM, Wong WH, Blow MJ, Visel A, Pennachio LA, Elnitski L, Petrykowska HM, Abyzov A, Aken B, Barrell D, Barson G, Berry A, Bignell A, Boychenko V, Bussotti G, Davidson C, Despacio-Reyes G, Diekhans M, Ezkurdia I, Frankish A, Gilbert J, Gonzalez JM, Griffiths E, Harte R, Hendrix DA, Hunt T, Jungreis I, Kay M, Khurana E, Leng J, Lin MF, Loveland J, Lu Z, Manthravadi D, Mariotti M, Mudge J, Mukherjee G, Notredame C, Pei B, Rodriguez JM, Saunders G, Sboner A, Searle S, Sisu C, Snow C, Steward C, Tapanari E, Tress ML, van Baren MJ, Washieti S, Wilming L, Zadissa A, Zhengdong Z, Brent M, Haussler D, Valencia A, Raymond A, Addleman N, Alexander RP, Auerbach RK, Bettinger K, Bhardwaj N, Boyle AP, Cao AR, Cayting P, Charos A, Cheng Y, Eastman C, Euskirchen G, Fleming JD, Grubert F, Habegger L, Hariharan M, Harmanci A, Iyenger S, Jin VX, Karczewski KJ, Kasowski M, Lacroute P, Lam H, Larnarre-Vincent N, Lian J, Lindahl-Allen M, Min R, Miotto B, Monahan H, Moqtaderi Z, Mu XJ, O'Geen H, Ouyang Z, Patacsil D, Raha D, Ramirez L, Reed B, Shi M, Slifer T, Witt H, Wu L, Xu X, Yan KK, Yang X, Struhl K, Weissman SM, Tenebaum SA, Penalva LO, Karmakar S, Bhanvadia RR, Choudhury A, Domanus M, Ma L, Moran J, Victorsen A, Auer T, Centarin L, Eichenlaub M, Gruhl F, Heerman S, Hoeckendorf B, Inoue D, Kellner T, Kirchmaier S, Mueller C, Reinhardt R, Schertel L, Schneider S, Sinn R, Wittbrodt B, Wittbrodt J, Jain G, Balasundaram G, Bates DL, Byron R, Canfield TK, Diegel MJ, Dunn D, Ebersol AK, Frum T, Garg K, Gist E, Hansen RS, Boatman L, Haugen E, Humbert R, Johnson AK, Johnson EM, Kutyavin TM, Lee K, Lotakis D, Maurano MT, Neph SJ, Neri FV, Nguyen ED, Qu H, Reynolds AP, Roach V, Rynes E, Sanchez ME, Sandstrom RS, Shafer AO, Stergachis AB, Thomas S, Vernot B, Vierstra J, Vong S, Weaver MA, Yan Y, Zhang M, Akey JA, Bender M, Dorschner MO, Groudine M, MacCoss MJ, Navas P, Stamatoyannopoulos G, Stamatoyannopoulos JA, Beal K, Brazma A, Flicek P, Johnson N, Lukk M, Luscombe NM, Sobral D, Vaquerizas JM, Batzoglou S, Sidow A, Hussami N, Kyriazopoulou-Panagiotopoulou S, Libbrecht MW, Schaub MA, Miller W, Bickel PJ, Banfai B, Boley NP, Huang H, Li JJ, Noble WS, Bilmes JA, Buske OJ, Sahu AO, Kharchenko PV, Park PJ, Baker D, Taylor J, Lochovsky L. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kozomara A, Griffiths-Jones S. Mirbase: Integrating microrna annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finnegan EF, Pasquinelli AE. Microrna biogenesis: Regulating the regulators. Crit Rev Biochem Mol Biol. 2013;48:51–68. doi: 10.3109/10409238.2012.738643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graves P, Zeng Y. Biogenesis of mammalian micrornas: A global view. Genomics Proteomics Bioinformatics. 2012;10:239–245. doi: 10.1016/j.gpb.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny rnas with probable regulatory roles in caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 15.Lee RC, Feinbaum RL, Ambros V. The c. Elegans heterochronic gene lin-4 encodes small rnas with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 16.Claverie JM. Fewer genes, more noncoding rna. Science. 2005;309:1529–1530. doi: 10.1126/science.1116800. [DOI] [PubMed] [Google Scholar]
- 17.Pasquinelli AE. Micrornas and their targets: Recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet. 2012;13:271–282. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- 18.Bartel DP. Micrornas: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, Subramaniam A, Propp S, Lollo BA, Freier S, Bennett CF, Bhanot S, Monia BP. Mir-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metabolism. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 20.Rayner KJ, Suarez Y, Davalos A, Parathath S, Fitzgerald ML, Tamehiro N, Fisher EA, Moore KJ, Fernandez-Hernando C. Mir-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328:1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zernecke A, Bidzhekov K, Noels H, Shagdarsuren E, Gan L, Denecke B, Hristov M, Koppel T, Jahantigh MN, Lutgens E, Wang S, Olson EN, Schober A, Weber C. Delivery of microrna-126 by apoptotic bodies induces cxcl12-dependent vascular protection. Science signaling. 2009;2:ra81. doi: 10.1126/scisignal.2000610. [DOI] [PubMed] [Google Scholar]
- 22.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Circulating micrornas as stable blood-based markers for cancer detection. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, Banham AH, Pezzella F, Boultwood J, Wainscoat JS, Hatton CS, Harris AL. Detection of elevated levels of tumour-associated micrornas in serum of patients with diffuse large b-cell lymphoma. British journal of haematology. 2008;141:672–675. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 24.Brooks-Wilson A, Marcil M, Clee SM, Zhang LH, Roomp K, van Dam M, Yu L, Brewer C, Collins JA, Molhuizen HO, Loubser O, Ouelette BF, Fichter K, Ashbourne-Excoffon KJ, Sensen CW, Scherer S, Mott S, Denis M, Martindale D, Frohlich J, Morgan K, Koop B, Pimstone S, Kastelein JJ, Genest J, Jr, Hayden MR. Mutations in abc1 in tangier disease and familial high-density lipoprotein deficiency. Nature genetics. 1999;22:336–345. doi: 10.1038/11905. [DOI] [PubMed] [Google Scholar]
- 25.Bodzioch M, Orso E, Klucken J, Langmann T, Bottcher A, Diederich W, Drobnik W, Barlage S, Buchler C, Porsch-Ozcurumez M, Kaminski WE, Hahmann HW, Oette K, Rothe G, Aslanidis C, Lackner KJ, Schmitz G. The gene encoding atp-binding cassette transporter 1 is mutated in tangier disease. Nature genetics. 1999;22:347–351. doi: 10.1038/11914. [DOI] [PubMed] [Google Scholar]
- 26.Lawn RM, Wade DP, Garvin MR, Wang X, Schwartz K, Porter JG, Seilhamer JJ, Vaughan AM, Oram JF. The tangier disease gene product abc1 controls the cellular apolipoprotein-mediated lipid removal pathway. The Journal of clinical investigation. 1999;104:R25–31. doi: 10.1172/JCI8119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rust S, Rosier M, Funke H, Real J, Amoura Z, Piette JC, Deleuze JF, Brewer HB, Duverger N, Denefle P, Assmann G. Tangier disease is caused by mutations in the gene encoding atp-binding cassette transporter 1. Nature genetics. 1999;22:352–355. doi: 10.1038/11921. [DOI] [PubMed] [Google Scholar]
- 28.Timmins JM, Lee JY, Boudyguina E, Kluckman KD, Brunham LR, Mulya A, Gebre AK, Coutinho JM, Colvin PL, Smith TL, Hayden MR, Maeda N, Parks JS. Targeted inactivation of hepatic abca1 causes profound hypoalphalipoproteinemia and kidney hypercatabolism of apoa-i. The Journal of clinical investigation. 2005;115:1333–1342. doi: 10.1172/JCI23915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chung S, Sawyer JK, Gebre AK, Maeda N, Parks JS. Adipose tissue atp binding cassette transporter a1 contributes to high-density lipoprotein biogenesis in vivo. Circulation. 2011;124:1663–1672. doi: 10.1161/CIRCULATIONAHA.111.025445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McGillicuddy FC, Reilly MP, Rader DJ. Adipose modulation of high-density lipoprotein cholesterol: Implications for obesity, high-density lipoprotein metabolism, and cardiovascular disease. Circulation. 2011;124:1602–1605. doi: 10.1161/CIRCULATIONAHA.111.058453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brunham LR, Kruit JK, Iqbal J, Fievet C, Timmins JM, Pape TD, Coburn BA, Bissada N, Staels B, Groen AK, Hussain MM, Parks JS, Kuipers F, Hayden MR. Intestinal abca1 directly contributes to hdl biogenesis in vivo. The Journal of clinical investigation. 2006;116:1052–1062. doi: 10.1172/JCI27352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brunham LR, Kruit JK, Pape TD, Parks JS, Kuipers F, Hayden MR. Tissue-specific induction of intestinal abca1 expression with a liver × receptor agonist raises plasma hdl cholesterol levels. Circ Res. 2006;99:672–674. doi: 10.1161/01.RES.0000244014.19589.8e. [DOI] [PubMed] [Google Scholar]
- 33.Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE, Naar AM. Microrna-33 and the srebp host genes cooperate to control cholesterol homeostasis. Science. 2010;328:1566–1569. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gerin I, Clerbaux LA, Haumont O, Lanthier N, Das AK, Burant CF, Leclercq IA, MacDougald OA, Bommer GT. Expression of mir-33 from an srebp2 intron inhibits cholesterol export and fatty acid oxidation. J Biol Chem. 2010;285:33652–33661. doi: 10.1074/jbc.M110.152090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marquart TJ, Allen RM, Ory DS, Baldan A. Mir-33 links srebp-2 induction to repression of sterol transporters. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:12228–12232. doi: 10.1073/pnas.1005191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horie T, Ono K, Horiguchi M, Nishi H, Nakamura T, Nagao K, Kinoshita M, Kuwabara Y, Marusawa H, Iwanaga Y, Hasegawa K, Yokode M, Kimura T, Kita T. Microrna-33 encoded by an intron of sterol regulatory element-binding protein 2 (srebp2) regulates hdl in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:17321–17326. doi: 10.1073/pnas.1008499107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davalos A, Goedeke L, Smibert P, Ramirez CM, Warrier NP, Andreo U, Cirera-Salinas D, Rayner K, Suresh U, Pastor-Pareja JC, Esplugues E, Fisher EA, Penalva LO, Moore KJ, Suarez Y, Lai EC, Fernandez-Hernando C. Mir-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:9232–9237. doi: 10.1073/pnas.1102281108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brown MS, Ye J, Goldstein JL. Medicine. Hdl mir-ed down by srebp introns. Science. 2010;328:1495–1496. doi: 10.1126/science.1192409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rayner K, et al. Inhibition of mir-33a/b in non-human primates raises plasma hdl and lowers vldl triglycerides. Nature. 2011;478:404–409. doi: 10.1038/nature10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rayner KJ, Esau CC, Hussain FN, McDaniel AL, Marshall SM, van Gils JM, Ray TD, Sheedy FJ, Goedeke L, Liu X, Khatsenko OG, Kaimal V, Lees CJ, Fernandez-Hernando C, Fisher EA, Temel RE, Moore KJ. Inhibition of mir-33a and b in non-human primates raises plasma hdl cholesterol and reduces vldl triglycerides. Nature. 2011 doi: 10.1038/nature10486. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Williams KJ, Feig JE, Fisher EA. Rapid regression of atherosclerosis: Insights from the clinical and experimental literature. Nat Clin Pract Cardiovasc Med. 2008;5:91–102. doi: 10.1038/ncpcardio1086. [DOI] [PubMed] [Google Scholar]
- 42.Goedeke L, Vales-Lara FM, Fenstermaker M, Cirera-Salinas D, Chamorro-Jorganes A, Ramirez CM, Mattison JA, de Cabo R, Suarez Y, Fernandez-Hernando C. A regulatory role for microrna 33* in controlling lipid metabolism gene expression. Mol Cell Biol. 2013;33:2339–2352. doi: 10.1128/MCB.01714-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mah SM, Buske C, Humphries RK, Kuchenbauer F. Mirna*: A passenger stranded in rna-induced silencing complex? Critical reviews in eukaryotic gene expression. 2010;20:141–148. doi: 10.1615/critreveukargeneexpr.v20.i2.40. [DOI] [PubMed] [Google Scholar]
- 44.Voellenkle C, Rooij J, Guffanti A, Brini E, Fasanaro P, Isaia E, Croft L, David M, Capogrossi MC, Moles A, Felsani A, Martelli F. Deep-sequencing of endothelial cells exposed to hypoxia reveals the complexity of known and novel micrornas. RNA. 2012;18:472–484. doi: 10.1261/rna.027615.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Graff JW, Dickson AM, Clay G, McCaffrey AP, Wilson ME. Identifying functional micrornas in macrophages with polarized phenotypes. J Biol Chem. 2012;287:21816–21825. doi: 10.1074/jbc.M111.327031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kang SM, Choi JW, Hong SH, Lee HJ. Up-regulation of microrna* strands by their target transcripts. Int J Mol Sci. 2013;14:13231–13240. doi: 10.3390/ijms140713231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramirez CM, Davalos A, Goedeke L, Salerno AG, Warrier N, Cirera-Salinas D, Suarez Y, Fernandez-Hernando C. Microrna-758 regulates cholesterol efflux through posttranscriptional repression of atp-binding cassette transporter a1. Arterioscler Thromb Vasc Biol. 2011;31:2707–2714. doi: 10.1161/ATVBAHA.111.232066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun D, Zhang J, Xie J, Wei W, Chen M, Zhao X. Mir-26 controls lxr-dependent cholesterol efflux by targeting abca1 and arl7. FEBS Lett. 2012;586:1472–1479. doi: 10.1016/j.febslet.2012.03.068. [DOI] [PubMed] [Google Scholar]
- 49.Kim J, Yoon H, Ramirez CM, Lee SM, Hoe HS, Fernandez-Hernando C. Mir-106b impairs cholesterol efflux and increases abeta levels by repressing abca1 expression. Exp Neurol. 2012;235:476–483. doi: 10.1016/j.expneurol.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramirez CM, Rotllan N, Vlassov AV, Davalos A, Li M, Goedeke L, Aranda JF, Cirera-Salinas D, Araldi E, Salerno A, Wanschel A, Zavadil J, Castrillo A, Kim J, Suarez Y, Fernandez-Hernando C. Control of cholesterol metabolism and plasma high-density lipoprotein levels by microrna-144. Circ Res. 2013;112:1592–1601. doi: 10.1161/CIRCRESAHA.112.300626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Aguiar Vallim TQ, Tarling EJ, Kim T, Civelek M, Baldan A, Esau C, Edwards PA. Microrna-144 regulates hepatic atp binding cassette transporter a1 and plasma high-density lipoprotein after activation of the nuclear receptor farnesoid × receptor. Circ Res. 2013;112:1602–1612. doi: 10.1161/CIRCRESAHA.112.300648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Aguiar Vallim T, Tarling E, Kim T, Civelek M, Baldan A, Esau C, Edwards P. Microrna-144 regulates hepatic abca1 and plasma hdl following activation of the nuclear receptor fxr. Circ Res. 2013 doi: 10.1161/CIRCRESAHA.112.300648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramirez CM, Rotllan N, Vlassov AV, Davalos A, Li M, Goedeke L, Aranda JF, Cirera-Salinas D, Araldi E, Salerno A, Wanschel AC, Zavadil J, Castrillo A, Jungsu K, Suarez Y, Fernandez-Hernando C. Control of cholesterol metabolism and plasma hdl levels by mirna-144. Circ Res. 2013 doi: 10.1161/CIRCRESAHA.112.300626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yamamoto S, Tanigawa H, Li X, Komaru Y, Billheimer JT, Rader DJ. Pharmacologic suppression of hepatic atp-binding cassette transporter 1 activity in mice reduces high-density lipoprotein cholesterol levels but promotes reverse cholesterol transport. Circulation. 2011;124:1382–1390. doi: 10.1161/CIRCULATIONAHA.110.009704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ouimet M. Autophagy in obesity and atherosclerosis: Interrelationships between cholesterol homeostasis, lipoprotein metabolism and autophagy in macrophages and other systems. Biochim Biophys Acta. 2013;1831:1124–1133. doi: 10.1016/j.bbalip.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 56.Le Guezennec X, Brichkina A, Huang YF, Kostromina E, Han W, Bulavin DV. Wip1-dependent regulation of autophagy, obesity, and atherosclerosis. Cell Metab. 2012;16:68–80. doi: 10.1016/j.cmet.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 57.Ouimet M, Franklin V, Mak E, Liao X, Tabas I, Marcel YL. Autophagy regulates cholesterol efflux from macrophage foam cells via lysosomal acid lipase. Cell Metab. 2011;13:655–667. doi: 10.1016/j.cmet.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Razani B, Feng C, Coleman T, Emanuel R, Wen H, Hwang S, Ting JP, Virgin HW, Kastan MB, Semenkovich CF. Autophagy links inflammasomes to atherosclerotic progression. Cell Metab. 2012;15:534–544. doi: 10.1016/j.cmet.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Robinet P, Ritchey B, Smith JD. Physiological difference in autophagic flux in macrophages from 2 mouse strains regulates cholesterol ester metabolism. Arterioscler Thromb Vasc Biol. 2013;33:903–910. doi: 10.1161/ATVBAHA.112.301041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen WX, Hu Q, Qiu MT, Zhong SL, Xu JJ, Tang JH, Zhao JH. Mir-221/222: Promising biomarkers for breast cancer. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2013 doi: 10.1007/s13277-013-0750-y. [DOI] [PubMed] [Google Scholar]
- 61.Chen Y, Liersch R, Detmar M. The mir-290-295 cluster suppresses autophagic cell death of melanoma cells. Scientific reports. 2012;2:808. doi: 10.1038/srep00808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Frankel LB, Wen J, Lees M, Hoyer-Hansen M, Farkas T, Krogh A, Jaattela M, Lund AH. Microrna-101 is a potent inhibitor of autophagy. The EMBO journal. 2011;30:4628–4641. doi: 10.1038/emboj.2011.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Korkmaz G, le Sage C, Tekirdag KA, Agami R, Gozuacik D. Mir-376b controls starvation and mtor inhibition-related autophagy by targeting atg4c and becn1. Autophagy. 2012;8:165–176. doi: 10.4161/auto.8.2.18351. [DOI] [PubMed] [Google Scholar]
- 64.Qased AB, Yi H, Liang N, Ma S, Qiao S, Liu X. Microrna-18a upregulates autophagy and ataxia telangiectasia mutated gene expression in hct116 colon cancer cells. Molecular medicine reports. 2013;7:559–564. doi: 10.3892/mmr.2012.1214. [DOI] [PubMed] [Google Scholar]
- 65.Tekirdag KA, Korkmaz G, Ozturk DG, Agami R, Gozuacik D. Mir181a regulates starvation- and rapamycin-induced autophagy through targeting of atg5. Autophagy. 2013;9:374–385. doi: 10.4161/auto.23117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu H, Wang F, Hu S, Yin C, Li X, Zhao S, Wang J, Yan X. Mir-20a and mir-106b negatively regulate autophagy induced by leucine deprivation via suppression of ulk1 expression in c2c12 myoblasts. Cellular signalling. 2012;24:2179–2186. doi: 10.1016/j.cellsig.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 67.Xu Y, An Y, Wang Y, Zhang C, Zhang H, Huang C, Jiang H, Wang X, Li X. Mir-101 inhibits autophagy and enhances cisplatin-induced apoptosis in hepatocellular carcinoma cells. Oncology reports. 2013;29:2019–2024. doi: 10.3892/or.2013.2338. [DOI] [PubMed] [Google Scholar]
- 68.Yang X, Zhong X, Tanyi JL, Shen J, Xu C, Gao P, Zheng TM, DeMichele A, Zhang L. Mir-30d regulates multiple genes in the autophagy pathway and impairs autophagy process in human cancer cells. Biochemical and biophysical research communications. 2013;431:617–622. doi: 10.1016/j.bbrc.2012.12.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu Y, Cao L, Yang L, Kang R, Lotze M, Tang D. Microrna 30a promotes autophagy in response to cancer therapy. Autophagy. 2012;8:853–855. doi: 10.4161/auto.20053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhai H, Song B, Xu X, Zhu W, Ju J. Inhibition of autophagy and tumor growth in colon cancer by mir-502. Oncogene. 2013;32:1570–1579. doi: 10.1038/onc.2012.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ucar A, Gupta SK, Fiedler J, Erikci E, Kardasinski M, Batkai S, Dangwal S, Kumarswamy R, Bang C, Holzmann A, Remke J, Caprio M, Jentzsch C, Engelhardt S, Geisendorf S, Glas C, Hofmann TG, Nessling M, Richter K, Schiffer M, Carrier L, Napp LC, Bauersachs J, Chowdhury K, Thum T. The mirna-212/132 family regulates both cardiac hypertrophy and cardiomyocyte autophagy. Nature communications. 2012;3:1078. doi: 10.1038/ncomms2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pan W, Zhong Y, Cheng C, Liu B, Wang L, Li A, Xiong L, Liu S. Mir-30-regulated autophagy mediates angiotensin ii-induced myocardial hypertrophy. PloS one. 2013;8:e53950. doi: 10.1371/journal.pone.0053950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Alvarez-Erviti L, Seow Y, Schapira AH, Rodriguez-Oroz MC, Obeso JA, Cooper JM. Influence of microrna deregulation on chaperone-mediated autophagy and alpha-synuclein pathology in parkinson's disease. Cell death & disease. 2013;4:e545. doi: 10.1038/cddis.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brest P, Lapaquette P, Mograbi B, Darfeuille-Michaud A, Hofman P. Risk predisposition for crohn disease: A “Menage a trios” Combining irgm allele, mirna and xenophagy. Autophagy. 2011;7:786–787. doi: 10.4161/auto.7.7.15595. [DOI] [PubMed] [Google Scholar]
- 75.Brest P, Lapaquette P, Souidi M, Lebrigand K, Cesaro A, Vouret-Craviari V, Mari B, Barbry P, Mosnier JF, Hebuterne X, Harel-Bellan A, Mograbi B, Darfeuille-Michaud A, Hofman P. A synonymous variant in irgm alters a binding site for mir-196 and causes deregulation of irgm-dependent xenophagy in crohn's disease. Nature genetics. 2011;43:242–245. doi: 10.1038/ng.762. [DOI] [PubMed] [Google Scholar]
- 76.Aiello RJ, Brees D, Bourassa PA, Royer L, Lindsey S, Coskran T, Haghpassand M, Francone OL. Increased atherosclerosis in hyperlipidemic mice with inactivation of abca1 in macrophages. Arterioscler Thromb Vasc Biol. 2002;22:630–637. doi: 10.1161/01.atv.0000014804.35824.da. [DOI] [PubMed] [Google Scholar]
- 77.van Eck M, Bos IS, Kaminski WE, Orso E, Rothe G, Twisk J, Bottcher A, Van Amersfoort ES, Christiansen-Weber TA, Fung-Leung WP, Van Berkel TJ, Schmitz G. Leukocyte abca1 controls susceptibility to atherosclerosis and macrophage recruitment into tissues. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:6298–6303. doi: 10.1073/pnas.092327399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Khera AV, Cuchel M, de la Llera-Moya M, Rodrigues A, Burke MF, Jafri K, French BC, Phillips JA, Mucksavage ML, Wilensky RL, Mohler ER, Rothblat GH, Rader DJ. Cholesterol efflux capacity, high-density lipoprotein function, and atherosclerosis. The New England journal of medicine. 2011;364:127–135. doi: 10.1056/NEJMoa1001689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rayner KJ, Sheedy FJ, Esau CC, Hussain FN, Temel RE, Parathath S, van Gils JM, Rayner AJ, Chang AN, Suarez Y, Fernandez-Hernando C, Fisher EA, Moore KJ. Antagonism of mir-33 in mice promotes reverse cholesterol transport and regression of atherosclerosis. J Clin Invest. 2011;121:2921–2931. doi: 10.1172/JCI57275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Allen RM, Marquart TJ, Albert CJ, Suchy FJ, Wang DQ, Ananthanarayanan M, Ford DA, Baldan A. Mir-33 controls the expression of biliary transporters, and mediates statin- and diet-induced hepatotoxicity. EMBO Mol Med. 2012;4:882–895. doi: 10.1002/emmm.201201228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wolf A, Bauer B, Hartz AM. Abc transporters and the alzheimer's disease enigma. Front Psychiatry. 2012;3:54. doi: 10.3389/fpsyt.2012.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Velagapudi VR, Hezaveh R, Reigstad CS, Gopalacharyulu P, Yetukuri L, Islam S, Felin J, Perkins R, Boren J, Oresic M, Backhed F. The gut microbiota modulates host energy and lipid metabolism in mice. J Lipid Res. 2010;51:1101–1112. doi: 10.1194/jlr.M002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, Britt EB, Fu X, Wu Y, Li L, Smith JD, Didonato JA, Chen J, Li H, Wu GD, Lewis JD, Warrier M, Brown JM, Krauss RM, Tang WH, Bushman FD, Lusis AJ, Hazen SL. Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. 2013;19:576–585. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang D, Xia M, Yan X, Li D, Wang L, Xu Y, Jin T, Ling W. Gut microbiota metabolism of anthocyanin promotes reverse cholesterol transport in mice via repressing mirna-10b. Circ Res. 2012;111:967–981. doi: 10.1161/CIRCRESAHA.112.266502. [DOI] [PubMed] [Google Scholar]
- 85.Wang D, Zou T, Yang Y, Yan X, Ling W. Cyanidin-3-o-beta-glucoside with the aid of its metabolite protocatechuic acid, reduces monocyte infiltration in apolipoprotein e-deficient mice. Biochem Pharmacol. 2011;82:713–719. doi: 10.1016/j.bcp.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 86.Wang L, Jia XJ, Jiang HJ, Du Y, Yang F, Si SY, Hong B. Micrornas 185, 96, and 223 repress selective high-density lipoprotein cholesterol uptake through posttranscriptional inhibition. Mol Cell Biol. 2013;33:1956–1964. doi: 10.1128/MCB.01580-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hu Z, Shen WJ, Kraemer FB, Azhar S. Micrornas 125a and 455 repress lipoprotein-supported steroidogenesis by targeting scavenger receptor class b type i in steroidogenic cells. Mol Cell Biol. 2012;32:5035–5045. doi: 10.1128/MCB.01002-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mohammadpour AH, Akhlaghi F. Future of cholesteryl ester transfer protein (cetp) inhibitors: A pharmacological perspective. Clin Pharmacokinet. 2013 doi: 10.1007/s40262-013-0071-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hewing B, Fisher EA. Rationale for cholesteryl ester transfer protein inhibition. Curr Opin Lipidol. 2012;23:372–376. doi: 10.1097/MOL.0b013e328353ef1d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rayner KJ, Hennessy EJ. Extracellular communication via microrna: Lipid particles have a new message. J Lipid Res. 2013;54:1174–1181. doi: 10.1194/jlr.R034991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Devaux Y, Vausort M, Goretti E, Nazarov PV, Azuaje F, Gilson G, Corsten MF, Schroen B, Lair ML, Heymans S, Wagner DR. Use of circulating micrornas to diagnose acute myocardial infarction. Clin Chem. 2012;58:559–567. doi: 10.1373/clinchem.2011.173823. [DOI] [PubMed] [Google Scholar]
- 92.Fiedler J, Thum T. Micrornas in myocardial infarction. Arterioscler Thromb Vasc Biol. 2013;33:201–205. doi: 10.1161/ATVBAHA.112.300137. [DOI] [PubMed] [Google Scholar]
- 93.Hergenreider E, Heydt S, Treguer K, Boettger T, Horrevoets AJ, Zeiher AM, Scheffer MP, Frangakis AS, Yin X, Mayr M, Braun T, Urbich C, Boon RA, Dimmeler S. Atheroprotective communication between endothelial cells and smooth muscle cells through mirnas. Nat Cell Biol. 2012;14:249–256. doi: 10.1038/ncb2441. [DOI] [PubMed] [Google Scholar]
- 94.Zhang Y, Liu D, Chen X, Li J, Li L, Bian Z, Sun F, Lu J, Yin Y, Cai X, Sun Q, Wang K, Ba Y, Wang Q, Wang D, Yang J, Liu P, Xu T, Yan Q, Zhang J, Zen K, Zhang CY. Secreted monocytic mir-150 enhances targeted endothelial cell migration. Molecular cell. 2010;39:133–144. doi: 10.1016/j.molcel.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 95.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. Micrornas are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wagner J, Riwanto M, Besler C, Knau A, Fichtlscherer S, Roxe T, Zeiher AM, Landmesser U, Dimmeler S. Characterization of levels and cellular transfer of circulating lipoprotein-bound micrornas. Arterioscler Thromb Vasc Biol. 2013;33:1392–1400. doi: 10.1161/ATVBAHA.112.300741. [DOI] [PubMed] [Google Scholar]
- 97.Hair P, Cameron F, McKeage K. Mipomersen sodium: First global approval. Drugs. 2013;73:487–493. doi: 10.1007/s40265-013-0042-2. [DOI] [PubMed] [Google Scholar]
- 98.Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, Zhou Y, Persson R, King BD, Kauppinen S, Levin AA, Hodges MR. Treatment of hcv infection by targeting microrna. The New England journal of medicine. 2013;368:1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 99.Feig JE, Parathath S, Rong JX, Mick SL, Vengrenyuk Y, Grauer L, Young SG, Fisher EA. Reversal of hyperlipidemia with a genetic switch favorably affects the content and inflammatory state of macrophages in atherosclerotic plaques. Circulation. 2011;123:989–998. doi: 10.1161/CIRCULATIONAHA.110.984146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Feig JE, Rong JX, Shamir R, Sanson M, Vengrenyuk Y, Liu J, Rayner K, Moore K, Garabedian M, Fisher EA. Hdl promotes rapid atherosclerosis regression in mice and alters inflammatory properties of plaque monocyte-derived cells. Proc Natl Acad Sci U S A. 2011;108:7166–7171. doi: 10.1073/pnas.1016086108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Horie T, Baba O, Kuwabara Y, Chujo Y, Watanabe S, Kinoshita M, Horiguchi M, Nakamura T, Chonabayashi K, Hishizawa M, Hasegawa K, Kume N, Yokode M, Kita T, Kimura T, Ono K. Microrna-33 deficiency reduces the progression of atherosclerotic plaque in apoe-/- mice. J Am Heart Assoc. 2012;1:e003376. doi: 10.1161/JAHA.112.003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rotllan N, Ramirez CM, Aryal B, Esau CC, Fernandez-Hernando C. Therapeutic silencing of microrna-33 inhibits the progression of atherosclerosis in ldlr-/- mice. Arterioscler Thromb Vasc Biol. 2013 doi: 10.1161/ATVBAHA.113.301732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Marquart TJ, Wu J, Lusis AJ, Baldan A. Anti-mir-33 therapy does not alter the progression of atherosclerosis in low-density lipoprotein receptor-deficient mice. Arterioscler Thromb Vasc Biol. 2013;33:455–458. doi: 10.1161/ATVBAHA.112.300639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wijesekara N, Zhang LH, Kang MH, Abraham T, Bhattacharjee A, Warnock GL, Verchere CB, Hayden MR. Mir-33a modulates abca1 expression, cholesterol accumulation, and insulin secretion in pancreatic islets. Diabetes. 2012;61:653–658. doi: 10.2337/db11-0944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hirsch-Reinshagen V, Maia LF, Burgess BL, Blain JF, Naus KE, McIsaac SA, Parkinson PF, Chan JY, Tansley GH, Hayden MR, Poirier J, Van Nostrand W, Wellington CL. The absence of abca1 decreases soluble apoe levels but does not diminish amyloid deposition in two murine models of alzheimer disease. J Biol Chem. 2005;280:43243–43256. doi: 10.1074/jbc.M508781200. [DOI] [PubMed] [Google Scholar]
- 106.Rader DJ, Tall AR. The not-so-simple hdl story: Is it time to revise the hdl cholesterol hypothesis? Nat Med. 2012;18:1344–1346. doi: 10.1038/nm.2937. [DOI] [PubMed] [Google Scholar]