Abstract
The emergence of vancomycin intermediate Staphylococcus aureus (VISA) and heterogeneous VISA (hVISA) is of major concern worldwide. Our objective was to investigate the prevalence, phenotypic and molecular features of hVISA strains isolated from bacteremic patients and to determine the clinical significance of the hVISA phenotype in patients with bacteremia. A total of 104 S. aureus blood isolates were collected from a teaching hospital of Argentina between August 2009 and November 2010. No VISA isolate was recovered, and 3 out of 92 patients (3.3%) were infected with hVISA, 2 of them methicillin-resistant S. aureus (MRSA) (4.5% of MRSA). Macro Etest and prediffusion method detected 3/3 and 2/3 hVISA respectively. Considering the type of bacteremia, the three cases were distributed as follows: two patients had suffered multiple episodes of bacteremia (both hVISA strains recovered in the second episode), while only one patient had suffered a single episode of bacteremia with hVISA infection. MRSA bloodstream isolates exhibiting the hVISA phenotype were related to HA-MRSA Cordobes clone (ST5-SCCmec I-spa t149) and MRSA Argentinean pediatric clone (ST100-SCCmec IVNV-spa t002), but not to CA-MRSA-ST30-SCCmec IV-spa t019 clone that was one of the most frequent in our country. Although still relatively infrequent in our hospital, hVISA strains were significantly associated with multiple episodes of bacteremia (p=0.037) and genetically unrelated.
Introduction
Staphylococcus aureus is a common pathogen associated with soft tissue infections, and more severe infections such as endocarditis, osteomyelitis, and bloodstream infections. The therapeutic choice in severe infections is often vancomycin, especially for methicillin-resistant S. aureus (MRSA).
Since 1997, when the first strains with reduced susceptibility to vancomycin intermediate S. aureus (VISA) were reported in Japan,17 there has been an increasing concern about the emergence of this phenotype around the world.
Heterogeneous VISA (hVISA) are defined as strains with minimal inhibitory concentrations (MICs) in the susceptible range (MIC ≤2 μg/ml), but containing subpopulations of cells in the vancomycin-intermediate range (VISA, MIC 4–8 μg/ml).6 These strains have been described for both MRSA and methicillin-susceptible S. aureus (MSSA) respectively.1,35
hVISA strains appear to be an early stage in the development of VISA and have been related to persistent bacteremia, greater rates of complications, and vancomycin treatment failure. Some authors consider that this phenotype could be responsible for treatment failure, whereas others have proposed that it has arisen as a consequence of treatment failure and prolonged vancomycin exposures.18
As hVISA strains enter the susceptible range by MIC conventional methods such as microdilution, clinical laboratory detection is very difficult. So far, the gold standard is the population analysis profile-area under the curve (PAP-AUC) method, but it is cost- and time consuming, and labor intensive. Other methods such as macro Etest and screening plates have been described but none of them has the sensitivity and specificity of the PAP-AUC method.20
The prevalence of hVISA varies depending on the geographic location, the population under study and the methodology applied.29,34,46 Because of the multiplicity of tests and the lack of standardized procedures, it is difficult to compare results between studies. There has been special interest in vancomycin MIC creep and treatment outcomes over the years.18,22,37 Recently, we have reported a case of endocarditis due to hVISA infection in the Hospital de Clínicas José de San Martín, a teaching hospital of Buenos Aires, Argentina.35 A few months later, in the same institution, another hVISA appeared, generating concern (described in XX-Congreso Latinoamericano de Microbiología; Montevideo-2010, unpublished data). The Hospital de Clínicas José de San Martín is a 500-bed general hospital, is the main hospital of the University of Buenos Aires, and receives patients from all over the country. Despite the few cases reported in Argentina,35,42,43 the prevalence, molecular epidemiology, and clinical significance of hVISA strains in our country are unknown because no standardized epidemiological studies have been performed. This information is necessary to decide the antibiotic therapy in patients with long-term vancomycin treatment. It is also necessary to have an appropriate method or a combination of methods suitable for accurate and easier detection of hVISA in clinical settings.
This study had three main objectives: (i) to define the prevalence and evaluate different methods to detect the hVISA phenotype among S. aureus bacteremia in a teaching hospital; (ii) to evaluate the genotypic characteristics of MRSA bloodstream isolates exhibiting the hVISA phenotype, and (iii) to assess the clinical significance of the hVISA phenotype in patients with bacteremia by S. aureus.
Materials and Methods
Isolates and study design
A prospective observational study was designed to evaluate the prevalence, phenotypic and molecular features of hVISA strains from all S. aureus isolates recovered from bloodstream infections at Hospital de Clínicas José de San Martín, between August 2009 and November 2010. Only one isolate from each patient was included in the analysis, but isolates from subsequent blood samples obtained from patients with multiple episodes of bacteremia were also stored for vancomycin susceptibility testing. Persistent bacteremia was defined as that lasting 7 days or more, despite administration of antibiotic to which the isolate was susceptible to in vitro, whereas recurrent bacteremia was defined as the return of S. aureus subsequent to documentation of negative blood cultures or clinical improvement, after completing a course of antistaphylococcal therapy. The study was approved by the Institutional Review Board.
All blood cultures were analyzed using BacT/ALERT® (BioMerieux, Inc., Durham, NC) and all S. aureus isolates were identified by standard methods.
Susceptibility testing
MIC to antimicrobial agents (oxacillin, cefoxitin, gentamicin, erythromycin, clindamycin, tetracycline, trimethoprim/sulfamethoxazole, ciprofloxacin, tigecycline, rifampicin, vancomycin, teicoplanin, and linezolid; all from Sigma-Aldrich, St Louis, MO) was determined by the agar dilution procedure using Mueller-Hinton agar (MHA; Difco BD, Sparks, MD) according to Clinical and Laboratory Standards Institute (CLSI) guidelines. Inducible clindamycin resistance in isolates displaying erythromycin resistance was detected by the D-test according to the recommendations of the CLSI.
Additionally, vancomycin MIC was determined by Etest® (AB BioMerieux, Solna, Sweeden) according to the manufacturer's instructions.
CLSI interpretation criteria were used for all antimicrobial agents except for tigecycline, for which FDA/EUCAST breakpoints were used.6
Screening for hVISA
All clinical isolates recovered in the study (n=104) were screened for hVISA and those positive by more than one method were confirmed by PAP-AUC. S. aureus Mu3 (hVISA), Mu50 (VISA), and ATCC 29213 (vancomycin-susceptible S. aureus [VSSA]) were used as control strains in all the experiments. Mu3 and Mu50 were obtained from the Network on Antimicrobial Resistance in S. aureus (NARSA) strain collection. The screening methods included were as follows:
(a) Screening plates: BHIV3, BHIV4, BHIV5, BHIV6 (Brain Heart Infusion agar [BHI; Difco BD] containing 3, 4, 5, and 6 μg/ml vancomycin respectively), and MHAT5 (MHA containing 5 μg/ml teicoplanin) were assayed as previously described using 10 μl26 but also 1 μl of bacterial suspensions equivalent to 0.5 McFarland for BHI screening plates, and equivalent to 2.0 McFarland for MHAT5 screening plates. Plates were incubated for 48 hr at 35°C and growth was considered as a positive result at any condition.
(b) Macro Etest: Inoculum was prepared from overnight growth on blood agar plate; 200 μl of bacterial suspension in saline solution equivalent to a 2.0 McFarland was plated and streaked onto BHI agar and vancomycin Etest strips® (AB BioMerieux) were applied. Plates were incubated for 48 hr at 35°C and values above or equal to 6 μg/ml were considered as a positive result.20
(c) Prediffusion method: Vancomycin 30 μg and teicoplanin 30 μg Neo-Sensitabs™ tablets (Rosco Diagnostica, Taastrup, Denmark) were prediffused for 2 hr on MHA plates, and after removing the tablets, the plates were maintained at room temperature for 18 hr. A bacterial suspension equivalent to a 0.5 McFarland inoculum was streaked. Plates were incubated for 24 hr at 35°C. The results were interpreted following the manufacturer's recommendation: vancomycin inhibition zone diameter ≤22 mm and/or teicoplanin inhibition zone diameter <20 mm were considered as a positive result.23,38
PAP-AUC method
PAP-AUC was performed for strains displaying at least two positive results on different screening assays/screening media. Briefly, serial 10-fold dilutions of cultures were plated onto increasing concentrations of vancomycin BHI agar (0, 0.25, 0.5, 1, 1.5, 2.0, 3.0, 4.0, 6.0, 8.0, and 10 μg/ml) (Difco BD). Colony growth at 48 hr was counted and viable count (log10 CFU/ml) was plotted against vancomycin concentration using GraphPad Prism Software. S. aureus ATCC 29213 (VSSA), Mu50 (VISA), and Mu3 (hVISA) were included. The AUC was measured for each sample and the ratio of test isolate AUC/mean Mu3 AUC was calculated. As described by Wootton et al., the criteria to define hVISA and VSSA were a PAP/AUC ratio ≥0.90 and a PAP/AUC ratio <0.90 respectively.49
PCR identification of mecA, lukS/F-PV, SCCmec, and agr types
Detection of mecA and PVL encoding genes (lukS/F-PV) was attempted after extraction of genomic DNA for all MRSA isolates as previously described.47 SCCmec types were determined by PCR with a simplified version of Kondo's typing system, including M-PCR-1 and M-PCR-2.25 In specific cases, SCCmec typing was performed as recommended by Oliveira and de Lencastre,33 and SCCmec IVNV characterization using primers described by Sola et al.43
The agr locus was genotyped by multiplex PCR, as previously described.15
Genetic analysis by pulsed-field gel electrophoresis, multilocus sequence typing, and spa typing
Genotyping analysis was conducted using pulsed-field gel electrophoresis (PFGE) with SmaI, as previously described.5 The PFGE fingerprints were compared by the unweighted pair-group method with arithmetic mean (UPGMA) clustering analysis, applying the Dice correlation coefficient. MRSA clones previously described in Argentina were included in the PFGE pattern analysis.8,13,41 Multilocus sequence typing (MLST) was performed for representative isolates of the main pulsotypes.7 Confirmed hVISA strains were also studied by spa typing16 and MLST.
Statistical analysis
p-Values were calculated by χ2 Yates test and Fisher's exact test for categorical variables. A p-value <0.05 was considered significant.
Results
Antimicrobial susceptibility and molecular characterization of isolates
Forty-eight out of the 92 (52.2%) patients with S. aureus bacteremia included in the study were infected by MSSA, whereas 44 (47.8%) were infected by MRSA. Table 1 compares the antimicrobial resistance rates of MSSA and MRSA among S. aureus blood isolates. MRSA isolates were more likely to be resistant to gentamicin, erythromycin/clindamycin, and ciprofloxacin (p<0.0001). As revealed by the D-test, in the MSSA group, 9/9 erythromycin-resistant isolates displayed inducible clindamycin resistance. By contrast, in the MRSA group, only 5/26 erythromycin-resistant isolates displayed the inducible phenotype, 1/26 was negative by the D-test, and the remaining isolates (20/26) showed constitutive clindamycin resistance. All isolates were susceptible to linezolid, tigecycline, vancomycin, and teicoplanin.
Table 1.
Antimicrobial Resistance Rates of Methicillin-Susceptible S. aureus and Methicillin-Resistant Staphylococcus aureus Among S. aureus Blood Isolates
| MSSA (n=48) | MRSA (n=44) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| (μg/ml) | (μg/ml) | ||||||||
| Antimicrobial agent | Range | MIC50 | MIC90 | %Ra | Range | MIC50 | MIC90 | %Ra | p-Value |
| Gentamicin | 0.063 to 128 | 0.25 | 32 | 12.5 | <0.063 to >512 | 32 | >512 | 61.4 | <0.0001 |
| Erythromycin | 0.125 to >512 | 0.25 | >512 | 18.75 | 0.125 to >512 | >512 | >512 | 59 | 0.0002 |
| Clindamycin | <0.063 to 0.125 | <0.063 | 0.063 | 18.75 | <0.063 to >512 | 0.063 | >512 | 56.8 | 0.0004 |
| Tetracycline | 0.063 to 1 | 0.25 | 0.5 | 0 | 0.063 to 64 | 0.125 | 0.5 | 4.5 | 0.23 |
| TMP-SMX | 0.25 to 1 | 1 | 1 | 0 | 0.25 to >512 | 1 | 2 | 4.5 | 0.23 |
| Ciprofloxacin | 0.063 to 8 | 0.25 | 1 | 8.3 | 0.063 to 256 | 8 | 64 | 59 | <0.0001 |
| Tigecycline | 0.125 to 1 | 0.25 | 0.5 | 0 | 0.063 to 1 | 0.25 | 1 | 0 | — |
| Rifampicin | <0.002 to 0.008 | 0.004 | 0.016 | 0 | <0.002 to 2 | 0.008 | 0.5 | 9.1 | 0.05 |
| Vancomycin | 0.5 to 2 | 1 | 1 | 0 | 0.5 to 2 | 1 | 1 | 0 | — |
| Teicoplanin | 0.25 to 4 | 0.5 | 1 | 0 | 0.125 to 2 | 1 | 1 | 0 | — |
| Linezolid | 1 to 4 | 2 | 2 | 0 | 1 to 2 | 2 | 2 | 0 | — |
%R, percentage of resistant isolates.
MIC, minimal inhibitory concentration; MRSA, methicillin-resistant S. aureus; MSSA, methicillin-susceptible S. aureus; TMP-SMX, trimethoprim/sulfamethoxazole.
The molecular characterization of the MRSA group revealed 43/44 isolates harboring the mecA gene. The most frequent SCCmec types were SCCmec I and IV (41.8% and 39.5% respectively) and the most frequent agr group among them was the agr group II (53.5%). Only 13/44 (29.5%) harbored the PVL coding genes (lukS/F-PV), and all PVL-positive isolates also harbored SCCmec IVc or SCCmec IVa.
PFGE analysis revealed that 14/18 MRSA isolates carrying SCCmec I corresponded to the Cordobés clone (pulsotype D-ST5-SCCmec I).12,41 Most isolates carrying SCCmec IV were related to the main community-acquired MRSA (CA-MRSA) clones described in Argentina: pulsotype C (ST30-SCCmec IVc) and pulsotype A (ST5-SCCmec IVa) (nine and five isolates respectively) (Table 2).8,11,13,31,43
Table 2.
Methicillin-Resistant S. aureus Clones Detected According to SCCmec Type, Pulsed-Field Gel Electrophoresis, and Multilocus Sequence Typing
| SCCmec type | No. of isolates | PFGE pulsotype | STa | Clonally related clone |
|---|---|---|---|---|
| I | 14 | D | 5 | Cordobés clone (ST5-I) |
| 4 | Other | NDb | ND | |
| IVc | 9 | C | 30 | Southwest Pacific clone (ST30-IV) |
| IVa | 5 | A | 5 | CAA clone (ST5-IV)b |
| IVNVc | 2 | B | 100 | Pediatric clone (ST100-IV) |
| IVvard | 1 | E | 72 | USA700 |
| V | 2 | Other | ND | ND |
| III | 1 | Other | ND | ND |
| NT | 5 | Other | ND | ND |
ST, sequence type. ST was determined for one representative isolate of each PFGE pulsotype.
CAA clone (ST5-IV), community-acquired MRSA described in Argentina (PFGE A, ST5-IV).13
IVNV, Tn4001 was detected integrated into the class B mec complex.
IVvar, tnp20 was detected integrated into the class B mec complex.
ND, not determined; NT, not typeable; PFGE, pulsed-field gel electrophoresis.
Vancomycin susceptibility and prevalence of hVISA
Vancomycin susceptibility analysis included 104 clinical isolates recovered from 92 patients with one or more episodes of bacteremia by S. aureus. Eleven patients had suffered multiple episodes of bacteremia (2 of them suffered persistent bacteremia and 9 recurrent bacteremia).
No VISA (vancomycin MIC from 4 to 8 μg/ml) or vancomycin-resistant (MIC ≥16 μg/ml) S. aureus strains were detected by agar dilution MIC or Etest. All isolates were susceptible to teicoplanin with MIC ≤4 μg/ml. By agar dilution, vancomycin MIC90 was 1 μg/ml (range 0.5–2 μg/ml), while by the Etest, MIC90 was 1.5 μg/ml (range 0.38–2 μg/ml).
Eleven isolates were considered to be hVISA because they were positive by at least two screening assays (including screening plates, Macro Etest, and prediffusion methods), and subsequently analyzed by the PAP-AUC method. Only three were confirmed as hVISA. Two hVISA strains were MRSA, while the other was a MSSA strain (Table 3). All hVISA isolates displayed Etest MIC >1 μg/ml.
Table 3.
Phenotypic and Genotypic Features of Presumable and Confirmed Heterogeneous Vancomycin Intermediate S. aureus
| Presumable hVISA | MIC (μg/ml) | Screening plates 48 hr (10 μl) | Prediffusion zone diameter (mm) | PAP-AUC | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Isolate | Genotypic features | VAN | TEI | Etest VAN (μg/ml) | Macro Etest VAN (μg/ml) | MHAT5 | BHIV3 | BHIV4 | BHIV5 | BHIV6 | VAN 30 μg | TEI 30 μg | PAP-AUC Mu3 | Interpretation |
| 55703 | SCCmec I agr II, luk-PV− |
1 | 1 | 1.5 | 6 | − | − | − | − | − | 22 | 25 | 1.05 | hVISA |
| 55576 | SCCmec IVNV agr II, luk-PV− |
1 | 1 | 2 | 6 | + | + | − | − | − | 24 | 20 | 0.96 | hVISA |
| 53079 | MSSA | 2 | 4 | 2 | 6 | − | + | − | − | − | 20 | 13 | 1.02 | hVISA |
| 53735 | SCCmec I agr II, luk-PV− |
1 | 2 | 1 | 4 | + | + | − | − | − | 24 | 10 | 0.49 | VSSA |
| 57748 | SCCmec V agr NT, luk-PV− |
1 | 2 | 1 | 4 | + | + | − | − | − | 25 | 11 | 0.37 | VSSA |
| 54467 | SCCmec I agr II, luk-PV− |
1 | 1 | 0.75 | 4 | + | + | − | − | − | 28 | 26 | 0.39 | VSSA |
| 54925 | MSSA | 1 | 1 | 0.75 | 3 | − | + | + | − | − | 28 | 27 | 0.49 | VSSA |
| 55133 | SCCmec I agr II, luk-PV− |
1 | 1 | 1 | 3 | + | + | − | − | − | 30 | 27 | 0.36 | VSSA |
| 56617 | SCCmec I agr II, luk-PV− |
1 | 0.5 | 0.5 | 4 | + | + | + | − | − | 28 | 27 | 0.36 | VSSA |
| 58269 | MSSA | 1 | 1 | 1 | 4 | + | + | − | − | − | 24 | 20 | 0.52 | VSSA |
| 55262 | SCCmec NT agr II, luk-PV− |
0.5 | 0.5 | 0.5 | 4 | + | + | − | − | − | 25 | 24 | 0.32 | VSSA |
hVISA, heterogeneous vancomycin intermediate S. aureus; PAP-AUC, population analysis profile-area under the curve; VSSA, vancomycin-susceptible S. aureus.
Twenty-seven isolates grew on at least one screening assay/medium. Screening plates were tested with two different inocula. Using 10 μl (0.5 McFarland for BHI screening plates and 2.0 McFarland for MHAT5 screening plates), we found 19 positive isolates with MHAT5 (one of them confirmed as hVISA), 15 positive isolates with BHIV3 (two of them confirmed as hVISA), and 2 positive isolates with BHIV4. On the other hand, using 1 μl, only one isolate was positive with MHA5T and three with BHIV3 (one of them confirmed as hVISA). No single isolate was positive with BHIV5 or BHIV6 at any condition.
The macro Etest successfully detected 3/3 hVISA isolates, whereas the prediffusion method detected four presumable hVISA, being only two confirmed by PAP-AUC (Table 3).
Clinical, microbiological, and molecular evaluation of hVISA bacteremias
Three out of the 92 (3.3%) patients suffering bacteremia at Hospital de Clínicas during 2009–2010 had hVISA. Considering the type of bacteremia, the three cases were distributed as follows: 2/11 patients had suffered multiple episodes of bacteremia (both hVISA strains recovered in the second episode), while only 1/81 patients had suffered a single episode of bacteremia with hVISA infection. These results indicate that hVISA isolation was significantly associated with multiple episodes of bacteremia (p=0.037).
In patient 1, isolates were recovered from persistent bacteremia. The patient suffered endocarditis secondary to a catheter-related bacteremia, and did not undergo surgery. The patient had no prior S. aureus infection and no history of treatment with vancomycin. Isolate 55703 (hVISA) was recovered after vancomycin treatment, and exhibited a higher PAP/AUC ratio than the first blood culture isolate (55603, VSSA). By contrast, patient 2 had prior S. aureus infection and history of vancomycin treatment. This case was a recurrent bacteremia and isolate 55343 was recovered from blood culture without a known source. Isolate 55576 (hVISA) was recovered after vancomycin treatment failure, and also exhibited a higher PAP/AUC than the first isolate (55343, VSSA). Both patients 1 and 2 died. The clonality of the isolates recovered before and after vancomycin treatment was demonstrated by PFGE (Fig. 1). In patient 3, the isolate recovered (53079) corresponded to a MSSA single bacteremia secondary to parotiditis, which, after treatment with β-lactams, had a good clinical outcome.
FIG. 1.
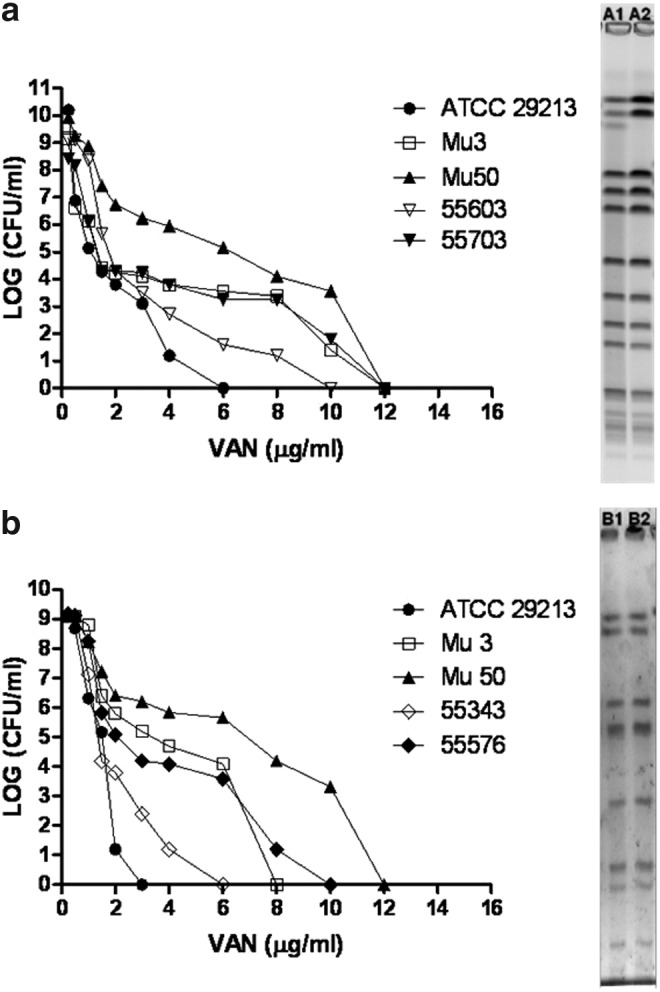
Population analysis profile and pulsed-field gel electrophoresis of the heterogeneous vancomycin intermediate Staphylococcus aureus (hVISA) strains recovered from multiple episodes of S. aureus bacteremia. ● ATCC 29213, □ Mu3, and ▲ Mu50. (a) Isolates recovered from patient 1: ▽ 55603 (first isolate, A1, population analysis profile-area under the curve [PAP-AUC] ratio: 0.73) and ▼ 55703 (isolate recovered after vancomycin treatment, A2, PAP-AUC ratio: 1.05). (b) Isolates recovered from patient 2: ◊55343 (first isolate, B1, PAP-AUC ratio: 0.47) and ♦ 55576 (isolate recovered after vancomycin treatment, B2, PAP-AUC ratio: 0.96).
As revealed by MLST, PFGE macrorestriction analyses, and spa typing, hVISA strains were genetically unrelated (Table 4). Isolate 55703 (ST5-SCCmec I-agr II, luk-PV negative) was characterized as spa t149-pulsotype D, revealing that this isolate is a subtype of the Cordobés clone, previously described as prevalent in this hospital.12 Isolate 55576 (ST100-SCCmec IVNV, -agr II-luk-PV negative) was spa t002-pulsotype B, had a Tn4001 insertion into the class B mec complex, and is related to the Argentine pediatric clone.43 Isolate 53079 was MSSA spa t267 and belonged to ST97, a sequence type predominant in bovines with mastitis and also described in Argentina causing osteomyelitis.28
Table 4.
Genotyping of Heterogeneous Vancomycin Intermediate S. aureus Isolates and Main Methicillin-Resistant S. aureus Clones Described in Argentina
| Strain | SCCmec | Allelic profile (arc-aro-glp-gmk-pta-tpi-yqi) | ST | CC | spa type |
|---|---|---|---|---|---|
| 55703 (hVISA) | I | 1-4-1-4-12-1-10 | 5 | 5 | t149 |
| 55576 (hVISA) | IVNV | 1-65-1-4-12-1-10 | 100 | 5 | t002 |
| 53079 (hVISA) | — | 3-1-1-1-1-5-3 | 97 | 97 | t267 |
| Cordobés clone | I | 1-4-1-4-12-1-10 | 5 | 5 | t149 |
| CAA clonea | IVa | 1-4-1-4-12-1-10 | 5 | 5 | t311 |
| Southwest Pacific clone | IVc | 2-2-2-2-6-3-2 | 30 | 30 | t019 |
CAA clone, community-acquired MRSA described in Argentina (PFGE A, ST5-IV).14
CC, clonal complex.
Discussion
In this study, we describe for the first time the prevalence and molecular characterization of hVISA isolates in a teaching hospital of Buenos Aires (Argentina) during 2009–2010. No VISA isolate was recovered, and the rate of hVISA was 3.3% of the S. aureus bloodstream infections (4.5% of MRSA). This is a low prevalence value when compared with results obtained by other researchers. In an Italian multicenter surveillance study conducted between 2007 and 2009, the frequency of hVISA was 19.5%.3 A higher rate (47.8% of MRSA) was reported by Horne at Austin Hospital (Australia) during 2005–2006 considering all clinical specimens.19 However, the rate found in our study is similar to that described for blood culture isolates in Taiwan.29
As reviewed by Howden et al., changes in hVISA can occur over time within specific institutions.20 In Argentina, hVISA strains were described in 2009 for the first time, one of them in our institution (Hospital de Clínicas).35,42 Considering the changing epidemiology of S. aureus and the impact of local antibiotic use politics it is difficult to predict future trends; so further surveillance epidemiological studies in this center as well as in other institutions of our country need to be conducted.
The selection of an optimal laboratory screening test for hVISA remains to be difficult. Difficulties are mainly based on the differences experienced by researchers in terms of specificity, sensitivity, and population under study. Different screening plates have been extensively studied in terms of specificity and sensitivity,20 but ultimately all presumable hVISA isolates should be confirmed by PAP-AUC. In our hands, none of the screening plates detected all three hVISA described in this study. BHIV3 detected two of them, but, as previously described, it has a high false positive rate.26 Two isolates were detected as presumable hVISA by MHAT5 and teicoplanin prediffusion tablets, but not by vancomycin-based screening tests, and PAP-AUC revealed that these isolates were not hVISA. Isolates exhibiting a reduction in teicoplanin susceptibility may not always display a reduction in vancomycin susceptibility.
Macro Etest has been proposed as an accurate method to detect hVISA. Surprisingly, while the manufacturer's breakpoint is 8 μg/ml, the highest value among all clinical isolates recovered in our study, including the three hVISA strains was 6 μg/ml. Only one isolate, which was not confirmed as hVISA by PAP-AUC, exhibited macro Etest result of 6 μg/ml, and was negative by other screening tests (data not shown). The prediffusion method has been described for the detection of colistin-resistance, daptomycin-nonsusceptible strains, and the hVISA/VISA phenotype.2,23,32 Although this screening method is not expensive and might represent a simpler alternative for hVISA detection in clinical laboratories, it has not been extensively evaluated. Despite the two false positives, we had acceptable results with the prediffusion method (2/3 hVISA isolates detected).
Our results show that macro Etest with a breakpoint ≥6 μg/ml (lower than the value proposed by the manufacturer) could be used to predict likelihood of hVISA in laboratories where PAP-AUC testing is not performed routinely, especially in cases of persistent/recurrent bacteremia. Alternatively, prediffusion method should be considered a very good option as the cost of the tablets is comparatively convenient.
The three hVISA isolates identified in this study presented vancomycin MIC values ≥1.5 μg/ml by the Etest. In agreement with various reports, hVISA isolates displayed vancomycin Etest MIC values close to the breakpoint (2 μg/ml) and higher than those determined by the agar dilution method.36,39 Additionally, two of the three cases were associated with mortality. The role of vancomycin in the treatment of high but susceptible vancomycin MIC isolates of MRSA has raised controversial discussions. Several studies have explored the clinical significance of vancomycin MIC in S. aureus infections and have demonstrated episodes of vancomycin treatment failure with a high MIC (≥1.5 μg/ml by the Etest).44,45,48 By contrast, others authors observed no association of mortality with increasing vancomycin MICs.27,40 However, treatment recommendations continue to support vancomycin for MRSA bacteriemia for isolates with vancomycin MIC values ≤2 μg/ml, and changes to therapy should be guided by the patient's clinical response.30
One MRSA isolate was mecA negative and mecC negative (data not shown), suggesting that the mechanism responsible for β-lactam resistance might be β-lactamase hyperproduction or changes in affinity of penicillin-binding proteins.
Although the hVISA phenotype has been described mainly from MRSA, it has also been reported from MSSA strains.1,14,35 Our results revealed that 2/3 cases were MRSA and 1/3 MSSA strains, all of them presenting different genotypes. Therefore, hVISA strains recovered during the study period are not clonally related, indicating that this emergence is the result of independent genetic events. By contrast, other surveillance studies have demonstrated hVISA clonal dissemination with higher prevalence values.14
The two MRSA hVISA strains belong to clonal complex 5, one of them related to HA-MRSA Cordobés clone (ST5-SCCmec I) and the other to the Argentine pediatric clone (ST100-SCCmec IVNV). Clonal complex 5 is widely distributed in Argentina and also around the world.4,9,10,43 It has been described that this genotype has an extraordinary genomic plasticity, which might explain the large proportion of hVISA, VISA and all the VRSA strains belonging to this lineage.21,24
Although the ST30-SCCmec IVc-pulsotype C clone, recently described as the main cause of CA-MRSA infections in Argentina,8,31 was the most frequent CA-MRSA clone recovered in this study, we found no hVISA strains belonging to this lineage. To our knowledge, only one hVISA strain has been described within Clonal Complex 30, carrying SCCmec I.21
Regarding the clinical significance of these strains, in this study we found a significant association between hVISA phenotype and multiple episodes of bacteremia. In addition, 2/3 hVISA were MRSA strains recovered from persistent/recurrent bacteremias and selected after vancomycin treatment. As reviewed by Howden et al.,20 the emergence of hVISA after vancomycin therapy has been reported by several authors. Therefore, the presence of this phenotype should be considered in face of patients with these clinical situations. However, one hVISA isolate was recovered from a MSSA bacteremia not treated with vancomycin. Related to this, in a retrospective study, Yamakawa et al. reported the emergence of hVISA before vancomycin introduction in Japan.50
Limitations of this work should be mentioned. The small number of hVISA isolated in this study are insufficient to generalize conclusions about the appropriate screening methods for the detection of this phenotype and our observations have to be corroborated with a higher number of strains by further surveillance studies in Argentinean clinical settings. Nevertheless, these results constitute an important contribution to phenotypic, genetic, and clinical knowledge of hVISA, especially considering that most of the published reports originate from developed nations, and data from Latin American countries are grossly underrepresented. Nonetheless, further epidemiological surveillance in Argentinean settings is required for a better understanding of this phenomenon and its evolution.
Conclusions
This study makes several key observations. We describe for the first time the prevalence and molecular characterization of hVISA isolates in a teaching hospital of Buenos Aires (Argentina). The hVISA phenotype occurred in 3.3% of the S. aureus bloodstream infections and no VISA isolate was recovered. Even if the prevalence is still relatively low compared to other studies, the clinical impact is concerning as a significant association between hVISA phenotype and multiple episodes of bacteremia isolates was determined. The majority of reported hVISA and VISA isolates evolved from MRSA strains, nevertheless in this study 1/3 hVISA were detected among MSSA isolates, consequently this result highlight the importance of screening hVISA phenotype in MSSA isolates as in MRSA.
The results of molecular typing indicate that the hVISA isolates were genetically diverse. The two methicillin-resistant hVISA isolates belong to MRSA clones previously disseminated in Argentina.
In addition, our results show that the combination of Etest MIC >1 μg/ml, macro Etest value ≥6 μg/ml, and the prediffusion method could be considered in future studies as useful strategies to predict likelihood of hVISA in laboratories where PAP-AUC testing is not performed routinely, especially in cases of persistent/recurrent bacteremia.
Acknowledgments
This work was supported in part by grants from University of Buenos Aires, Argentina (20020100100510 and 20020100100296, 2011–2014) to M.M. and A.F., Agencia Nacional de Promoción Científica y Tecnológica (PICT 1634 and 2362) and Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) (PIP Nro 11220110100707CO) to M.M. M.M. is member of “Carrera del Investigador” of CONICET. S.D.G. is a doctoral fellow of CONICET.
Disclosure Statement
No competing financial interests exist.
References
- 1.Bobin-Dubreux S., Reverdy M.E., Nervi C., Rougier M., Bolmstrom A., Vandenesch F., and Etienne J.2001. Clinical isolate of vancomycin-heterointermediate Staphylococcus aureus susceptible to methicillin and in vitro selection of a vancomycin-resistant derivative. Antimicrob. Agents Chemother. 45:349–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyen F., Vangroenweghe F., Butaye P., De Graef E., Castryck F., Heylen P., Vanrobaeys M., and Haesebrouck F.2010. Disk prediffusion is a reliable method for testing colistin susceptibility in porcine E. coli strains. Vet. Microbiol. 144:359–362 [DOI] [PubMed] [Google Scholar]
- 3.Campanile F., Bongiorno D., Falcone M., Vailati F., Pasticci M.B., Perez M., Raglio A., Rumpianesi F., Scuderi C., Suter F., et al. 2012. Changing Italian nosocomial-community trends and heteroresistance in Staphylococcus aureus from bacteremia and endocarditis. Eur. J. Clin. Microbiol. Infect. Dis. 31:739–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chatterjee S.S., and Otto M.2013. Improved understanding of factors driving methicillin-resistant Staphylococcus aureus epidemic waves. Clin. Epidemiol. 5:205–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung M., de Lencastre H., Matthews P., Tomasz A., Adamsson I., Aires de Sousa M., Camou T., Cocuzza C., Corso A., Couto I., et al. 2000. Molecular typing of methicillin-resistant Staphylococcus aureus by pulsed-field gel electrophoresis: comparison of results obtained in a multilaboratory effort using identical protocols and MRSA strains. Microb. Drug Resist. 6:189–198 [DOI] [PubMed] [Google Scholar]
- 6.CLSI. 2012. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically. Approved Standard M7-A7. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 7.Enright M.C., Knox K., Griffiths D., Crook D.W., and Spratt B.G.2000. Molecular typing of bacteria directly from cerebrospinal fluid. Eur. J. Clin. Microbiol. Infect. Dis. 19:627–630 [DOI] [PubMed] [Google Scholar]
- 8.Fernandez S., de Vedia L., Lopez Furst M.J., Gardella N., Di Gregorio S., Ganaha M.C., Prieto S., Carbone E., Lista N., Rotrying F., et al. 2013. Methicillin-resistant Staphylococcus aureus ST30-SCCmec IVc clone as the major cause of community-acquired invasive infections in Argentina. Infect. Genet. Evol. 14:401–405 [DOI] [PubMed] [Google Scholar]
- 9.Gardella N., Fernandez S., Di Gregorio S., Cuirolo A., Gutkind G., and Mollerach M. [Comparative study of clones from isolates methicillin-resistant Staphylococcus aureus prevalent in Argentina]. Rev. Panam. Salud Publica 30:665–666 [DOI] [PubMed] [Google Scholar]
- 10.Gardella N., Fernandez S., Gregorio S.D., Cuirolo A., Gutkind G., and Mollerach M.2011. Estudio comparativo de los clones de aislamientos de Staphylococcus aureus resistente a meticilina prevalentes en la Argentina. Rev. Panam. Salud Pública 30:665–666 [DOI] [PubMed] [Google Scholar]
- 11.Gardella N., Murzicato S., Di Gregorio S., Cuirolo A., Desse J., Crudo F., Gutkind G., and Mollerach M.2011. Prevalence and characterization of methicillin-resistant Staphylococcus aureus among healthy children in a city of Argentina. Infect. Genet. Evol. 11:1066–1071 [DOI] [PubMed] [Google Scholar]
- 12.Gardella N., Picasso R., Predari S.C., Lasala M., Foccoli M., Benchetrit G., Famiglietti A., Catalano M., Mollerach M., and Gutkind G.2005. Methicillin-resistant Staphylococcus aureus strains in Buenos Aires teaching hospitals: replacement of the multidrug resistant South American clone by another susceptible to rifampin, minocycline and trimethoprim-sulfamethoxazole. Rev. Argent. Microbiol. 37:156–160 [PubMed] [Google Scholar]
- 13.Gardella N., von Specht M., Cuirolo A., Rosato A., Gutkind G., and Mollerach M.2008. Community-associated methicillin-resistant Staphylococcus aureus, eastern Argentina. Diagn. Microbiol. Infect. Dis. 62:343–347 [DOI] [PubMed] [Google Scholar]
- 14.Garnier F., Chainier D., Walsh T., Karlsson A., Bolmstrom A., Grelaud C., Mounier M., Denis F., and Ploy M.C.2006. A 1 year surveillance study of glycopeptide-intermediate Staphylococcus aureus strains in a French hospital. J. Antimicrob. Chemother. 57:146–149 [DOI] [PubMed] [Google Scholar]
- 15.Gilot P., Lina G., Cochard T., and Poutrel B.2002. Analysis of the genetic variability of genes encoding the RNA III-activating components Agr and TRAP in a population of Staphylococcus aureus strains isolated from cows with mastitis. J. Clin. Microbiol. 40:4060–4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harmsen D., Claus H., Witte W., Rothganger J., Turnwald D., and Vogel U.2003. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 41:5442–5448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hiramatsu K., Hanaki H., Ino T., Yabuta K., Oguri T., and Tenover F.C.1997. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J. Antimicrob. Chemother. 40:135–136 [DOI] [PubMed] [Google Scholar]
- 18.Holmes N.E., Johnson P.D., and Howden B.P.2012. Relationship between vancomycin-resistant Staphylococcus aureus, vancomycin-intermediate S. aureus, high vancomycin MIC, and outcome in serious S. aureus infections. J. Clin. Microbiol. 50:2548–2552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horne K.C., Howden B.P., Grabsch E.A., Graham M., Ward P.B., Xie S., Mayall B.C., Johnson P.D., and Grayson M.L.2009. Prospective comparison of the clinical impacts of heterogeneous vancomycin-intermediate methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-susceptible MRSA. Antimicrob. Agents Chemother. 53:3447–3452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howden B.P., Davies J.K., Johnson P.D., Stinear T.P., and Grayson M.L.2010. Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin-intermediate strains: resistance mechanisms, laboratory detection, and clinical implications. Clin. Microbiol. Rev. 23:99–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Howe R.A., Monk A., Wootton M., Walsh T.R., and Enright M.C.2004. Vancomycin susceptibility within methicillin-resistant Staphylococcus aureus lineages. Emerg. Infect. Dis. 10:855–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacob J.T., and Diazgranados C.A.2013. High vancomycin minimum inhibitory concentration and clinical outcomes in adults with methicillin-resistant Staphylococcus aureus infections: a meta-analysis. Int. J. Infect. Dis. 17:e93–e100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katz B.D., Luperchio S.A., and Thorne G.M.2008. Detection of daptomycin-nonsusceptible strains using the Neo-Sensitab prediffusion method. Diagn. Microbiol. Infect. Dis. 61:315–320 [DOI] [PubMed] [Google Scholar]
- 24.Kobayashi S.D., Musser J.M., and DeLeo F.R.2012. Genomic analysis of the emergence of vancomycin-resistant Staphylococcus aureus. MBio 3:e00170–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kondo Y., Ito T., Ma X.X., Watanabe S., Kreiswirth B.N., Etienne J., and Hiramatsu K.2007. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob. Agents Chemother. 51:264–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kosowska-Shick K., Ednie L.M., McGhee P., Smith K., Todd C.D., Wehler A., and Appelbaum P.C.2008. Incidence and characteristics of vancomycin nonsusceptible strains of methicillin-resistant Staphylococcus aureus at Hershey Medical Center. Antimicrob. Agents Chemother. 52:4510–4513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lalueza A., Chaves F., San Juan R., Daskalaki M., Otero J.R., and Aguado J.M.2010. Is high vancomycin minimum inhibitory concentration a good marker to predict the outcome of methicillin-resistant Staphylococcus aureus bacteremia?. J. Infect. Dis. 201:311–312; author reply 312–313 [DOI] [PubMed] [Google Scholar]
- 28.Lattar S.M., Tuchscherr L.P., Centron D., Becker K., Predari S.C., Buzzola F.R., Robinson D.A., and Sordelli D.O.2012. Molecular fingerprinting of Staphylococcus aureus isolated from patients with osteomyelitis in Argentina and clonal distribution of the cap5(8) genes and of other selected virulence genes. Eur. J. Clin. Microbiol. Infect. Dis. 31:2559–2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin S.Y., Chen T.C., Chen F.J., Chen Y.H., Lin Y.I., Siu L.K., and Lu P.L.2012. Molecular epidemiology and clinical characteristics of hetero-resistant vancomycin intermediate Staphylococcus aureus bacteremia in a Taiwan Medical Center. J. Microbiol. Immunol. Infect. 45:435–441 [DOI] [PubMed] [Google Scholar]
- 30.Liu C., Bayer A., Cosgrove S.E., Daum R.S., Fridkin S.K., Gorwitz R.J., Kaplan S.L., Karchmer A.W., Levine D.P., Murray B.E., et al. 2011. Clinical practice guidelines by the infectious diseases society of america for the treatment of methicillin-resistant Staphylococcus aureus infections in adults and children: executive summary. Clin. Infect. Dis. 52:285–292 [DOI] [PubMed] [Google Scholar]
- 31.Lopez Furst M.J., de Vedia L., Fernandez S., Gardella N., Ganaha M.C., Prieto S., Carbone E., Lista N., Rotryng F., Morera G.I., et al. 2013. Prospective multicenter study of community-associated skin and skin structure infections due to methicillin-resistant Staphylococcus aureus in Buenos Aires, Argentina. PLoS One 8:e78303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nielsen S.V., and Casals J.B.2005. Detection of decreased susceptibility to glycopeptides in S. aureus using tablet (disc) prediffusion. In15th European Congress of Clinical Microbiology and Infectious Diseases Copenhagen/Denmark, April2–5, 2005 [Google Scholar]
- 33.Oliveira D.C., and de Lencastre H.2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 46:2155–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park K.H., Kim E.S., Kim H.S., Park S.J., Bang K.M., Park H.J., Park S.Y., Moon S.M., Chong Y.P., Kim S.H., et al. 2012. Comparison of the clinical features, bacterial genotypes and outcomes of patients with bacteraemia due to heteroresistant vancomycin-intermediate Staphylococcus aureus and vancomycin-susceptible S. aureus. J. Antimicrob. Chemother. 67:1843–1849 [DOI] [PubMed] [Google Scholar]
- 35.Perazzi B., Bello N., Mollerach M., Vay C., Lasala M.B., and Famiglietti A.2011. Endocarditis caused by methicillin-susceptible Staphylococcus aureus with reduced susceptibility to vancomycin: a case report. J. Med. Case Rep. 5:555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prakash V., Lewis J.S., 2nd, and Jorgensen J.H.2008. Vancomycin MICs for methicillin-resistant Staphylococcus aureus isolates differ based upon the susceptibility test method used. Antimicrob. Agents Chemother. 52:4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rojas L., Bunsow E., Munoz P., Cercenado E., Rodriguez-Creixems M., and Bouza E.2012. Vancomycin MICs do not predict the outcome of methicillin-resistant Staphylococcus aureus bloodstream infections in correctly treated patients. J. Antimicrob. Chemother. 67:1760–1768 [DOI] [PubMed] [Google Scholar]
- 38.Rosco Diagnostica A/S T.D. 2010. Neo-Sensitabs™. User's Guide. Susceptibility testing EUCAST and CLSI potency Neo-Sensitabs™. Document 1.5.0. Prediffussion Method
- 39.Sader H.S., Rhomberg P.R., and Jones R.N.2009. Nine-hospital study comparing broth microdilution and Etest method results for vancomycin and daptomycin against methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 53:3162–3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schweizer M.L., Furuno J.P., Sakoulas G., Johnson J.K., Harris A.D., Shardell M.D., McGregor J.C., Thom K.A., and Perencevich E.N.2011. Increased mortality with accessory gene regulator (agr) dysfunction in Staphylococcus aureus among bacteremic patients. Antimicrob. Agents Chemother. 55:1082–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sola C., Gribaudo G., Vindel A., Patrito L., and Bocco J.L.2002. Identification of a novel methicillin-resistant Staphylococcus aureus epidemic clone in Cordoba, Argentina, involved in nosocomial infections. J. Clin. Microbiol. 40:1427–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sola C., Lamberghini R.O., Ciarlantini M., Egea A.L., Gonzalez P., Diaz E.G., Huerta V., Gonzalez J., Corso A., Vilaro M., et al. 2011. Heterogeneous vancomycin-intermediate susceptibility in a community-associated methicillin-resistant Staphylococcus aureus epidemic clone, in a case of Infective Endocarditis in Argentina. Ann. Clin. Microbiol. Antimicrob. 10:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sola C., Paganini H., Egea A.L., Moyano A.J., Garnero A., Kevric I., Culasso C., Vindel A., Lopardo H., and Bocco J.L.2012. Spread of epidemic MRSA-ST5-IV clone encoding PVL as a major cause of community onset Staphylococcal infections in Argentinean children. PLoS One 7:e30487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soriano A., Marco F., Martinez J.A., Pisos E., Almela M., Dimova V.P., Alamo D., Ortega M., Lopez J., and Mensa J.2008. Influence of vancomycin minimum inhibitory concentration on the treatment of methicillin-resistant Staphylococcus aureus bacteremia. Clin. Infect. Dis. 46:193–200 [DOI] [PubMed] [Google Scholar]
- 45.Takesue Y., Nakajima K., Takahashi Y., Ichiki K., Ishihara M., Wada Y., Tsuchida T., Uchino M., and Ikeuchi H.2011. Clinical characteristics of vancomycin minimum inhibitory concentration of 2 μg/ml methicillin-resistant Staphylococcus aureus strains isolated from patients with bacteremia. J. Infect. Chemother. 17:52–57 [DOI] [PubMed] [Google Scholar]
- 46.van Hal S.J., Wehrhahn M.C., Barbagiannakos T., Mercer J., Chen D., Paterson D.L., and Gosbell I.B.2011. Performance of various testing methodologies for detection of heteroresistant vancomycin-intermediate Staphylococcus aureus in bloodstream isolates. J. Clin. Microbiol. 49:1489–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.von Specht M., Gardella N., Tagliaferri P., Gutkind G., and Mollerach M.2006. Methicillin-resistant Staphylococcus aureus in community-acquired meningitis. Eur. J. Clin. Microbiol. Infect. Dis. 25:267–269 [DOI] [PubMed] [Google Scholar]
- 48.Wang J.L., Wang J.T., Sheng W.H., Chen Y.C., and Chang S.C.2010. Nosocomial methicillin-resistant Staphylococcus aureus (MRSA) bacteremia in Taiwan: mortality analyses and the impact of vancomycin, MIC=2 mg/L, by the broth microdilution method. BMC Infect. Dis. 10:159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wootton M., Howe R.A., Hillman R., Walsh T.R., Bennett P.M., and MacGowan A.P.2001. A modified population analysis profile (PAP) method to detect hetero-resistance to vancomycin in Staphylococcus aureus in a UK hospital. J. Antimicrob. Chemother. 47:399–403 [DOI] [PubMed] [Google Scholar]
- 50.Yamakawa J., Aminaka M., Okuzumi K., Kobayashi H., Katayama Y., Kondo S., Nakamura A., Oguri T., Hori S., Cui L., et al. 2012. Heterogeneously vancomycin-intermediate Staphylococcus aureus (hVISA) emerged before the clinical introduction of vancomycin in Japan: a retrospective study. J. Infect. Chemother. 18:406–409 [DOI] [PubMed] [Google Scholar]


