To address the issue of an initial pathological event in Fanconi anemia-related bone marrow failure (FA-BMF), induced pluripotent stem cells (iPSCs) were established from fibroblasts of six patients with FA and FANCA mutations. Results indicated that the hematopoietic consequences in FA patients originate from the early hematopoietic stage and highlight the potential usefulness of iPSC technology for elucidating the pathogenesis of FA-BMF.
Keywords: Induced pluripotent stem cells, Fanconi anemia, Hematopoietic progenitors, Differentiation, Transcription factors
Abstract
Fanconi anemia (FA) is a disorder of genomic instability characterized by progressive bone marrow failure (BMF), developmental abnormalities, and an increased susceptibility to cancer. Although various consequences in hematopoietic stem/progenitor cells have been attributed to FA-BMF, the quest to identify the initial pathological event is still ongoing. To address this issue, we established induced pluripotent stem cells (iPSCs) from fibroblasts of six patients with FA and FANCA mutations. An improved reprogramming method yielded iPSC-like colonies from all patients, and iPSC clones were propagated from two patients. Quantitative evaluation of the differentiation ability demonstrated that the differentiation propensity toward the hematopoietic and endothelial lineages is already defective in early hemoangiogenic progenitors. The expression levels of critical transcription factors were significantly downregulated in these progenitors. These data indicate that the hematopoietic consequences in FA patients originate from the early hematopoietic stage and highlight the potential usefulness of iPSC technology for elucidating the pathogenesis of FA-BMF.
Introduction
Fanconi anemia (FA) is a disorder of genomic instability characterized by progressive bone marrow failure (BMF), developmental abnormalities, and an increased susceptibility to cancer [1–3]. The responsible genes form a common DNA repair network referred to as the FA pathway [4]. The FA pathway has been considered to be involved in repair of DNA interstrand cross-links (ICLs) [1–4]. The FA pathway is activated by ICL damage, leading to the monoubiquitination of Fanconi anemia, complementation group D2 (FANCD2) and Fanconi anemia, complementation group I (FANCI) proteins (ID complex) by FA core E3 ligase complex. The monoubiquitinated ID complex is then loaded on damaged chromatin and mediates homologous recombination and translesion synthesis. Although accelerated apoptosis and cell cycle arrest in hematopoietic stem/progenitor cells have been associated with BMF in patients with FA [5], precisely how and when the initial event that causes this consequence occurs have been unclear. Recent reports have indicated that the fate of hematopoietic progenitors has already been determined during fetal liver hematopoiesis both in humans and in mouse models of FA [5, 6]. However, tracing the earlier developmental events to capture the initial event is both technically and ethically impossible at present in humans.
Induced pluripotent stem cells (iPSCs) established from patients with FA may be useful to address these issues because, in combination with a proper hematopoietic differentiation system, iPSCs can enable evaluation of the human developmental stages [7, 8]. In this study, we established iPSCs from patients with FA and found that their differentiation propensity toward the hematopoietic lineage was already defective in the early hemoangiogenic progenitor stage. This report shows the possibility that the hematopoietic consequences in patients with FA originate at the earliest hematopoietic stage.
Materials and Methods
Detailed methods are included in the supplemental online data.
Study Ethics
This study was approved by the ethics committees of Kyoto University and Tokai University, and informed consent was obtained from the patients’ guardians in accordance with the Declaration of Helsinki.
Establishment and Differentiation of iPSCs From Patients With FA
Fibroblasts obtained from the six patients with FA (detail shown in supplemental online Table 1) were reprogrammed with episomal vectors encoding OCT3/4, SOX2, KLF4, LIN28, and L-MYC and a short hairpin RNA (shRNA) encoding a p53 knockdown sequence, as described previously [9]. Plasmids were kindly provided by Dr. Keisuke Okita (Kyoto University). For hematopoietic differentiation, a two-dimensional hematopoietic differentiation system was used, as described previously [10, 11].
Results and Discussion
We tried to reprogram fibroblasts obtained from six patients with FA and mutations in complementation group A (FA-A; patients with mutations in the Fanconi anemia, complementation group A [FANCA] gene) (supplemental online Table 1) by introducing the previously described reprogramming factors (OCT3/4, SOX2, KLF4, c-MYC) with retroviral [7] or Sendai viral vectors [12] under hypoxic conditions (5% O2). Consistent with the previous reports [13], no iPSC-like colonies emerged. Recently, several groups reported that using a combination of highly efficient methods, including improved vectors such as a polycistronic OSKM cassette or additional reprograming factors under hypoxic culture conditions, could overcome the reprogramming resistance of FA cells [14–16]. Hence, we tried to establish FA patient-specific iPSCs by using episomal vectors encoding OCT3/4, SOX2, KLF4, LMYC, LIN28, and an shRNA-mediated p53 knockdown construct [9] under hypoxic conditions (5% O2). As a result, iPSC-like colonies arose from all FA patient-derived fibroblasts (supplemental online Table 1); however, we could pick up and maintain only the TKFA02 and TKFA07 patient-derived iPSC (FA-iPSC) lines. The colonies arising from the TKFA03, FA44, FA45, and FA46 patients could not be picked up and propagated. Consequently, we selected one and three FA-iPSC lines of the TKFA02 and TKFA07 cells, respectively (Fig. 1A).
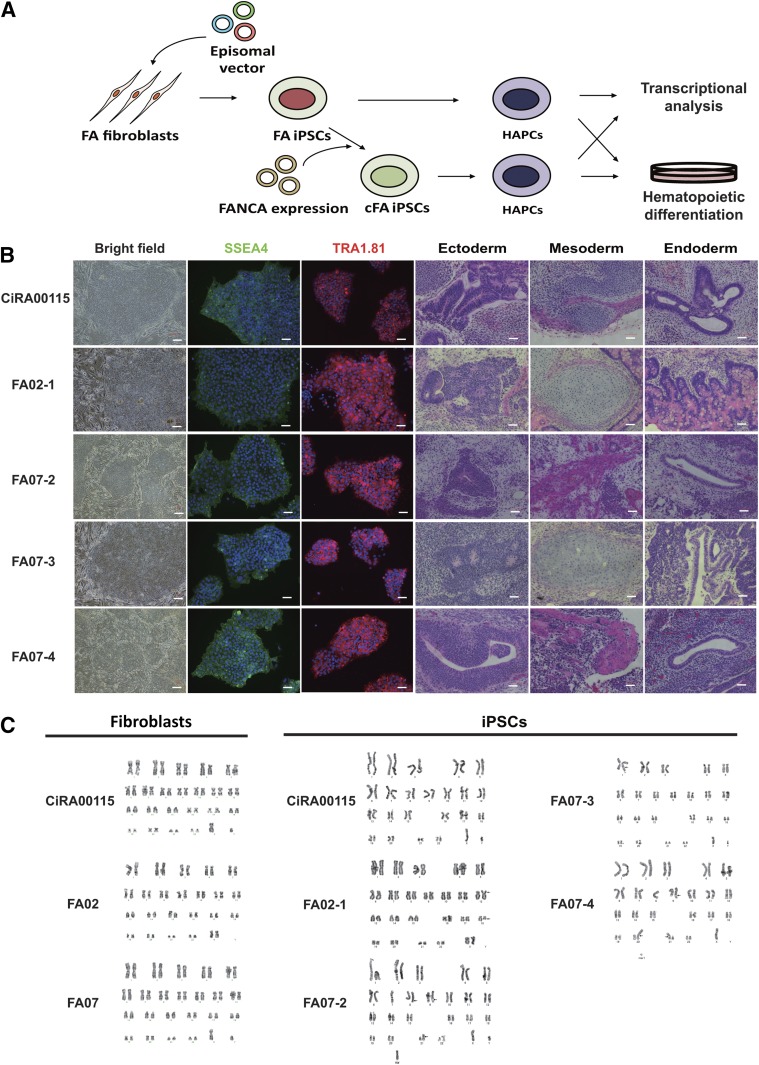
Figure 1.
Establishment of FA-iPSC lines from patients with FA and mutations in FANCA. CiRA00115 is a control iPSC line. (A): The experimental scheme. (B): The morphology and immunostaining of pluripotent markers of FA-iPSCs and teratoma formation by the FA-iPSC lines (ectoderm: neural rosette; mesoderm: cartilage; endoderm: gut-like structure). Scale bars indicate 100 μm. (C): The results of karyotype analysis of fibroblasts and iPSCs from FA patients. The karyotypes of the fibroblasts were 46XX and 46XY for FA02 and FA07, respectively. The karyotype of FA-iPSCs from FA02 was summarized as 46, XX, t(12;18)(p11.2;p11.2) [20 cells]. The karyotypes of FA-iPSCs from FA07 were as follows: for FA07-2, 46, XY, add(1)(q32) [14 cells] and 46, XY [6 cells]; for FA07-3, 42-46, XY, add(8)(q24.1),add(11)(q23),del(11)(q23),add(18)(q21) [17 cells] and 42-46, XY, add(8)(q24.1),add(9)(q34),add(21)(p11.2) [3 cells]; for FA07-4, 45-46, XY, add(8)(q24.1),add(9)(q34),add(21)(p11.2) [20 cells]. Twenty metaphases were analyzed for each sample. Abbreviations: cFA, FANCA-complemented FA-iPSC clone; FA, Fanconi anemia; HAPC, hemoangiogenic progenitor cell; iPSC, induced pluripotent stem cell.
The FA-iPSC lines showed human pluripotent cell-like morphology, expressed pluripotent markers, and differentiated into three germ layers in the teratoma formation assay (Fig. 1B). A short tandem repeated analysis confirmed that the patient identity was conserved throughout the reprogramming process (supplemental online Fig. 1). The FA-iPSC lines had residual transgene expressions, as reported previously [16] (supplemental online Fig. 2A). Consistent with this, these FA-iPSC clones, including their complemented counterparts (discussed below), showed reduced p53 expression (supplemental online Fig. 2B). Although the fibroblasts from both patients had normal karyotypes, both FA-iPSC lines had aberrant karyotypes (Fig. 1C), which is compatible with a previous report [16] and indicates that the FA pathway has an important role in determining chromosomal stability through reprogramming events. Collectively, by combining the reprogramming factors with modulation of the p53 pathway, we were able to develop a robust reprogramming strategy that enabled us to establish iPSC-like colonies from FA patient-derived somatic cells.
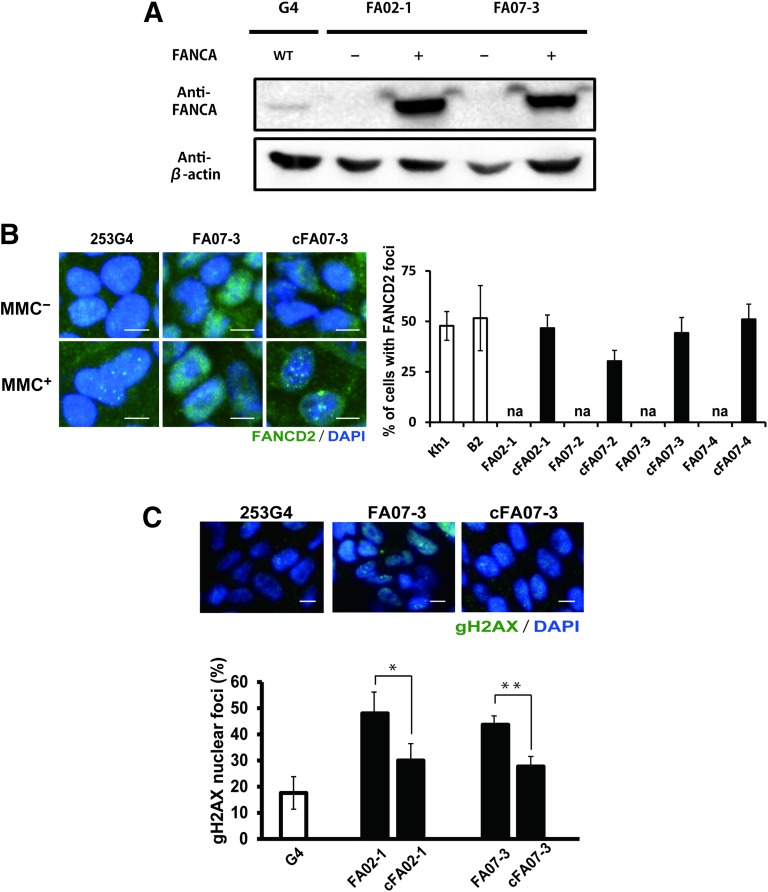
To establish isogenic FANCA-complemented clones, we introduced exogenous wild-type FANCA cDNA into each clone (Fig. 2A; supplemental online Fig. 3). Mitomycin C-induced FANCD2 foci formation was restored in these FANCA-complemented FA-iPSC clones (designated as cFA-iPSCs) (Fig. 2B). During maintenance of the iPSCs, the FA-iPSCs showed an increased DNA double-strand break rate, as evaluated by the frequency of nuclear foci of phosphorylated H2AX compared with the complemented counterparts (Fig. 2C), but the distribution of the cells in the different phases of the cell cycle was not significantly different (supplemental online Fig. 4). Consequently, the complementation of the FA pathway recovers the in vitro phenotype of FA-iPSCs.
Figure 2.
Validation of FA pathway in FA-iPSC lines. KhES1 is a control embryonic stem cell line, and 409B2 and 253G4 are control iPSC lines. (A): Immunoblotting for FANCA. (B): The formation of FANCD2 foci (green) in the nuclei (stained with DAPI; blue) of mitomycin C-treated FA-iPSCs (FA07) and cFA-iPSCs (cFA07) (left). The percentage of nuclei positive for FANCD2 foci (right). (C): Representative immunofluorescent staining (upper) and quantification (lower) of the nuclear foci of phosphorylated H2AX. The y-axis in the graph indicates the percentage of cells with three or more nuclear phosphorylated H2AX foci. All data are presented as mean ± SD and are representative of three independent experiments. All p values were determined by Student's t test. ∗, p < .05; ∗∗, p < .01 (n = 3). Scale bars indicate 10 μm. Abbreviations: cFA, FANCA-complemented FA-iPSC clone; DAPI, 4′,6-diamidino-2-phenylindole; FA, Faconi anemia; iPSC, induced pluripotent stem cell; MMC, mitomycin C; WT, wild type.
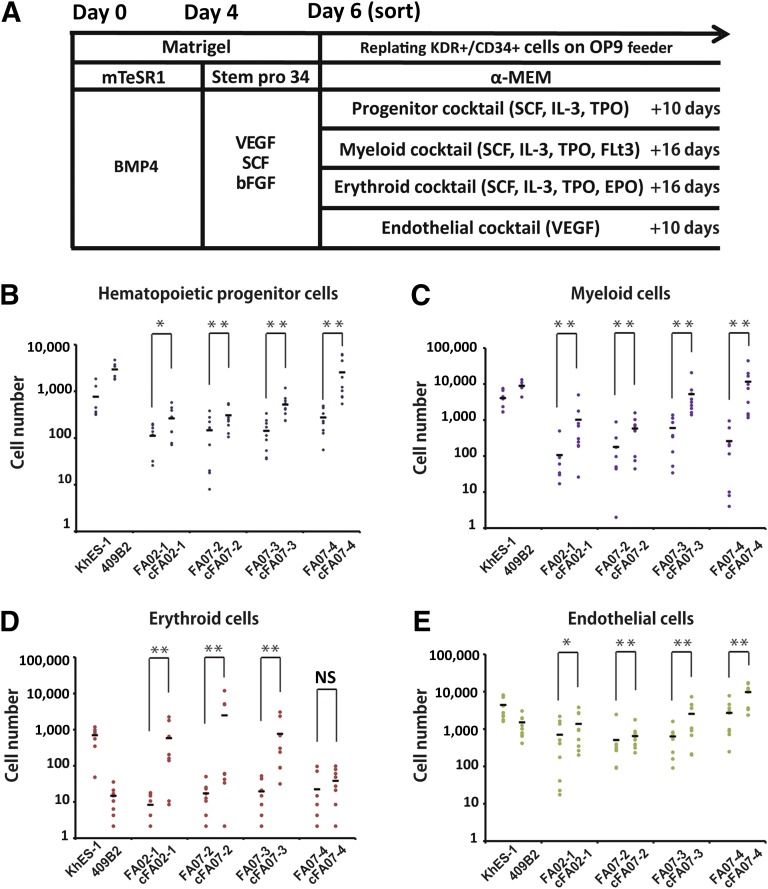
To determine whether the hematopoietic differentiation is affected by disruption of the FA pathway, we next differentiated the FA-iPSC lines into hematopoietic progenitor cells through the previously reported serum- and feeder-free monolayer culture system [10, 11]. Under this protocol, the progenitor cells committed to the hematopoietic lineage are obtained as KDR+CD34+ early hemoangiogenic progenitor cells (HAPCs), which give rise to both endothelial and hematopoietic cells. To evaluate the differentiation propensity of HAPCs quantitatively, we sorted the KDR+CD34+ HAPCs on day 6 and cultured them on OP9 feeder cells with lineage-specific cytokine cocktails (Fig. 3A; supplemental online Fig. 5). The FA-iPSC lines showed a significant reduction of CD34+CD45+ hematopoietic precursors compared with cFA-iPSC lines (Fig. 3B). Subsequently, the myeloid and erythroid lineage hematopoietic cells were significantly reduced (Fig. 3C, 3D). The number of CD31+ endothelial cells from FA-iPSC-derived HAPCs (FA-HAPCs) was also reduced compared with that in the cFA-iPSC lines (Fig. 3E). The distribution of the cell cycle in FA-iPSC-derived KDR+CD34+ HAPCs was comparable to that of the complemented counterparts (supplemental online Fig. 6). We also confirmed that KDR+CD34+ HAPCs were not apoptotic (supplemental online Fig. 7). Although the expression level of p53 reduced and varied among the FA- or cFA-iPSC lines (supplemental online Fig. 2B), the ability of these cells to undergo hematopoietic differentiation was dependent on the FANCA status but not on the p53 level, indicating that, at least in this study, the expression level of p53 has no obvious impact on the differentiation propensity of HAPCs. Taken together, these findings indicate that FA-HAPCs showed a defective propensity to differentiate toward both hematopoietic and endothelial lineages.
Figure 3.
The defective differentiation propensity in Fanconi anemia induced pluripotent stem cell-derived hemoangiogenic progenitors. KhES1 and 409B2 are control embryonic and induced pluripotent stem cell lines, respectively. (A): A schematic diagram of hematopoietic differentiation. (B–E): The number of differentiated cells derived from 5,000 sorted KDR+CD34+ cells. (B): CD34+CD45+ hematopoietic progenitors. (C): CD33+CD45+ myeloid cells. (D): CD45−CD235α+ erythroid cells. (E): CD31+CD34+ endothelial cells. All data are presented as mean ± SD and are representative of three independent experiments. All p values were determined by Wilcoxon rank sum test. ∗, p < .05; ∗∗, p < .01; ∗∗∗, p < .001 (n = 3). Abbreviations: α-MEM, alpha Minimum Essential Medium; bFGF, basic fibroblast growth factor; EPO, erythropoietin; IL-3, interleukin 3; NS, not significant; SCF, stem cell factor; TPO, thyroid peroxidase; VEGF, vascular endothelial growth factor.
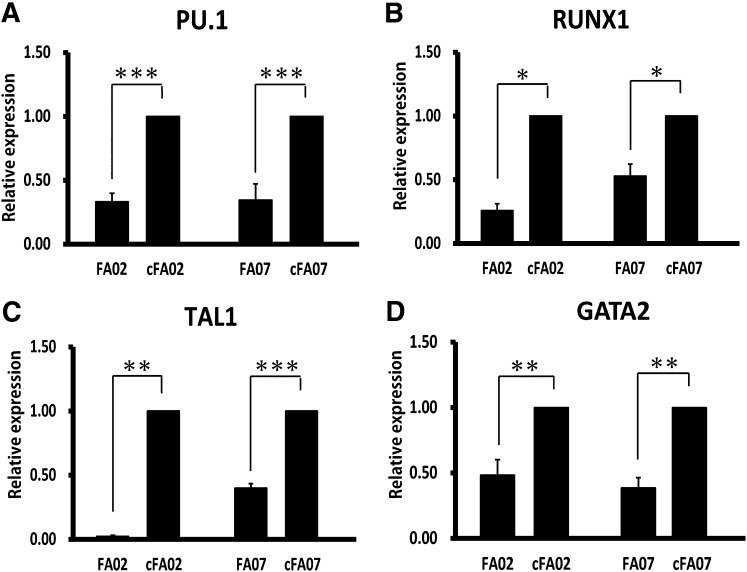
To further elucidate the mechanism underlying the defective hematopoietic differentiation, we next quantified the expression levels of critical transcription factors requird for hematopoietic differentiation in HAPCs [17–20]. HAPCs from FA-iPSC lines showed significant downregulation of these transcription factors (Fig. 4A–4D), indicating that the FA pathway might be involved in maintaining the transcriptional network critical for determining the differentiation propensity of HAPCs. We also compared the global expression profiles of HAPCs from FA- and cFA-iPSCs (supplemental online Fig. 8A, 8B). We identified that 227 genes were significantly upregulated and 396 genes were significantly downregulated in FA-HAPCs (supplemental online Fig. 8C; supplemental online Table 2). A gene ontology enrichment analysis revealed that genes associated with mesodermal differentiation, vascular formation, and hematopoiesis were extensively downregulated in FA-HAPCs (supplemental online Fig. 8D).
Figure 4.
FANCA-deficient hemoangiogenic progenitor cells show the decreased expression of hematopoietic marker genes. (A–D): The results of the quantitative polymerase chain reaction analysis for hematopoietic marker genes in KDR+CD34+ hemoangiogenic progenitors (day 6). The relative gene expression (y-axis) was calculated relative to the expression in corresponding cFA induced pluripotent stem cells after normalization to the expression of glyceraldehyde-3-phosphate dehydrogenase. All data are presented as mean ± SD and are representative of three independent experiments. All p values were determined by Student's t test. ∗, p < .05; ∗∗, p < .01; ∗∗∗, p < .001 (n = 3). Abbreviation: cFA, FANCA-complemented Fanconi anemia induced pluripotent stem cell clone.
Conclusion
In summary, we successfully established FA-iPSCs from FA-A patients. We found that an early pathological phenotype was detected in HAPCs as a defective differentiation propensity into both hematopoietic and endothelial lineages. Interestingly, during hematopoiesis, the expression of FANCA and FANCC were specifically upregulated in KDR+CD34+ HAPCs (supplemental online Fig. 9), indicating the stage-specific requirement of these genes in HAPCs. Because the expression of hematopoietic and angiogenic genes was affected, FANCA may have an important role in regulating these genes in FA-HAPCs. Conducting a comprehensive analysis of patient-derived affected progenitors is not feasible without iPSC technology, which provides an unprecedented opportunity to gain further insight into the pathogenesis of BMF in patients with FA.
Supplementary Material
Acknowledgments
We thank M. Yamane and S. Benno for technical assistance, H. Watanabe for administrative assistance, and M. Yoshida and K. Isoda for performing the teratoma assay. Funding was provided by grants from the Ministry of Health, Labor, and Welfare to T.N.; a grant from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) to T.N.; grants from the Leading Project of MEXT to T.N.; a grant from the Funding Program for World-Leading Innovative Research and Development on Science and Technology (FIRST Program) of the Japan Society for the Promotion of Science (JSPS) to T.N.; grants from the JSPS to T.N. and M.K.S.; a grant-in-aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (Grant 20591262) to M.Y.; the Program for Intractable Diseases Research Utilizing Disease-Specific Induced Pluripotent Stem (iPS) Cells from the Japan Science and Technology Agency (JST) to T.N.; the grant for Core Center for iPS Cell Research of Research Center Network for Realization of Regenerative Medicine from JST to T.N. and M.K.S.; and a Research Grant for Intractable Diseases (H-21-061) from the Japanese Ministry of Health, Labor, and Welfare to M.Y.
Author Contributions
N.M.S.: collection and/or assembly of data, data analysis and interpretation, and manuscript writing; A.N., C.O., N.A., and A.W.: collection and/or assembly of data, data analysis and interpretation; M.Y.: provision of study material or patients; A.H.: data analysis and interpretation; K.-I.W. and T.H.: provision of study material or patients; M.T.: data analysis and interpretation, provision of study material or patients, conception and design; T.N.: conception and design, data analysis and interpretation; M.K.S.: conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript.
Disclosure of Potential Conflicts of Interest
C.O. is an uncompensated employee of the Mitsubishi Space Software Co., Ltd., with which CiRA entered into an agreement of bioinformatics analysis service. The other authors indicated no potential conflicts of interest.
References
- 1.Deans AJ, West SC. DNA interstrand crosslink repair and cancer. Nat Rev Cancer. 2011;11:467–480. doi: 10.1038/nrc3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kottemann MC, Smogorzewska A. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature. 2013;493:356–363. doi: 10.1038/nature11863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim H, D'Andrea AD. Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Genes Dev. 2012;26:1393–1408. doi: 10.1101/gad.195248.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takata M, Yamamoto K, Matsushita N, et al. The Fanconi anemia pathway promotes homologous recombination repair in DT40 cell line. Subcell Biochem. 2006;40:295–311. doi: 10.1007/978-1-4020-4896-8_17. [DOI] [PubMed] [Google Scholar]
- 5.Ceccaldi R, Parmar K, Mouly E, et al. Bone marrow failure in Fanconi anemia is triggered by an exacerbated p53/p21 DNA damage response that impairs hematopoietic stem and progenitor cells. Cell Stem Cell. 2012;11:36–49. doi: 10.1016/j.stem.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kamimae-Lanning AN, Goloviznina NA, Kurre P. Fetal origins of hematopoietic failure in a murine model of Fanconi anemia. Blood. 2013;121:2008–2012. doi: 10.1182/blood-2012-06-439679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 8.Zhu Z, Huangfu D. Human pluripotent stem cells: An emerging model in developmental biology. Development. 2013;140:705–717. doi: 10.1242/dev.086165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okita K, Matsumura Y, Sato Y, et al. A more efficient method to generate integration-free human iPS cells. Nat Methods. 2011;8:409–412. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- 10.Niwa A, Heike T, Umeda K, et al. A novel serum-free monolayer culture for orderly hematopoietic differentiation of human pluripotent cells via mesodermal progenitors. PLoS ONE. 2011;6:e22261. doi: 10.1371/journal.pone.0022261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yanagimachi MD, Niwa A, Tanaka T, et al. Robust and highly-efficient differentiation of functional monocytic cells from human pluripotent stem cells under serum- and feeder cell-free conditions. PLoS ONE. 2013;8:e59243. doi: 10.1371/journal.pone.0059243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seki T, Yuasa S, Oda M, et al. Generation of induced pluripotent stem cells from human terminally differentiated circulating T cells. Cell Stem Cell. 2010;7:11–14. doi: 10.1016/j.stem.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 13.Raya A, Rodriguez-Piza I, Guenechea G, et al. Disease-corrected haematopoietic progenitors from Fanconi anaemia induced pluripotent stem cells. Nature. 2009;460:53–59. doi: 10.1038/nature08129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muller LU, Milsom MD, Harris CE, et al. Overcoming reprogramming resistance of Fanconi anemia cells. Blood. 2012;119:5449–5457. doi: 10.1182/blood-2012-02-408674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muller LU, Schlaeger TM, DeVine AL, et al. Induced pluripotent stem cells as a tool for gaining new insights into Fanconi anemia. Cell Cycle. 2012;11:2985–2990. doi: 10.4161/cc.21109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yung SK, Tilgner K, Ledran MH, et al. Brief report: Human pluripotent stem cell models of Fanconi anemia deficiency reveal an important role for Fanconi anemia proteins in cellular reprogramming and survival of hematopoietic progenitors. Stem Cells. 2013;31:1022–1029. doi: 10.1002/stem.1308. [DOI] [PubMed] [Google Scholar]
- 17.Scott EW, Simon MC, Anastasi J, et al. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265:1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- 18.Okuda T, van Deursen J, Hiebert SW, et al. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 19.Shivdasani RA, Mayer EL, Orkin SH. Absence of blood formation in mice lacking the T-cell leukaemia oncoprotein tal-1/SCL. Nature. 1995;373:432–434. doi: 10.1038/373432a0. [DOI] [PubMed] [Google Scholar]
- 20.Tsai FY, Keller G, Kuo FC, et al. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature. 1994;371:221–226. doi: 10.1038/371221a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.